Abstract
Tomato flower abscises at the anatomically distinct abscission zone that separates the pedicel into basal and apical portions. During abscission, cell separation occurs only at the abscission zone indicating distinctive molecular regulation in its cells. We conducted a transcriptome analysis of tomato pedicel tissues during ethylene promoted abscission. We found that the abscission zone was the most active site with the largest set of differentially expressed genes when compared with basal and apical portions. Gene Ontology analyses revealed enriched transcription regulation and hydrolase activities in the abscission zone. We also demonstrate coordinated responses of hormone and cell wall related genes. Besides, a number of ESTs representing homologs of key Arabidopsis shoot apical meristem activity genes were found to be preferentially expressed in the abscission zone, including WUSCHEL (WUS), KNAT6, LATERAL ORGAN BOUNDARIES DOMAIN PROTEIN 1(LBD1), and BELL-like homeodomain protein 1 (BLH1), as well as tomato axillary meristem genes BLIND (Bl) and LATERAL SUPPRESSOR (Ls). More interestingly, the homologs of WUS and the potential functional partner OVATE FAMILIY PROTEIN (OFP) were subsequently down regulated during abscission while Bl and AGL12 were continuously and specifically induced in the abscission zone. The expression patterns of meristem activity genes corroborate the idea that cells of the abscission zone confer meristem-like nature and coincide with the course of abscission and post-abscission cell differentiation. Our data therefore propose a possible regulatory scheme in tomato involving meristem genes that may be required not only for the abscission zone development, but also for abscission.
Introduction
Organ abscission is a ubiquitous physiological process in the plant kingdom. While seasonal senescence and foliage abscission set the enchanted autumn scenery, the dispersal of seeds and the efficiency of grain threshing and fruit picking are highly relevant to agriculture. Abscission occurs at specific sites called abscission zones. Up to date, a vast number of genes, particularly hydrolytic enzymes, such as polygalacturonases (PGs), xyloglucan endotransglucosylase/hydrolase (XTH), β-1,4-glucanase (cellulase), and expansins have been shown to be implicated in abscission for the degradation of cell wall and middle lamella at the abscission zones that eventually result in the shedding of distal organs [1], [2], [3], [4], [5], [6].
During this process, ethylene is known to be an efficient accelerator for organ abscission, although the initiation of abscission is considered to be timed or repressed by auxin [6], [7], [8], [9]. In this regard, abscission zones are made up with so-called type II cells whose expansive growth can be enhanced by ethylene, but not auxin [7]. Despite the dominant role of ethylene in plant organ abscission, genes whose function in abscission is independent of ethylene are increasingly reported, especially in Arabidopsis. For instance, HAESA and HSL2 (HAESA-like 2) are receptors of the small peptide ligand IDA (INFLORESCENCE DEFICIENT IN ABSCISSION) and together modulate the downstream MAPK cascade to induce floral organ shedding in an ethylene-independent manner [10]. Other genes contributing to the abscission zone development and its capacity are transcription factors (TFs) including MADS-box genes AGL15 and AGL18 [11], KNAT/BP [12], and AtZFP2 (ZINC FINGER PROTEIN2) [13]. Ethylene-independent mechanisms are proposed to be involved in the establishment of the abscission zone that is subsequently responsive to ethylene signals, either through guiding the correct architecture formation of the abscission zone itself, or by facilitating the efficacy of ethylene signaling pathways [14], [15], [16]. Thus, an intrinsic relation between the development of the abscission zone and its function is plausible.
The tomato flower abscission zone is located in the middle of the pedicel dividing it into two distinct sections: the basal and the apical portions. The abscission zone is composed of a few layers of smaller cells lacking vacuoles and any aspect of maturation [17], indicating that cell growth and differentiation is arrested at an early stage of development [18], [19]. In this regard, the persistent expression of KNOX family genes in the abscission zone were suggested [20]. In tomato, a few prominent genes have been found to affect the abscission zone development, including JOINTLESS (J), LATERAL SUPPRESSOR (Ls), and JOINTLESS2 (J2) [21], [22], [23]. BLIND (Bl), encoding a R2R3-class Myb transcription factor, genetically interacts with J [24], [25], [26], suggesting a possible role of this gene in abscission. In Arabidopsis, the Bl homolog REGULATOR OF AXILLARY MERISTEMS1 (RAX1) is also required for lateral meristem initiation [27], [28]. Furthermore, mutations of RAX genes affect the expression domains of the meristem activity gene SHOOTMERISTEMLESS (STM) which works with KNAT6, both KNOX-family genes, in shoot apical meristem maintenance and organ separation [29]. Recently, KNAT6, together with KNAT1 and 2 were shown to act downstream in the IDA-HAE/HSL2 signaling pathway to regulate floral organ abscission [30]. Therefore, a picture of regulatory network involving meristem activity genes in abscission is emerging.
In tomato, two recent transcriptome analyses were performed using pedicels as materials. Meir et al. (2010) compared the transcriptome differences between the abscission zone and the basal portion of pedicel in a time course after flower removal with or without the treatment of 1-methylcyclopropene (1-MCP), an effective inhibitor of ethylene action, while Nakano et al. (2012) compared the whole pedicel of the wild type and the jointless mutant or MACROCYLYX (MC) knock-down transgenic lines at pre-abscission stage under normal growth conditions. Although it has long been observed that application of exogenous ethylene accelerates tomato flower abscission, a comparative analysis of the transcriptome profiles in abscission zone relative to the neighboring tissues (the basal and apical portions) in abscission is still lacking. We performed a microarray assay on three pedicel tissues at 0, 3, 6 h time points of ethylene treatment. Our results showed that homologs of meristem activity genes were preferentially expressed in abscission zone when compared with the neighboring tissues. We also show that some of these genes were subsequently repressed in the abscising tissues. The expression patterns of meristem activity genes support the idea that they may play a role in maintaining meristem-like status of abscission zone cells, correlating with their performance before and after abscission.
Results
The Experimental Design
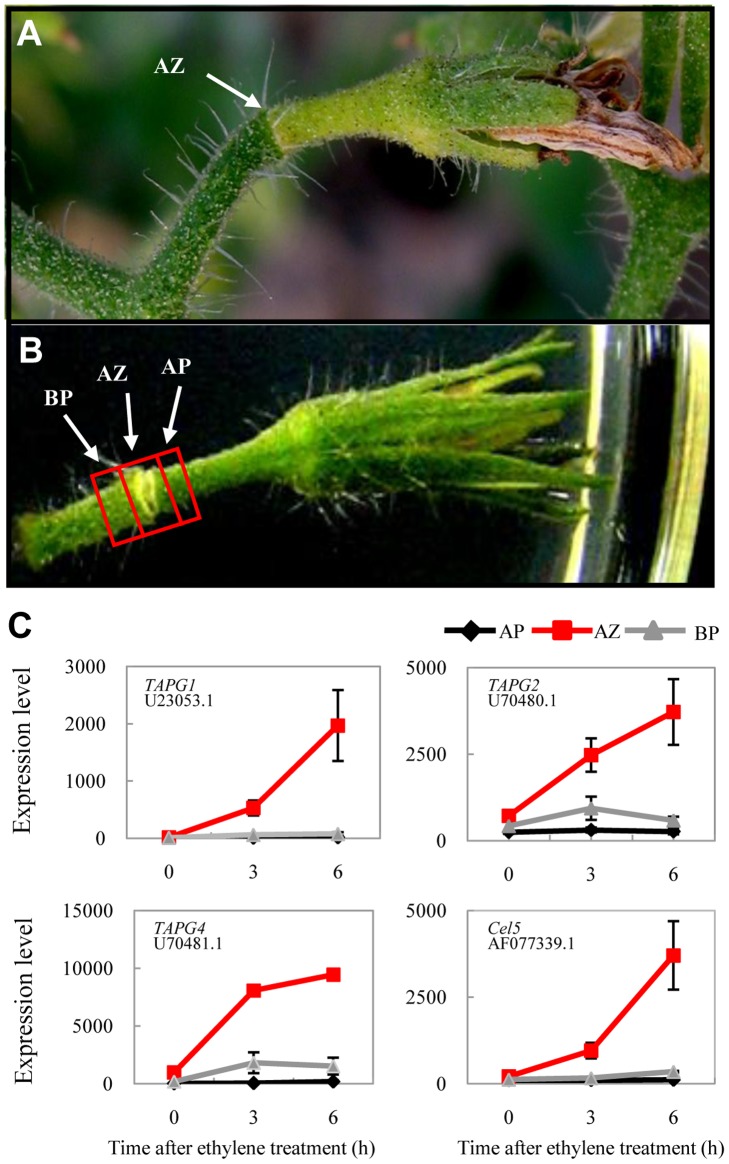
During the natural senescence of a tomato flower, the abscission zone and the apical portion become yellowish, withered, and eventually separated along the abscission zone whereas the basal portion often remains green (Fig.1A; [16], [31]). It has been shown that application of 10 µl/L ethylene can initiate abscission of floral explants within 6 hour (h) while in the absence of ethylene abscission does not occur for up to 5 days [32]. We adopted a higher ethylene concentration (50 µl/L) to treat pre-anthesis floral explants (see Materials and Methods). Under our condition, the abscission zone started to lose green color after about 6 h. To study the early molecular events during tomato pedicel abscission, we manually collected ∼1.5 mm of the abscission zone and neighboring tissues at 0, 3, and 6 h ethylene treatment and froze them immediately in the liquid nitrogen (Fig. 1B). RNAs were extracted and applied in microarray analyses using Affymatrix GeneChip Tomato Genome Array that consists more than 10,000 tomato probe sets from around 9,000 ESTs (or genes). Initial assessment showed that previously characterized abscission zone-specific genes, such as tomato abscission polygalacturonase 1 (TAPG1, U23053.1), TAPG2 (U70480.1), TAPG4 (U70481.1), and the cellulase gene Cel5 (AF077339.1) were specifically induced in the abscission zone under our experimental condition (Fig. 1C), indicating that application of exogenous ethylene had indeed caused changes in internal gene expressions. Twenty five genes with various expression patterns in the three tissues as detected by microarrays were further validated using RT-PCR assays. Except two genes (AI777697 and BT012857.1), all the other 23 genes showed similar expression patterns as detected by the microarray, giving a consistency rate of >92% (Table S1). To assess the effects of wounding incurred during the preparation of flower explants and the subsequent experiment, we chose 68 genes and tested their expression patterns under 6 h air treatment using real time PCR. As shown in Table S2, it seems that wounding to some extent affects expressions of some of the genes under air condition. However, considering the high exogenous ethylene concentration applied, the effect of wounding can be largely ignored. In the following sections, analyses were performed on differentially expressed genes with a fold change ≥2 and a false discovery rate q ≤0.01 at either 3 h and/or 6 h ethylene-treatment when compared with their expression levels at 0 h or between respective pedicel tissues.
Figure 1. A tomato flower in abscission.
A, The morphological change of a tomato pedicel during natural flower senescence; Note the color changes at abscission zone (AZ) and its apical portion while the basal portion remains green. B, An abscising floral explant on the ½ MS media under ethylene treatment, showing the portions of tissues collected for microarray analysis. BP, basal portion; AP, apical portion. C, Induction of cell wall degradation marker genes from microarray analysis in the tomato abscission zone during ethylene-promoted abscission. Marker genes include TAPG1, TAPG2, TAPG4, and Cel5, with corresponding GenBank accession number indicated below. Error bars derived from three biological replicates.
Differential Transcriptome Responses of Three Tomato Pedicel Tissues during Abscission
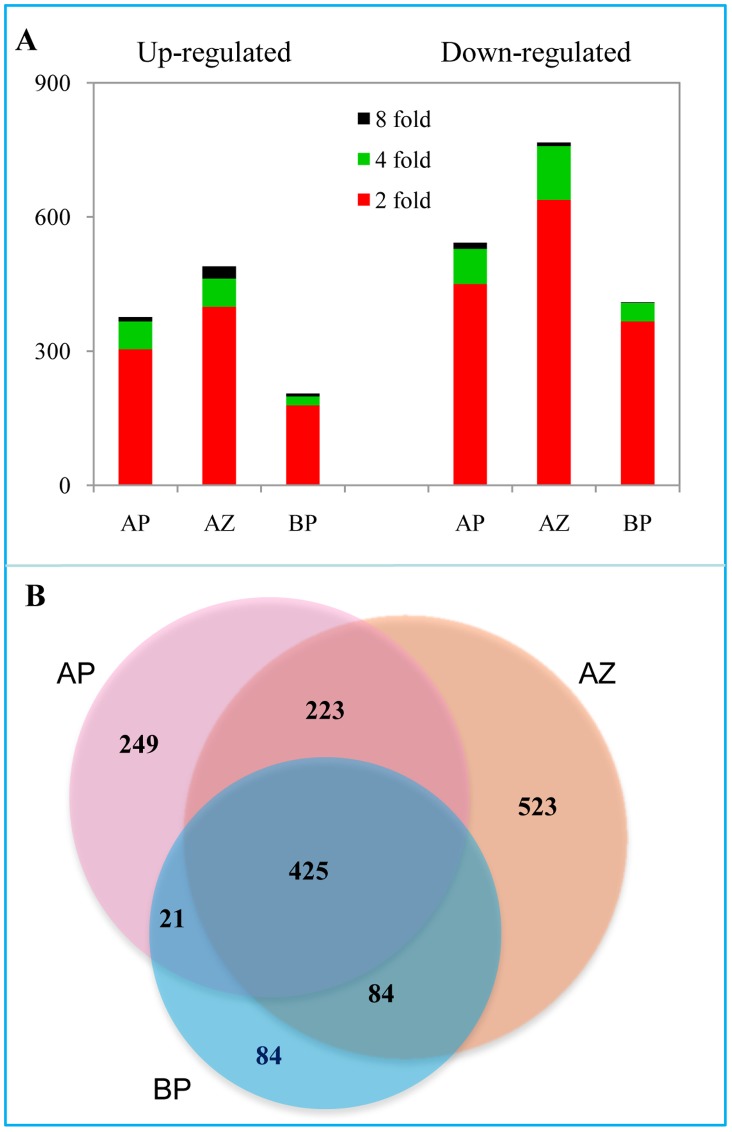
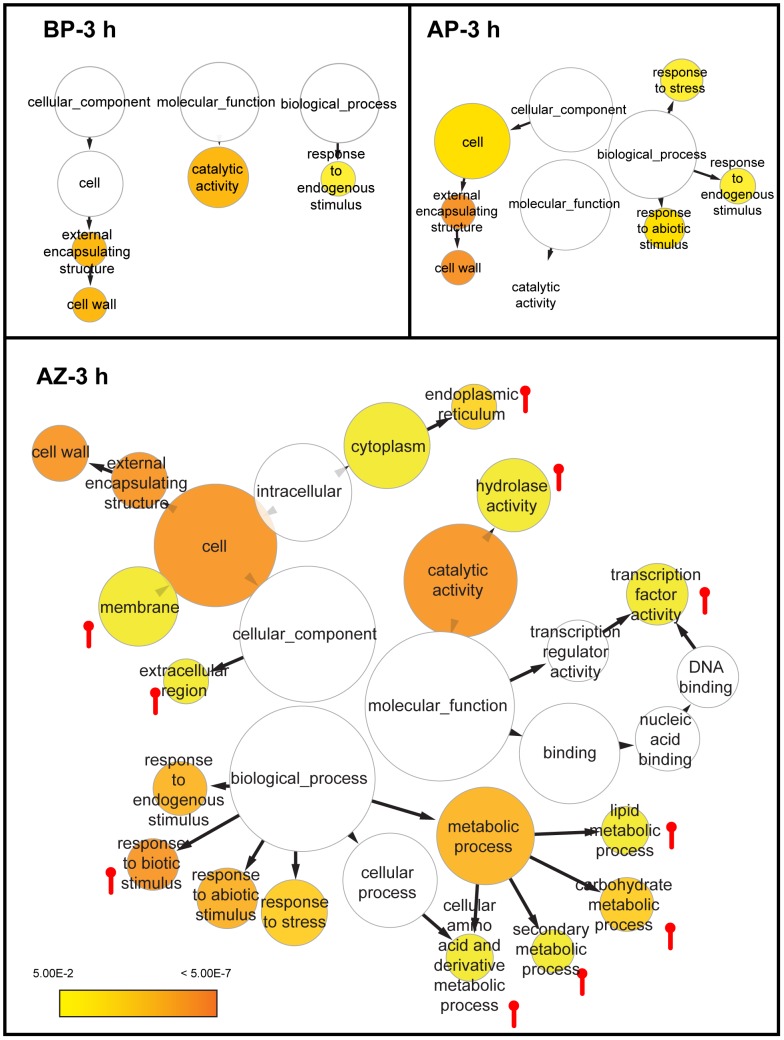
A total of 1,255 ESTs (or in general “genes”) were found to be differentially expressed in the abscission zone, much more than those in the basal portion (614 ESTs) and the apical portion (918 ESTs; Table S3,S4,S5). In all three tissues, more genes were down regulated than up regulated, mostly at the range of 2 fold change (Fig. 2A). A total of 425 differentially expressed genes were shared in all three tissues that may represent the common ethylene responsive genes (Fig. 2B). More genes (41% of the total) were specifically regulated in the abscission zone than in the apical portion (27%) and the basal portion (13.7%) respectively. These numbers demonstrate that the abscission zone is the most active site in gene expression during abscission. Gene Ontology (GO) enrichment analyses showed that the Cellular Component (CC) term “cell wall” and the Molecular Function (MF) term “catalytic activity” were enriched among differentially expressed genes of all three tissues, suggesting a general effect of ethylene on cell wall structures and catalytic enzymes, while the Biological Process (BP) term “response to abiotic stimulus” was shared only by genes from the abscission zone and the apical portion (Fig. 3). The abscission zone conferred several specifically enriched GO terms, including CC terms “membrane”, “extracellular region”, and “endoplasmic reticulum”; MF terms “hydrolase activity” and “transcription factor”; and BP terms “response to biotic stimulus” and various metabolic processes. These observations indicate that the abscission zone is distinctively more sensitive to ethylene than its neighboring tissues during abscission.
Figure 2. Transcriptome responses of different tissues of tomato pedicels during ethylene-promoted abscission.
A, The total numbers of genes differentially expressed (fold changes ≥2, 4, 8; p<0.05) at 3 h and/or 6 h after ethylene treatment. B, Venn diagram showing the overlapping of differentially expressed genes in the abscission zone and flanking tissues (the basal portion and the apical portion). Lists of genes in each tissue are presented in Tables S3,S4,S5.
Figure 3. Enriched GO terms among genes differentially expressed in abscission zone after 3 h ethylene treatment.
A Cytoscape view of enriched GO terms in BinGO where categories in GoSlimPlants (Maere et al., 2005) were used to simplify the analysis. The color bar shows the statistical significance, with enrichment significance level P<0.05, and the false discovery rate FDR <0.05. The size of the node is proportional to the number of genes in the GO category. The pin-like symbols indicate enriched GO terms specifically in the abscission zone relative to the basal portion and the apical portion.
Clustering Analysis of Ethylene Responsive Genes in the Abscission Zone
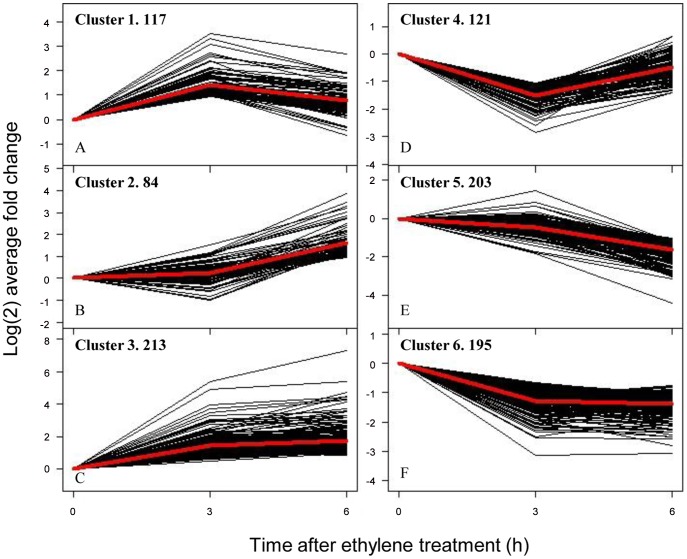
Based on the similarity of the kinetic expression patterns we classified 933 non-redundant differential expression genes of the abscission zone into six clusters. Cluster 1 comprises 117 genes that were transiently induced at 3 h and were then repressed to the initial expression level at 6 h (Fig. 4A). MapMan classification revealed genes involved in amino acid metabolism, development and transcription, and ethylene signaling genes (Table S6). The tomato AGL15 homolog (BG725110) was significantly induced at 3 h in both the abscission zone and the apical portion and was slightly up regulated in the basal portion (Fig. S1). Cluster 2 contains 84 genes that were not induced until 6 h after ethylene treatment (Fig. 4B). Hormone related genes were featured in this cluster, indicating a delay in non-ethylene hormone responses during abscission. Meanwhile, pectate lyases (2), expansins (2), and peroxidases (4) seem to become the major cell wall modification proteins. Cluster 3 genes (213) were induced immediately at 3 h and most maintained high expression levels at 6 h (Fig. 4C). It was the largest group among the six clusters comprising nearly half of all up regulated genes. This cluster was featured by genes encoding TFs, biotic/abiotic responses, ethylene signaling/biogenesis, and cell wall degradation enzymes. The varieties of TFs include C2C2 zinc finger protein (5 genes), AP2 domain (2), WRKYs (2), as well as the MADS-box gene AGL12 homolog (AY098737.2) and Bl. The presence of two ethylene signaling genes (homologs of Arabidopsis EIN4 and AtERF-1) and three synthesis-degradation genes (X58885.1, AW223067, AJ715790.1) indicates rapid responses of ethylene related genes.
Figure 4. K means clustering of 933 non-redundant differentially expressed genes.
Genes in clusters 1–3 are generally induced while genes in clusters 4–6 are repressed.
Clusters 4, 5, and 6 contain genes that were largely suppressed during abscission (Fig. 4D–F). Again, genes associated with “RNA transcription” were predominant in all three clusters. In cluster 4 (121 genes), for instance, there were 10 RNA transcription associated genes, including Arabidopsis LBD4 (LATERAL ORGAN BOUNDARIES domain protein 4; AI774397) and AS1 (ASYMMETRIC LEAVES 1; AF148934.1) homologs. In tomato pedicel, AS1 was clearly down regulated at the abscission zone during abscission, much significantly than in the apical portion and the basal portion (Table S6). Cluster 4 also contains 10 cell wall modification genes, including four genes similar to Arabidopsis expansins A6, A8, A10, and A13. Cluster 5 genes (203) were not repressed until 6 h. There were 21 photosynthesis related genes, 17 of which were for the light reaction in the light harvest center whereas the other seven were enzymes for the Calvin cycle. The steady and profound repression of photosynthesis related genes explains the pale color of the abscission zone tissues during abscission when the basal portion and the apical portion still remain green (Fig. 1B). Cluster 6 consists of 195 genes that were characterized with 14 RNA transcription/regulation genes, including the putative ortholog of Arabidopsis SHOOT MERISTEMLESS (STM, AF000141.1) and the tomato KNOTTED 2 gene (U76408), a homolog of KNAT6. The rapid repression of the KNOX genes may imply possible involvement of meristem gene regulation in tomato pedicel abscission [29].
Responses of Hormone and Cell Wall Related Genes
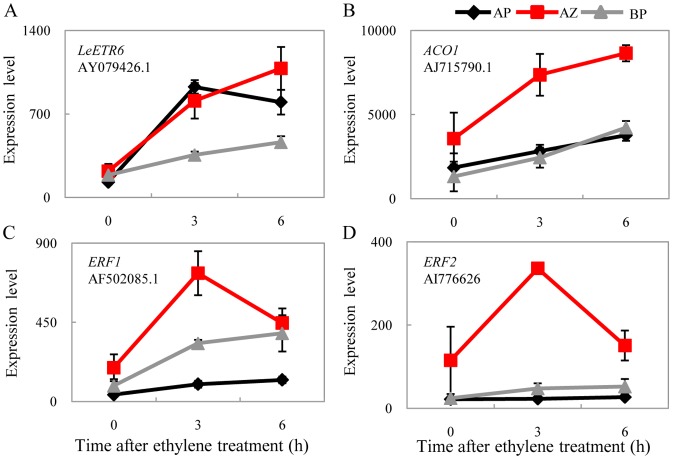
To distinguish the effect caused by endogenous ethylene concentration changes over the general responses to exogenous ethylene treatment, we mostly focused on genes with expression patterns distinguishable among the three tissues. We found that a third (12 out of 37) of differentially expressed hormone genes in the abscission zone was for ethylene, but most of them also differentially expressed in the basal portion and/or the apical portion. There were two ethylene receptors. LeETR6 (AY079426.1) was continuously induced by ethylene in the abscission zone, from 3 h to 6 h (3.67 and 4.89 fold respectively; Fig. 5A). It was more significantly induced in the apical portion (7.18 and 6.19 fold at 3 h and 6 h respectively) and was marginally induced (1.89 and 2.46 fold respectively) in the basal portion. In contrast, LeETR2 (AF043085.1) was repressed in the abscission zone (0.38 and 0.48 fold at 3 h and 6 h respectively; Table S6). Interestingly, LeETR4, which exhibited significant induction in the flower removal experiment [8], was not significantly regulated in our study. In spite of this, one 1-aminocyclopropane-1-carboxylate oxidase gene (ACO1; AJ715790.1) was specifically induced in the abscission zone (Fig. 5B), together with two ERFs (ethylene response factor; AF502085.1, AI776626; Fig. 5C & D), indicating that the specificity of ethylene functions is probably achieved by downstream effective genes.
Figure 5. Response of ethylene related genes.
Induction of (A) the ethylene receptor LeETR6, (B) ethylene biosynthesis gene 1-aminocyclopropane-1-carboxylate oxidase gene (ACO1), and (C-D) two ERF transcription factors during ethylene-promoted abscission.
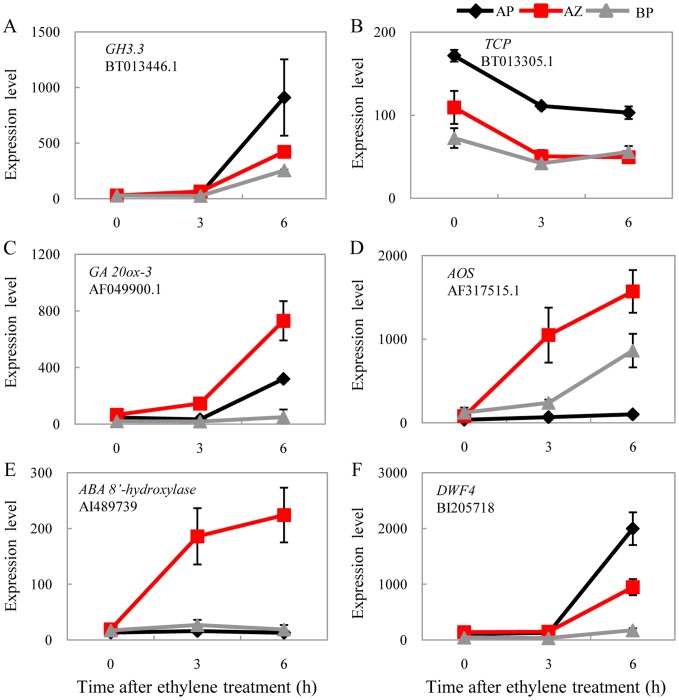
Similar tissue specific patterns were also found for genes related to auxin, the functionally antagonistic hormone of ethylene during abscission which is considered to be generated in the flower and transported along the pedicel [8]. The homolog of the Arabidopsis auxin homeostasis gene GH3.3 (BT013446.1) that encodes the indole-3-acetic acid amido synthetase for IAA-amino acid conjugation was induced 33, 14, and 9 fold in the apical portion, the abscission zone, and the basal portion respectively after 6 h ethylene treatment. Such a distal to proximal expression gradient along the pedicel indicates a negative regulation on auxin levels in the corresponding tissues (Fig. 6A), consistent with the repression patterns of the auxin regulated TCP transcription factor (BT013305.1; Fig. 6B). Except for the induction of ABA 8′-hydroxylase CYP707A3 gene homolog (AI489739) that may promote increased ABA breakdown specifically in the abscission zone, abscission induced differential responses in rate limiting genes were found for gibberellin (gibberellin 20-oxidase-3, AF049900.1), jasmonic acid (ALLENE OXIDE SYNTHASE, or AOS; AF317515.1), and brassinosteroid (DWF4, BI205718) in the three pedicel tissues (Fig. 6C–F). It has to be noted that ACO1 and DWF4 were also strongly induced by wounding (Table S2).
Figure 6. Coordinated expression of auxin and other hormone related genes.
Note the formation of distal to proximal gradient expression pattern for GH3.3 (A), TCP (B), and DWF4 (F). GA 20ox-3 (C), AOS (D), and ABA 8′-hydoxylase (E) genes were most highly expressed in the abscission zone during abscission.
Cell wall and middle lamella degradation is one of the major targets of abscission. Despite the overall complexity in expression patterns for cell wall modification genes, three PGs and one cellulase were specifically induced in abscising abscission zones (Fig. 1C), while three XTHs were induced with two of them XTH8 (X82684.1) and SlXTH (AY497476.1) only transiently at 3 h (Fig. S2). Two expansins, EXP11 (AJ560646.1) and the Arabidopsis AtEXPa4 homolog (AF548376.1) were later induced significantly at 6 h, together with one pectin lyase gene (BT012714.1) that is involved in cell wall enlargement and pectin re-modification [33], [34], [35]. Thus, our data confirmed previous reports how cell wall modification genes respond and corroborated with the functions of ethylene in abscission.
Abscission Zone-preferential Expression of Key Meristem Protein Complex Genes
Expression domains may indicate unique functions of a gene in a particular tissue. A total of 107 genes were specifically expressed in the abscission zone relative to their expression levels in the apical portion and the basal portion (Table 1 & Table S7). Among them, WUSCHEL (WUS), Bl, and Ls also displayed differential expressions between wild type and mc mutant (and hence jointless) pedicels [36]. What was unexpected was the abscission zone specific expression patterns for all three homologs of Arabidopsis KNAT6 (U76408), BELL-like homeodomain protein 1 (BLH1, AF375967), and OFP (OVATE FAMILY PROTEIN, AY140893) whose family members can potentially form ternary complexes critical for meristem activities [37], [38], [39], [40]. Homologs of a LATERAL ORGAN BOUNDARIES DOMAIN PROTEIN 1(LBD1) and Myb78 were also highly expressed in the abscission zone. In addition, the expression of the homolog for the second key meristem gene STM was the highest in the abscission zone, but was also expressed in the basal portion and the apical portion (Table S7). Since these genes have been shown to interact with each other in Arabidopsis shoot apical meristem and axillary meristem, their preferential expressions in the abscission zone support the idea that meristem activity genes play roles in maintaining the undeveloped status of tomato abscission zone cells [16]. On the contrary, the expression of two MADS-box genes TAG1 (the tomato AGAMOUS) and TDR8 (the Arabidopsis SHATTERPROOF 2 homolog) were excluded from the abscission zone (Table 1). And so was a SQUAMOSA-promoter binding protein gene LeSPL3 (BI929558) that could potentially regulate the abscission development gene MC. The sequestration of TAG1 and TDR8 expressions in the abscission zone, in contrast to the requirement of two additional MADS-box genes J and MC, demonstrates diverse roles of MADS-box genes in abscission zone functions.
Table 1. List of abscission zone-specific transcription factors.
| Probe Set ID | GenBank# | Annotation | Arabidopsis homolog1 | Fold change | Raw readings | |||
| AZ:AP | AZ:BP | AP | AZ | BP | ||||
| Preferentially expressed in AZ | ||||||||
| Les.3693.1.S1_at | AF426174 | BLIND | 24.79 | 9.73 | 14.9 | 368.41 | 37.88 | |
| LesAffx.62669.2.A1_at | AW442297 | LOB domain protein 1 | LBD1(AT1G07900) | 20.35 | 3.15 | 34.2 | 696.58 | 221.31 |
| Les.4136.1.S1_at | AJ538329 | WUSCHEL | 16.62 | 15.32 | 61.2 | 1016.3 | 66.33 | |
| Les.3571.1.S1_at | U76408 | KNOTTED 2 protein | KNAT6 (AT1G23380) | 10.86 | 2.05 | 51.4 | 558.11 | 271.83 |
| Les.4309.1.S1_at | AF375967 | BELL-like homeodomainprotein 4 | BLH1(AT2G02850) | 10.61 | 4.29 | 30.5 | 323.88 | 75.47 |
| Les.69.1.S1_at | AF098674 | LATERAL SUPPRESSOR | 8 | 7.73 | 17 | 135.83 | 17.58 | |
| Les.3819.1.S1_at | AY140893 | OVATE | AT2G18500 | 4.3 | 3.34 | 260 | 1120.7 | 335.53 |
| LesAffx.62138.1.S1_at | AW737374 | MYP Cpm10 | MYB78 (AT5G49620) | 2.63 | 2.7 | 110 | 289.36 | 107.14 |
| Preferentially expressed in AP and BP | ||||||||
| Les.46.1.S1_at | BT014391 | TDR8 | SHP2 (AT2G42830) | 0.49 | 0.3 | 148 | 71.98 | 238.13 |
| Les.3065.1.S1_at | BI929558 | SQUAMOSA PROMOTER-BINDING PROTEIN LIKE protein | SPL3 (AT2G33810) | 0.32 | 0.5 | 965 | 309.75 | 618.07 |
| LesAffx.53591.1.S1_at | AI899018 | MYB, methyl jasmonateinduced | MYB13 (AT1G06180) | 0.31 | 0.5 | 390 | 121.56 | 242.3 |
| Les.3620.1.S1_at | L26295 | TAG1 | 0.31 | 0.47 | 335 | 102.7 | 220.81 | |
Best Arabidopsis homolog if not functional studied in tomato; See text for tomato gene details.
Transcription Repression of Key Meristem Activity Genes in Abscission Zone during Abscission
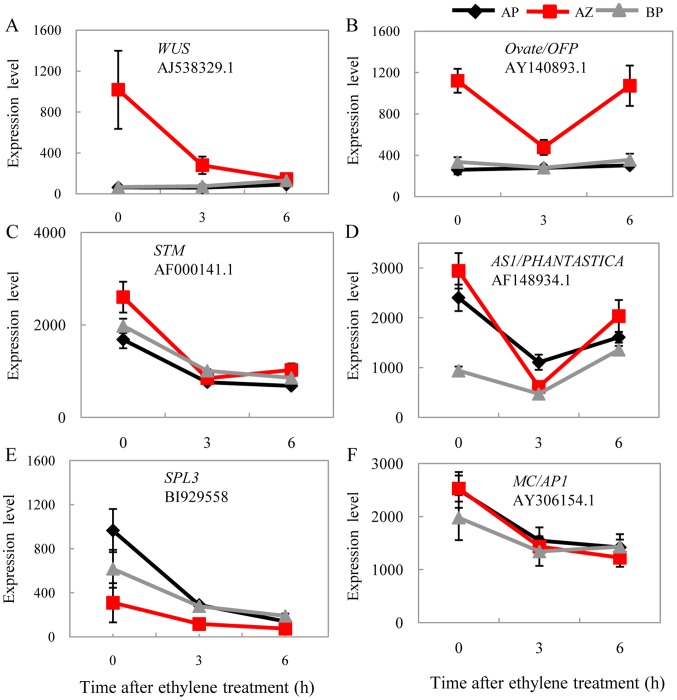
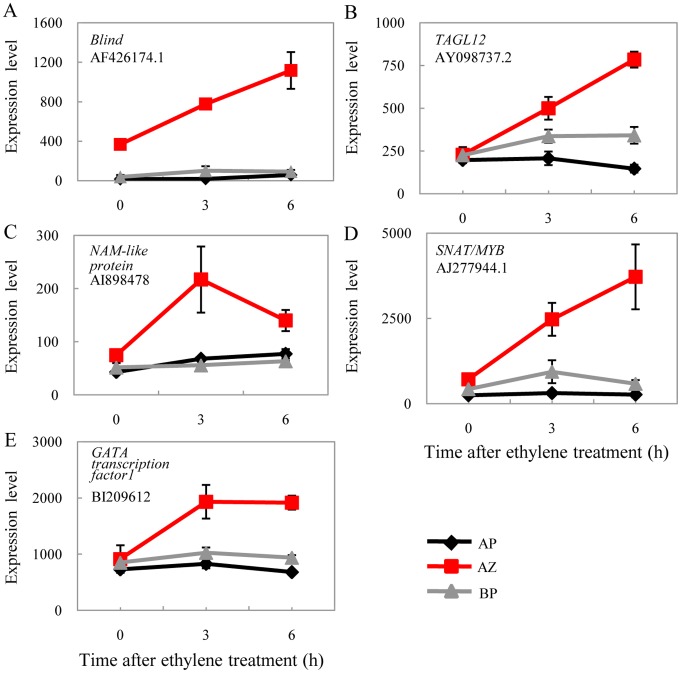
Despite their preferred expressions in the abscission zone, homologs of WUS and the putative interacting partner OFP were clearly repressed during abscission (Fig. 7A & B), while their expressions in the basal portion and the apical portion remained unchanged. The tomato WUS displayed strong tissue-specific expression in the abscission zone, 15 fold higher than both the basal portion and the apical portion. The expression of WUS was reduced to the basal level as in the basal portion and the apical portion after 6 h ethylene-promoted abscission. As a matter of fact, STM was also significantly repressed during abscission, but in all three of the tissues. And so was the AS1 (ASYMETRIC LEAVES1; Fig. 7C & D). In Arabidopsis, all these genes were found to be functionally related in the shoot apical meristem and the axillary meristem that regulate (or being regulated by) KNOX genes [38]. The co-regulation of these genes in tomato pedicel abscission zones and during abscission support the under developed meristem status of the abscission zone cells [16] and may be also necessary for post-abscission cell differentiation [41]. Except for the KNOX genes, the expression of the SPL3 homolog was also further repressed in all three tissues, together with MC, the APETALA1 (AP1) and SQUAMOSA (SQUA) orthologs and potential regulating target [42], but not in the abscission zone-specific manner (Fig. 7E & F). In monocots, a SPL gene LIGULELESS 1 (LG1) and its orthologs have been found to be required for the proper development of auricles of grass plants (maize, [43]; barley, [44]; rice, [45]), which define regions equivalent to tomato abscission zones. These data therefore prompt speculation of a common SPL regulatory pathway for the abscission zone in both monocots and eudicots, which deserves further investigation. In contrast, TAGL12 and Bl were continuously and specifically induced in the abscission zone during ethylene treatment, together with three other transcription factors (Fig. 8). Expression patterns of these genes coincide with abscission genes such as those for hormone biogenesis and signaling and cell wall modifications. The repression of meristem genes and induction of various transcription factors may indicate a certain regulatory relationship among them.
Figure 7. Repression of meristem activity genes during abscission.
Homologs of WUS (A) and OVATE (B) are specifically repressed in the abscission zone. STM (C) and AS1 (D) were repressed in the abscission zone, but also in one or more additional tissues. E–F show the repression of the SPL3 homolog and the tomato MC gene, a putative ortholog of the Antirrhinum SQUAMOSA gene.
Figure 8. Induction of transcription factor genes during abscission.
(A) Blind is specifically expressed in the abscission zone and is further induced significantly during the course of abscission. (B-E) De novo significant induction of four transcription factors.
Discussion
Organ abscission is one of the key steps in the plant life cycle. Both promoted abscission, such as in cotton defoliation and mechanical citrus harvest, and delayed abscission, such as in most vegetables and horticultural crops, are desirable for farmers to provide higher quality products [15]. The effect of ethylene on regulating plant organ abscission has long been established [46] and applied in horticulture and agriculture practices. But comparative transcriptome profiling of the three pedicel tissues is still not available. The better understanding of the molecular events associated with the abscission zone development and physiology may provide new insights in the basic biology of abscission and assist its manipulation in an agriculturally favorable manner.
Experimental Approaches to Study Abscission Zone-specific Gene Profiles
The distinct location of the tomato flower abscission zone makes it one of the most convenient materials for abscission study. Our work, together with Meir et al. (2010) and Nakano et al. (2012), demonstrate that results derived from manually collected abscission zone tissues were reliable and repeatable. However, despite a large number of genes were differentially expressed in each experiment, only a portion of them were co-regulated under both conditions, indicating that different strategies reveal different biological scenarios. One difference is in material preparation. For instance, we used pedicel explants with flower attached while Meir et al. (2010) used pedicels with flowers removed. Similarly, Meir et al. (2010) studied a 14-h time course and we are interested in the early molecular events at 6 h with relatively high ethylene concentration. These differences should contribute to the discrepancies in gene expression patterns. Despite this, our approach was substantiated by the expected expression patterns of the abscission zone marker genes such as TAPG2 and TAPG4 that have been experimentally characterized [8], [47]. Particularly, most abscission zone differentially expressed genes in our study showed unique expression patterns when compared with its expressions in the basal and apical portions, demonstrating that responses of different tissues during abscission are underlined by different molecular mechanisms.
Differential Gene Expressions among Tomato Pedicel Tissues
Despite its simplicity in cell type (small, undifferentiated), our transcriptome analyses showed that the tomato pedicel abscission zone conferred a dominant number of differentially expressed genes over its neighboring tissues in terms of both tissue specificity and abscission. Consistent with previous work, a variety of genes were found in the abscission zone. Major categories include those for the degradation and synthesis of cell wall polysaccharides (PGs. XTHs, CELs, EXPs), hormone metabolism (ethylene, jasmonic acid, auxin, abscisic acid, brassinosteroid, gibberellin), defense and interaction with the environment (ROS, PRs, LRRs), secondary metabolism (flavonoids, isoprenoids, phenylpropanoids), PS, signaling (kinases, calcium-binding proteins, G-proteins), and transcription (TFs). Generally, genes involved in stress and amino acid metabolism were largely induced while those for PS, secondary metabolism, cell wall, and transport were significantly suppressed, indicating a slow down of biochemical reactions inside the abscission zone and the activation of the stress response and protection systems during abscission.
The interplay of plant hormones has been widely reported. Similar to previously reported, the induction of ethylene biosynthesis and signaling genes were characterized among differentially expressed genes during abscission, although none of the three ethylene receptors exhibited abscission zone-specific patterns. As expected, auxin regulating genes were largely suppressed, mostly in non-tissue-specific manners, displaying an antagonistic interaction between these two major players in abscission. The expressions of auxin related genes, such as the auxin homeostasis gene GH3.3, often form steep gradient from distal to proximal along the pedicel, consistent with the proposed polar transportation of auxin from the flower to the base of the pedicel [7], [8], [48]. For other hormones, the induction of rate limiting enzyme genes for gibberellin, jasmonic acid, and brassinosteroid suggests a coordinated regulatory modes among these hormone related genes, while enhanced expression of the ABA catabolism gene 8′-hydroxylase CYP707A3 [49] may indicate increased abscisic acid breakdown during abscission. It has to be noted that the immediate responses of cell wall modification genes in the abscission zone, especially those encoding TAPGs and Cels should provide additional evidence for the effectiveness and specificity of our experimental approach.
Transcription Regulation in the Tomato Flower Abscission Zone: From Development to Abscission
Abscission is generally considered to be achieved through four major steps: the development of the abscission zone cells, the acquisition of competence to respond to abscission signals, the activation of abscission, and the differentiation of a protective layer at the distal end of the basal portion [50]. This last step requires (1) the abscission zone cells to confer meristem-like nature and (2) the meristem status be removed by repressing the expression of meristem maintenance genes, such as WUS and STM. Our genome-wide tissue specific expression analysis of tomato pedicels supports such a notion. We found that key meristem maintenance genes were preferentially expressed in the abscission zone, including homologs of Arabidopsis WUS, KNAT6, LBD1, LH1and OFP, as well as the axillary meristem initiation genes Bl and Ls. Our findings are consistent with the recent discoveries in Arabidopsis where shoot apical meristem and axillary meristem genes work together for lateral organ development including leaf and flower, in which the formation of the abscission zone is an integrative step [51], [52]. Unexpectedly, homologs of WUS and its potential functional partner OFP appear to be specifically repressed in the abscission zone during abscission when their expression level in the basal portion and the apical portion remained largely unchanged. In fact, STM, as well as its closely interacting gene AS1, was also significantly repressed in the abscission zone, but not in an abscission zone specific manner. These data together corroborate the idea that reduction of meristem activity gene expression is necessary for additional cell differentiation in the abscission zone during or after abscission. Finally, the modulation of the LeSPL3 homolog could add another layer of regulation of abscission zone functions by microRNAs [53]. Since similar genes are found for grass auricle development, the regulatory scheme proposed in our work may represent a common scheme for both monocot and eudicot abscission.
Materials and Methods
Plant Materials, Growth Conditions, and Ethylene Treatment
Tomato plants, Solanum lycopersicon accession LA3021, were grown under greenhouse conditions with a 16-h photoperiod and 25°C constant temperatures. Floral explants were inserted into a petri dish with 1/2 MS agar located inside in a Plexiglass box in a tray filled with a layer of water. Ethylene gas from an airtight vessel was conducted into the box with a soft tube through the water and reached the final concentration of 50 ul/L. Three duplicates were prepared so that tissues can be obtained at 0, 3, and 6 h once treatment started.
Flower Dissection and RNA Extraction
Pedicels of pre-anthesis flowers were dissected using a sharp razor blade into three segments (abscission zone, distal, and proximal tissues; Fig. 1B), each about 1.5 mm, and were put into liquid nitrogen immediately. RNA was isolated using TRIZOL (Invitrogen) according to manufacturer’s instructions. Three independent biology repeats were performed.
Microarray Analysis
Microarray hybridization and initial data subtraction were performed by Bo-ao company (Beijing) using the Affymatrix platform. We used Robust Multichip Analysis [54] in the Affymatrix package for initially probe signal summarization, normalization, and background subtraction with default parameters. Data comparison of different time points and tissues were performed using the SAM software [55]. The approach of [56] was applied to generate adjusted P values (q values) so as to control the level of false discoveries due to multiple comparisons. Three biology replicates were performed per treatment. Differentially expressed genes were screened using threshold fold change ≥2 and FDR (false discovery rate) ≤0.01. Microarray probe sequences were further annotated by comparison with Arabidopsis proteins using BlastX. Redundant ESTs that were annotated to be the same Arabidopsis genes were removed. Non-redundant dataset were imported into the MapMan program for functional classification. Gene ontology enrichment analysis was performed in the BinGO website [57]. The mean normalized signal intensity for each probe set was calculated from three biological replications. Signal intensities of each gene were standardized so that each gene (row) would have a mean of zero and an SD of 1. Six clusters were chosen by the gap statistic [58]. The K-mean clustering method was used to assign membership of each probe set [59].
Semi-quantitative and Real Time RT-PCR
RNA samples were reverse transcribed using superscript II reverse transcriptase (Invitrogen). cDNA then diluted 10 times for semi-quantitative RT-PCR reaction. To ensure equal amounts of cDNA, a control reaction was performed with the tomato ACTIN gene. Quantitative real time RT-PCR reaction was performed in triplicate using QuantiTect SYBR Green PCR Master Mix (Qiagen) on an Iq5 Real Time PCR Detection System (Bio-Rad). Average threshold cycle (CT) values were calculated for each gene of interest, and were normalized and used to calculate relative transcript levels of the transcript using the 2−ΔΔCT method described by [60]. The GAPDH (glyceraldehyde-3-phosphate dehydrogenase) from tomato was used as an internal standard for normalization.
Supporting Information
Modulation of additional transcription factors in tomato pedicel tissues during ethylene-promoted abscission.
(TIF)
Induction of additional cell wall modification genes in tomato pedicel tissues during ethylene-promoted abscission.
(TIF)
RT-PCR validation of differentially expressed genes. RNA concentrations were adjusted using the actin gene.
(PDF)
Change in gene expression induced by ethylene compared to air for 68 genes.
(PDF)
Differentially expressed genes after 3 h or 6 h ethylene treatment in the abscission zone.
(PDF)
Differentially expressed genes in the basal portion after 3 h or 6 h ethylene treatment.
(PDF)
Differentially expressed genes in the apical portion after 3 h or 6 h of ethylene treatment.
(PDF)
MapMan classification of differentially expressed non-redundant genes in abscising abscission zone.
(PDF)
The full list of the abscission zone-preferentially expressed genes.
(PDF)
Acknowledgments
We thank the handling editor and the two anonymous reviewers for very constructive suggestions.
Funding Statement
This work is supported in part by Natural Science Foundation of China (number 31070260) and National Transgenic Key project of MOA of China (number 2013ZX08009-001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lashbrook CC, Gonzalez-Bosch C, Bennett AB (1994) Two divergent endo-beta-1,4-glucanase genes exhibit overlapping expression in ripening fruit and abscising flowers. Plant Cell 6: 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. del Campillo E, Bennett AB (1996) Pedicel breakstrength and cellulase gene expression during tomato flower abscission. Plant Physiol 111: 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cho HT, Cosgrove DJ (2000) Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci U S A 97: 9783–9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Taylor, Whitelaw C (2001) Signals in abscission. New Phytol 151: 323–339. [Google Scholar]
- 5. Ogawa M, Kay P, Wilson S, Swain SM (2009) ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21: 216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roberts JA, Elliott KA, Gonzalez-Carranza ZH (2002) Abscission, dehiscence, and other cell separation processes. Annu Rev Plant Biol 53: 131–158. [DOI] [PubMed] [Google Scholar]
- 7. McManus MT (2008) Further examination of abscission zone cells as ethylene target cells in higher plants. Ann Bot (Lond) 101: 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meir S, Philosoph-Hadas S, Sundaresan S, Selvaraj KS, Burd S, et al. (2010) Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol 154: 1929–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osborne DJ, McManus MT (2005) Hormones, signals and target cells in plant development.: Cambridge University Press. 252 p.
- 10. Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, et al. (2008) Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci U S A 105: 15629–15634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Adamczyk BJ, Lehti-Shiu MD, Fernandez DE (2007) The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J 50: 1007–1019. [DOI] [PubMed] [Google Scholar]
- 12. Wang X, Xu W, Ma L, Fu Z, Deng X, et al. (2006) Requirement of KNAT1/BP for the Development of Abscission Zones in Arabidopsis thaliana. J Integr Plant Biol 48: 15–26. [Google Scholar]
- 13. Cai S, Lashbrook CC (2008) Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiol 146: 1305–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patterson SE, Bleecker AB (2004) Ethylene-dependent and -independent processes associated with floral organ abscission in Arabidopsis. Plant Physiol 134: 194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Binder BM, Patterson SE (2009) Ethylene-dependent and -independent regulation of abscission. Stewart Postharvest Review 5: 1–10. [Google Scholar]
- 16. van Nocker S (2009) Development of the abscission zone. Stewart Postharvest Review 1: 1–6. [Google Scholar]
- 17. Osborne DJ (1989) Abscission. Crit Rev in Plant Sci 8: 103–129. [Google Scholar]
- 18. Gawadi AG, Avery GSJ (1950) Leaf abscission and the so-called “abscission layer”. Am J Bot 37: 172–180. [Google Scholar]
- 19. Rolland-Lagan AG (2008) Vein patterning in growing leaves: axes and polarities. Curr Opin Genet Dev 18: 348–353. [DOI] [PubMed] [Google Scholar]
- 20.van Nocker S, Ek-Ramos J (2007) Control of flowering time. In Regulation of transcription in plants. Grasser KD Ed. Plant Reviews (Blackwell Publishing, Ltd.).
- 21. Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K (1999) The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci U S A 96: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mao L, Begum D, Chuang HW, Budiman MA, Szymkowiak EJ, et al. (2000) JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 406: 910–913. [DOI] [PubMed] [Google Scholar]
- 23. Yang TJ, Lee S, Chang SB, Yu Y, de Jong H, et al. (2005) In-depth sequence analysis of the tomato chromosome 12 centromeric region: identification of a large CAA block and characterization of pericentromere retrotranposons. Chromosoma 114: 103–117. [DOI] [PubMed] [Google Scholar]
- 24. Schmitz G, Tillmann E, Carriero F, Fiore C, Cellini F, et al. (2002) The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc Natl Acad Sci U S A 99: 1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Szymkowiak EJ, Irish EE (2006) JOINTLESS suppresses sympodial identity in inflorescence meristems of tomato. Planta 223: 646–658. [DOI] [PubMed] [Google Scholar]
- 26. Quinet M, Kinet JM, Lutts S (2011) Flowering response of the uniflora:blind:self-pruning and jointless:uniflora:self-pruning tomato (Solanum lycopersicum) triple mutants. Physiol Plant 141: 166–176. [DOI] [PubMed] [Google Scholar]
- 27. Muller D, Schmitz G, Theres K (2006) Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18: 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keller T, Abbott J, Moritz T, Doerner P (2006) Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18: 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Belles-Boix E, Hamant O, Witiak SM, Morin H, Traas J, et al. (2006) KNAT6: an Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 18: 1900–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Butenko MA, Shi CL, Aalen RB (2012) KNAT1, KNAT2 and KNAT6 act downstream in the IDA-HAE/HSL2 signaling pathway to regulate floral organ abscission. Plant Signal Behav 7: 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Addicott FT (1982) Abscission. Berkeley, CA: University of California Press.
- 32. Roberts JA, Schindler CB, Tucker GA (1984) Ethylene-promoted tomato flower abscission and the possible involvement of an inhibitor. Planta 160: 159–163. [DOI] [PubMed] [Google Scholar]
- 33. Cosgrove DJ, Li LC, Cho HT, Hoffmann-Benning S, Moore RC, et al. (2002) The growing world of expansins. Plant Cell Physiol 43: 1436–1444. [DOI] [PubMed] [Google Scholar]
- 34. Zenoni S, Fasoli M, Tornielli GB, Dal Santo S, Sanson A, et al. (2011) Overexpression of PhEXPA1 increases cell size, modifies cell wall polymer composition and affects the timing of axillary meristem development in Petunia hybrida. New Phytol 191: 662–677. [DOI] [PubMed] [Google Scholar]
- 35. Lee Y, Choi D, Kende H (2001) Expansins: ever-expanding numbers and functions. Curr Opin Plant Biol 4: 527–532. [DOI] [PubMed] [Google Scholar]
- 36. Nakano T, Kimbara J, Fujisawa M, Kitagawa M, Ihashi N, et al. (2012) MACROCALYX and JOINTLESS Interact in the transcriptional regulation of tomato fruit abscission zone development. Plant Physiol 158: 439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hackbusch J, Richter K, Muller J, Salamini F, Uhrig JF (2005) A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc Natl Acad Sci U S A 102: 4908–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hake S, Smith HM, Holtan H, Magnani E, Mele G, et al. (2004) The role of knox genes in plant development. Annu Rev Cell Dev Biol 20: 125–151. [DOI] [PubMed] [Google Scholar]
- 39. Hamant O, Pautot V (2010) Plant development: A TALE story. Comptes Rendus Biologies 333: 371–381. [DOI] [PubMed] [Google Scholar]
- 40. Li E, Wang S, Liu Y, Chen JG, Douglas CJ (2011) OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J 67: 328–341. [DOI] [PubMed] [Google Scholar]
- 41. Bleecker AB, Patterson SE (1997) Last exit: senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9: 1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Preston JC, Hileman LC (2010) SQUAMOSA-PROMOTER BINDING PROTEIN 1 initiates flowering in Antirrhinum majus through the activation of meristem identity genes. Plant J 62: 704–712. [DOI] [PubMed] [Google Scholar]
- 43. Moreno MA, Harper LC, Krueger RW, Dellaporta SL, Freeling M (1997) liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev 11: 616–628. [DOI] [PubMed] [Google Scholar]
- 44. Rossini L, Vecchietti A, Nicoloso L, Stein N, Franzago S, et al. (2006) Candidate genes for barley mutants involved in plant architecture: an in silico approach. Theor Appl Genet 112: 1073–1085. [DOI] [PubMed] [Google Scholar]
- 45. Lee J, Park JJ, Kim SL, Yim J, An G (2007) Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol Biol 65: 487–499. [DOI] [PubMed] [Google Scholar]
- 46. Jackson MB, Osborne DJ (1970) Ethylene, the natural regulator of leaf abscission. Nature 225: 1019–1022. [DOI] [PubMed] [Google Scholar]
- 47. Hong SB, Sexton R, Tucker ML (2000) Analysis of gene promoters for two tomato polygalacturonases expressed in abscission zones and the stigma. Plant Physiol 123: 869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Osborne DJ, Sargent JA (1976) The positional differentiation of abscission zones during the development of leaves of Sambucus nigra and the response of the cells to auxin and ethylene. Planta 132: 197–204. [DOI] [PubMed] [Google Scholar]
- 49. Umezawa T, Okamoto M, Kushiro T, Nambara E, Oono Y, et al. (2006) CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J 46: 171–182. [DOI] [PubMed] [Google Scholar]
- 50. Patterson SE (2001) Cutting loose. Abscission and dehiscence in Arabidopsis. Plant Physiol 126: 494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rutjens B, Bao D, van Eck-Stouten E, Brand M, Smeekens S, et al. (2009) Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J 58: 641–654. [DOI] [PubMed] [Google Scholar]
- 52. Majer C, Hochholdinger F (2011) Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci 16: 47–52. [DOI] [PubMed] [Google Scholar]
- 53. Chen X, Zhang Z, Liu D, Zhang K, Li A, et al. (2010) SQUAMOSA Promoter-Binding Protein-Like Transcription Factors: Star Players for Plant Growth and Development. J Integr Plant Biol 52: 946–951. [DOI] [PubMed] [Google Scholar]
- 54. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- 55. Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57: 289–300. [Google Scholar]
- 57. Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449. [DOI] [PubMed] [Google Scholar]
- 58. Tibshirani R, Walther G, Hastie T (2001) Estimating the number of clusters in a data set via the gap statistic. J R Statist Soc B 63: 411–423. [Google Scholar]
- 59.Kaufman L, Rousseeuw P (1990) Finding Groups in Data: An Introduction to Cluster Analysis. New York: Wiley.
- 60. Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Modulation of additional transcription factors in tomato pedicel tissues during ethylene-promoted abscission.
(TIF)
Induction of additional cell wall modification genes in tomato pedicel tissues during ethylene-promoted abscission.
(TIF)
RT-PCR validation of differentially expressed genes. RNA concentrations were adjusted using the actin gene.
(PDF)
Change in gene expression induced by ethylene compared to air for 68 genes.
(PDF)
Differentially expressed genes after 3 h or 6 h ethylene treatment in the abscission zone.
(PDF)
Differentially expressed genes in the basal portion after 3 h or 6 h ethylene treatment.
(PDF)
Differentially expressed genes in the apical portion after 3 h or 6 h of ethylene treatment.
(PDF)
MapMan classification of differentially expressed non-redundant genes in abscising abscission zone.
(PDF)
The full list of the abscission zone-preferentially expressed genes.
(PDF)