Abstract
Mammalian circadian clocks restrict cell proliferation to defined time windows, but the mechanism and consequences of this interrelationship are not fully understood. Previously we identified the multifunctional nuclear protein NONO as a partner of circadian PERIOD (PER) proteins. Here we show that it also conveys circadian gating to the cell cycle, a connection surprisingly important for wound healing in mice. Specifically, although fibroblasts from NONO-deficient mice showed approximately normal circadian cycles, they displayed elevated cell doubling and lower cellular senescence. At a molecular level, NONO bound to the p16-Ink4A cell cycle checkpoint gene and potentiated its circadian activation in a PER protein-dependent fashion. Loss of either NONO or PER abolished this activation and circadian expression of p16-Ink4A and eliminated circadian cell cycle gating. In vivo, lack of NONO resulted in defective wound repair. Because wound healing defects were also seen in multiple circadian clock-deficient mouse lines, our results therefore suggest that coupling of the cell cycle to the circadian clock via NONO may be useful to segregate in temporal fashion cell proliferation from tissue organization.
Keywords: keratinocyte, p54nrb, RNA-binding protein, paraspeckle protein
The circadian clock adapts organisms to their daily surroundings both behaviorally and physiologically. In animals, not only are complex behaviors such as sleep and mood governed by this oscillator, but also different body functions such as digestion, circulation, and respiration (1). The basic mechanism of this clock is cell-autonomous in all studied species possessing a circadian clock. In mammals, individual clocks in most cells are synchronized by a brain “master clock” in the suprachiasmatic nucleus of the hypothalamus to orchestrate all rhythmic physiology (2). On a cellular level, circadian physiology extends even to processes such as proliferation (3–7), apoptosis (8), and DNA damage repair (6, 9), which are thought to play important roles in cancer control (8, 10).
In individual cells, the circadian clock mechanism consists of oscillating feedback loops of transcription of “core” oscillator genes and posttranslational modifications of their protein products that regulate protein stability, activity, and/or localization. For example, in mammals the transcription of periods (Per) and cryptochomes (Cry) are activated by BMAL1:CLOCK heterodimers at cis-acting elements called E-boxes, and their protein products form complexes that repress their own transcription (11). We originally identified the RNA-binding protein NONO (also called p54nrb) biochemically as a new member of this circadian transcriptional repressor complex in mice, and mutation of its ortholog NonA in flies resulted in severe attenuation of circadian rhythmicity (12). However, apart from its interaction with this circadian repressor complex, NONO’s mechanism of action within the clock remains unknown.
The mechanism of the cell cycle has been reviewed extensively elsewhere (13, 14). Rather than having a fixed duration, its period is tightly regulated by cellular processes via checkpoint proteins that gate cell cycle progression. The circadian clock is one such regulatory process, and it has been shown that the circadian clock can directly time cell division (3, 4), although not necessarily in all cell types (15, 16). The exact mechanism of this regulation is at the moment only partially understood and may involve circadian regulation of cell cycle checkpoint genes like wee1 (3) and p21-Waf1 (17), or interaction of clock factors with CSK-homologous kinase 1/2 (CHK1/2) proteins (18, 19). In addition, a limited number of accessory factors have been assigned roles in both processes, including the mammalian Timeless protein (TIM), which interacts with both the circadian CRY2 protein and the cell cycle CHK1 protein (18), and p54nrb/NONO, which interacts with proteins involved in DNA damage repair (20, 21).
To understand the role of the NONO protein in the circadian clock and its relationship to the cell cycle, we created NONO-deficient mice. Although these mice showed only minor circadian defects (22), circadian gating by the cell cycle was lost in fibroblasts from these mice, allowing us to probe both the mechanism and the physiological relevance of the connection between the two processes. Our investigations not only uncover a unique role for NONO in the circadian transcriptional regulation of the p16-Ink4A cell cycle checkpoint gene, but also suggest that circadian gating of the cell cycle by NONO plays an unsuspected role in tissue regeneration.
Results
Fibroblasts from Nonogt Mice Show Increased Proliferation and Decreased Senescence.
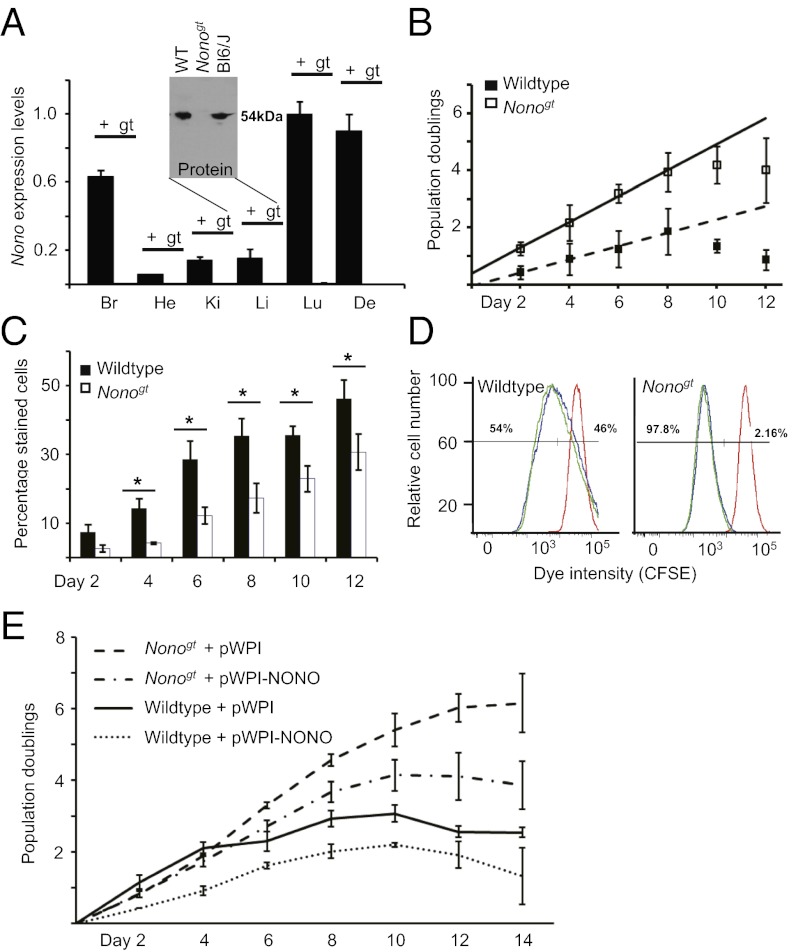
To better understand the function of NONO, we used a “genetrapped” embryonic stem cell line (23) to generate Nonogt mice that completely lack coding Nono transcript and NONO protein in all tissues tested (Fig. 1A). In addition to a circadian phenotype of slight period shortening in animal behavior and cellular gene expression that was expected from our previous studies (22), we noticed that the primary fibroblasts taken to characterize these mice showed more robust division than their WT counterparts. To quantify this effect, we serially split cells from WT and Nonogt littermates at a predetermined frequency so that confluence was not reached and counted cells at each passage. Nonogt cells indeed showed a markedly increased rate of population doubling relative to WT cells (Fig. 1B). In principle, such an effect could have arisen either because cells divided faster or because they reached senescence—the normal postproliferative arrest of cell division in adult tissues—at a lower rate. To examine senescence, we stained the same cells for senescence-associated β-galactosidase (SA-βgal) activity (24) at each passage from their initial isolation until their complete senescence. Nonogt cells exhibited a roughly twofold decreased proportion of senescent cells at every passage (Fig. 1C).
Fig. 1.
Mice deficient for NONO show increased cell division and reduced senescence. (A) NONO RNA expression measured by qPCR in various tissues taken from WT (+) and Nonogt animals (gt, not detectable). y axis, expression levels relative to maximum observed expression. Br, brain; De, dermis; He, heart; Ki, kidney; Li, liver; Lu, lung (n = 4 animals). (Inset) NONO protein measured in liver nuclear extract pooled from two representative animals, as well as in unrelated C57-Bl6J mice (Bl6/J). (B) WT and Nonogt primary fibroblasts were counted and passaged every 2 d and a constant number of cells plated to a new dish. Total cell number over time is plotted relative to initial cell number as population doublings. (Student t test for significant difference of doubling rates, P = 0.05.) (C) Cells from each passage in B were stained for SA-βgal activity, and the percentages of total cells expressing this marker were recorded. In this and all subsequent figures, *P < 0.05 and **P < 0.01 (Student t test). (D) Duplicate nonconfluent plates of WT and Nonogt primary fibroblasts were stained with CFSE and allowed to divide for 4 additional days. Dye intensity was then measured by flow cytometry (duplicate plates of cells in green and blue). As a control, other plates of the same cells were treated with mitomycin C to inhibit cell division immediately after staining and then treated in parallel (red). Numbers near curves reflect the percentage overlap between the green/blue curves and red curve. (E) WT and Nonogt primary fibroblasts were infected with lentivirus WPI (expressing GFP) or WPI-NONO (expressing NONO) and allowed to proliferate via serial passaging as in A. Relative cell number for each cell type ±SD was plotted at each passage (i.e., every 2 d).
If Nonogt cells had reduced senescence rather than an increased division rate, then fewer cells should remain nondividing in cultures of equivalent age. We tested this hypothesis by staining dividing Nonogt and WT cells from the same passage with the permanent cytoplasmic stain CFSE (carboxyfluorescein diacetate, succinimidyl ester) and then determining dye content by flow cytometry 4 d later. This dye remains trapped within the cells but is diluted with each cytokinesis. Hence it provides a quantitative analysis of the percentage of a cell population that has divided (25). According to this experiment, all Nonogt cells had divided at least once, whereas 40% of the WT cells had not divided (Fig. 1D). Reintroduction of NONO into primary Nonogt fibroblasts via lentiviral transduction slowed division and increased senescence, and addition of NONO to WT cells slowed division even further (Fig. 1E), confirming the dose-dependent role of NONO in restraining cell proliferation and pointing to a probable role for this protein in the cell cycle.
NONO Is a Transcriptional Activator of the p16-INK4A Cell Cycle Checkpoint Gene.
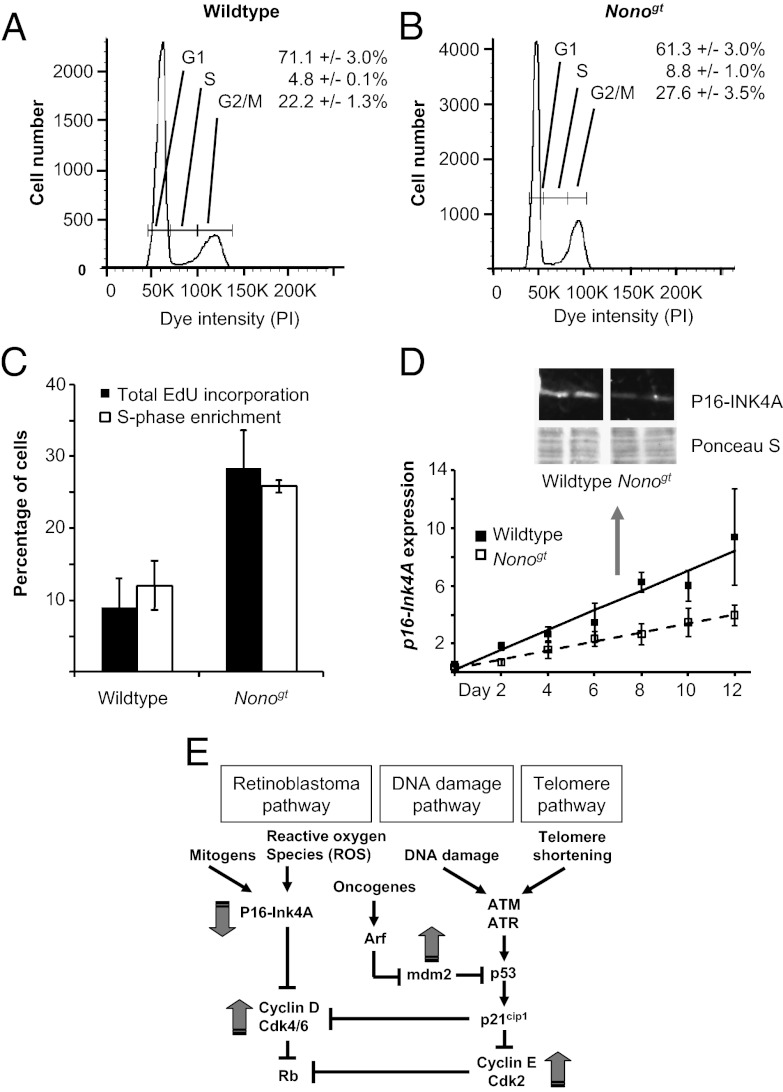
To identify the cell cycle stage at which NONO has a role, we fixed actively dividing fibroblasts from WT and Nonogt animals and labeled them with propidium iodide, a fluorescent DNA-binding dye (26). Subsequent flow cytometry experiments allowed us to quantify the proportion of cells in different stages of the cell cycle in each population. Our results demonstrated a twofold increase of cells in synthesis phase (S phase) of the cell cycle when NONO was absent. NONO may thus act as a regulator of exit from the G1 phase (Fig. 2 A and B). To confirm the effects of NONO upon S phase, we also pulse-labeled cells with 5-ethynyl-2´-deoxyuridine (EdU), an immunohistochemically detectable nucleotide analog incorporated during DNA replication in S phase. By comparing signals from EdU with those from propidium iodide in the same cells, we were able to verify that more cells were indeed in S phase in Nonogt fibroblasts than in WT ones (Fig. 2C).
Fig. 2.
NONO-deficient mice show an increased proportion of cells in S phase and reduced levels of p16-Ink4A. (A) Dividing cultures of primary fibroblasts from WT mice were stained with propidium iodide and analyzed by flow cytometry to quantify cellular DNA content. (B) Identical experiments were performed with Nonogt primary fibroblasts. (C) Percentage of cells in S phase in dividing cultures of primary fibroblasts from WT and Nonogt mice, as measured in a separate experiment by pulse-labeling with EdU. (P < 0.001, difference between genotypes.) Experiment is shown both by total EdU incorporation (black bars) and in cells previously sorted for S phase via propidium iodide incorporation (white bars). (D) Total RNA was harvested from each passage of WT (black squares) and Nonogt (white squares) primary fibroblasts passaged serially as in Fig 1B. p16-Ink4A transcript levels were determined by qPCR for each passage. (Expression is shown relative to WT at day 0, Student t test for significant difference of p16 accumulation rates, P = 0.01.) (Inset) Western blot showing levels of p16-INK4A protein from duplicate cultures of dividing WT and Nonogt fibroblasts. To verify equal loading of protein, Ponceau-S staining of the same blot section is depicted beneath. (E) Total RNA was harvested from cells under conditions as in A and subjected to qPCR array analysis. A selection of genes relative to senescence pathways is shown. Genes regulated in Nonogt cells are shown with gray arrows.
In most other systems, NONO has been identified as a transcriptional cofactor regulating either gene expression or RNA processing (27). Therefore, we considered it likely that NONO would exert its effects upon senescence and the cell cycle via transcriptional regulation of cell cycle genes. On the basis of the knowledge that NONO acted upon both senescence and the exit from the G1 phase checkpoint, we considered the p16-Ink4A locus as a likely target. P16-INK4A has been implicated previously as a regulator of the mitogen-responsive retinoblastoma pathway and is one of the key cellular components regulating senescence. It is known to repress the cyclin D-dependent kinases 4 and 6 (Cdk4 and Cdk6), resulting in a G1 arrest that slows cell division and promotes senescence (28, 29). To investigate whether the p16-Ink4A gene is systematically misregulated in Nonogt fibroblasts, we serially passaged cells from both WT and Nonogt mice and examined the expression levels of p16-Ink4A at each passage. Indeed, quantitative RT-PCR confirmed that RNA abundance of p16-Ink4A was systematically down-regulated in Nonogt cells at each passage, and this change in expression was also reflected in levels of p16-INK4A protein (Fig. 2D).
Because p16-INK4A negatively regulates cell division and positively regulates senescence, its repression is consistent with the phenotype observed in Nonogt fibroblasts. As a control, we also looked at key regulators of other pathways known to control senescence: the DNA damage-responsive p53 locus, and tankyrase, a downstream regulator of telomere length. No differences were observed in the levels of tankyrase mRNA between WT and knockout cells, and p53 was twofold lower, a direction inconsistent with the phenotype that we observe. In addition, for all known upstream regulators of p16-Ink4A—the Ets1, Ets2, and Id1 loci—transcript levels in Nonogt cells were unchanged or inconsistent with the observed down-regulation of p16-Ink4A (Fig. S1 A–E), supporting our hypothesis that p16-Ink4A is a possible direct regulatory target of NONO. More broadly, we compared expression of cell cycle genes in Nonogt fibroblasts and their WT counterparts using a cell cycle gene RT-PCR array, and not surprisingly found transcriptional misregulation of multiple other cell cycle genes (Fig. 2E and Table S1), confirming the effects of NONO upon the cell cycle in general and the extensive coregulation of cell cycle processes.
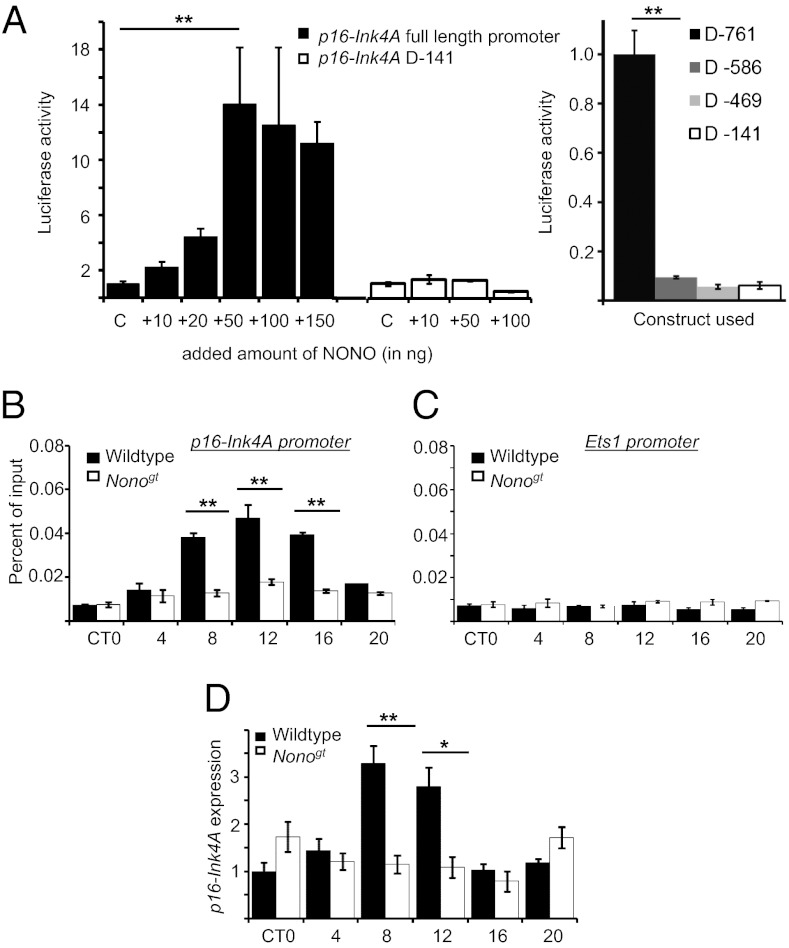
To examine further whether NONO might directly regulate the p16-Ink4A locus transcriptionally, we first cotransfected 3T3 fibroblast cells with a vector expressing NONO and with a luciferase reporter for p16-Ink4A promoter activity. The addition of NONO resulted in a dose-dependent increase up to 14-fold in luciferase signal compared with the levels obtained with the reporter alone (Fig. 3A), whereas a minimal promoter of p16-Ink4A (D-141) was not activated, suggesting that NONO functions as a transcriptional coactivator of the p16-Ink4A locus. By contrast, transcription of a reporter containing the CMV promoter driving expression of a hybrid p16-Ink4A-luciferase transcript including the entire 3′ untranslated region was unchanged (Fig. S2). Therefore, the effect of NONO on the p16-Ink4A locus is likely on the transcriptional rather than the posttranscriptional level.
Fig. 3.
NONO binds to the p16-Ink4A promoter to activate transcription. (A, Left) 3T3 cells were transfected with a luciferase reporter plasmid driven by 0.8 kb of the p16-Ink4A promoter (D-761), as well as indicated amounts of a plasmid expressing NONO. After 2 d, relative bioluminescence in cellular extracts was determined (solid bars; data plotted in arbitrary units, signal from reporter alone = 1). Comparable experiments were performed using a reporter construct containing only a minimal p16-Ink4A promoter, 0.14kb (D-141) (open bars). (A, Right) equivalent experiments were performed using constructs containing deletions of the p16-Ink4A promoter containing the indicated number of nucleotides upstream of the transcription start site. (B) Liver chromatin was harvested from WT and Nonogt mice at different times of day and subjected to immunoprecipitation using an anti-NONO antibody. Genomic DNA was purified from the precipitate, and DNA from the p16-Ink4A promoter was quantified by qPCR. (C) Bar graph quantifying DNA from the Ets1 promoter, using the same ChIP reactions as in B. (D) RNA was harvested from the livers in B, and p16-Ink4A transcript levels were quantified by qPCR.
NONO and PER Proteins Bind to the p16-Ink4A Promoter to Direct Its Circadian Transcription in Vivo.
To investigate whether NONO also acts directly as a transcriptional coactivator of p16-Ink4A in vivo, we performed ChIP of endogenous NONO protein with the p16-Ink4A promoter. In murine liver NONO indeed binds to the p16-Ink4A promoter region (Fig. 3B). As a control, we verified that NONO does not bind to the promoter of the Ets1 gene, an upstream regulator of p16-Ink4A (Fig. 3C). Importantly, NONO binds to p16-Ink4A in circadian fashion (Fig. 3B), implying that it might confer a circadian expression pattern upon this important cell cycle checkpoint gene.
To test this hypothesis, we examined the expression of p16-Ink4A directly in mouse liver at different times of day. As predicted, p16-Ink4A mRNA levels were indeed circadian in their abundance. In livers from Nonogt mice, this circadian regulation was lost, and p16-Ink4A transcript levels were nonrhythmic and low (Fig. 3D). Therefore, circadian regulation by NONO is indeed responsible for the circadian gene expression of p16-Ink4A.
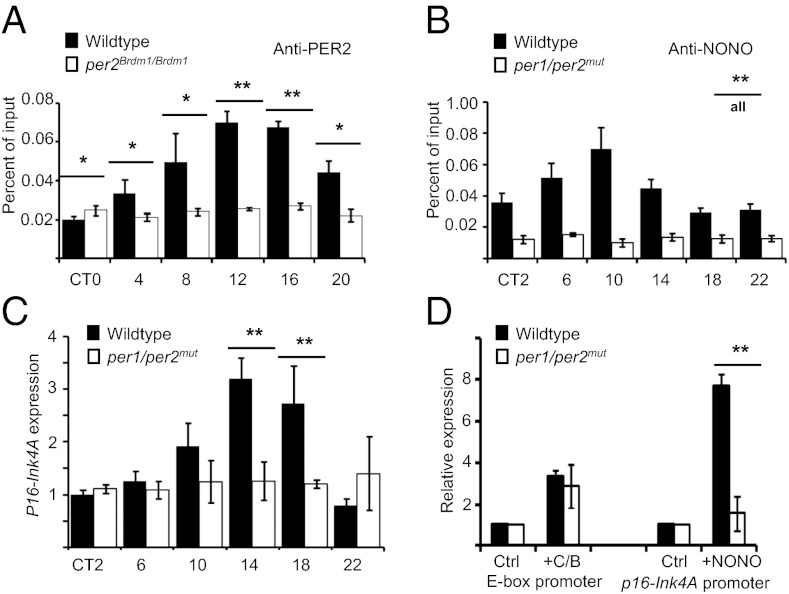
We have shown previously that overall NONO transcript and protein levels are constant throughout the day but that one particular NONO complex (also containing the PER proteins of the circadian oscillator) shows circadian variations in abundance (12). Because the p16-Ink4A promoter was occupied by NONO in circadian fashion, we also tested to determine whether PER proteins bound there. ChIP analyses from mouse liver demonstrated that PER2 proteins are present at the p16-Ink4A promoter with the same kinetics as NONO. As expected, no PER2 binding was observed in a per2Brdm1/Brdm1 strain lacking this protein (Fig. 4A). To determine whether PER proteins were necessary for the effects of NONO upon the p16-Ink4A promoter, we examined the levels of mRNA from this locus in per1Brdm1/Brdm1per2Brdm1/Brdm1 mutant mice (henceforth designated per1/per2mut for simplicity), which lack both proteins (30). In livers from these mice, the binding of NONO to the p16-Ink4A promoter was noncircadian and low or absent (Fig. 4B), and p16-Ink4A transcription itself was also constantly low (Fig. 4C).
Fig. 4.
Period proteins are necessary for binding and transcriptional activation of p16-Ink4A by NONO. (A) Liver chromatin was harvested from WT and per2Brdm1/Brdm1 mice at different times of day and subject to immunoprecipitation using an anti-PER2 antibody. Genomic DNA was purified from the precipitate, and DNA from the p16-Ink4A promoter was quantified by qPCR. (B) Bar graph showing ChIP experiments identical to those in A but using WT and per1/per2mut mice and immunoprecipitated with an anti-NONO antibody (P ≤ 0.001 for all pairs.) (C) RNA was harvested from the livers in B, and p16-Ink4A transcript levels were quantified by qPCR. (D) Transfection experiments identical to those in Fig. 2C were performed using primary fibroblasts isolated from WT mice and per1/per2mut mice.
These results suggest that a PER–NONO complex is responsible for the effects of NONO upon the p16-Ink4A promoter. To test this hypothesis explicitly and determine whether PER proteins are required for NONO to activate p16-Ink4A, we compared the ability of NONO to activate transcription of a p16-Ink4A reporter in transient transfections in primary fibroblasts from WT and per1/per2mut mice. Transcriptional activation by the circadian transcriptional activator complex BMAL1/CLOCK was achieved in both WT and per1/per2mut cells to the same extent (Fig. 4D). Whereas activation by NONO was observed in WT cells, no activation was observed in mutant cells (Fig. 4D), indicating that PER proteins are required for NONO activity (or vice versa).
NONO is Necessary for Circadian Gating of the Cell Cycle.
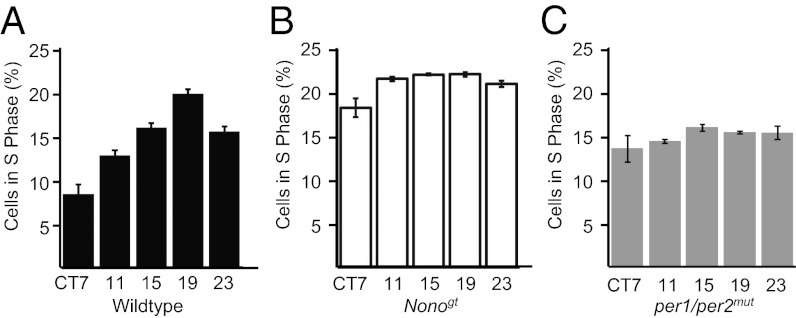
Circadian transcriptional activation of p16-Ink4A via NONO has predictable consequences for cellular physiology. In numerous previous studies in a variety of contexts, increased expression of p16-INK4A suppressed cell division, and reduced expression of this protein augmented it (31). Therefore, if NONO regulates p16-Ink4a in circadian fashion, we reasoned that NONO might be one of the unknown regulators that couple the circadian clock to cell division. To test this hypothesis, we synchronized circadian rhythms in duplicate plates of WT and Nonogt fibroblasts using dexamethasone (32). Subsequently these plates were fixed at different times after synchronization and then stained with propidium iodide and analyzed by flow cytometry to quantify DNA content. WT cells showed marked circadian variations in cell division, with 2.6-fold variation in the proportion of cells in S phase at midday and midnight (Fig. 5A). In per1/per2mut cells, no such variations were observed (Fig. 5C). This lack of circadian gating was equally observed in Nonogt cells, which divided throughout the day at a level equal to the peak of WT cells—consistent with the derepression of p16-INK4A that normally would negatively regulate cell division (Fig. 5B). Thus the NONO protein is necessary for diurnal cell cycle gating in these cells, and its absence likely leads to a disinhibition of the G1–S transition at specific circadian phases due to low levels of p16-INK4A.
Fig. 5.
NONO protein is necessary for circadian gating of the cell cycle in primary fibroblasts. Dividing primary fibroblasts were clock-synchronized with dexamethasone and then harvested at different times of day, fixed, and stained with propidium iodide. The percentage of dividing cells (i.e., in S phase) from (A) WT, (B) per1/per2mut, and (C) Nonogt cultures was then quantified by FACS analysis. (Times indicated are relative to synchronization.)
Absence of NONO Results in Defective Dermal Wound Healing.
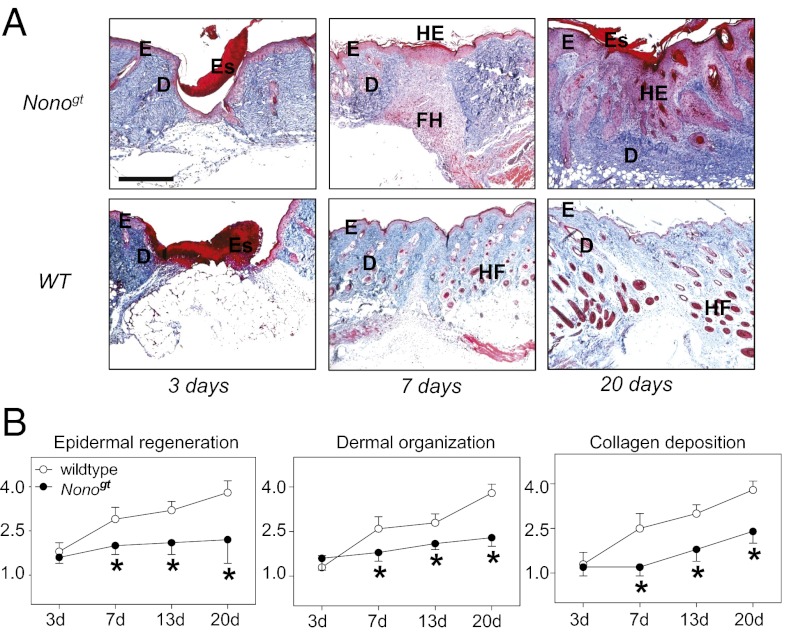
Despite this dramatic phenotype in primary cells, Nonogt mice develop normally. Hence, any importance for cell division is probably confined to adult animals. One situation in which cell division in adult animals plays an important role is during wound repair. To test whether NONO is required for normal wound repair, we wounded the skin of adult WT and Nonogt mice, closed the resulting full-thickness incisional wound with Steri-strips, and followed the wound healing process after 3, 7, 13, and 20 d by histological wound healing scores. Before wounding, skin structure appeared indistinguishable among WT and Nonogt animals (Fig. S3A), and shortly after wounding, scab formation, inflammation, and angiogenesis were normal (Fig. 6A, Left). However, later after wounding, histological analysis revealed profound alterations in both dermal and epidermal regeneration. WT wounds exhibited good reepithelialization and granulation tissue organization. Granulation tissue consisted of oval or spindle fibroblasts (dark red) embedded in a dense fascicular system of collagen fibers (gray; Fig. 6A, Center). The epidermal layer was well structured and gave, together with the well organized granulation tissue, rise to new skin that reestablished tissue integrity (Fig. 6A, Right). In Nonogt mice, immature granulation tissue was characterized by continued fibroblast proliferation, occupying most of the wound area (red, Fig. 6A, Right) by day 20. These plumped and round-to-polyhedral fibroblasts were distributed within a loose matrix with hardly any collagen production. Similarly, the keratinocyte layer (stained in pink) was characterized by hyperproliferation and little epidermal organization (Fig. 6A, Center and Right). Thus, even as collagen-secreting fibroblasts hyperproliferated in wounded Nonogt animals, collagen secretion was dramatically diminished (Fig. 6B). The dividing cells could form only an immature dysfunctional epidermal layer resting on disorganized granulation tissue, which prevented healing (Fig. 6B and Fig. S3).
Fig. 6.
NONO protein is necessary for correct dermal wound healing. (A) WT and Nonogt mice were incisionally wounded and then killed 3, 7, or 20 d later. Representative Masson-Goldner trichrome-stained paraffin sections from the center of these wounds are shown. (B) Wound healing subscores of normal healing WT mice compared with Nonogt mice on day 3, 7, 13, and 20 after incisional wounding in the dorsal skinfold. Data represent means ± SD (n = 5).
Circadian Clock-Deficient Mice Also Show Defects in Wound Healing.
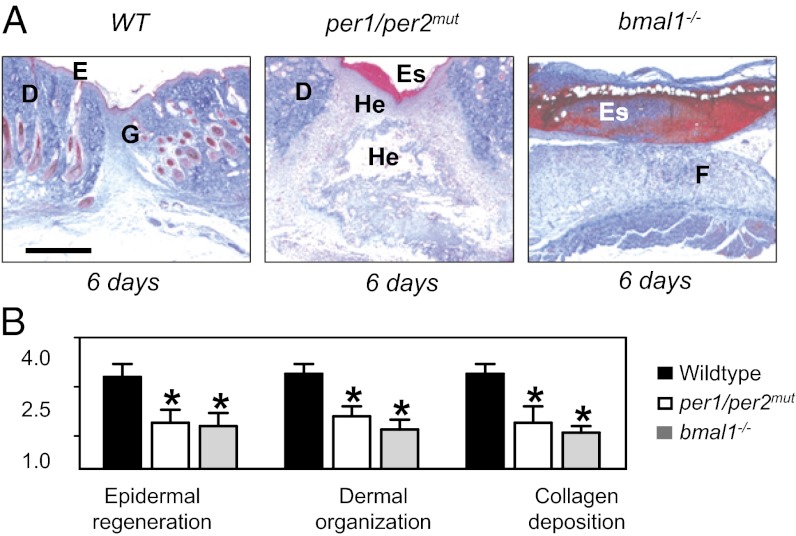
In principle, the defect in wound healing observed in Nonogt mice might be linked to the circadian cell cycle phenotype described above, or arise from another unrelated function of NONO. To distinguish between these hypotheses, we also tested wound healing in two strains of mice lacking functional circadian clocks: per1/per2mut mice and bmal1−/− mice. Both strains also showed defective wound healing. Consistent with the requirement of PER proteins for NONO activity at the p16-Ink4a promoter, the per1/per2mut mouse showed a thick layer of immature granulation tissue that was dominated by an excess of fibroblasts as well as polymorphonuclear cells. Bmal1−/− mice also showed severe wound healing defects, although this time marked by lack of epithelial coverage and a highly disorganized granulation tissue throughout the observation period. Most wounds in bmal1−/− mice consisted mainly of an inflammatory fibrin clot with hardly any fibroblast or keratinocyte proliferation (Fig. 7 A and B). Taken together, our data suggest that a functional circadian clock is important for wound healing and that its effects are likely mediated by NONO.
Fig. 7.
Functional circadian clock is necessary for correct dermal wound healing. (A) Masson-Goldner trichrome staining of 6-d wounds from WT (Left), per1/per2mut (Center), and bmal1−/− mice (Right). D, dermis; E, epidermis; Es, Eschar; F, fibrin; FH, fibroblast hyperproliferation; G, granulation tissue; He, hemorrhage; HE, hyperproliferative epithelium; HF, hair follicle. (Scale bar, 1 mm.) (B) Wound healing subscores of normal healing WT mice compared with per1/per2mut and bmal1−/− mice on day 6 after incisional wounding in the dorsal skinfold. Data represent means ± SD (n = 6).
Discussion
In this article we show that the NONO protein—which we have identified previously as a component of the basic circadian oscillator in Drosophila and in mouse cells (12)—plays an equally important role to gate circadian cell division in fibroblasts. Elimination of NONO entirely abrogated circadian-cell cycle coupling in fibroblasts, suggesting that NONO serves as a necessary link between these two processes, at least in these cells. This uncoupling allowed us to probe possible functions of this link.
NONO as a Circadian Effector.
NONO was initially identified by its homology to splicing factors (33) and as a Drosophila factor involved in courtship songs (34). Since then, diverse studies have described it as a protein with pleiotropic functions mainly involved in RNA processing and transport, as well as a transcription factor (27). It has been implicated in diverse pathways, such as nuclear receptor signaling (35), DNA repair (36, 37), and viral infection (38). Here we have shown that it can directly activate the p16-Ink4A locus, an important regulator of the G1 exit checkpoint of the cell cycle. The detailed mechanism of its coactivator function remains unclear. In the case of transducer of regulated CREB-binding protein (TORC)-mediated coactivation by NONO, Amelio et al. (39) suggested that NONO directly bridges interactions between RNA polymerase II and other activators. Other groups have shown that NONO regulates nuclear retention of RNAs (40, 41) or transcription termination (42), so it is possible that NONO couples transcription initiation and downstream RNA processing.
NONO itself is not expressed in circadian fashion. Nevertheless, it binds to PER proteins and has been shown to antagonize their repressive activity (12). Recently, a protein highly homologous to NONO called splicing factor, proline- and glutamine-rich (SFPQ) has been shown to bind to PER proteins as well, but to assist in their repressive activity by recruiting the mSIN3A histone deacetylase (43). Consistent with these results, we have previously demonstrated that all three members of the DBHS protein family [Drosophila Behavior Human Splicing, of which NONO, SFPQ, and paraspeckle component 1 (PSPC1) are members] likely play redundant roles via cis-acting E-box elements at circadian genes (22). Here we show that NONO plays a strong activating role, also circadian, at the p16-Ink4A promoter. However, this activation is dependent upon a particular 175-bp sequence within the promoter region of this gene that does not contain an E-box. Thus, multiple sequences likely direct activities of DBHS proteins, and these can be activating or repressing. DBHS proteins themselves seem to have no sequence-specific DNA-binding activity, although structure-specific binding (DNA triplexes, RNA loops) has been postulated (44, 45). We propose that NONO and homologous factors might act as circadian output effectors, linking circadian regulation via PER proteins to activating and repressing moities to confer circadian expression upon diverse aspects of physiology and gene expression. The exact mechanism of this regulation will doubtless constitute a fascinating subject of future investigations.
NONO as a Link Between the Circadian Clock and the Cell Cycle.
In keeping with NONO’s proposed role as a circadian output effector, we present here data that show that NONO directly activates the p16-INK4A cell cycle checkpoint gene in circadian fashion to regulate cell cycle progression. Multiple circadian influences upon the cell cycle have already been documented, acting at multiple different checkpoints. For example, it has been suggested that transcriptional control of cell cycle genes such as p21-waf1 and wee1 might play important roles. Both Wee1 transcript and protein levels show circadian fluctuation probably governed by cis-acting E-box elements, and one of the targets of the WEE1 kinase, CDC2, also shows circadian phosphorylation (3). A similar story has been constructed for p21-waf1, whose circadian transcription is governed by REV-ERBα response element (RRE) elements within the promoter that respond to REV-ERBα/β and retinoid-related orphan receptor (ROR) proteins (17). Another class of clock factors, the PER proteins, themselves function as tumor suppressors (46, 47). Similarly, TIM—shown by some groups to play a role in the mammalian circadian clock (48)—has been shown to bind to the CHK1 and CHK2 cell cycle proteins (19). Thus, in total, five different sites of action of the circadian clock upon the cell cycle have been proposed. One of the principal difficulties in evaluating the roles of these independent mechanisms in different tissues lies in the importance of cell cycle checkpoint proteins, which precludes meaningful loss-of-function experiments. What is particularly interesting about NONO is that its disruption completely abrogates circadian gating of the cell cycle in fibroblasts without severely disrupting either the circadian clock or the cell cycle. Importantly for our conclusions about circadian regulation of the cell cycle, even if the NONO homolog NON-A is essential for circadian function in flies (12), NONO-deficient mice possess essentially normal clocks in all tissues examined. This discrepancy likely arises because of functional redundancy between NONO and homologous proteins SFPQ and PSPC1 within the circadian oscillator in mammals (22). Thus, the Nonogt mouse provides a mouse model in which the cell cycle is uncoupled from a functional circadian clock.
Circadian Control and Wound Repair.
Because Nonogt fibroblasts show increased proliferation and decreased senescence, it is perhaps to be expected that dermal wound repair is disrupted in Nonogt animals. Nevertheless, because collagen is secreted by fibroblasts, a dramatic lack of collagen in wounds in Nonogt animals is not what we anticipated in the fibroblast-hyperproliferated wounds of these animals. It might hint, however, at the underlying function of the gating of the cell cycle by NONO. We hypothesize that regulated cell division might allow for organized cycles of division and tissue building and thereby facilitate the organization of complex tissue structures. By contrast, when cell division occurs constantly and randomly, such organization never occurs, resulting in the overproliferation and under-epithelialization we have observed. Our argument is supported by the fact that two different types of circadian clock-deficient animals (bmal1−/− mice and per1/per2mut mice) also show defects in wound healing. The requirement for clocks in wound healing seems to be conserved in other vertebrates: very recently, a role for the circadian clock in zebrafish fin healing was also documented (49). Other recent research points to a network of clock and clock-controlled genes whose expression is important for proliferation of fibroblasts and keratinocytes, including the clock genes Clock (5) and Bmal1 (6) and the circadian output effector Klf9 (7). Hence the effects that we observe are unlikely to be caused by an activity of the NONO protein unrelated to the circadian clock. The loosely “opposite” phenotypes of different circadian mutant mice further support the idea that circadian gating per se is important for dermal healing. Bmal1−/− mice, defective in the “positive limb” of the circadian oscillator, show excessive fibrin with reduced fibroblast number; and per1/per2mut mice defective in the “negative limb” show increased fibroblasts and reduced fibrin.
Of course, correct wound healing is not simply a matter of cell division but also of differentiation. Prekeratinocyte and other stem cells first augment in number and then differentiate to their final nonproliferative types. It has been suggested that the circadian clock in hair root follicles plays an important role in generating heterogenous populations of cells necessary for hair growth (50), and a similar role in skin regeneration is not only possible, but to the authors likely. Through its role in replicative senescence, the p16-INK4A protein plays a key role in mesenchymal stem cell differentiation (51), so it is possible that circadian regulation of this protein via NONO would affect not only cell division but also stem cell populations. However, these processes are unseparable and intertwined: our observation of overproliferation of keratinocytes at wounds in Nonogt animals also implies an overrepresentation of prekeratinocyte stem cells at these times, and the overproliferation of fibroblasts equally shows direct control of cell division itself. The key to both observations—and also the key conclusion of this study—is that circadian control of the p16-INK4A checkpoint via NONO is necessary for correct dermal wound healing, and lack of this control results in loss of circadian cell cycle gating in vitro and tissue overproliferation in vivo. The exact downstream mechanism of this control and its consequences for tissue structure will provide a fascinating and medically relevant subject for future investigations. It should be noted, however, that tissue formation during development in both Nonogt and clock-disrupted mice seems normal. Therefore, it is likely that such control is either unnecessary or redundant earlier in development.
As more is known about the feedback loops that control circadian clock mechanism, increasing numbers of laboratories have begun to address the questions of how and why the circadian oscillator communicates timing to all of the physiological and cellular process that it governs. In these studies a pattern has begun to emerge: basic clock components can themselves directly regulate genes involved in the “output pathways” that control diurnal function (52). In this article we show that the NONO protein plays an essential intermediary role. More generally, coopting the circadian clock as a synchronizing timekeeper—completely independent of its role in diurnal adaptation—might provide an important future paradigm for clock function at a cellular level.
Materials and Methods
Plasmids.
The bioluminescence reporter construct pBmal1-Luciferase and the NONO-targeting siRNA6 have been described previously (12). For NONO overexpression, a commercially obtained pSPORT construct was used (clone ID 360A935 from Open Biosystems). p16-promoter-luciferase plasmid (D-761) and minimal p16 promoter plasmid (D-141) (53) were obtained from B. A. Mock and S. Zhang (Laboratory of Genetics, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD).
Animal Husbandry.
Chimeric mice were obtained from Nonogt ES cells (C57Bl6 genotype) via standard blastocyst injection into SV129 mice by the University of California, Davis. Individual chimeric mice were back-crossed 4–10 generations against C57Bl6. All experiments were performed with littermates. Animal housing and experimental procedures are in agreement with veterinary law of the canton of Zurich. Genotyping was done as described at www.mmrrc.org/strains/38/ctr_protocol.pdf using a NONO-specific primer set (sense 5′-TTA GGG GGC CGA ACT ACT TGA ATT G-3′, antisense 5′-GGG CCG GGC AGA TTT ACT AGT TTT T-3′. Quantitative PCR (qPCR) primer sequences are listed in the quantitative real-time PCR section, below).
Primary Cell Isolation and Culture.
Primary adult dermal fibroblasts (ADFs) were taken from a 0.5-cm piece of mouse tail that was cut into several small pieces by using a razor blade. Digestion occurred in 1.8 mL DMEM containing 20% FBS, 1% penicillin/streptomycin, and 1% amphotericin B supplemented with 0.7 U Liberase blendzyme (Roche), at 37 °C and 5% CO2 for 8 h (all concentrations are vol/vol). After centrifugation in 1× PBS, the pellet was resuspended in DMEM containing 20% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2.5 μg/mL amphotericin B and kept at 37 °C and 5% CO2. The next day, medium was exchanged, and remaining tail pieces were removed. Another medium exchange was done 3 d later. After 1 wk the medium was exchanged for medium without amphotericin B. Subsequently, ADFs were cultured at 37 °C and 5% CO2 in DMEM supplemented with 20% FBS and 1% penicillin/streptomycin.
cDNA Production, Quantitative Real-Time PCR, and PCR Arrays.
RNA was extracted as described in (12). Five hundred nanograms of total RNA was transcribed to cDNA with SuperScript II (Invitrogen) using oligo (dT) primers according to the manufacturer’s instructions. For quantitative real-time PCR 20 ng of cDNA was used, and transcript levels of genes were detected by Taqman probes used with the Taqman PCR mix protocol (Roche) using the AB7900 thermocycler. Primers used for detection of specific genes are listed in Table S2. PCR arrays to quantify cell cycle components were performed according to the manufacturer’s instructions using 500 ng of total RNA (SABiosciences; array name, PAMM-020E). RNA was extracted from duplicate plates of either WT or Nonogt primary fibroblasts 14 d after isolation.
Western Blotting and Immunohistochemistry.
Western blotting was performed using standard procedures (Current Protocols in Molecular Biology, Wiley). Equal loading and size detection using protein ladder was verified by Ponceau-S staining of membranes before probing. The probing of the primary anti-NONO antibody (polyclonal antibody raised by Charles River Labs using bacterially produced NONO protein, affinity-purified against the antigen used for injection) was done at a 1:200 dilution. The probing of the secondary antibody was done at a 1:10,000 dilution for anti-rabbit IgG coupled to horseradish peroxidase (Sigma) and 1:1,000 for anti-mouse HRP (Sigma), respectively.
Cellular Senescence and Cytometric Measurements.
For growth curves, primary fibroblasts from WT and Nonogt animals were split every other day, plating each time 1 × 106 cells so that confluence was never reached. Within these plates, a coverslip was laid for senescence-associated β-galactosidase staining, and nonplated cells were saved for RNA isolation. Population doublings were calculated as the logarithm of the number of cells counted at the current passage divided by the number of cells at the first passage, adjusted for plating ratios. Senescence-associated β-galactosidase staining was performed using a senescence β-galactosidase Staining Kit (Cell Signaling Technology) following the protocol of the manufacturer.
To measure cell cycle progression at different circadian times, circadian clocks in identical plates of WT and Nonogt primary fibroblasts were synchronized with 100 nM dexamethasone for 30 min as previously described (32) at day 10 of serial splitting. Seven hours after synchronization cells from duplicate plates were fixed every 4 h. Fixation and flow cytometric measurements of DNA content via propidium iodide staining were performed according to Current Protocols in Cell Biology, chapter 8.4.4 (54). Cell cycle stages were quantified using the Watson cell cycle model of the FlowJo software.
For EdU pulse labeling the commercially available Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit from Invitrogen was used, following the manufacturer’s instructions. Flow cytometry data were analyzed with the FlowJo software package.
For CFSE staining, cells were trypsinized, resuspended in complete cell medium, and then washed with 1× PBS. From this, 1 × 106 cells were transferred and spun down. The pellet of cells was then resuspended in 0.2 mL 1× PBS supplemented with 5 μM CFSE and incubated for 10 min at 37 °C. By adding ice-cold complete cell medium the reaction was stopped and the cells were washed with 1× PBS. The pellet of cells was resuspended in complete cell medium, plated, and allowed to grow until analysis. These cells were compared with a plate that was mitomycin C treated to arrest cell division and then stained in the same way as described above with CFSE. In brief, 10 mL of a 10 μg/mL mitomycin C (SIGMA, M4287) solution were added to a 10-cm dish of cells and incubated for 3 h at 37 °C with 5% CO2. Afterward cells were washed three times with 1× PBS supplemented with 5% of FBS and then normal growth medium was added and cells were incubated overnight. The next day the plate was split 1:1 to remove dead cells and cell debris, and then CFSE treated as above. After 4 d both arrested and dividing cells were trypsinized, washed three times with 1× PBS, and then resuspended in 1× PBS supplemented with 1% FBS for FCSE measurement using a BD FacsCanto II machine.
Transient Transfections.
For p16Ink4A reporter transfection studies Lipofectamine LTX with PLUS reagent (Invitrogen) was used according to the manufacturer’s instructions, cultivating cells in 24-well plates and transfecting them with a total of 650 ng DNA, of which 50 ng were the promoter luciferase reporter construct. Varying amounts of pCMV-NONO plasmid were “balanced” by the addition of pKS(+) to a total of 600 ng. Cells were harvested after 60 h by washing once with 1× PBS and extracting with the supplied buffer from a luciferase assay kit (Promega). Quantification was done by scintillation counting, and normalized against total protein amount.
ChIP.
Chromatin from mouse liver and tissue culture cells was obtained as described previously (55). Equal amounts of precleared chromatin were incubated overnight at 4 °C with 1 μL of anti-NONO antibody or anti-PER2 antibody. The capture of the DNA:protein complexes, the washing conditions, and the purifiction of the DNA fragments before qPCR, as well the control antibodies have been described previously (56). The region-specific primer/probe pairs are listed in Table S2.
Histological Wound Scoring.
WT and Nonogt mice were anesthetized i.p. with a mixture of 90 mg/kg body weight (BW) ketamine hydrochloride (Ketavet; Parke Davis) and 25 mg/kg BW xylazine hydrochloride (Rompun; Bayer). The dorsal region was shaved and treated with a depilatory agent (Pilca Perfect; Stafford-Miller Continental). Three full-thickness incisions (6 mm) perpendicular to the dorsal midline were made at one anterior and two posterior dorsal sites, and the skin margins were closed with Steri-Strips (Steri-Strip S Surgical Skin Closure; 3M). Mice were killed on day 3, 7, 13, and 20 after wounding and the wounds embedded according to standard procedures and stained with Masson-Goldner trichrome and hematoxylin eosin. A total of 16 animals (8 WT and 8 NONOgt, n = 2 for each timepoint) were wounded and killed on the corresponding time points, and two additional WT and NONOgt mice without any incisions served as controls. Subsequently, identical experiments were performed on three each of WT, bmal1−/−, and per1/per2mut mice, with killing on day 6 after wounding. Histological wound scoring has been described previously (57) and was based on the quality of dermal organization, epidermal regeneration, collagen deposition, cellular content, and wound vascularity. The criteria used as histological scores of wound healing are summarized in Table S3.
Statistical Methods.
In bar graphs, Student’s t test was used to determine significant differences between control and experimental groups. In all figures, n = 3–4 independent experiments, with each experiment conducted in technical duplicate. Data are plotted ±SEM. In line graphs, for each experimental data set, linear regression was conducted to determine best-fit line describing the data from each independent experiment. Overall significance of differences in doubling (e.g., Fig. 1A) or in accumulation rate (e.g., Fig. 2A) were then determined using a Student two-tailed t test of slopes of the regression lines from each data set (n = 2–4).
Supplementary Material
Acknowledgments
We thank Claudia Dumrese at the University of Zürich, Center for Microscopy and Image Analysis for flow cytometry assistance, Pia Fuchs (Unispital Zürich) for histology assistance, Roland Dürr for assistance with statistical analysis of data, R. Dallmann (University of Zurich) for a critical reading of the manuscript, and B. Mock (Harvard University) for the kind gift of plasmids containing the p16-Ink4a promoter. This work was funded by Swiss National Science Foundation grants (to S.A.B., J.A.R., and U.A.). Further support was provided by the Zürich Neurozentrum and Molecular Life Sciences graduate programs of the Life Sciences Zürich graduate school.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 1565.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213317110/-/DCSupplemental.
References
- 1.Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U. The mammalian circadian timing system: From gene expression to physiology. Chromosoma. 2004;113(3):103–112. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- 2.Kowalska E, Brown SA. Peripheral clocks: Keeping up with the master clock. Cold Spring Harb Symp Quant Biol. 2007;72:301–305. doi: 10.1101/sqb.2007.72.014. [DOI] [PubMed] [Google Scholar]
- 3.Matsuo T, et al. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302(5643):255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 4.Nagoshi E, et al. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119(5):693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Miller BH, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci USA. 2007;104(9):3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geyfman M, et al. Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc Natl Acad Sci USA. 2012;109(29):11758–11763. doi: 10.1073/pnas.1209592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spörl F, et al. Krüppel-like factor 9 is a circadian transcription factor in human epidermis that controls proliferation of keratinocytes. Proc Natl Acad Sci USA. 2012;109(27):10903–10908. doi: 10.1073/pnas.1118641109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gery S, et al. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22(3):375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 9.Gaddameedhi S, Selby CP, Kaufmann WK, Smart RC, Sancar A. Control of skin cancer by the circadian rhythm. Proc Natl Acad Sci USA. 2011;108(46):18790–18795. doi: 10.1073/pnas.1115249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filipski E, et al. Host circadian clock as a control point in tumor progression. J Natl Cancer Inst. 2002;94(9):690–697. doi: 10.1093/jnci/94.9.690. [DOI] [PubMed] [Google Scholar]
- 11.Ripperger J, Brown SA. In: The Circadian Clock. Albrecht U, editor. Vol 12. New York: Springer; 2009. pp. 37–78. [Google Scholar]
- 12.Brown SA, et al. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308(5722):693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- 13.Vermeulen K, Van Bockstaele DR, Berneman ZN. The cell cycle: A review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003;36(3):131–149. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: Genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9(2):115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 15.Pendergast JS, Yeom M, Reyes BA, Ohmiya Y, Yamazaki S. Disconnected circadian and cell cycles in a tumor-driven cell line. Commun Integr Biol. 2010;3(6):536–539. doi: 10.4161/cib.3.6.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeom M, Pendergast JS, Ohmiya Y, Yamazaki S. Circadian-independent cell mitosis in immortalized fibroblasts. Proc Natl Acad Sci USA. 2010;107(21):9665–9670. doi: 10.1073/pnas.0914078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gréchez-Cassiau A, Rayet B, Guillaumond F, Teboul M, Delaunay F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J Biol Chem. 2008;283(8):4535–4542. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- 18.Unsal-Kaçmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25(8):3109–3116. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X, Wood PA, Hrushesky WJ. Mammalian TIMELESS is required for ATM-dependent CHK2 activation and G2/M checkpoint control. J Biol Chem. 2010;285(5):3030–3034. doi: 10.1074/jbc.M109.050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Proteau A, et al. The multifunctional nuclear protein p54nrb is multiphosphorylated in mitosis and interacts with the mitotic regulator Pin1. J Mol Biol. 2005;346(4):1163–1172. doi: 10.1016/j.jmb.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 21.Stier S, et al. Identification of p54(nrb) and the 14-3-3 Protein HS1 as TNF-alpha-inducible genes related to cell cycle control and apoptosis in human arterial endothelial cells. J Biochem Mol Biol. 2005;38(4):447–456. doi: 10.5483/bmbrep.2005.38.4.447. [DOI] [PubMed] [Google Scholar]
- 22.Kowalska E, et al. Distinct roles of DBHS family members in the circadian transcriptional feedback loop. Mol Cell Biol. 2012;32(22):4585–4594. doi: 10.1128/MCB.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skarnes WC, et al. International Gene Trap Consortium A public gene trap resource for mouse functional genomics. Nat Genet. 2004;36(6):543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Litaker JR, et al. Expression profile of senescence-associated beta-galactosidase and activation of telomerase in human ovarian surface epithelial cells undergoing immortalization. Int J Oncol. 1998;13(5):951–956. doi: 10.3892/ijo.13.5.951. [DOI] [PubMed] [Google Scholar]
- 25.Lyons AB, Parish CR. Determination of lymphocyte division by flow cytometry. J Immunol Methods. 1994;171(1):131–137. doi: 10.1016/0022-1759(94)90236-4. [DOI] [PubMed] [Google Scholar]
- 26.Krishan A. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J Cell Biol. 1975;66(1):188–193. doi: 10.1083/jcb.66.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO—multi-functional nuclear proteins. FEBS Lett. 2002;531(2):109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- 28.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366(6456):704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 29.Ohtani N, Yamakoshi K, Takahashi A, Hara E. The p16INK4a-RB pathway: Molecular link between cellular senescence and tumor suppression. J Med Invest. 2004;51(3-4):146–153. doi: 10.2152/jmi.51.146. [DOI] [PubMed] [Google Scholar]
- 30.Zheng B, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105(5):683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 31.Huschtscha LI, Reddel RR. p16(INK4a) and the control of cellular proliferative life span. Carcinogenesis. 1999;20(6):921–926. doi: 10.1093/carcin/20.6.921. [DOI] [PubMed] [Google Scholar]
- 32.Balsalobre A, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289(5488):2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 33.Dong B, Horowitz DS, Kobayashi R, Krainer AR. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res. 1993;21(17):4085–4092. doi: 10.1093/nar/21.17.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rendahl KG, Jones KR, Kulkarni SJ, Bagully SH, Hall JC. The dissonance mutation at the no-on-transient-A locus of D. melanogaster: Genetic control of courtship song and visual behaviors by a protein with putative RNA-binding motifs. J Neurosci. 1992;12(2):390–407. doi: 10.1523/JNEUROSCI.12-02-00390.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong X, et al. p54nrb is a transcriptional corepressor of the progesterone receptor that modulates transcription of the labor-associated gene, connexin 43 (Gja1) Mol Endocrinol. 2009;23(8):1147–1160. doi: 10.1210/me.2008-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S, et al. Involvement of p54(nrb), a PSF partner protein, in DNA double-strand break repair and radioresistance. Nucleic Acids Res. 2009;37(20):6746–6753. doi: 10.1093/nar/gkp741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salton M, Lerenthal Y, Wang SY, Chen DJ, Shiloh Y. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle. 2010;9(8):1568–1576. doi: 10.4161/cc.9.8.11298. [DOI] [PubMed] [Google Scholar]
- 38.Zolotukhin AS, et al. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol Cell Biol. 2003;23(18):6618–6630. doi: 10.1128/MCB.23.18.6618-6630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amelio AL, et al. A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proc Natl Acad Sci USA. 2007;104(51):20314–20319. doi: 10.1073/pnas.0707999105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123(2):249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106(4):465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 42.Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 2007;21(14):1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duong HA, Robles MS, Knutti D, Weitz CJ. A molecular mechanism for circadian clock negative feedback. Science. 2011;332(6036):1436–1439. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson LD, et al. Triplex DNA-binding proteins are associated with clinical outcomes revealed by proteomic measurements in patients with colorectal cancer. Mol Cancer. 2012;11(1):38. doi: 10.1186/1476-4598-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng R, et al. PSF and p54nrb bind a conserved stem in U5 snRNA. RNA. 2002;8(10):1334–1347. doi: 10.1017/s1355838202022070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gery S, Koeffler HP. Circadian rhythms and cancer. Cell Cycle. 2010;9(6):1097–1103. doi: 10.4161/cc.9.6.11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111(1):41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 48.Barnes JW, et al. Requirement of mammalian Timeless for circadian rhythmicity. Science. 2003;302(5644):439–442. doi: 10.1126/science.1086593. [DOI] [PubMed] [Google Scholar]
- 49.Idda ML, et al. Circadian timing of injury-induced cell proliferation in zebrafish. PLoS One. 2012;7(3):e34203. doi: 10.1371/journal.pone.0034203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janich P, et al. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature. 2011;480(7376):209–214. doi: 10.1038/nature10649. [DOI] [PubMed] [Google Scholar]
- 51.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460(7259):1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14(6):679–689. [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang S, et al. p16 INK4a gene promoter variation and differential binding of a repressor, the ras-responsive zinc-finger transcription factor, RREB. Oncogene. 2003;22(15):2285–2295. doi: 10.1038/sj.onc.1206257. [DOI] [PubMed] [Google Scholar]
- 54.Darzynkiewicz Z, Juan G, Bedner E. Determining cell cycle stages by flow cytometry. In: Bonifacino JS, Dasso M, Harford JB, Lippincott-Schwartz J, Yamada KM, editors. Current Protocols in Cell Biology. New York: John Wiley & Sons; 2001. pp. 8.4.1–8.4.18. [DOI] [PubMed] [Google Scholar]
- 55.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38(3):369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 56.Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24(4):345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Altavilla D, et al. Inhibition of lipid peroxidation restores impaired vascular endothelial growth factor expression and stimulates wound healing and angiogenesis in the genetically diabetic mouse. Diabetes. 2001;50(3):667–674. doi: 10.2337/diabetes.50.3.667. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.