Abstract
Background
Low-molecular-weight glutenin subunits (LMW-GS) strongly influence the bread-making quality of bread wheat. These proteins are encoded by a multi-gene family located at the Glu-A3, Glu-B3 and Glu-D3 loci on the short arms of homoeologous group 1 chromosomes, and show high allelic variation. To characterize the genetic and protein compositions of LMW-GS alleles, we investigated 16 Aroona near-isogenic lines (NILs) using SDS-PAGE, 2D-PAGE and the LMW-GS gene marker system. Moreover, the composition of glutenin macro-polymers, dough properties and pan bread quality parameters were determined for functional analysis of LMW-GS alleles in the NILs.
Results
Using the LMW-GS gene marker system, 14–20 LMW-GS genes were identified in individual NILs. At the Glu-A3 locus, two m-type and 2–4 i-type genes were identified and their allelic variants showed high polymorphisms in length and nucleotide sequences. The Glu-A3d allele possessed three active genes, the highest number among Glu-A3 alleles. At the Glu-B3 locus, 2–3 m-type and 1–3 s-type genes were identified from individual NILs. Based on the different compositions of s-type genes, Glu-B3 alleles were divided into two groups, one containing Glu-B3a, B3b, B3f and B3g, and the other comprising Glu-B3c, B3d, B3h and B3i. Eight conserved genes were identified among Glu-D3 alleles, except for Glu-D3f. The protein products of the unique active genes in each NIL were detected using protein electrophoresis. Among Glu-3 alleles, the Glu-A3e genotype without i-type LMW-GS performed worst in almost all quality properties. Glu-B3b, B3g and B3i showed better quality parameters than the other Glu-B3 alleles, whereas the Glu-B3c allele containing s-type genes with low expression levels had an inferior effect on bread-making quality. Due to the conserved genes at Glu-D3 locus, Glu-D3 alleles showed no significant differences in effects on all quality parameters.
Conclusions
This work provided new insights into the composition and function of 18 LMW-GS alleles in bread wheat. The variation of i-type genes mainly contributed to the high diversity of Glu-A3 alleles, and the differences among Glu-B3 alleles were mainly derived from the high polymorphism of s-type genes. Among LMW-GS alleles, Glu-A3e and Glu-B3c represented inferior alleles for bread-making quality, whereas Glu-A3d, Glu-B3b, Glu-B3g and Glu-B3i were correlated with superior bread-making quality. Glu-D3 alleles played minor roles in determining quality variation in bread wheat. Thus, LMW-GS alleles not only affect dough extensibility but greatly contribute to the dough resistance, glutenin macro-polymers and bread quality.
Keywords: Triticum aestivum, Glu-3 alleles, Bread-making quality
Background
The unique viscoelastic properties conferred by gluten proteins in bread wheat are the basis of the flexible processing qualities in producing a wide range of food products for a large proportion of the world population. Gluten proteins, also named prolamins, are classically divided into gliadins and glutenins, based on different solubilities in an alcohol/water mixture [1]. The gliadins are generally monomeric proteins, divided into three groups, α/β-, γ- and ω-gliadins, based on their electrophoretic mobilities at low pH [2]. Glutenins form polymeric proteins stabilized by interchain disulfide bonds. Based on different molecular weights, glutenins can be classified into two groups, high-molecular-weight glutenin subunits (HMW-GS) and low-molecular-weight glutenin subunits (LMW-GS) [2,3]. LMW-GS are further divided into B-, C- and D-group subunits according to their mobilities in sodium dodecyl sulphate polyacrylamide-gel electrophoresis (SDS-PAGE) [4].
In bread wheat, HMW-GS are encoded by genes at the orthologous Glu-1 loci on the long arms of chromosomes 1A, 1B and 1D (Glu-A1, Glu-B1 and Glu-D1). Each locus possesses two paralogous genes encoding one x- and one y-type subunit [5]. LMW-GS genes are located at the Glu-A3, Glu-B3 and Glu-D3 loci on the short arms of group 1 chromosomes. The LMW-GS genes at the Glu-3 loci and the gliadin genes at the Gli-1 loci are tightly linked and form gene clusters covering several centimorgans (cMs) [6-8]. Moreover, unlike the simple composition of HMW-GS, LMW-GS are encoded by a complex multigene family without the information of the exact number of genes [9,10]. A large number of genes and abundant allelic variations at Glu-3 loci and their tight linkage with gliadin genes make it difficult to elucidate the composition and function of LMW-GS genes in bread wheat [4].
SDS-PAGE is widely used to investigate the abundant seed storage proteins in bread wheat. Based on the mobility of proteins in SDS-PAGE gels, whole seed proteins are divided into four groups, HMW-GS, D-group, B-group, and C-group. D-group proteins are mainly composed of ω-gliadin proteins, whereas B-group mostly consists of LMW-GS proteins, and C-group comprise α, β and γ-type gliadins and several LMW-GS proteins [4]. Based on the different electrophoretic patterns, LMW-GS protein alleles encoded by Glu-3 loci are designated alphabetically (e.g., Glu-A3a) [11]. However, identification of the LMW-GS composition in breeding programs remains a significant challenge because determination of LMW-GS alleles with SDS-PAGE needs much experience. This is why the functions of LMW-GS alleles are not well characterized. Gene-specific markers for Glu-A3 and Glu-B3 alleles were developed to identify different LMW-GS alleles. However, molecular markers for Glu-D3 alleles are still not available due to the slight differences among alleles [12-17]. Using BAC library screening and proteomics methods, LMW-GS genes in Norin 61 (Glu-A3d, Glu-B3i and Glu-D3c), Glenlea (Glu-A3g, Glu-B3g and Glu-D3c) and Xiaoyan 54 (Glu-A3d, Glu-B3d and Glu-D3c) were identified and characterized [10,18-20]. These studies greatly improved our understanding of the unique genes encoding different LMW-GS alleles in bread wheat. Recently, based on the conserved and polymorphic structure of LMW-GS genes, we developed a LMW-GS gene marker system and a full-length gene cloning method [21,22]. They were successfully used to identify and characterize more than 16 LMW-GS genes in individual wheat varieties [21]. Both methods are helpful in elucidating the composition of Glu-3 alleles in LMW-GS genes of bread wheat.
The effects of glutenin alleles on dough properties and processing qualities were mostly studied in two types of populations: structured populations (e.g., recombinant inbred lines (RILs) and doubled haploid lines) derived from biparental crosses, and non-structured populations, general collections of varieties and breeding lines. Due to the simple composition and easy identification of allelic variants of HMW-GS, the contributions of HMW-GS to dough properties and end-use quality were well investigated and widely used in breeding programs [23]. However, HMW-GS alone could not explain the variation in quality among wheat varieties, as LMW-GS also contributed to dough properties [24-31]. For example, LMW-GS alleles made a slightly larger contribution than HMW-GS to dough extensibility [32,33]. Compared with HMW-GS, LMW-GS formed highly polymorphic protein complex and contain abundant allelic variation. Using SDS-PAGE and allele-specific primers, LMW-GS alleles were identified in wheat collections or structured populations, and their effects on processing quality were analyzed and discussed. However, controversies were common in regard to different kinds of populations or collections. For example, Cane et al. [34] and Eagles et al. [33] reported that Glu-A3e was correlated with inferior dough resistance and extensibility, whereas Zheng et al. [35] showed that Glu-A3e was a favorable allele for dough-mixing properties. Due to the complex composition of LMW-GS alleles and difficulties in distinguishing LMW-GS from gliadins in SDS-PAGE gels, the molecular genetic mechanisms behind the functional differences of LMW-GS alleles are not well investigated.
In the present study, a set of near-isogenic lines (NILs) containing five Glu-A3 alleles, eight Glu-B3 alleles and five Glu-D3 alleles, was used to study the effects of LMW-GS on the composition of glutenin macro-polymers (GMP), dough properties, and pan bread making quality. These NILs were investigated using SDS-PAGE, the LMW-GS gene marker system and two-dimensional gel electrophoresis (2D-PAGE) for identifying the composition of LMW-GS genes and proteins in each LMW-GS allele, and analyzing their association with dough properties and bread-making quality.
Results
Separation of LMW-GS proteins in Aroona NILs using SDS-PAGE
The glutenin alleles in the flour of 16 Aroona NILs were separated by SDS-PAGE (Figure 1a). Aroona possessed five HMW-GS proteins, viz., 1, 7 + 9 and 2 + 12, encoded by genes at Glu-A1, Glu-B1 and Glu-D1 loci, respectively. All NILs had the same HMW-GS as Aroona, and their unique LMW-GS and gliadin bands were labeled in Figure 1a. Four Glu-A3 NILs possessed unique LMW-GS bands at the B-group region from Aroona (Glu-A3c). Aroona-Glu-A3d and Aroona-Glu-A3e also contained specific gliadin bands (Figure 1a). Among eight Glu-B3 NILs, Aroona (Glu-B3b), Aroona-Glu-B3a, B3f and B3g shared similar B-group proteins, whereas Aroona-Glu-B3c, B3d, B3h and B3i showed another group of electrophoretic patterns (Figure 1a). Among the latter four NILs, Aroona-Glu-B3c possessed the lowest quantity of B-group LMW-GS proteins (Additional file 1: Figure S1), especially the protein with the largest molecular weight. Four Glu-D3 NILs and Aroona shared the same B-group LMW-GS proteins (Figure 1a). Aroona-Glu-D3b and D3d had the same protein bands and contained one unique LMW-GS from Aroona. Except for one gliadin protein, the electrophoretic pattern of Aroona-Glu-D3a was the same as those of Aroona-Glu-D3b and D3d. Aroona-Glu-D3f produced a quite different protein pattern in the C-group region compared with other Glu-D3 NILs. Generally, the Glu-3 loci were tightly linked with Gli-1 loci [6-8], and it was difficult to break this linkage through genetic recombination in conventional crosses. Thus, each Glu-3 NIL not only possessed unique LMW-GS but also contained 1 or 2 specific gliadins.
Figure 1.
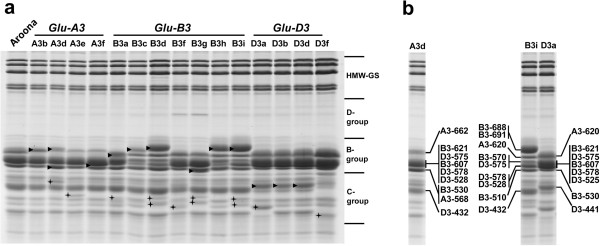
a) Separation of glutenin proteins from flour of Aroona NILs. Arrowheads mark unique LMW-GS protein bands in individual NILs, and asterisks label specific gliadin bands. b) Identification of LMW-GS bands in Aroona-Glu-A3d, Aroona-Glu-B3i and Aroona-Glu-D3a.
Dissecting genes encoding LMW-GS alleles in the Aroona NILs
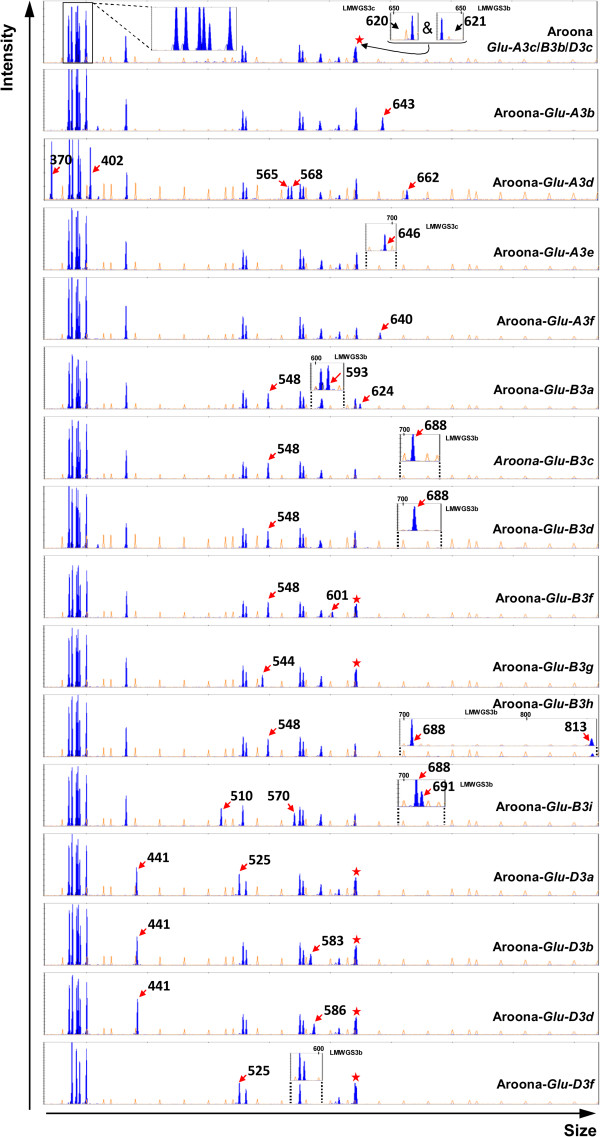
To analyze the genes encoding LMW-GS alleles in Aroona NILs, we investigated the 16 Aroona NILs using a previously developed LMW-GS gene marker system, that was efficient in separating LMW-GS genes in bread wheat (Figure 2; Table 1) [21,22]. Eighteen LMW-GS genes were identified in Aroona, including four Glu-A3 genes (A3-391, A3-400, A3-502-2 and A3-620), four Glu-B3 genes (B3-530-2, B3-578, B3-607 and B3-621-1), and eight Glu-D3 genes (D3-385, D3-393, D3-394, D3-432, D3-528, D3-575, D3-578-1 and D3-591) (Table 1). The other two novel genes corresponding to DNA fragments 388 and 410 present in all 16 lines (Table 1) were pseudogenes with premature termination codons in the CDS regions. Among 16 Aroona NILs, Aroona-Glu-D3f possessed 14 unique genes, with the least number of LMW-GS genes, whereas Aroona-Glu-A3d had the largest number of LMW-GS genes (20 genes). Comparison of the LMW-GS genes among NILs indicated that each NIL differed from Aroona at only one Glu-3 locus. With the help of the complete gene sequences available in Aroona NILs and some other wheat varieties [10,13,14,17-19,21], the composition of LMW-GS genes in each Aroona NIL was well characterized.
Figure 2.
Separation of LMW-GS genes in Aroona NILs using the LMW-GS gene molecular marker system. The marker system contained three sets of primers, LMWGS1, LMW-GS2 and LMWGS3. The main data were derived from primer set LMWGS1, and the added figures were from the primer LMWGS3b or LMWGS3c. The peaks marked with the asterisk comprise two genes, A3-620 and B3-621. The arrows indicate unique genes in each Aroona NIL.
Table 1.
LMW-GS genes identified in 16 Aroona NILs using the LMW-GS gene molecular marker system
| Line | Genes at Glu-A3 locus | Genes at Glu-B3 locus | Genes at Glu-D3 locus | New genes | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aroona(Glu-A3c, B3b, D3c) |
391 |
400 |
502-2 |
|
|
620a |
530-2 |
|
|
578 |
607 |
621-1 |
|
385 |
393 |
394 |
432 |
528 |
575 |
578-1 |
591 |
388 |
410 |
| Aroona-Glu-A3b |
391 |
400 |
502-1 |
|
|
643 |
530-2 |
|
|
578 |
607 |
621-1 |
|
385 |
393 |
394 |
432 |
528 |
575 |
578-1 |
591 |
388 |
410 |
| Aroona-Glu-A3d |
370 |
402 |
484 |
565 |
568 |
662 |
530-2 |
|
|
578 |
607 |
621-1 |
|
385 |
393 |
394 |
432 |
528 |
575 |
578-1 |
591 |
388 |
410 |
| Aroona-Glu-A3e |
391 |
400 |
502-3 |
|
|
646 |
530-2 |
|
|
578 |
607 |
621-1 |
|
385 |
393 |
394 |
432 |
528 |
575 |
578-1 |
591 |
388 |
410 |
| Aroona-Glu-A3f |
391 |
400 |
502-4 |
|
573 |
640 |
530-2 |
|
|
578 |
607 |
621-1 |
|
385 |
393 |
394 |
432 |
528 |
575 |
578-1 |
591 |
388 |
410 |
| Aroona-Glu-B3a |
391 |
400 |
502-2 |
|
|
620 |
530-1 |
548 |
|
578 |
593 |
624 |
|
385 |
393 |
394 |
432 |
528 |
575 |
578-1 |
591 |
388 |
410 |
| Aroona-Glu-B3c |
391 |
400 |
502-2 |
|
|
620 |
530-1 |
548 |
|
|
|
688-1 |
|
385 |
393 |
394 |
432 |
528 |
575 |
578-1 |
591 |
388 |
410 |
| Aroona-Glu-B3d |
391 |
400 |
502-2 |
|
|
620 |
530-1 |
548 |
|
|
|
688-2 |
|
385 |
393 |
394 |
432 |
528 |
575 |
578-1 |
591 |
388 |
410 |
| Aroona-Glu-B3f |
391 |
400 |
502-2 |
|
|
620 |
530-3 |
548 |
|
578 |
601 |
621-1 |
|
385 |
393 |
394 |
432 |
528 |
575 |
578-1 |
591 |
388 |
410 |
| Aroona-Glu-B3g |
391 |
400 |
502-2 |
|
|
620 |
530-3 |
|
|
578 |
544 |
621-2 |
|
385 |
393 |
394 |
432 |
528 |
575 |
578-1 |
591 |
388 |
410 |
| Aroona-Glu-B3h |
391 |
400 |
502-2 |
|
|
620 |
530-2 |
548 |
|
|
|
688-3 |
813 |
385 |
393 |
394 |
432 |
528 |
575 |
578-1 |
591 |
388 |
410 |
| Aroona-Glu-B3i |
391 |
400 |
502-2 |
|
|
620 |
510 |
|
570 |
|
|
688-4 |
691 |
385 |
393 |
394 |
432 |
528 |
575 |
578-1 |
591 |
388 |
410 |
| Aroona-Glu-D3a |
391 |
400 |
502-2 |
|
|
620 |
530-2 |
|
|
578 |
607 |
621-1 |
|
385 |
393 |
394 |
441 |
525 |
575 |
578-2 |
591 |
388 |
410 |
| Aroona-Glu-D3b |
391 |
400 |
502-2 |
|
|
620 |
530-2 |
|
|
578 |
607 |
621-1 |
|
385 |
393 |
394 |
441 |
528 |
575 |
578-2 |
583 |
388 |
410 |
| Aroona-Glu-D3d |
391 |
400 |
502-2 |
|
|
620 |
530-2 |
|
|
578 |
607 |
621-1 |
|
385 |
393 |
394 |
441 |
528 |
575 |
578-2 |
586 |
388 |
410 |
| Aroona-Glu-D3f | 391 | 400 | 502-2 | 620 | 530-2 | 578 | 607 | 621-1 | -- | -- | 394 | -- | 525 | 575 | 578 | -- | 388 | 410 | |||||
a Bold numbers indicate active LMW-GS genes.
For Glu-A3 alleles, 4–6 genes were identified in Aroona (Glu-A3c) and four Glu-A3 NILs (Table 1; Figure 2). Except for Aroona-Glu-A3d, these NILs possessed LMW-GS genes A3-391 and A3-400 (Table 1; Figure 2), which were m-type pseudogenes with premature termination codons. The other genes were i-type genes. The A3-502 gene contained four allelic variants with unique SNPs and InDels, i.e., A3-502-1, A3-502-2, A3-502-3 and A3-502-4. All A3-502 allelic variants and the A3-640 gene were pseudogenes, containing premature termination codons in the coding sequences. The i-type genes, A3-643, A3-620, A3-646 and A3-573, were the only active genes in Aroona-Glu-A3b, A3c, A3e and A3f, respectively. Sequence alignments showed that A3-620 and A3-643 shared high identity (> 99%) differing only in one InDel and one SNP, whereas A3-573 and A3-646 were greatly different from each other and the other i-type genes (Additional file 1: Figure S2). A3-573 was much shorter than the other i-type proteins due to deletion of three repetitive units and several glutamine residues in repetitive regions, and lacking GTFLQPH in the C-terminal domain (Additional file 1: Figure S2). Aroona-Glu-A3d contained six unique LMW-GS genes (Table 1; Figure 2). For two m-type genes, A3-370 was a pseudogene, whereas A3-402 contained an intact coding sequence that could be expressed in the developing grain [18]. The other four genes, A3-484, A3-565, A3-568 and A3-662 were i-type genes, and among them A3-568 and A3-662 were active. Both genes contained unique SNPs in the C-terminal domain and different lengths of repetitive region compared to other i-type genes (Additional file 1: Figure S2). Thus, compared to only one LMW-GS gene expressed in the other Glu-A3 alleles, Glu-A3d contained three active genes, one m-type (A3-402) and two i-type (A3-568 and A3-662) LMW-GS genes. Except for A3-402, the active genes at the Glu-A3 locus were i-type genes. These i-type genes encoded B-group LMW-GS, producing different electrophoretic patterns in the B-group region among Glu-A3 NILs (Figure 1).
For Glu-B3 alleles, 3–5 LMW-GS genes were identified in Aroona (Glu-B3b) and seven Glu-B3 NILs. Gene B3-530 was present in all Aroona NILs and four allelic variants (B3-530-1, -2, -3 and B3-510) were identified (Table 1). Sequence alignment showed that the deduced protein sequences of the three B3-530 allelic variants differed in only four amino acids, whereas B3-510 contained a deletion in the repetitive domain and differed by seven amino acids from the B3-530 proteins (Additional file 1: Figure S3). B3-544, B3-593, B3-601 and B3-607 were allelic variants, sharing over 99% of identity and their deduced protein sequences differed only in the repetitive domain with the deletion of glutamine residues or one repetitive unit (Additional file 1: Figure S4). B3-621-1, B3-621-2 and B3-624 genes were highly conserved and their proteins differed only in one InDel of a glutamine residue in the repetitive domain and one amino acid at the C terminal region (Additional file 1: Figure S5). The B3-688 gene was quite distinct, differing from B3-621 and B3-624 in several SNPs and InDels (Additional file 1: Figure S5). Moreover, B3-688 varied among Aroona-Glu-B3c, B3d, B3h and B3i, and the protein products differed in seven amino acids and two InDels (Additional file 1: Figure S5). Among Glu-B3 genes, B3-548, B3-578 and B3-813 were pseudogenes with premature termination codons, whereas the other genes contained intact ORFs (Open Reading Frame). Sequence analysis showed that B3-530, B3-510, B3-548 and B3-570 were m-type genes and the others were s-type genes. Based on the differences in s-type genes, Glu-B3 NILs were classified into two groups. Aroona-Glu-B3a, B3b, B3f and B3g contained B3-578, B3-544/593/601/607, and B3-621/624, and the others had longer sequences of genes, B3-688 and B3-691/813/Null (Table 1). This classification was consistent with the different electrophoretic patterns contrasting the two groups (Figure 1a).
Aroona-Glu-B3a, B3b, B3f and B3g had similar LMW-GS compositions, but each of them possessed unique allelic variants (Table 1; Figure 2). Both Aroona-Glu-B3f and B3g contained B3-530-3, but they differed in B3-544, B3-601, B3-621-1 and B3-621-2 genes (Table 1; Additional file 1: Figures S4 and S5).Aroona-Glu-B3f and Aroona (Glu-B3b) shared the same B3-621-1, but possessed different genes B3-601/B3-530-3 and B3-607/B3-530-2, respectively (Table 1; Additional file 1: Figures S3 and S4). On the other hand, Aroona-Glu-B3c, B3d, B3h, and B3i also differed in active genes and their allelic variants. Aroona-Glu-B3c and B3d contained B3-530-1, but their B3-688 proteins differed in five amino acids (Table 1; Additional file 1: Figure S5). Aroona-Glu-B3i was unique, containing four active LMW-GS genes B3-510, B3-570, B3-688-4 and B3-691 (Table 1; Figure 2). B3-570 was only present in Glu-B3i, encoding an m-type LMW-GS with the N-terminal domain METSQIPGLEKPS. Although B3-688-4 and B3-691 were different genes, they shared about 99% identity and B3-691 is formally reported for the first time.
At the Glu-D3 locus, eight genes were identified from individual NILs except Aroona-Glu-D3f (Table 1). D3-393 and D3-583/586/591 had premature termination codons. All the other genes were widely reported and were expressed in developing grains [10,17,18,20]. D3-385, D3-394 and D3-575 were highly conversed among the Aroona NILs. D3-432 and D3-441 were allelic variants with over 99% identity, and differed in only two SNPs and two InDels. Allelic variants D3-525 and D3-528 were identical, except for one InDel. Two allelic variants, D3-578-1 and D3-578-2 differed by eight SNPs (Table 1). Compared with the active Glu-D3 genes in Aroona (Glu-D3c), D3-441 and D3-578-2 were present in Aroona-Glu-D3b and D3d, which possessed the same active LMW-GS genes (Table 1; Figure 2). Besides D3-441 and D3-578-2, Aroona-Glu-D3a contained unique gene D3-525 in contrast to Aroona. In Aroona-Glu-D3f, only four LMW-GS genes were identified; the other four genes might be absent due to deletion, substitution or other mutations (Table 1; Figure 2).
Each Aroona NIL possessed unique LMW-GS alleles. The gene compositions of these LMW-GS alleles were dissected, all the LMW-GS gene sequences were characterized, and the proteins sequences were deduced (Table 1; Figure 2). Based on the size of the deduced LMW-GS and the LMW-GS 2D-PAGE spots identified from Xiaoyan 54, Jing 411 and Norin 61 [18,19], LMW-GS genes were assigned to protein bands in SDS-PAGE gels (Figure 1b). The active i-type genes at the Glu-A3 locus, all the active genes at Glu-B3 and the genes D3-525/528, D3-575 and D3-578 at Glu-D3 encoded B-group LMW-GS, whereas the protein products of the other active genes belonged to the C-group. Moreover, B3-621/624, B3-593/601/607, D3-575 and D3-578 encoded proteins with similar molecular weights (38–40 kDa), and ran synchronously on SDS-PAGE, forming the thickest protein bands in Aroona-Glu-B3a, B3b and B3f. B3-688 genes encoding the longest protein conferred the unique protein pattern on Aroona-GluB3d, B3h and B3i. (Figure 1).
Comparison of whole proteins among LMW-GS NILs using 2D-PAGE
The data from SDS-PAGE and the LMW-GS marker system showed that each Glu-3 allele contained several unique LMW-GS and gliadin genes. To analyze the composition of LMW-GS and gliadin proteins in individual NILs, we performed 2D-PAGE analysis of the whole flour proteins (Figures 3, 4 and 5). In the previous studies, using 2D-PAGE coupled with MS or N-terminal sequencing technology, LMW-GS proteins in Xiaoyan 54, Jing 411, Norin 61 and Butte 86 were successfully identified [18,19,36]. These data greatly contributed to the identification of flour proteins in the Aroona NILs.
Figure 3.
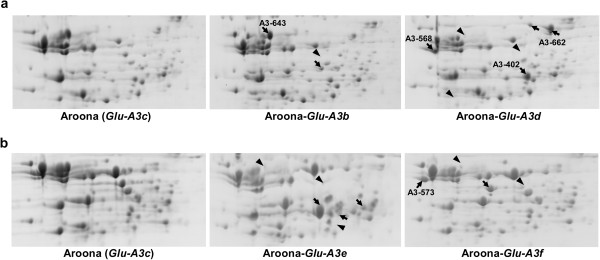
Separation of flour proteins in Glu-A3 NILs using two-dimensional gel electrophoresis (2D-PAGE). a) Comparison of storage proteins from Aroona, Aroona-Glu-A3b and Aroona-Glu-A3d. b) Comparison of storage proteins from Aroona, Aroona-Glu-A3e and Aroona-Glu-A3f. The arrows indicate the unique protein spots in each NIL, and the arrowheads show the protein spots present in Aroona but absent in other NILs. The high molecular weight glutenin subunit protein spots are the same and not shown here due to limited space.
Figure 4.
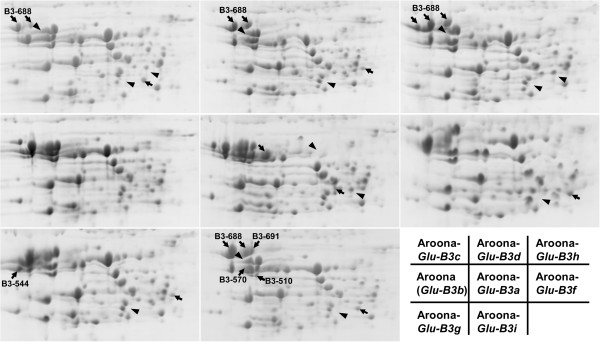
Separation of flour proteins in eight Glu-B3 NILs using two-dimensional gel electrophoresis (2D-PAGE). Arrows indicate the unique protein spots in each NIL, and the arrowheads show protein spots present in Aroona but absent in other NILs.
Figure 5.
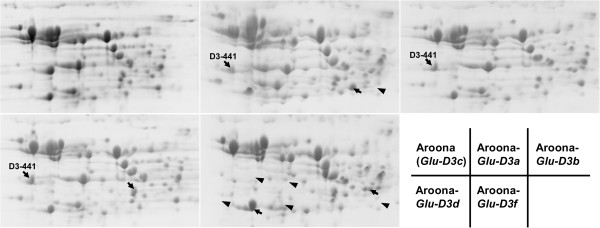
Separation of flour proteins in five Glu-D3 NILs using two-dimensional gel electrophoresis (2D-PAGE). The arrows indicate the unique protein spots in each NIL, and the arrowheads show the protein spots present in Aroona but absent in other NILs.
Among five Glu-A3 NILs, Aroona-Glu-A3b and Aroona (Glu-A3c) shared the same protein patterns, except for one gliadin spot, which was in agreement with the similar composition of LMW-GS genes and the high identity between A3-620 and A3-643 (Figure 3a). Compared with Aroona, Aroona-Glu-A3d contained three unique LMW-GS, A3-402, A3-568 and A3-662, and two gliadin spots were absent (Figure 3a). For Aroona-Glu-A3e, the protein product of the unique i-type gene A3-646 was too little to be detected, but three unique gliadin spots were present in 2D-PAGE (Figure 3b). Aroona-Glu-A3f also contained one unique i-type protein (A3-573) and one gliadin spot different from Aroona (Figure 3b). Thus, accompanying the unique LMW-GSs in individual Glu-A3 NILs, generally 1–2 gliadin spots were different among them. The only exception was Aroona-Glu-A3e which possessed the highest number of gliadins and the lowest quantity (none) of i-type LMW-GS among the Glu-A3 NILs (Figure 3b).
Among eight Glu-B3 NILs, Aroona-Glu-B3c, B3d, B3h and B3i shared similar protein spot patterns on 2D-PAGE, consistent with their similar electrophoretic patterns on SDS-PAGE (Figures 1a and 4). Compared with the protein spot pattern of Aroona (Glu-B3b), Aroona-Glu-B3c, B3d, B3h and B3i contain the unique B3-688 proteins (Figure 4). The B3-688 protein spot in Aroona-Glu-B3c was much weaker than those in the other three NILs (Additional file 1: Figure S6). Except for LMW-GS proteins, only one or two small gliadin spot differences were identified among the four NILs (Figure 4). The other three Aroona NILs (i.e., Aroona-Glu-B3a, B3f and B3g) had similar protein spot patterns with Aroona (Glu-B3b; Figure 4). Among these four NILs, a unique LMW-GS protein B3-544 was detected in Aroona-Glu-B3g, whereas the other three alleles shared the same LMW-GS protein spot patterns (Figure 4). Thus, the 2D-PAGE patterns of Aroona and seven Glu-B3 NILs suggested that their different proteins mainly were unique LMW-GS proteins rather than gliadins.
For the Glu-D3 NILs, the protein spots encoded by the D3-441 gene in Aroona-Glu-D3a, D3b and D3d had larger molecular weights and pIs than its allelic variant D3-432 in Aroona (Figure 5). No other different LMW-GS spots were detected among these Glu-D3 NILs, and Aroona-Glu-D3a and D3d, each contained only one unique gliadin spot (Figure 5). The Aroona-Glu-D3f allele was unique, lacking two LMW-GS proteins (i.e., D3-385 and D3-441) and at least three medium gliadin spots (Figure 5). The absence of four LMW-GS genes was also observed using the LMW-GS gene marker system (Table 1). Thus, except for Aroona-Glu-D3f, only one or two different protein spots were detected among Glu-D3 NILs, and the proteins encoded by genes at the Glu-D3 locus were highly conserved.
Quality properties of the LMW-GS NILs
Dough properties such as Zeleny-sedimentation value (ZSV), Farinograph and Extensograph parameters, GMP parameters, and pan bread quality parameters, were measured on the Aroona NILs (Table 2). The 16 genotypes showed significant differences (P < 0.05 or P < 0.01) in most quality parameters, suggesting that genotype had an important influence on variation in wheat quality properties (Additional file 2: Table S1). Significant differences in some parameters (e.g., kernel protein, ZSV, Farinograph water absorption, glutenin and gliadin contents, and external bread color and structure) were also observed between the two locations (Additional file 2: Table S1). However, no significant interaction effects between genotypes and locations were detected for most quality parameters, except for external color of bread and Farinograph development time (Additional file 2: Table S1). Analysis of variance (ANOVA) and multiple comparisons of all quality parameters were performed among all NILs within each Glu-3 group (Table 2, Additional file 2: Tables S2, S3 and S4).
Table 2.
F values of one way ANOVA of wheat quality parameters within each Glu-3 groupa
| Group | LV | LVS | EC | Sha | IC | Smo | Spr | Str | TF | PBTS |
|---|---|---|---|---|---|---|---|---|---|---|
| Aroona-Glu-A3 |
5.31* |
2.33 |
0.7 |
3.35 |
1.14 |
12.14** |
0.67 |
6.45* |
2 |
4.75* |
| Aroona-Glu-B3 |
2.85* |
2.07 |
0.8 |
1.57 |
1.28 |
2.09 |
0.75 |
5.09** |
2 |
5.78** |
| Aroona-Glu-D3 |
3.74 |
2.29 |
1.71 |
2.57 |
1 |
1 |
0.33 |
0.52 |
0.12 |
0.68 |
|
Group |
KP |
ZSV |
ST |
DT |
Wab |
Rmax |
EA |
Ext |
|
|
| Aroona-Glu-A3 |
3.3 |
59.35** |
6.55* |
11.57** |
27.98** |
10.34** |
13.46** |
4.51* |
|
|
| Aroona-Glu-B3 |
2.54 |
43.89** |
6.19** |
8.47** |
5.4** |
9.97** |
10.06** |
2.34 |
|
|
| Aroona-Glu-D3 |
3.01 |
0.85 |
2.47 |
3.05 |
4.04* |
2.58 |
1.18 |
2.33 |
|
|
|
Group |
Glutenin |
Gliadin |
Gli/Glu |
EPP |
UPP |
%UPP |
|
|
|
|
| Aroona-Glu-A3 |
0.28 |
3.13 |
4.62* |
2.87 |
7.1** |
9.58** |
|
|
|
|
| Aroona-Glu-B3 |
0.55 |
8.81** |
4.65** |
2.14 |
3.15* |
3.63* |
|
|
|
|
| Aroona-Glu-D3 | 3.64 | 0.32 | 6.55* | 1.51 | 1.53 | 1.48 |
* Significant at P < 0.05; ** Significant at P < 0.01.
a LV: Loaf volume (cm3), LVS: Loaf volume score, EC: External color, Sha: Shape, IC: Inner color, Smo: Smoothness, Spr: Springiness, Str: Structure, TF: Taste flavor, PBTS: Pan bread total score, KP: Kernel protein (%, 14% m.b.), ZSV: Zeleny sedimentation value (mL), ST: Farinograph stability time (min), DT: Farinograph development time (min), Wab: Farinograph water absorption (%), Rmax: Extensograph maximum resistance (B.U.), EA: Energy area (cm2), Ext: Extensograph extensibility (mm), EPP: Extractable glutenin polymeric protein; UPP: Unextractable glutenin polymeric protein; Gli/Glu: Ratio of gliadin to glutenin; %UPP = UPP/(UPP + EPP) × 100.
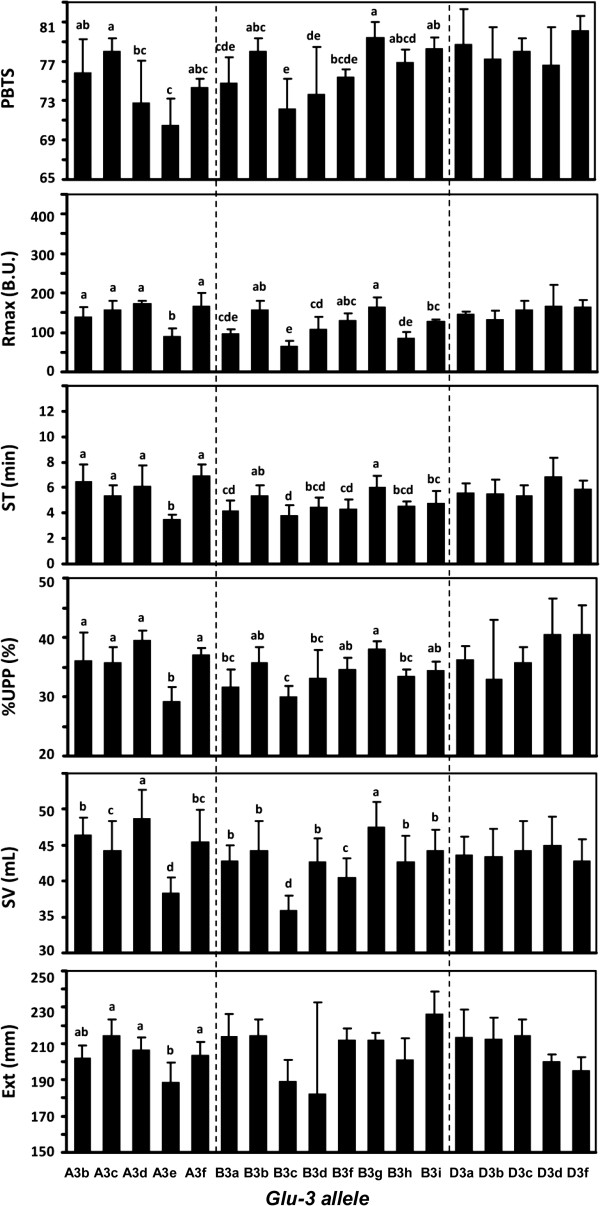
ANOVA showed that the Glu-A3 NILs were significantly different in most quality parameters (Table 2). This suggested that Glu-A3 alleles affected the bread-making quality of bread wheat. Subsequent multiple comparisons indicated that Aroona-Glu-A3e always produced the worst quality parameters, e.g., ZSV, Farinograph stability time (ST), Extensograph maximum resistance (Rmax), Extensograph extensibility (Ext), percentage of SDS-unextractable fraction in total polymeric protein (%UPP), and pan bread total score (PBTS), whereas the other five Glu-A3 NILs had similar values for Rmax, ST, %UPP and Ext (Figure 6). Although Aroona-Glu-A3d had the largest ZSV, it showed only moderate PBTS (Figure 6).
Figure 6.
Comparison of wheat quality properties among Glu-A 3NILs, Glu-B3 NILs and Glu-D3 NILs. Statistical analysis of the quality parameters of Aroona NILs for each glutenin locus by ANOVA and Duncan LSR (P < 0.05). The bars are labeled by different letters or letter combinations based on multiple statistical comparisons. No statistical significance exists between alleles labeled by one or more identical letters. PBTS, Pan bread total score; Rmax, Extensograph maximum resistance; ST, Farinograph stability time; %UPP, Percent of SDS-unextractable fraction in total polymeric protein; ZSV, Zeleny-sedimentation value; Ext, Extensograph extensibility.
Glu-B3 NILs showed large differences in the most important quality parameters (Table 2), indicating that Glu-B3 alleles were also significantly associated with bread-making quality. Among eight Glu-B3 NILs, Aroona-Glu-B3g produced the highest PBTS, ZSV, ST, Rmax, %UPP and some other parameters (Figure 6, Additional file 2: Tables S2, S3 and S4). Similarly, Aroona (Glu-B3b) and Aroona-Glu-B3i had higher quality parameters than the other Glu-B3 NILs, excluding Aroona-Glu-B3g. In contrast, Aroona-Glu-B3c performed the lowest in ZSV, ST, Rmax, %UPP, PBTS, and some other parameters (Figure 6; Additional file 2: Tables S2, S3 and S4), suggesting that Glu-B3c was an undesirable allele for bread-making quality. The other Glu-B3 NILs (Aroona-Glu-B3a, B3d, B3f and B3h) possessed similar moderate quality parameters, without significant differences.
Five Glu-D3 NILs produced similar quality parameters, without significant differences, except for Farinograph water absorption and the ratio of gliadin to glutenin (Table 2). This was consistent with the similar composition of LMW-GS genes and proteins among Glu-D3 NILs.
Discussion
In the present study, SDS-PAGE, 2D-PAGE and LMW-GS gene molecular marker system were used for characterization of LMW-GS genes and proteins in 16 Aroona NILs. Each NIL contained a unique LMW-GS allele. The genetic and protein compositions of each LMW-GS allele were dissected. In addition, we analyzed the functional differences among the LMW-GS alleles in bread-making quality using these NILs. The molecular mechanisms behind the functional differences were discussed based on the characterization of LMW-GS genes and proteins in the NILs.
Characterization of LMW-GS genes and proteins in the NILs
SDS-PAGE is a useful method to accurately identify HMW-GS in bread wheat, but is inefficient for separating LMW-GS alleles, because of the presence of large numbers of LMW-GS proteins with similar mobilities with each other and with gliadins (Figure 1b). In the present study, the LMW-GS gene marker system was successfully used to separate 14–20 LMW-GS from each NIL, and also distinguished 16 Glu-3 NILs, except for Aroona-Glu-B3c and B3d without the length polymorphisms among B3-688-1 and B3-688-2 (Table 1; Figure 2). These results indicated that the LMW-GS gene marker system was efficient and accurate not only in separating members of the LMW-GS gene family [21], but also in distinguishing allelic variants of individual LMW-GS genes. Recently, allele-specific markers were widely used in distinguishing Glu-A3 and Glu-B3 alleles [12-14]. However, the high conservation among Glu-D3 allelic variants made it difficult to develop allele-specific markers for discriminating different allelic variants. Because these allelic variants showed DNA sequence polymorphisms in length, the LMW-GS gene marker system worked well in dissecting the complex genes and allelic variants at the Glu-D3 locus (Table 1). On the other hand, compared with only one gene in individual alleles identified with gene-specific primers [13,14], almost all the genes in Glu-3 alleles were displayed using the LMW-GS gene marker system (Table 1). Identification of all genes in each allele will greatly contribute to an understanding of the molecular mechanism determining functional differences among Glu-3 alleles in bread wheat.
In the present study, Aroona and its NILs contained most of the Glu-3 allelic variations identified from the worldwide wheat germplasm in a cooperative program [12]. Each LMW-GS allele was encoded by several linked genes or haplotypes [13,14,16,17,21,22]. The dissection of LMW-GS genes in Aroona NILs would facilitate the haplotype analysis of LMW-GS genes in common wheat. At the Glu-A3 locus, besides two m-type genes, 2–4 i-type genes were identified from individual Glu-A3 alleles. Unlike the conserved m-type genes, i-type genes were completely different among five alleles. These i-type genes might be tightly linked, forming unique haplotype in each Glu-A3 allele, e.g. A3-484/A3-565/A3-568/A3-662 in Glu-A3d (Table 1) [13,18,19,21]. At the Glu-B3 locus, 1–2 m-type gene(s) and 1–3 s-type gene(s) were characterized in individual alleles. Generally, these s-type genes could be divided into two groups, one containing B3-578/B3-544/593/601/607/B3-621/624, the other having B3-688/B3-691/813/N (Table 1, Figure 2). These s-type genes in each group might cosegregate and form unique haplotypes in bread wheat [14]. Among four allelic variants of m-type gene B3-530, B3-530-2 was present in both Aroona (Glu-B3b) and Aroona-Glu-B3h, containing difference s-type haplotypes B3-578/B3-607/B3-621-1 and B3-688/B3-813, respectively. Thus, at the Glu-B3 locus, the m- and s-type genes appeared not to be tightly linked (Table 1; Figure 2) [6,18]. All the Glu-D3 alleles except Glu-D3f contained eight LMW-GS genes (Table 1). Only one or two allelic variants (D3-432/441, D3-525/528 and D3-578-1/-2 were identified for each LMW-GS gene and only a few SNPs were detected among allelic variants. Thus, Glu-D3 genes were highly conserved among wheat varieties [16,17], wherease Glu-A3 and Glu-B3 genes showed high diversities, which were derived from multiple allelic variations of i- and s-type genes, respectively.
To analyze differences in seed storage proteins among LMW-GS NILs, flour proteins were displayed using 2D-PAGE (Figures 3, 4 and 5). LMW-GS proteins shared some similar characteristics, such as mostly located in the B-group region and larger pIs than gliadins. Although it is difficult to detect small variation in molecular weight and quantity of each protein spot in 2D-PAGE, the composition and allelic variations of seed storage proteins for each Glu-3 allele were successfully characterized. These results were in agreement with the 2D protein patterns of Xiaoyan 54, Jing 411, Norin 61, Butte 86 and some other varieties identified with LC-MS or N-terminal sequencing [18,19,36]. For example, the Glu-A3d and Glu-B3d alleles showed similar electrophoretic patterns to Xiaoyan 54 and Norin 61 in i-type (A3-568 and A3-662) and s-type (B3-688) proteins, respectively [18,19], and Glu-B3g shared the same spot pattern of B3-544 with Glenlea [19]. Moreover, comparison of 2D-PAGE patterns showed that, except for the different LMW-GS proteins, only 1–3 unique small gliadin spots were present in individual Aroona NILs (Figures 3, 4 and 5). These results suggested that the differences among the Aroona NILs were mainly derived from allelic variation of LMW-GS genes.
LMW-GS alleles and bread-making quality
The NIL population was used to study the effects of LMW-GS on dough properties, GMP parameters, and pan bread quality parameters. The results confirmed that the LMW-GS played important roles in determining variation in wheat quality properties. Among the three Glu-3 loci, alleles Glu-A3 and Glu-B3 were of major importance in determining differences in processing qualities among the NILs (Table 2).
Glu-A3 alleles
Among five Glu-A3 alleles, Glu-A3e (A3-391, A3-400, A3-502-3 and A3-646) performed the poorest in almost all quality properties (Table 1, Figure 6; Additional file 2: Tables S2, S3 and S4). This was consistent with previous reports in which Glu-A3e was associated with lower extensibility and Rmax than Glu-A3d, A3b and A3c[33]. The negative effect of Glu-A3e on dough rheological properties was reported in several studies previously [24,33,37,38]. No unique i-type protein band was detected from the Glu-A3e allele using SDS-PAGE (Figure 1a) [39]. The protein product of the A3-646 gene was also not identified in 2D-PAGE although the i-type gene A3-646 in Aroona-Glu-A3e contained the intact ORF (Figure 3b, Additional file 1: Figure S2). Less i-type proteins and more gliadins in Glu-A3e increased the ratio of gliadin to glutenin and greatly reduced %UPP (Figure 6; Additional file 2: Table S4), resulting in the worst performance of the Glu-A3e genotype in dough resistance and extensibility and pan bread total score [39]. The other Glu-A3 NILs produced similar quality parameters, including Rmax, ST, Ext and %UPP. These data indicated that Glu-A3a, A3c, A3d and A3f all had equivalent positive effects on UPP content and dough resistance and extensibility (Figure 6). Among them, Glu-A3d had a significant effect on high ZSV, which was consistent with results from Xiaoyan 54 and Jing 411 RILs [18]. Some other studies also reported that the Glu-A3d allele had a superior effect on dough strength [40-42]. Compared with only one active gene at the Glu-A3 locus in the other alleles, the Glu-A3d allele possessed three active LMW-GS genes and produced the highest ZSV, Rmax, and %UPP (Figure 6). The large number of active genes in Glu-A3d might be the basis of the superior performance in wheat quality properties [18].
Glu-B3 alleles
Our study confirmed the important contribution of the Glu-B3 locus to quality variation of bread wheat varieties. Among eight Glu-B3 alleles, Glu-B3c (B3-530-1, B3-548 and B3-688-1) produced the lowest quality parameters, including ZSV, ST, Rmax, %UPP, and PBTS (Figure 6). The inferior effects of Glu-B3c allele were also reported in previous studies [24,33,43,44]. Glu-B3c and Glu-B3d (B3-530-1, B3-548 and B3-688-2) shared an identical LMW-GS composition, except for five amino acid differences between B3-688-1 and B3-688-2 (Table 1; Figure 2, Additional file 1: Figure S5), but produced significantly different Rmax and ZSV (Figure 6). B3-688-2 appeared to produce better quality properties than B3-688-1. On the other hand, Glu-B3c and Glu-B3h (B3-530-2, B3-548, B3-688-3 and B3-813) also contained similar active LMW-GS genes and had only a few SNPs in the B3-530 and B3-688 allelic variants, but Glu-B3h had significantly more positive effects on PBTS and ZSV than Glu-B3c (Figure 6). The 2D-PAGE showed that Glu-B3c had a smaller quantity of B3-688-1 protein than Glu-B3d and B3h (Figure 4, Additional file 1: Figure S6). This suggested that the unique SNPs in B3-688-1 might result in lower expression or difficulty in translation and polymerization. The small amount of B3-688-1 might directly cause the higher ratio of gliadin to glutenin and the lower %UPP for the Glu-B3c allele, finally resulting in its inferior effects on wheat quality (Additional file 2: Tables S2, S3 and S4). Compared to two active genes in the three Glu-B3 alleles above, Glu-B3i contained four active genes, m-type haplotype B3-510/B3-570 and s-type haplotype B3-688/B3-691. Both haplotypes provided the highest number of active genes and lead to the best quality performance among the four alleles [18].
The Glu-B3a, B3b, B3f and B3g alleles belonged to the same group of s-type haplotypes. They shared similar electrophoretic patterns and each possessed three active LMW-GS genes, but had significantly different effects on quality properties (Figure 6). Glu-B3g (B3-530-3, B3-548, B3-544 and B3-621-2) produced the best quality parameters, including ZSV, Rmax, ST, %UPP, LV and PBTS (Figure 6), in agreement with previous reports [40,42,45]. Glu-B3b (B3-530-2, B3-548, B3-607 and B3-621-1) also had positive effects on most quality parameters (Figure 6), again consistent with the previous studies [46,47]. The SE-HPLC parameters indicated that the low content of gliadin and the high content of UPP might be the main reasons for the superior performance of Glu-B3b and B3g (Figure 6; Additional file 2: Table S4). And the high content of UPP might be derived from the SNPs or InDels in the nucleotide sequences of three active genes (B3-530-2/3, B3-544/607 and B3-621-1/-2), which enhanced the ability of the protein products to form large glutenin macropolymers. More evidence was obtained by comparing Glu-B3f and Glu-B3g. Both alleles contained the same m-type gene, B3-530-3, but had different s-type haplotypes and significant differences in ZSV, ST, UPP, and PBTS. The s-type haplotype B3-578/B3-544/B3-621-2 formed more UPP and produced better bread-making quality than B3-578/B3-601/B3-621-1.
Glu-D3 alleles
The five Glu-D3 alleles produced similar values for almost all quality properties in the present study (Figure 6; Table 2), which confirmed previous reports that Glu-D3 alleles produced similar Rmax and extensibilities among large collections of wheat varieties [24,33,48], although some studies indicated different effects of Glu-D3 alleles on dough strength or mixing properties [27,29,38,49]. Glu-D3a, D3b, D3c and D3d contained similar 2D-PAGE spot patterns, and all six active genes were highly conserved among the alleles (> 99% identities). Their similarity in LMW-GS genes and whole proteins was consistent with their equivalent effects on all quality properties. Although Glu-D3f allele lacks two LMW-GS proteins and three gliadin spots, it produced similar quality properties to the other Glu-D3 alleles (Figure 5; Table 2). These results suggested that D3-385, D3-432 and the three missing gliadins were not related to quality improvement. However, the lack of active genes D3-394 and D3-528 at the Glu-D3 locus in Jing 411 (Glu-D3l) showed significant negative effects on ZSV [18]. Thus, except for Glu-D3l, Glu-D3 alleles appeared to play only minor roles in determining quality variation among bread wheat varieties, and they should be given the lowest priority among LMW-GS alleles in selecting for improved bread-making quality in wheat.
Conclusion
In the present study, we dissected the genetic and protein composition of 16 LMW-GS NILs, measured the dough property and bread-making quality properties of individual NILs, and performed functional analyses for each allele. Among five Glu-A3 alleles, Glu-A3e (i-type haplotype A3-502-3/A3-646) was inferior with negative effects on all quality properties. Among eight Glu-B3 alleles, Glu-B3b (m-type gene B3-530-2 and s-type haplotype B3-578/B3-607/B3-621-1), Glu-B3g (m-type gene B3-530-3 and s-type haplotype B3-578/B3-544/B3-621-2) and Glu-B3i (m-type haplotype B3-510/B3-570 and s-type haplotype B3-688-4/B3-691) were correlated with superior bread-making quality, whereas Glu-B3c (m-type gene B3-530-1 and s-type haplotype B3-688-1/N) produced inferior quality properties. Among five Glu-D3 alleles, there were no significant differences in all quality parameters measured in the present study. Moreover, all alleles with superior dough properties and pan bread quality also possessed high contents of UPP and %UPP. Thus, it is possible that LMW-GS alleles determine dough viscoelasticity by modifying the size distribution of glutenin polymers and aggregative properties of glutenins [50]. These results significantly enhance our understanding of the composition of LMW-GS, confirm the strong effects of LMW-GS on not only dough extensibility but dough strength, and provide useful information for quality improvement in bread wheat.
Methods
Plant materials
The wheat variety Aroona and 15 near isogenic lines (NILs) were kindly provided by Dr. Marie Appelbee and Prof. Ken Shepherd, SARDI Grain Quality Research Laboratory, South Australia. Each NIL contains a unique LMW-GS allele from a donor variety added to Aroona (Additional file 2: Table S5). They were planted at the Xinjiang Academy of Agri-Reclamation Sciences, Shihezi, and Xinjiang Academy of Agricultural Sciences, Urumqi, Xinjiang province, in randomized complete blocks with two replications during the 2010 cropping season.
Analysis of LMW-GS genes
Genomic DNA of 16 Aroona NILs was extracted from young leaves of seedlings following Saghai-Maroof et al. [51]. The LMW-GS genes were separated by the LMW-GS gene molecular marker system [22]. With the help of the LMW-GS genes available from the Aroona NILs and some other wheat varieties [10,13,14,17,18], LMW-GS genes were characterized using Lasergene software (DNAStar; http://www.dnastar.com/), ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/), and MEGA 5 software [52].
Isolation and separation of LMW-GS proteins
Glutenin extraction was performed according to the method described by Singh et al. [11]. These proteins were separated by SDS-PAGE using the method described by Sunbrook and Russell [53]. Whole seed proteins were isolated from wheat flour based on the SDS/Phenol method [54] with some modifications. Briefly, proteins in 0.12 g flour were precipitated with 10% TCA/acetone at −20°C overnight. After centrifuging at 20,000 g at 4°C for 15 min, the pellet was washed three times with 80% acetone, then dried at 50°C. Whole proteins were extracted with SDS/Phenol buffer (50% Tris-phenol pH 8.0, 30% sucrose, 2% SDS, 0.1 M Tris–HCl pH 8.0 and 2% DTT). The upper phenol phase was transferred into a new 2 mL tube. A fivefold volume of methanol containing 0.1 M ammonium acetate was added to the tube. The proteins were deposited at −20°C for 10 min or overnight. After centrifuging at 20,000 g for 5 min at 4°C, the pellet was washed once with 100% methanol and twice with 80% acetone before briefly drying in air. The proteins were dissolved in isoelectric focusing (IEF) sample extraction solution and used in 2D-PAGE analysis according to Dong et al. [18]. The images of SDS-PAGE were analyzed using NIH ImageJ software program (http://rsb.info.nih.gov/ij/).
Quality testing and evaluation of pan bread
Measures of grain hardness, protein content, Zeleny sedimentation values, Farinograph and Extensograph parameters, and pan bread qualities were performed by methods reported in He et al. [55]. The glutenin macro-polymer compositions were measured following Zhang et al. [56].
Statistical analysis
The SAS statistical package (SAS Institute, Cary, NC) was used for data analysis. All statistical analyses were based on averaged data from two locations.
Abbreviations
2D-PAGE: Two-dimensional gel electrophoresis (IEF × SDS-PAGE); %UPP: UPP/(UPP + EPP) × 100; ANOVA: Analysis of variance; CIMMYT: International maize and wheat improvement center; cMs: Centimorgans; DT: Farinograph development time (min); DTT: Dithiothreitol; EA: Energy area (cm2); EC: External color; EPP: Extractable glutenin polymeric protein; Ext: Extensograph extensibility (mm); Gli/Glu: Ratio of gliadin to glutenin; GMP: Glutenin macro-polymers; HMW-GS: High-molecular-weight glutenin subunits; HPLC: High-performance liquid chromatography; IC: Inner color; IEF: Isoelectric focusing; kDa: Kilodalton; KP: Kernel protein (%, 14% m.b.); LMW-GS: Low-molecular-weight glutenin subunits; LV: Loaf volume (cm3); LVS: Loaf volume score; NIL: Near-isogenic lines; ORFs: Open reading frame; PBTS: Pan bread total score; PCR: Polymerase chain reaction; pI: Isoelectric points; RILs: Recombinant inbred lines; Rmax: Extensograph maximum resistance (B.U.); SDS: Sodium dodecyl sulfate; SDS-PAGE: Sodium dodecyl sulphate-polyacrylamide gel electrophoresis; Sha: Shape; Smo: Smoothness; Spr: Springiness; ST: Farinograph stability time (min); Str: Structure; TCA: Trichloroacetic acid; TF: Taste flavor; Tris–HCl: Tris (hydroxymethyl) aminomethane hydrochloride; UPP: Unextractable glutenin polymeric protein; Wab: Farinograph water absorption (%); ZSV: Zeleny sedimentation value (mL).
Authors’ contributions
XZ carried out SDS-PAGE, 2D-PAGE and LMW-GS gene marker system analyses and drafted the manuscript. HJ measured glutenin macro-polymers, dough properties and pan bread quality parameters and performed statistical analysis. YZ participated in the measurement of processing quality parameters. DL participated in the identification of LMW-GS genes and proteins with SDS-PAGE, 2D-PAGE and LMW-GS gene marker system and revised the manuscript. GL participated in the design of the study and quality parameter analysis. XX participated in the design of the study and revised the manuscript. ZH conceived of the study and participated in its design, coordinated the various research groups, and revised the manuscript. AZ conceived of the study and participated in its design, and revised the manuscript. All authors read and approved the final manuscript.
Financial competing interests
· In the past five years have you received reimbursements, fees, funding, or salary from an organization that may in any way gain or lose financially from the publication of this manuscript, either now or in the future? Is such an organization financing this manuscript (including the article-processing charge)? If so, please specify.
No.
· Do you hold any stocks or shares in an organization that may in any way gain or lose financially from the publication of this manuscript, either now or in the future? If so, please specify.
No.
· Do you hold or are you currently applying for any patents relating to the content of the manuscript? Have you received reimbursements, fees, funding, or salary from an organization that holds or has applied for patents relating to the content of the manuscript? If so, please specify.
No.
· Do you have any other financial competing interests? If so, please specify.
No.
Non-financial competing interests
Are there any non-financial competing interests (political, personal, religious, ideological, academic, intellectual, commercial or any other) to declare in relation to this manuscript? If so, please specify.
No.
Supplementary Material
Quantitative gel densitometry measurements of B-group proteins in Glu-B3 NILs as determined using NIL ImageJ software program. Data points were normalized with respect to the Aroona value. Figure S2: Sequence alignments of i-type proteins identified in Aroona NILs. Figure S3: Sequence alignments of the B3-530 protein and its allelic variants identified in Aroona NILs. Figure S4: Sequence alignments of the B3-544/593/601/607 proteins identified in Aroona NILs. Figure S5: Sequence alignments of the B3-621/624 and B3-688 proteins identified in Aroona NILs. Figure S6: The differential display of B3-688 spots among Aroona-Glu-B3c, B3d and B3h. Spot volume values are expressed in percentages (%vol) of the total proteome.
F values of one way ANOVA of wheat quality properties by locations and genotypes. Table S2: Comparison of pan bread quality properties among Aroona NILs. Table S3: Comparison of dough properties among Aroona NILs. Table S4: Comparison of glutenin macro-polymer properties among Aroona NILs. Table S5: Composition of glutenin alleles in Aroona and its NILs.
Contributor Information
Xiaofei Zhang, Email: xfzhangcas@gmail.com.
Hui Jin, Email: jinhui1986913@yahoo.com.cn.
Yan Zhang, Email: zhangyan2012@caas.net.cn.
Dongcheng Liu, Email: dcliucas@gmail.com.
Genying Li, Email: LGY111@126.com.
Xianchun Xia, Email: xiaxianchun1963@163.com.
Zhonghu He, Email: zhhecaas@163.com.
Aimin Zhang, Email: amzhang@genetics.ac.cn.
Acknowledgement
The authors are grateful to Prof. Robert McIntosh, University of Sydney, for reviewing this manuscript. We thank Drs Marie Appelbee and Ken Shepherd, SARDI Grain Quality Research Laboratory, Adelaide, South Australia, for seed of the spring wheat variety Aroona and near-isogenic derivatives. This study was supported by the National Basic Research Program (2009CB118300), the National Science Foundation of China (30830072), International Collaboration Project from Ministry of Agriculture (2011-G3), and China Agriculture Research System (CARS-3-1-3).
References
- Osborne TB. The vegetable proteins. London: Longmans Green and Co; 1924. p. 154. [Google Scholar]
- Wasik RJ, Bushuk W. Sodium dodecyl sulfate-polyacrylamide gel-electrophoresis of reduced glutenin of durum wheats of different spaghetti-making quality. Cereal Chem. 1975;52(3):328–334. [Google Scholar]
- Payne PI. Genetics of wheat storage proteins and the effect of allelic variation on bread-making quality. Ann Rev Plant Physiol. 1987;38:141–153. doi: 10.1146/annurev.pp.38.060187.001041. [DOI] [Google Scholar]
- D'Ovidio R, Masci S. The low-molecular-weight glutenin subunits of wheat gluten. J Cereal Sci. 2004;39(3):321–339. doi: 10.1016/j.jcs.2003.12.002. [DOI] [Google Scholar]
- Shewry PR, Tatham AS, Barro F, Barcelo P, Lazzeri P. Biotechnology of breadmaking: unraveling and manipulating the multi-protein gluten complex. Nat Biotechnol. 1995;13(11):1185–1190. doi: 10.1038/nbt1195-1185. [DOI] [PubMed] [Google Scholar]
- Gao S, Gu YQ, Wu J, Coleman-Derr D, Huo N, Crossman C, Jia J, Zuo Q, Ren Z, Anderson OD. et al. Rapid evolution and complex structural organization in genomic regions harboring multiple prolamin genes in the polyploid wheat genome. Plant Mol Biol. 2007;65(1–2):189–203. doi: 10.1007/s11103-007-9208-1. [DOI] [PubMed] [Google Scholar]
- Pogna NE, Autran JC, Mellini F, Lafiandra D, Feillet P. Chromosome 1B encoded gliadins and glutenin subunits in durum wheat: genetics and relationship to gluten strength. J Cereal Sci. 1990;11(1):15–34. doi: 10.1016/S0733-5210(09)80178-1. [DOI] [Google Scholar]
- Ruiz M, Carrillo JM. Linkage relationships between prolamin genes on chromosome 1A and chromosome 1B of durum wheat. Theor Appl Genet. 1993;87(3):353–360. doi: 10.1007/BF01184923. [DOI] [PubMed] [Google Scholar]
- Cassidy BG, Dvorak J, Anderson OD. The wheat low-molecular-weight glutenin genes: characterization of six new genes and progress in understanding gene family structure. Theor Appl Genet. 1998;96(6–7):743–750. [Google Scholar]
- Huang XQ, Cloutier S. Molecular characterization and genomic organization of low molecular weight glutenin subunit genes at the Glu-3 loci in hexaploid wheat (Triticum aestivum L.) Theor Appl Genet. 2008;116(7):953–966. doi: 10.1007/s00122-008-0727-1. [DOI] [PubMed] [Google Scholar]
- Singh NK, Shepherd KW, Cornish GB. A simplified SDS-PAGE procedure for separating LMW subunits of glutenin. J Cereal Sci. 1991;14(3):203–208. doi: 10.1016/S0733-5210(09)80039-8. [DOI] [Google Scholar]
- Liu L, Ikeda TM, Branlard G, Pena RJ, Rogers WJ, Lerner SE, Kolman MA, Xia X, Wang L, Ma W. et al. Comparison of low molecular weight glutenin subunits identified by SDS-PAGE, 2-DE. MALDI-TOF-MS and PCR in common wheat. BMC Plant Biol. 2010;10:124. doi: 10.1186/1471-2229-10-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Li GY, Pena RJ, Xia XC, He ZH. Development of STS markers and establishment of multiplex PCR for Glu-A3 alleles in common wheat (Triticum aestivum L.) J Cereal Sci. 2010;51(3):305–312. doi: 10.1016/j.jcs.2010.01.005. [DOI] [Google Scholar]
- Wang LH, Zhao XL, He ZH, Ma W, Appels R, Pena RJ, Xia XC. Characterization of low-molecular-weight glutenin subunit Glu-B3 genes and development of STS markers in common wheat (Triticum aestivum L.) Theor Appl Genet. 2009;118(3):525–539. doi: 10.1007/s00122-008-0918-9. [DOI] [PubMed] [Google Scholar]
- Zhang W, Gianibelli MC, Rampling LR, Gale KR. Characterisation and marker development for low molecular weight glutenin genes from Glu-A3 alleles of bread wheat (Triticum aestivum. L) Theor Appl Genet. 2004;108(7):1409–1419. doi: 10.1007/s00122-003-1558-8. [DOI] [PubMed] [Google Scholar]
- Zhao XL, Xia XC, He ZH, Gale KR, Lei ZS, Appels R, Ma W. Characterization of three low-molecular-weight Glu-D3 subunit genes in common wheat. Theor Appl Genet. 2006;113(7):1247–1259. doi: 10.1007/s00122-006-0379-y. [DOI] [PubMed] [Google Scholar]
- Zhao XL, Xia XC, He ZH, Lei ZS, Appels R, Yang Y, Sun QX, Ma W. Novel DNA variations to characterize low molecular weight glutenin Glu-D3 genes and develop STS markers in Common Wheat. Theor Appl Genet. 2007;114(3):451–460. doi: 10.1007/s00122-006-0445-5. [DOI] [PubMed] [Google Scholar]
- Dong LL, Zhang XF, Liu DC, Fan HJ, Sun JZ, Zhang ZJ, Qin HJ, Li B, Hao ST, Li ZS. et al. New insights into the organization, recombination, expression and functional mechanism of low molecular weight glutenin subunit genes in bread wheat. PLoS One. 2010;5(10):e13548. doi: 10.1371/journal.pone.0013548. doi:10.1371/journal.pone.0013548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda TM, Araki E, Fujita Y, Yano H. Characterization of low-molecular-weight glutenin subunit genes and their protein products in common wheats. Theor Appl Genet. 2006;112(2):327–334. doi: 10.1007/s00122-005-0131-z. [DOI] [PubMed] [Google Scholar]
- Ikeda TM, Nagamine T, Fukuoka H, Yano H. Identification of new low-molecular-weight glutenin subunit genes in wheat. Theor Appl Genet. 2002;104(4):680–687. doi: 10.1007/s001220100756. [DOI] [PubMed] [Google Scholar]
- Zhang XF, Liu DC, Jiang W, Guo XL, Yang WL, Sun JZ, Ling HQ, Zhang AM. PCR-based isolation and identification of full-length low-molecular-weight glutenin subunit genes in bread wheat (Triticum aestivum L.) Theor Appl Genet. 2011;123(8):1293–1305. doi: 10.1007/s00122-011-1667-8. [DOI] [PubMed] [Google Scholar]
- Zhang XF, Liu DC, Yang WL, Liu KF, Sun JZ, Guo XL, Li YW, Wang DW, Ling HQ, Zhang AM. Development of a new marker system for identifying the complex members of the low-molecular-weight glutenin subunit gene family in bread wheat (Triticum aestivum L.) Theor Appl Genet. 2011;122(8):1503–1516. doi: 10.1007/s00122-011-1550-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne PI, Nightingale MA, Krattiger AF, Holt LM. The relationship between HMW glutenin subunit composition and the bread-making quality of British grown wheat varieties. J Sci Food Agr. 1987;40(1):51–65. doi: 10.1002/jsfa.2740400108. [DOI] [Google Scholar]
- Branlard G, Dardevet M, Saccomano R, Lagoutte F, Gourdon J. Genetic diversity of wheat storage proteins and bread wheat quality. Euphytica. 2001;119(1–2):59–67. [Google Scholar]
- Brett GM, Mills ENC, Tatham AS, Fido RJ, Shewry PR, Morgan MRA. Immunochemical identification of LMW subunits of glutenin associated with bread-making quality of wheat flours. Theor Appl Genet. 1993;86(4):442–448. doi: 10.1007/BF00838559. [DOI] [PubMed] [Google Scholar]
- Flaete NES, Uhlen AK. Association between allelic variation at the combined Gli-1, Glu-3 loci and protein quality in common wheat (Triticum aestivum L.) J Cereal Sci. 2003;37(2):129–137. doi: 10.1006/jcrs.2001.0447. [DOI] [Google Scholar]
- Gupta RB, Bekes F, Wrigley CW. Prediction of physical dough properties from glutenin subunit composition in bread wheats - correlation studies. Cereal Chem. 1991;68(4):328–333. [Google Scholar]
- Gupta RB, Paul JG, Cornish GB, Palmer GA, Bekes F, Rathjen AJ. Allelic variation at glutenin subunit and gliadin loci, Glu-1, Glu-3 and Gli-1, of Common Wheat. 1. Its additive and interaction effects on dough properties. J Cereal Sci. 1994;19(1):9–17. doi: 10.1006/jcrs.1994.1003. [DOI] [Google Scholar]
- Luo C, Griffin WB, Branlard G, McNeil DL. Comparison of low- and high molecular-weight wheat glutenin allele effects on flour quality. Theor Appl Genet. 2001;102(6–7):1088–1098. [Google Scholar]
- Nietotaladriz MT, Perretant MR, Rousset M. Effect of gliadins and HMW and LMW subunits of glutenin on dough properties in the F-6 recombinant inbred lines from a bread wheat cross. Theor Appl Genet. 1994;88(1):81–88. doi: 10.1007/BF00222398. [DOI] [PubMed] [Google Scholar]
- Nagamine T, Kai Y, Takayama T, Yanagisawa T, Taya S. Allelic variation at the Glu-1 and Glu-3 loci in southern Japanese wheats, and its effects on gluten properties. J Cereal Sci. 2000;32(2):129–135. doi: 10.1006/jcrs.2000.0323. [DOI] [Google Scholar]
- Bekes F, Kemeny S, Morell M. An integrated approach to predicting end-product quality of wheat. Eur J Agron. 2006;25(2):155–162. doi: 10.1016/j.eja.2006.04.008. [DOI] [Google Scholar]
- Eagles HA, Hollamby GJ, Gororo NN, Eastwood RF. Estimation and utilisation of glutenin gene effects from the analysis of unbalanced data from wheat breeding programs. Aust J Agr Res. 2002;53(4):367–377. doi: 10.1071/AR01074. [DOI] [Google Scholar]
- Cane K, Sharp PJ, Eagles HA, Eastwood RF, Hollamby GJ, Kuchel H, Lu M, Martin PJ. The effects on grain quality traits of a grain serpin protein and the VPM1 segment in southern Australian wheat breeding. Aust J Agr Res. 2008;59(10):883–890. doi: 10.1071/AR08114. [DOI] [Google Scholar]
- Zheng SS, Byrne PF, Bai GH, Shan XY, Reid SD, Haley SD, Seabourn BW. Association analysis reveals effects of wheat glutenin alleles and rye translocations on dough-mixing properties. J Cereal Sci. 2009;50(2):283–290. doi: 10.1016/j.jcs.2009.06.008. [DOI] [Google Scholar]
- Dupont FM, Vensel WH, Tanaka CK, Hurkman WJ, Altenbach SB. Deciphering the complexities of the wheat flour proteome using quantitative two-dimensional electrophoresis, three proteases and tandem mass spectrometry. Proteome Sci. 2011;9:10. doi: 10.1186/1477-5956-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RB, Singh NK, Shepherd KW. The cumulative effect of allelic variation in LMW and HMW glutenin subunits on dough properties in the progeny of two bread wheats. Theor Appl Genet. 1989;77(1):57–64. doi: 10.1007/BF00292316. [DOI] [PubMed] [Google Scholar]
- Park CS, Kang C-S, Jeung J-U, Woo S-H. Influence of allelic variations in glutenin on the quality of pan bread and white salted noodles made from Korean wheat cultivars. Euphytica. 2011;180(2):235–250. doi: 10.1007/s10681-011-0385-2. [DOI] [Google Scholar]
- Gupta RB, Macritchie F. Allelic variation at glutenin subunit and gliadin loci, Glu-1, Glu-3 and Gli-1 of Common Wheats. 2. Biochemical basis of the allelic effects on dough properties. J Cereal Sci. 1994;19(1):19–29. doi: 10.1006/jcrs.1994.1004. [DOI] [Google Scholar]
- Ito M, Fushie S, Maruyama-Funatsuki W, Ikeda TM, Nishio Z, Nagasawa K, Tabiki T, Yamauchi H. Effect of allelic variation in three glutenin loci on dough properties and bread-making qualities of winter wheat. Breeding Sci. 2011;61(3):281–287. doi: 10.1270/jsbbs.61.281. [DOI] [Google Scholar]
- Liu L, He ZH, Yan J, Zhang Y, Xia XC, Pena RJ. Allelic variation at the Glu-1 and Glu-3 loci, presence of the 1B.1R translocation, and their effects on mixographic properties in Chinese bread wheats. Euphytica. 2005;142(3):197–204. doi: 10.1007/s10681-005-1682-4. [DOI] [Google Scholar]
- Meng XG, Xie F, Shang XW, An LZ. Association between allelic variations at the Glu-3 loci and wheat quality traits with Lanzhou Alkaline Stretched Noodles quality in northwest China spring wheats. Cereal Res Commun. 2007;35(1):109–118. doi: 10.1556/CRC.35.2007.1.13. [DOI] [Google Scholar]
- Bekes F, Gras PW, Anderssen RS, Appels R. Quality traits of wheat determined by small-scale dough testing methods. Aust J Agr Res. 2001;52(11–12):1325–1338. [Google Scholar]
- Ma W, Appels R, Bekes F, Larroque O, Morell MK, Gale KR. Genetic characterisation of dough rheological properties in a wheat doubled haploid population: additive genetic effects and epistatic interactions. Theor Appl Genet. 2005;111(3):410–422. doi: 10.1007/s00122-005-2001-0. [DOI] [PubMed] [Google Scholar]
- Martinez-Cruz E, Espitia-Rangel E, Villasenor-Mir HE, Molina-Galan JD, Benitez-Riquelme I, Santacruz-Varela A, Pena-Bautista RJ. Dough rheology of wheat recombinant lines in relation to allelic variants of Glu-1 and Glu-3 loci. Cereal Res Commun. 2011;39(3):386–393. doi: 10.1556/CRC.39.2011.3.8. [DOI] [Google Scholar]
- Li Y, Zhou R, Branlard G, Jia J. Development of introgression lines with 18 alleles of glutenin subunits and evaluation of the effects of various alleles on quality related traits in wheat (Triticum aestivum L.) J Cereal Sci. 2010;51(1):127–133. doi: 10.1016/j.jcs.2009.10.008. [DOI] [Google Scholar]
- Tabiki T, Ikeguchi S, Ikeda TM. Effects of high-molecular-weight and low-molecular-weight glutenin subunit alleles on common wheat flour quality. Breeding Sci. 2006;56(2):131–136. doi: 10.1270/jsbbs.56.131. [DOI] [Google Scholar]
- Liu L, He ZH, Ma WJ, Liu JJ, Xia XC, Pena RJ. Allelic variation at the Glu-D3 locus in Chinese bread wheat and effects on dough properties, pan bread and noodle qualities. Cereal Res Commun. 2009;37(1):57–64. doi: 10.1556/CRC.37.2009.1.7. [DOI] [Google Scholar]
- Gobaa S, Bancel E, Branlard G, Kleijer G, Stamp P. Proteomic analysis of wheat recombinant inbred lines: Variations in prolamin and dough rheology. J Cereal Sci. 2008;47(3):610–619. doi: 10.1016/j.jcs.2007.07.001. [DOI] [Google Scholar]
- Popineau Y, Cornec M, Lefebvre J, Marchylo B. Influence of high M(r) glutenin subunits on glutenin polymers and rheological properties of glutens and gluten subfractions of near-isogenic lines of wheat Sicco. J Cereal Sci. 1994;19(3):231–241. doi: 10.1006/jcrs.1994.1030. [DOI] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Nat Acad Sci USA. 1984;81(24):8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9(4):299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Wang W, Vignani R, Scali M, Cresti M. A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis. 2006;27(13):2782–2786. doi: 10.1002/elps.200500722. [DOI] [PubMed] [Google Scholar]
- He ZH, Liu L, Xia XC, Liu JJ, Pena RJ. Composition of HMW and LMW glutenin subunits and their effects on dough properties, pan bread, and noodle quality of Chinese bread wheats. Cereal Chem. 2005;82(4):345–350. doi: 10.1094/CC-82-0345. [DOI] [Google Scholar]
- Zhang PP, He ZH, Zhang Y, Xia XC, Chen DS, Zhang Y. Association between % SDS-unextractable polymeric protein (%UPP) and end-use quality in Chinese bread wheat cultivars. Cereal Chem. 2008;85(5):696–700. doi: 10.1094/CCHEM-85-5-0696. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantitative gel densitometry measurements of B-group proteins in Glu-B3 NILs as determined using NIL ImageJ software program. Data points were normalized with respect to the Aroona value. Figure S2: Sequence alignments of i-type proteins identified in Aroona NILs. Figure S3: Sequence alignments of the B3-530 protein and its allelic variants identified in Aroona NILs. Figure S4: Sequence alignments of the B3-544/593/601/607 proteins identified in Aroona NILs. Figure S5: Sequence alignments of the B3-621/624 and B3-688 proteins identified in Aroona NILs. Figure S6: The differential display of B3-688 spots among Aroona-Glu-B3c, B3d and B3h. Spot volume values are expressed in percentages (%vol) of the total proteome.
F values of one way ANOVA of wheat quality properties by locations and genotypes. Table S2: Comparison of pan bread quality properties among Aroona NILs. Table S3: Comparison of dough properties among Aroona NILs. Table S4: Comparison of glutenin macro-polymer properties among Aroona NILs. Table S5: Composition of glutenin alleles in Aroona and its NILs.