Abstract
DNA polymerases select for the incorporation of deoxyribonucleotide triphosphates (dNTPs) using amino acid side-chains that act as a “steric-gate” to bar improper incorporation of rNTPs. An additional factor in the selection of nucleotide substrates resides in the preferred geometry for the furanose moiety of the incoming nucleotide triphosphate. We have probed the role of sugar geometry during nucleotide selection by model DNA polymerases from Sulfolobus solfataricus using fixed conformation nucleotide analogues. North-methanocarba-dATP (N-MC-dATP) locks the central ring into a RNA-type (C2′-exo, North) conformation near a C3′-endo pucker and South-methanocarba-dATP (S-MC-dATP) locks the central ring system into a (C3′-exo, South) conformation near a C2′-endo pucker. Dpo4 preferentially inserts N-MC-dATP and in the crystal structure of Dpo4 in complex with N-MC-dAMP, the nucleotide analogue superimposes almost perfectly with Dpo4 bound to unmodified dATP. Biochemical assays indicate that the S. solfataricus B-family DNA polymerase Dpo1 can insert and extend from both N-MC-dATP and S-MC-dATP. In this respect, Dpo1 is unexpectedly more tolerant of substrate conformation than Dpo4. The crystal structure of Dpo4 bound to S-MC-dADP shows that poor incorporation of the Southern pucker by the Y-family polymerase results from a hydrogen bond between the 3′-OH group of the nucleotide analogue and the OH group of the steric gate residue, Tyr12, shifting the S-MC-dADP molecule away from the dNTP binding pocket and distorting the base pair at the primer-template junction. These results provide insights into substrate specificity of DNA polymerases, as well as molecular mechanisms that act as a barrier against insertion of rNTPs.
Keywords: DNA polymerase, nucleotide selection, X-ray crystallography
INTRODUCTION
Barriers to accurate replication of the genome are many and diverse in nature (1). Evolution has retained overlapping DNA damage and replication stress responses that rely upon complex signalling cascades in order to maintain genomic integrity (2). Residing within these cellular processes are molecular checks against aberrant genomic replication that depend upon the structure and function of certain nucleic acid metabolizing enzymes. In this respect, DNA polymerases are an important part of the barrier against genomic instability, as they provide a means of selecting the appropriate substrate from a pool of nucleotide triphosphates to be inserted opposite a given base, whether the base is damaged or not (3,4). Outside of covalent modification to canonical nucleic acids (i.e. DNA damage), one of the most common threats to genomic stability is the aberrant incorporation of ribonucleotides in place of 2′-deoxyribonucleotides (5,6). The addition of a 2′-hydroxyl (OH) group to the furanose moiety of nucleic acids has dramatic and biologically important ramifications on the topology of the nucleic acid helix, protein-nucleic acid interactions and the overall stability of the macromolecule. Moreover, rNTP levels are higher than the corresponding dNTPs inside eukaryotic cells (7). The higher concentration of rNTPs inside of cells is a barrier to accurate DNA replication since it creates sub-optimal conditions for selection of dNTPs. It is, therefore, of great importance that we understand mechanisms that promote substrate specificity by DNA polymerases.
DNA polymerases discriminate against mis-insertion of ribonucleotide triphosphates (rNTPs) but the stringency of selecting deoxyribonucleotides (dNTPs) over rNTPs can vary several orders of magnitude from less than 10-fold to >106-fold depending upon the polymerase under investigation and the identity of the base pair (base pair) being examined (7). To a large degree, the difference in nucleotide selectivity can be attributed to structural differences between DNA polymerase families and several studies have elucidated the active site residues responsible for sugar selection during DNA synthesis (8–13). The primary means of selecting dNTPs over rNTPs lies in the use of so-called “steric gate” mechanisms, which rely upon amino acid side-chains or backbone atoms that are usually found in the palm domain of the polymerase. Different polymerase families utilize variable steric-gate residues. For example, the Y-family polymerases from bacteria to humans use either a tyrosine or a phenylalanine side-chain to help prevent mis-insertion of rNTPs (10,13). Included amongst these enzymes is the model Y-family DNA polymerase Dpo4 from Sulfolobus solfataricus, which employs Tyr12 in the palm domain to select against rNTPs (13). While mutation of the steric-gate residue in Dpo4 to alanine does abrogate sugar selectivity approximately 2,000-fold (14), dNTP insertion by the Dpo4 Y12A mutant is still preferred over rNTP insertion, which is suggestive of a role for factors beyond the steric-gate during furanose selection by Dpo4.
The covalent linkages between heterocyclic bases, the phosphate backbone and a central furanose moiety define the topology of nucleic acid structure. Furanose geometry within the context of nucleic acids has been studied in great detail and there are clearly established preferred conformations for deoxyribonucleotides and ribonucleotides (15). The conformation of the five-membered ring of furanose can be characterized by the values P and νmax in the pseudorotation cycle (Figure 1A). The diagrammatic representation of these values is often used as a way to quickly ascertain the nature of the molecules under investigation. In this regard, the “Northern” hemisphere represents furanose structures that are most often associated with C3′-endo ribonucleotides (RNA-like A-form helices) and the “Southern” hemisphere is most closely associated with C2′-endo deoxynucleotides (DNA-like B-form helices). The conformation of the furanose moiety in RNA and DNA can oscillate between “puckers” because the free-energy barrier allows relatively facile conversion between species (16). The study of furanose selectivity in the active site of DNA polymerases requires high-resolution crystal structures to identify conformational preferences. Recently, an elegant series of time-resolved crystal structures revealed the mechanism of phosphodiester bond formation by human DNA polymerase eta (hpol η) with unprecedented detail (17). The reported structures reveal that the incoming dATP molecule retains a C3′-endo (A-form) pucker throughout the hpol η catalytic cycle, while the primer terminus shifts from the C2′-endo (B-form) in the ground state to a C3′-endo conformation in the product state. These results suggest that the Y-family polymerases, like other DNA polymerases, have evolved to preferentially incorporate more RNA-like sugar puckers and must rely upon the steric-gate residue to prevent mis-insertion of rNTPs. Indeed, recent work has shown that the bacterial Y-family member, UmuC, displays proficient insertion of rNTPs and that mutation of the UmuC steric-gate residue converts the protein into a primer-dependent RNA polymerase (18). Y-family polymerases are enzymes that are involved in DNA damage tolerance and replication stress response mechanisms (19). Deciphering the factors involved in substrate selection by these enzymes is an important part of understanding how the unique active site structure participates in genomic maintenance.
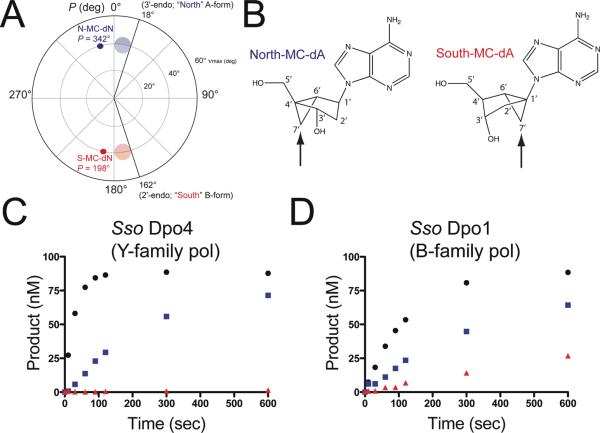
Figure 1.
Fixed-conformation nucleotide analogues probe the role of the furanose geometry during polymerase catalysis. (a) A pseudorotational cycle is shown to compare the N/S-MC conformations with the clustering of C3′-endo (semi-transparent blue circle) and C2′-endo sugar puckers (semi-transparent red circle) most commonly observed in RNA and DNA helices, respectively. (b) The chemical structures of N-MC-dA and S-MC-dA are shown with atoms in the ring system numbered. The arrows point to the different positioning of the cyclopropane ring relative to the cyclopentane ring. (c) Single-nucleotide insertion experiments were performed for Dpo4 (5 nM) and primer/template DNA (100 nM) with 10 μM dATP (black circles) N-MC-dATP (blue squares) and S-MC-dATP (red triangles). (d) Single-nucleotide insertion experiments were performed for Dpo1 (5 nM) and primer/template DNA (100 nM) with 10 μM dATP (black circles), N-MC-dATP (blue squares) and S-MC-dATP (red triangles).
Chemical modification of natural substrates is a classical approach to understanding how form defines function. An alternative way to study the relative influence of different sugar puckers on the catalytic properties of DNA polymerases is to covalently modify the furanose ring in such a way that the geometry becomes “locked” or “fixed” in a certain conformation. The use of a bicyclo[3.1.0]hexane scaffold can produce furanose mimics that are locked in either the North or South sugar pucker (20). The use of fixed-conformation nucleotide analogues to study DNA polymerase mechanism allows one to investigate the molecular features associated with dNTP selection over rNTPs (or other nucleotide analogues) by separating the influence of the sugar pucker for the incoming nucleotide triphosphate from contributions associated with protein interactions with the 2′-OH group. Previous results have shown that most polymerases and reverse transcriptases studied to date preferentially incorporate the more RNA-like North-methano-carba nucleotide triphosphates (N-MC-dNTPs) (21–23). The South-methanocarba dNTPs (S-MC-dNTPs) are generally not incorporated effectively by polymerases. We have utilized North and South oriented methanocarba-deoxyadenosine triphosphate (N-MC-dATP and S-MC-dATP, respectively) analogues to study furanose selection by two DNA polymerases from the crenarchaeote Sulfolobus solfataricus P2. We provide structural and functional data probing the role of sugar puckering during nucleotide selection by Dpo4 and compare these findings to experiments with the B-family replicative polymerase Dpo1. Our results provide new insights into the molecular features that contribute to the selection of nucleotides by DNA polymerases.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were of sufficient purity for crystallization studies. Unlabeled dNTPs were purchased from GE Healthcare Life Sciences (Piscataway, NJ). [γ-32P]-ATP was purchased from PerkinElmer Life Sciences (Boston, MA). N-MC-dATP and S-MC-dATP were synthesized as described previously (24) and converted to triphosphate form by TriLink BioTechnologies (San Diego, CA). DNA oligonucleotides used in the polymerase assays and for crystallization were purchased from Integrated DNA technologies (Coralville, IA). The oligonucleotides were purified by the manufacturer and analyzed by mass spectrometry. Dpo4 was expressed and purified as described previously (25). Dpo1 was expressed and purified as described previously (26).
Single-Nucleotide Insertion Polymerase Assays
For single-nucleotide insertion assays, a primer-template DNA substrate was prepared by radio-labeling. The 18-mer primer oligonucleotide sequence used was 5'-GGG GGC TCG TAA GGA TCC -3' and the 23-mer template oligonucleotide sequence was 5'-GCA CTG GAT CCT TAC GAG CCC CC-3'. 32P-labeled primer was annealed to template oligonucleotide by heating a 1:2 molar ratio of primer:template DNA to 95 °C for 5 min and then slowly cooling to room temperature. The primer:template DNA substrate was then incubated with either Dpo4 or Dpo1 polymerase (pol) prior to extension. Each reaction was initiated by adding dNTP•MgCl2 (0.5 mM dNTP and 5 mM MgCl2) solution to a pre-incubated polymerase•DNA complex (2 nM pol and 100 nM DNA). For some experiments discussed in the text, higher amounts of Dpo4 (100 nM) were required to compare N-MC-dATP and S-MC-dATP insertion rates. Unless otherwise stated, all enzymatic reactions were carried out at 37 °C in 50 mM HEPES buffer (pH 7.5) containing 60 mM KCl, 5 mM dithiothreitol (DTT), 100 μg mL−1 bovine serum albumin (BSA), and 10% (v/v) glycerol. At the indicated time points, 4 μL aliquots were quenched with 16 μL of a 95% formamide (v/v)/20 mM EDTA/0.1% bromophenol blue (w/v) solution and were separated by electrophoresis on a 16% polyacryamide (w/v)/7 M urea gel. The products were then visualized using a Typhoon imager (GE Healthcare Life Sciences) and quantified using ImageQuant™ software (GE Healthcare Life Sciences).
Full-length Extension Polymerase Assays
The same 18/23-mer primer:template DNA substrate used in the single-nucleotide insertion assays was used in the full-length extension experiments. The 18/23-mer primer:template DNA was then incubated with either Dpo4 or Dpo1 polymerase (pol) prior to extension. Each reaction was initiated by adding dNTP•MgCl2 (0.5 mM of each dNTP and 5 mM MgCl2) solution to a pre-incubated polymerase•DNA complex (5 nM pol and 100 nM DNA). At the indicated time points, 4 μL aliquots were quenched with 16 μL of a 95% formamide (v/v)/20 mM EDTA/0.1% bromophenol blue (w/v) solution and were separated by electrophoresis on a 16% polyacryamide (w/v)/7 M urea gel. The products were then visualized using a Typhoon imager (GE Healthcare Life Sciences) and quantified using ImageQuant™ software (GE Healthcare Life Sciences).
Steady-State Kinetics
To measure Michaelis-Menten kinetics, a 14/18-mer primer:template DNA substrate was used. A fluorescein-labeled 14 nucleotide primer strand (5'-FAM-GGG GGA AGG ATT CC-3') was annealed to a 18 nt template strand (5'-TCA TGG AAT CCT TCC CCC-3') with a 1:1 molar ratio of each oligonucleotide to generate a 14/18-mer primer/template DNA by heating to 95 °C for five minutes and then slow cooling to room temperature. Single-nucleotide incorporation by Dpo4 was measured over a range of dNTP concentrations. Each reaction was initiated by adding dNTP•MgCl2 (0.1 to 250 μM dNTP and 5 mM MgCl2) solution to a pre-incubated polymerase•DNA complex (2 nM pol and 100 nM DNA for Dpo4; 5 nM pol and 100 nM DNA for Dpo4). The products were analyzed as described above for full-length extension assays. The initial portion of the velocity curve was fit to a linear equation in the program GraphPad Prism (GraphPad Software, Inc., San Diego, CA). The resulting velocity was plotted as a function of dNTP concentration and then fit to a hyperbolic equation, correcting for enzyme concentration, to obtain estimates for the turnover number (kcat) and Michaelis constant (KM,dNTP).
Crystallization of Dpo4, X-ray Diffraction Data Collection and Processing
Crystallization of Dpo4 was performed essentially as described previously (27). A 14 nucleotide primer strand (5'-GGG GGA AGG ATT CC-3') was annealed to a 18 nt template strand (5'-TCA TGG AAT CCT TCC CCC-3') with a 1:1 molar ratio of each oligonucleotide to generate a 14/18-mer primer/template DNA by heating to 95 °C for five minutes and then slow cooling to room temperature. The 14/18-mer DNA was used in both the N-MC-dATP and the S-MC-dATP complexes. Dpo4 was mixed with DNA (1:1.2 molar ratio) in 20 mM Tris-HCl buffer (pH 8.0, 25 °C) containing 100 mM NaCl, 4% glycerol (v/v), and 5 mM β-mercaptoethanol and then placed on ice for 1 h prior to incubation with 5 mM MgCl2 and 0.5 mM of either N-MC-dATP or S-MC-dATP. The final Dpo4 concentration was ~ 10 mg mL−1. Crystals were grown using the sitting drop vapor-diffusion method with the reservoir solution containing 20 mM Tris-HCl (pH 8.0 at 25 °C), 15% polyethylene glycol 3350 (w/v), 60 mM NaCl, 5 mM MgCl2, and 4% glycerol (v/v). Droplets consisted of a 1:1 (v/v) mixture of the Dpo4:DNA:Mg2+:N/S-MC-dATP complex and the reservoir solutions and were equilibrated against the reservoir solutions. Crystals were soaked in mother liquor containing an additional 25% polyethylene glycol 3350 (w/v) and 15% ethylene glycol (v/v) and then swiped through paratone-N (Hampton Research, Aliso Viejo, CA) and flash frozen in liquid nitrogen. Data sets were collected at the Advanced Photon Source (Argonne, IL) on beamline ID24-E maintained by the Northeastern Collaborative Acess Team (NE-CAT). Scaling and indexing were performed using HKL2000 (28).
Structure Determination and Refinement
The structure was solved by the method of molecular replacement using the Phaser program in the Phenix software suite (29). For the Dpo4•North structure containing N-MC-dATP, we used the protein and DNA molecules from a previously reported Dpo4 ternary complex (PDB ID 2J6S) as the search model. For the Dpo4•South structure containing S-MC-dATP, we used the protein and DNA molecules from a previously reported Dpo4 ternary complex (PDB ID 3T5J) as the search model. Several rounds of refinement in Phenix and model building in Coot (30) resulted in the final structure. Each structure displayed good stereochemistry with 94.5% and 94.4% of the residues in the favored region of the Ramachandran plot. Pymol was used to create the figures representing the crystal structures (31).
RESULTS
Dpo4 preferentially incorporates fixed-conformation nucleotide analogues with RNA-like sugar geometry, while Dpo1 tolerates both N-MC-dATP and S-MC-dATP puckers
Two fixed-conformation nucleotide analogues were prepared as a means of probing the role of the sugar pucker in polymerase catalysis (Figure 1B). The N-MC-dATP conformation mimics the geometry normally preferred by Y-family DNA polymerases during nucleotide insertion, whereas the S-MC-dATP molecule is not effectively utilized by most DNA polymerases tested to date (21–23). In order to test the conformational preferences for sugar selection by two of the four DNA polymerases from S. solfataricus, we performed single-nucleotide insertion experiments with fixed-conformation analogues alongside unmodified dATP (Figure 1). Similar to results reported previously for the DinB-homologue human DNA polymerase kappa (hpol κ) (22), the archaeal Y-family DNA polymerase Dpo4 only utilized the RNA-like N-MC-dATP to any measurable degree during the time course tested (Figure 1C). The rate of nucleotide insertion was 1.9 ± 0.3 nM min−1 and 0.26 ± 0.01 nM min−1 for dATP and N-MC-dATP, respectively (Figure 1C). Dpo4 was not able to insert S-MC-dATP under these experimental conditions. Relative to dATP insertion, the rate of N-MC-dATP insertion by Dpo4 was inhibited 7.3-fold by substitution of the furanose moiety with the bicyclo[3.1.0]hexane ring system but there was a clear preference for the RNA-like N-MC-dATP over S-MC-dATP.
Surprisingly, the B-family DNA polymerase Dpo1 can utilize both the RNA-like N-MC-dATP and the DNA-like S-MC-dATP with relatively modest differences between the two molecules (Figure 1D). The rate of nucleotide insertion was 0.45 ± 0.03 nM min−1, 0.18 ± 0.03 nM min−1, and 0.051 ± 0.007 nM min−1 for dATP, N-MC-dATP and S-MC-dATP, respectively (Figure 1D). Nucleotide insertion by the high-fidelity B-family polymerase Dpo1 was inhibited 2.5-fold by N-MC-dATP and 8.8-fold by S-MC-dATP. In other words, Dpo1 inserts S-MC-dATP at a rate that is diminished about 3.50fold relative to the rate observed for N-MC-dATP. These results are similar to those reported for the Y-family member human DNA polymerase eta (hpol η) and represent only the second report of a polymerase using S-MC-dATP with any degree of efficiency (22). The initial time course experiments revealed that the high-fidelity polymerase Dpo1 is unexpectedly more tolerant of covalently modified dNTPs than the translesion polymerase Dpo4.
Extension from a North pucker is inhibited for both enzymes but Dpo1 shows greater tolerance by extending from either N-MC-dA or S-MC-dA
Next, we performed full-length extension experiments to compare the ability of Dpo1 and Dpo4 to insert and extend from the two fixed-conformation nucleotide analogues. A single template thymidine exists in the unpaired portion of the template strand of the substrate used in the full-length extension assay. As such, the polymerase should only utilize the fixed-conformation MC-dATP molecule once in the extension assay, namely at the initial insertion step. Extension from the modified residue should then proceed with the polymerase using only unmodified dNTPs. The resulting gels clearly revealed that Dpo4 inserts N-MC-dATP but that extension from the modified nucleotide is inhibited (Figure 2). A mechanistic rationale for inhibited extension from N-MC-dA may be found upon consideration of the recent time-resolved hpol η structures, where the primer terminus clearly adopts a DNA-like C2′-endo, South pucker in the ground-state and reactant state (17). It is only in the product state that the primer terminus adopts a C3′-endo pucker to avoid a clash with the non-bridging oxygen of the a-phosphate for the nucleotide being inserted by hpol η. In our experiment with the fixed-conformation N-MC-dATP, the incorporated N-MC-dA moiety would be unable to adjust to such a B-form type of sugar pucker in the early steps of the Dpo4 catalytic cycle and this may contribute to the inhibitory nature of extension from the fixed North pucker. Dpo1-catalyzed insertion and extension from N-MC-dA is inhibited relative to dATP but there is clearly more extended product formed by Dpo1 than that observed for Dpo4 (Figure 2, compare N-MC-dATP lanes for each enzyme).
Figure 2.

Analysis of full-length extension by Dpo1 and Dpo4 in the presence of fixed-conformation MC-dATPs. Both Dpo1 and Dpo4 (5 nM) were incubated with a primer/template DNA substrate (100 nM) possessing a single templating thymidine at the initial insertion point. The template sequence was designed so that only a single dATP will be inserted during accurate DNA synthesis. The reaction was initiated by the addition of a mixture of MgCl2 (1 mM) and dNTPs containing dCTP, dTTP, dGTP and either dATP, N-MC-dATP or S-MC-dATP (20 μM each). Following quenching, the products were separated by 20% polyacrylamide (w/v)/7 M urea and imaged. Time points are shown underneath each lane.
We then tested the ability of the two polymerases to insert and extend from a DNA-like sugar pucker by including S-MC-dATP in the reaction mixture. Dpo4 fails to utilize the more DNA-like S-MC-dATP as a substrate for polymerization, consistent with the single-nucleotide insertion results. Unlike Dpo4, the B-family pol, Dpo1, can use the S-MC-dATP molecule at the insertion step and extension from the modified S-MC-dA appears to progress one step beyond (Figure 2). However, extension from S-MC-dA by Dpo1 does not go any farther indicating that processive extension from the S-MC-dA moiety is inhibited relative to extension from N-MC-dA. Still, these results are consistent with single-nucleotide insertion experiments and suggest that Dpo1 is more tolerant than Dpo4 during insertion of fixed conformation nucleotide analogues mimicking the different sugar puckers.
Quantitative analysis of Dpo4-catalyzed insertion of N-MC-dATP and S-MC-dATP
Steady-state kinetic analysis of Dpo4-catalyzed insertion of N-MC-dATP opposite dT was performed in order to quantify the change in polymerase activity during insertion of the RNA-like fixed conformation nucleotide. We also measured the steady-state kinetic parameters defining Dpo1-catalyzed insertion of dATP, N-MC-dATP and S-MC-dATP. Determination of the specificity constant (kcat/Km,dNTP) revealed that though Dpo4 prefers the North-oriented geometry during insertion, the bicyclo[3.1.0]hexane modification inhibits polymerization approximately 2.6-fold (Table 1). The reduction in the specificity constant was primarily related to a decrease in the turnover number (kcat), which is decreased ~3.8-fold relative to dATP. We were unable to determine the steady-state kinetic parameters for S-MC-dATP using conditions similar to those for N-MC-dATP (data not shown). However, we were able to obtain some quantitative comparison between N-MC-dATP and S-MC-dATP insertion by increasing the concentration of enzyme in the reaction mixture to equimolar ratio with DNA substrate. Under these conditions, we observe Dpo4-catalyzed product formation for both fixed-conformation nucleotides (Figure 3). The measured rate of product formation was 0.26 ± 0.09 nM min−1 for N-MC-dATP and 0.010 ± 0.002 nM min−1 for S-MC-dATP. Therefore, the rate of DNA synthesis by Dpo4 is inhibited 26-fold by inclusion of the B-form type pucker in S-MC-dATP. This is in contrast to the results with Dpo1 where there was only a 3.5-fold difference in the rates observed for N-MC-dATP and S-MC-dATP (Figure 1). Steady-state kinetic analysis of Dpo1 insertion of N-MC-dATP and S-MC-dATP opposite template dT reveals that, like Dpo4, the reduction in catalytic efficiency results from a reduction in kcat, which is decreased ~1.6-fold and ~2.6-fold for N-MC-dATP and S-MC-dATP, respectively, relative to dATP. These results are consistent with the notion that Dpo1 is less perturbed by changes in the geometry of the central ring system of the incoming dNTP.
Table 1.
Steady-state kinetic parameters for Dpo1- and Dpo4-catalyzed insertion of dATP and N-MC-dATP
| kcat (min−1) | KM,dNTP (μM) | kcat/KM,dNTP (min−1 μM−1) | |
|---|---|---|---|
| Dpo1 | |||
| dATP | 1.4 ± 0.1 | 1.4 ± 0.6 | 1.00 |
| N-MC-dATP | 0.86 ± 0.24 | 1.2 ± 0.5 | 0.72 |
| S-MC-dATP | 0.53 ± 0.03 | 1.3 ± 0.3 | 0.41 |
| Dpo4 | |||
| dATP | 18.0 ± 1.3 | 10.0 ± 3.3 | 1.80 |
| N-MC-dATP | 4.8 ± 0.4 | 8.7 ± 3.0 | 0.55 |
| S-MC-dATP | n.d. | n.d. | n.d. |
n.d. = not determined.
Figure 3.

Comparison of Dpo4-catalyzed insertion of N-MC-dATP and S-MC-dATP using high concentrations of enzyme. (a) Dpo4 (100 nM) was incubated with primer/template DNA (100 nM) and the reaction was initiated by the addition of MgCl2 (5 mM) and either dATP (10 μM) or N-MC-dATP (10 μM) for up-to five minutes. The rate of product formation was measured by fitting the linear portion of the velocity curve. The observed rate was 1.7 ± 0.3 nM sec−1 for dATP and 0.12 ± 0.01 nM sec−1 for N-MC-dATP (b) Dpo4 (100 nM) was incubated with primer/template DNA (100 nM) and the reaction was initiated by the addition of MgCl2 (5 mM) and S-MC-dATP (10 μM and 50 μM) for up-to forty minutes. The rate of product formation was measured by fitting the linear portion of the velocity curve. The observed rate was 6.6 × 10−3 ± 0.5 nM sec−1 for 10 μM S-MC-dATP and 10.9 ± 0.9 nM sec−1 for 50 μM S-MC-dATP.
Structural comparison of Dpo4 in complex with N-MC-dATP and S-MC-dATP
We then sought to understand the molecular features associated with the relative intolerance of Dpo4 to different sugar puckers by solving the crystal structure of the protein in ternary complex with an incoming North and South MC-dATP molecules. We were able to successfully crystallize Dpo4 in complex with both fixed conformation nucleotide analogues. Crystals for each complex diffracted to 2.9 Å with good signal to noise and completeness (Table 2). We refer to the ternary complexes as Dpo4•North and Dpo4•South for the crystals grown in the presence of N-MC-dATP and S-MC-dATP, respectively. The Dpo4•North complex crystallized in the primitive orthorhombic P 21212 space group with a single protein•nucleic acid molecule in the asymmetric unit, while the Dpo4•South complex crystallized in the primitive monoclinic P 21 space group, which possessed two protein•nucleic acid molecules in the asymmetric unit. As described in the materials and methods, the structures were solved by molecular replacement using the protein and DNA molecules from previously reported Dpo4 ternary complexes. Each complex contained a 14 nt primer strand annealed to a 18 nt template with a thymidine set to pair with the incoming dNTP. Superimposition of Dpo4•North and the two protein molecules in the Dpo4•South structure reveals no dramatic alterations in protein structure between complexes (Figure 4A; R.M.S.D. = 0.64 and 0.76 Å for superimposition of Dpo4•North with Dpo4•South molecule A and B, respectively). The initial difference maps show clear positive density for the incoming nucleotide, as well as density near the metal ion binding sites. The metal ions were refined as calcium. A few minor changes are observed when comparing the primer-template DNA in the two structures. First, the dC:dG base pair at the far end of the duplex (i.e. the part of the DNA distant from the polymerase active site and exposed to solvent) is disordered in the Dpo4•South structure (Figure 4A). Also, the 3-dA located at the +1 position in the template strand adopts distinct conformations when comparing Dpo4•North and Dpo4•South (Figure 4A, right panel). In the Dpo4•North structure, 3-dA is found rotated towards the active site of Dpo4. With the exception of the active site changes discussed below, the conformation of the remainder of the primer-template DNA is largely conserved for the two structures. Analysis of the helical parameters using 3DNA (32,33) shows that the primer-template DNA in both structures adopts a B-form helix, even near the polymerase active site. The dC residue at the primer terminus is found to adopt a C2′-endo conformation in all three protein•nucleic acid complexes. As expected the most interesting structural alterations are observed in and around the active site of Dpo4.
Table 2.
Crystal data and refinement parameters
| Parameter | Dpo4 North | Dpo4 South |
|---|---|---|
| X-ray source | APS | APS |
| Beamline | ID24-E | ID24-E |
| Detector | ADSC Q315 | ADSC Q315 |
| Wavelength (Å) | 0.979 | 0.979 |
| Temperature (K) | 110 | 110 |
| No. of Crystals | 1 | 1 |
| Space group | P21212 | P21 |
| Unit Cell (a, b, c; Å) | 93.6,103.4,52.5 | 52.7,101.1,100.0 |
| Resolution range (Å) | 46.8–2.90 | 47.5–2.89 |
| Highest resolution shella | (3.00–2.90) | (2.96–2.89) |
| No. of measurements | 267168 | 262183 |
| No. of unique reflections | 11041 (1130) | 21258 (1829) |
| Redundancy | 3.6 (3.5) | 3.2 (3.0) |
| Completeness (%) | 97.8 (84.2) | 96.0 (63.0) |
| R-mergeb (%) | 8.8 (70.4) | 14.0 (78.8) |
| Signal to noise (I/sI) | 9.3 (1.9) | 12.7 (1.8) |
| Model Composition | ||
| No. of amino acid residues | 341 | 682 |
| No. of water molecules | 46 | 65 |
| No. of Ca2+ ions | 4 | 9 |
| No. of template nucleotides | 16 | 29 |
| No. of primer nucleotides | 14 | 26 |
| No. of MC-dATP | 1 | 2 |
| Rfc (%) | 20.2 | 20.6 |
| R-freed (%) | 27.6 | 26.5 |
| Temperature factors | ||
| From Wilson plot (Å2) | 75.1 | 60.8 |
| r.m.s. standard deviation from ideal values | ||
| Bond lengths (Å) | 0.008 | 0.010 |
| Bond angles (°) | 1.3 | 1.4 |
| Dihedral angles (°) | 20.6 | 20.8 |
Values in parentheses correspond to the highest resolution shells.
R-merge = ΣhklΣj=1,N|〈Ihkl〉−Ihkl,j|/ΣhklΣj=1,N|Ihkl,j|, where the outer sum (hkl) is taken over unique the reflections.
Rf= Σhkl∥Fo,hkl|−k|Fc,hkl∥/Σhkl|Fo,hkl|, where |Fo,hkl| and |Fc,hkl| are the observed and calculated structure factors respectively.
Rfree idem, for the set of reflections (10% of the total) omitted from the refinement process.
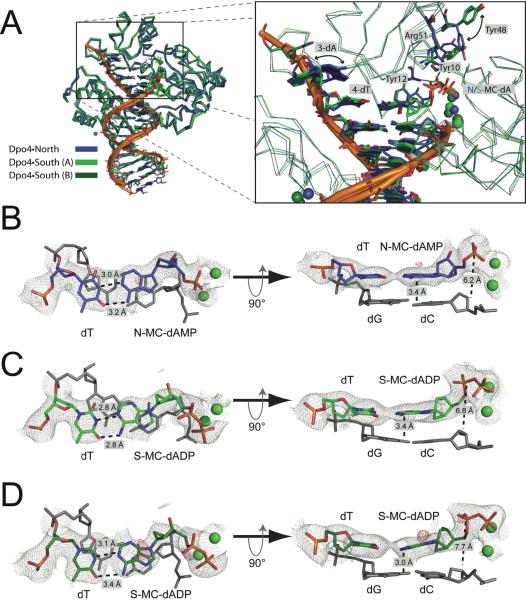
Figure 4.
The crystal structure of Dpo4 in complex with fixed-conformation nucleotide analogues. (a) The Dpo4•North structure (blue carbons) is shown superimposed upon both molecules A (light green carbons) and B (dark green carbons) from the asymmetric unit of the Dpo4•South structure. The active site is shown in the panel to the right to illustrate several changes surrounding the nascent base pair. The side chains for Tyr10, Tyr48 and Arg51 are shown to illustrate conformational differences between the Dpo4•North and Dpo4•South structures. There is no change in conformation for the steric-gate residue, Tyr12, when comparing the two structures. (b) The active site base pairs and calcium ions from the Dpo4•North structure are shown. The N-MC molecule has been hydrolyzed to the monophosphate form (N-MC-dAMP) and is base paired with the template dT. The base pair at the −1 position is shown (gray). Hydrogen bonding distances are noted in the left panel. The base stacking distance and the distance between the α-phosphate and the 3'-OH group are indicated in the right panel. The 2Fo-Fc Fourier sum electron density is shown contoured to the 1σ level (gray wire mesh) and the 2Fo-Fc difference map is shown contoured at 3σ (green wire mesh) and −3σ (red wire mesh). (c) The active site base pairs and calcium ions from molecule A of the Dpo4•South structure are shown. The distances noted are the same as those shown in panel b, as are the electron density representations. (d) The active site base pairs and calcium ions from molecule B of the Dpo4•South structure are shown. The distances noted are the same as those shown in panel b, as are the electron density representations.
First, the incoming nucleotide triphosphate has been hydrolyzed to the monophosphate in the Dpo4•North structure (Figure 4B), while both molecules in the Dpo4•South structure are found in the diphosphate form (S-MC-dADP) (Figure 4C and 4D). Both N-MC-dAMP and S-MC-dADP are paired with template dT in a Watson-Crick fashion and two calcium ions are bound in the active site of each complex (Figure 4B, 4C and 4D). Residues in the thumb domain of each protein molecule coordinate a third calcium ion. An additional calcium ion is observed between the DNA substrate and the little finger domain, specifically coordinated with the side-chain of Asp294 and the phosphate moiety 10-dA in the primer strand. The base-stacking interactions observed for the nascent base pair are intriguing when one compares Dpo4•North to Dpo4•South. An average base-stacking distance of 3.4 Å between the incoming N-MC-dAMP and the dC:dG base pair at the −1 position is similar to molecule A from the Dpo4•South structure. However, molecule B of the Dpo4•South structure exhibits a severely compressed base stacking interaction, with an average distance of around 3.0 Å between S-MC-dADP:dT and the −1 dC:dG base pair (Figure 4D). In both Dpo4•South structures there is a slight buckling of the −1 base pair that results from the positioning of the incoming S-MC-dADP (Figure 4C and 4D, right panel). The S-MC-dADP molecules are both shifted towards the major groove side of the growing helix, which avoids clashes with the steric gate residue, Tyr12 (Figure 5A). In fact, the 3′-OH group of S-MC-dADP appears to form a hydrogen bond with the OH group on Tyr12, which may help stabilize the non-productive binding of the nucleotide analogue. The distance between the α-phosphate and the 3′-OH group of the primer terminus extends from 6.2 Å in the Dpo4•North structure to 6.8 Å and 7.7 Å in molecules A and B of the Dpo4•South structure. Additionally, the position of the terminal 3′-OH group on the primer is moved inwards, away from the incoming nucleotide, for the S-MC-dADP containing complexes (Figure 5B). Overall, the Dpo4•North structure possesses features indicative of a more productive catalytic complex than either molecule in the Dpo4•South structure. Indeed, the active sites of the two molecules in the Dpo4•South structure appear to represent a gradient in terms of the deviation from Dpo4•North structure, with Dpo4•South molecule A serving as an intermediate between Dpo4•North and Dpo4•South molecule B. Additional structural observations and comparisons are discussed in detail below.
Figure 5.
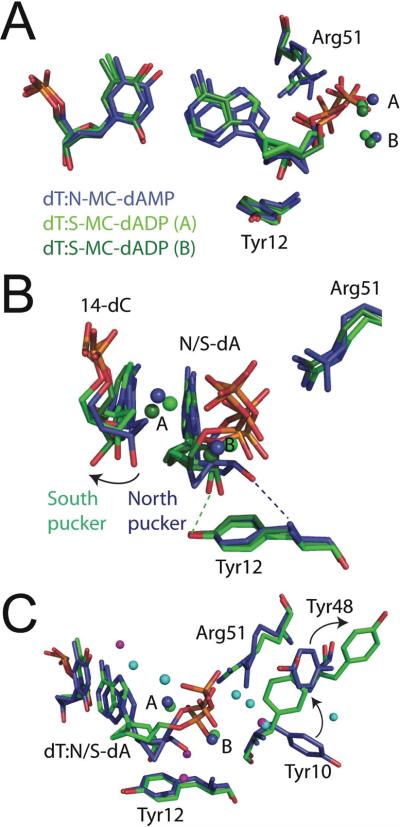
Comparative view of Dpo4 bound to fixed-conformation nucleotides and unmodified nucleotides. (a) Superimposition of Dpo4•North (blue carbons) and the two molecules in the Dpo4•South asymmetric unit (molecule A, light green carbons; molecule B, dark green carbons) reveals distinct differences in both the nascent base pairs and in the conformations of amino acid side-chains near the dNTP binding cleft of Dpo4. The active site metal ions are shown as spheres and are labelled A and B for the proposed catalytic and coordinating ions, respectively. The side-chains for Tyr12 and Arg51 are shown to provide their relative orientations for the subsequent panels. (b) Compared to the Dpo4•North structure, the primer terminus in Dpo4•South is forced inward by the movement of the S-MC-dADP molecules away from the steric gate, which is due to the constricted geometry of the central bicyclo[3.1.0]hexane ring system. The dashed lines denote hydrogen bonding between the 3′-OH group of the incoming nucleotide analogues and the steric-gate residue (Tyr12). (c) Additional conformational changes in the amino acid side-chains are shown near the Dpo4 active site. A rotation of Tyr48 from the position adopted in the Dpo4•North structure is accompanied by the rotation of Tyr10 in both molecules of the Dpo4•South structure. The active site water molecules for Dpo4•North and Dpo4•South (molecule A) structures are shown in cyan and magenta, respectively.
DISCUSSION
The ability of nucleic acid polymerases to select specific substrates from a pool of highly similar molecules is one the most fascinating aspects of nucleic acid synthesis and a foundation for the accurate propagation of genomic material. In the case of selecting either dNTPs or rNTPs, polymerases must distinguish between substrates that differ in a single atom housed in the furanose ring. Importantly, the insertion of rNTPs during replication of DNA is a biologically relevant source of genomic instability (6). Estimates in Saccharomyces cerevisiae place the number of rNTPs inserted by the replicative DNA polymerases near 10,000 per cell division (7), which is comparable to estimates for abasic site formation and oxidative DNA lesions in eukaryotic cells (34). Moreover, ribonucleotide concentrations are higher than the corresponding deoxyribonucleotides inside cells (35), which further adds to the need for strict selection of the appropriate dNTP during genomic replication. Therefore, a precise understanding of molecular determinants that select against mis-insertion of rNTPs is of great importance.
The addition of a single hydroxyl group at the C2′ position of nucleotides alters the preferred geometry of the molecule and influences subsequent interactions with the polymerase active site. Deoxyribonucleotides preferentially adopt C2′-endo (South) conformations, while ribonucleotides most commonly reside in a C3′-endo (North) pucker. Comparing the pucker of the incoming dNTP observed in crystal structures of DNA polymerases from different organisms and families reveals that these enzymes almost universally prefer North sugar puckers for the incoming dNTP. Moreover, all DNA polymerases studied to date rely upon a steric exclusion, or steric “gate”, mechanism of one sort or another to prevent insertion of rNTP substrates into the growing primer strand. While the steric gate confers a large portion of the selective power for DNA polymerases, other properties intrinsic to nucleotide geometry also play a role in determining catalytic preferences. Separating the role of the steric gate residue during substrate selection from intrinsic conformational preferences in the nucleotide triphosphate is difficult. The fixed-conformation nucleotides used in our study restrict the central ring system in a geometry that resembles either a North, RNA-like pucker or a South, DNA-like pucker (Figure 1). These molecules have been used previously to study how enzymes of various classes (e.g. kinases, DNA methyl transferases, polymerases/reverse transcriptases) utilize puckers from two antipodal regions of the pseudorotational cycle (20), as well as in the study of nucleic acid structural properties (36). We sought to understand how these modified nucleotides with a fixed sugar pucker interact with two DNA polymerases from S. solfataricus, the B-family replicative pol, Dpo1, and the Y-Family translesion pol, Dpo4, in order to ascertain how discrete puckers affect the activity of these enzymes. In this way, we were able to dissect the catalytic response of the Dpo1 and Dpo4 active sites to specific types of sugar puckering.
We first tested the functional capacity of Dpo1 and Dpo4 to insert N-MC-dATP and S-MC-dATP. Dpo4 shows a strong preference for insertion of the more RNA-like N-MC-dATP compared to S-MC-dATP in single-nucleotide insertion experiments (Figure 1C). This was not surprising, as it has been shown for Dpo4 and other polymerases that the incoming dNTP is found in a C3′-endo, North-type pucker (13). The Y-family polymerase Dpo4 does not insert the South-oriented S-MC-dATP to any measurable extent under steady-state conditions used here. We were able to observe and quantify Dpo4-catalyze insertion of S-MC-dATP by increasing the enzyme concentration (Figure 3). Under these conditions, Dpo4 catalysis was slowed nearly 9-fold by S-MC-dATP when compared to the RNA-like pucker of N-MC-dATP. These results are similar to those obtained previously with HIV-1 Reverse Transcriptase (HIV-1 RT) (21) and for human DNA polymerase kappa (hpol κ) (22). In contrast to Dpo4, the B-family member, Dpo1, can catalyze insertion of both N-MC-dATP and S-MC-dATP opposite template dT with relatively modest levels of inhibition by either nucleotide analogue (Figure 1D). While both fixed conformation nucleotides are inserted at a slower rate than dATP, there is only a 3.5-fold reduction in the rate for S-MC-dATP compared to N-MC-dATP, indicating that Dpo1 can use a South-oriented dNTP with relative ease compared to Dpo4. It is strange, in this instance, to consider the translesion polymerase Dpo4 to be the less tolerant of the two enzymes tested, as Dpo4 has been shown to bypass bulky DNA adducts in the template strand with greater efficiency than Dpo1 (26). The relative tolerance of Dpo1 for the modified nucleotide analogues was intriguing.
Full-length extension assays were then employed to determine if Dpo1 and Dpo4 could extend from the fixed conformation nucleotide analogues (Figure 2). Again, Dpo4 only uses N-MC-dATP under the conditions tested and the enzyme is apparently inhibited at the extension step once N-MC-dA has been incorporated into the growing primer. In this respect, Dpo4 more closely resembles HIV-1 RT in that neither enzyme is terribly proficient at extending from the inserted N-MC-dA moiety, where as hpol κ can insert and extend beyond N-MC-dA with relative ease. The inability of Dpo4 to extend from the 3′-OH group of the incorporated N-MC-dA is perhaps not surprising since the residue at the end of the primer strand in consistently found in a South C2′-endo pucker in crystal structures of Dpo4, including the current structures. Recently reported time-resolved X-ray crystal structures of the Y-family member hpol η show that the primer terminus adopts the DNA-like C2′-endo pucker in the early steps in polymerase catalysis (i.e. the ground-state and reactant-state), but like other polymerase families, a North C3′-endo pucker is observed once phosphoryl transfer has occurred (17). A key step in nucleotidyl transfer by polymerases is the deprotonation of the 3′-OH group, which relies upon binding of the catalytic metal ion and a transfer of the proton from the 3′-OH group to the transient water molecule. A North-oriented C3′-endo pucker at the primer terminus clashes with the position of the transient water molecule in the hpol η structural study (17). Inhibition of extension from N-MC-dA by Dpo4 would be expected since the 3′-OH group in the fixed-conformation nucleotide blocks productive positioning of the transient water molecule in the reactant state. Consistent with the single-nucleotide insertion assays, Dpo1 appears to insert and extend from both N-MC-dATP and S-MC-dATP (Figure 2). Extension of the fixed-conformation nucleotides by Dpo1 is not as efficient as that observed for dATP, with the extension from S-MC-dA being largely limited to one nucleotide. The structural basis for greater tolerance by Dpo1 is unknown and further investigation will be required to dissect the molecular determinants associated with Dpo1- catalyzed insertion of South-oriented puckers.
The biochemical assays reported here suggest that the B-family DNA polymerase is better able to accommodate variations in the geometry of the central ring system of incoming nucleotide triphosphates when compared to Dpo4. We were intrigued by these results and set out to solve the crystal structures of Dpo4 bound to both N-MC-dATP and S-MC-dATP in order to better understand the relative intolerance observed in the functional assays. We were successful in determining the structure of Dpo4 in complex with both fixed conformation nucleotide analogues. Superimposition of Dpo4•North structure with each of the two molecules in the asymmetric unit of the Dpo4•South structure reveals essentially no change in the overall protein structure. Each incoming nucleotide was hydrolyzed from the triphosphate to either the monophosphate in the case of N-MC-dAMP or the diphosphate for the South-oriented molecules (Figure 4). Comparing the relative positioning of the incoming N-MC-dAMP and S-MC-dADP molecules to one another and to previously reported structures with dATP or ADP reveals much concerning the selection of sugar puckering by Dpo4.
Both N-MC-dAMP and S-MC-dAMP are paired with the template dT in a canonical Watson-Crick hydrogen bonding pattern. However, close inspection of the two structures reveals distinct and most likely, functionally relevant alterations. Relative to N-MC-dATP, the purine ring system for S-MC-dADP is shifted away from the Dpo4 active site residues (Figure 5A). A hydrogen bond is formed between the 3′-OH group of S-MC-dADP and the OH group on the steric gate residue, Tyr12 (Figure 5A). Movement of S-MC-dADP opens up a space for a water molecule to bind in the position occupied by the 3′-OH group of N-MC-dAMP, which forms a hydrogen bond with the backbone amide of Tyr12 in the Dpo4•North structure (Figure 5C). The steric gate adopts the same position in both Dpo4 structures reported here (Figure 5). Since the fixed conformation S-MC moiety will not readily oscillate to a conformation that is tolerated by the position of Tyr12, the S-MC-dADP molecule is forced towards the −1 base pair in a manner that buckles the primer-template junction and places the α-phosphate in a position that is less favourable for transfer. These observations provide a structural basis for why Dpo4 inserts North-oriented sugar puckers over nucleotides that adopt a South-pucker in the polymerase active site.
While the overall fold of the protein does not deviate between structures, there are several interesting changes in the amino acids side chains surrounding the dNTP binding cleft of Dpo4. There are only minor side-chain differences observed for the catalytic Asp7, Asp105 and Glu106 residues when comparing the two structures. An alteration in the side-chain of Arg51 is observed but is most likely due to a passive “tracking” of the phosphates for N-MC-dAMP and S-MC-dADP and not as a result of active repositioning the nucleotides. Two residues (Tyr10 and Tyr48) are found in markedly distinct conformations when comparing Dpo4•North and Dpo4•South (Figure 5C). In the Dpo4•North structure, a water-mediated hydrogen bond between Tyr48 and the α-phosphate moiety appears to assist in the stabilization of the incoming N-MC-dAMP. Such an interaction is not observed in the Dpo4•South structure (Figure 5C). When dATP occupies the active site of Dpo4 (PDB 2AGQ), Tyr10 and Tyr48 are found in the same orientation as that observed for Dpo4•North. In the previously reported structure the hydroxyl group of Tyr48 forms a hydrogen bond with the γ-phosphate of dATP (37). Thus, Tyr48 appears to act as a “back-gate” to dNTP binding that does not close when S-MC-dADP is bound to Dpo4.
The Dpo4•North structure very closely resembles a previously reported complex of Dpo4 bound to dATP paired with dT (37). In the previous structure, dATP adopts the slightly “Northwestern” C2′-exo pucker (P = 344°). The N-MC-dAMP molecule is also found in a C2′-exo Northwestern pucker (P = 334°). Indeed, N-MC-dAMP and dATP superimpose almost perfectly, with the exception of the phosphates (Figure 6), which is consistent with a preferential insertion of North-oriented puckering modes for Dpo4. By way of contrast, a recent study reported the crystal structure of the Dpo4 steric-gate mutant Y12A with an incoming ADP (13). As stated previously, Dpo4 relies upon stacking between the aromatic ring of Tyr12 and a hydrogen bond between the 3′-OH group of the incoming dNTP and a backbone amide group to select against ribonucleotides. The 2′-OH group of ADP is only accommodated when Tyr12 is mutated to alanine (Figure 6B) (13). The structure of the Dpo4 Y12A steric-gate mutant bound to ADP shows that the ribonucleotide is shifted down to occupy the space normally filled by Tyr12 and that the sugar is observed in a Northeastern C4′-exo pucker (P = 45°). Comparison of our Dpo4•North structure to the Dpo4(Y12A)•ADP complex shows the expected difference in positioning of ADP because of the loss of Tyr12 (Figure 6B). Comparison of the three molecules (N-MC-dAMP, dATP and ADP) reveals a graded positioning of the α-phosphate that correlates nicely with the rate of Dpo4-catalyzed insertion (dATP > N-MC-dATP > ATP).
Figure 6.
Dpo4 binds N-MC-dAMP in a manner that closely resembles unmodified dATP. (a) The Dpo4•North structure is shown superimposed with wild-type Dpo4 in complex with dATP (PDB code 2AGQ, light red carbons) and a steric-gate mutant form of Dpo4 (Y12A) bound to ADP (PDB code 3PR5, yellow carbons). (b) The active site base pairing modes in panel a are shown rotated approximately 90° to illustrate the orientation relative to the primer terminus. The side-chains at position 12 and 51 of the Dpo4 proteins are shown.
The fixed-conformation nucleotide analogues used in our study rely upon a bicyclo[3.1.0]hexane scaffold to lock the geometry of the central ring system. As such, one might suppose that the cyclopropane group could, in itself, alter the kinetic properties of the polymerases through steric effects during insertion of the modified nucleotide. However, several points argue against such a scenario. First, the central ring system of N-MC-dATP bound to Dpo4 superimposes quite nicely with the deoxy-ribose moiety of dATP reported previously (Figure 6). There are no protein contacts near the cyclopropane ring and yet we observe a diminished turnover number for Dpo4-catalyzed insertion of N-MC-dATP (Table 1). The main structural difference observed between N-MC-dATP and dATP is the position of the α-phosphate, which is placed 2.2 Å closer to the primer terminus when dATP is bound in the Dpo4 active site. Similarly, the cyclopropane moiety of S-MC-dADP does not interact with the protein. The perturbations associated with S-MC-dADP binding to Dpo4 result from interactions with the 3′-OH group of the nucleotide and the steric gate residue, which wedges the incoming S-MC-dADP into the primer terminus and positions the phosphates poorly for catalysis. Recently we showed that another Y-family member, human DNA polymerase iota (hpol ι) utilizes N-MC-dATP more efficiently than the unmodified nucleotide (38). Again, there were no protein contacts near the cyclopropane ring with either N-MC-dATP or S-MC-dATP bound to the hopl ι active site. The primary effect of the bicyclo[3.1.0]hexane moiety was to alter the syn/anti equilibrium such that the adenine group of N-MC-dATP preferentially adopted the anti conformation. As covalent modifications go, the cyclopropane moiety is a relatively innocuous addition that tucks in under the five-membered ring system of the modified dNTP. Thus, it would appear that any inhibitory effect imparted upon polymerase structure and function by the added size of the cyclopropane group is minimal.
The role of the furanose geometry in DNA polymerase catalysis has been studied in great detail (10,12). Sugar selection by DNA polymerases can have important ramifications upon genomic stability. Notably, the ability to discriminate against rNTP insertion varies several orders of magnitude depending upon which polymerase is under investigation (10). Similar steric-gate mechanisms are employed in general terms by all DNA polymerases to prevent mis-insertion of rNTPs. The best selectivity against rNTPs appears to reside in the A- and B-family DNA polymerases. X-family polymerases, such as pol β and pol μ, are clearly the worst polymerase family at selecting against rNTPs, where as Y-family polymerases represent intermediate selectivity against rNTPs. The preferential pucker of dNTPs and rNTPs in the active site of polymerases can contribute to how effectively these enzymes select from a pool of very similar substrates. The current study was an attempt to dissect the relative influence of the sugar pucker upon DNA polymerase activity by using fixed-conformation nucleotide analogues that mimic either a North or a South sugar pucker. Based on our functional assays, insertion of N-MC-dATP should represent an intermediate between efficient insertion of unmodified dATP and ineffective utilization of S-MC-dATP and indeed this is what the structures reveal. Insertion of South-oriented nucleotides is impaired by interactions with the steric-gate residue, which positions the incoming S-MC-dADP molecule in a non-productive orientation and also disrupts the primer-template junction. N-MC-dAMP adopts a position that is almost identical to that observed for Dpo4 in complex with dATP. Finally, we report the interesting observation that the B-family polymerase from S. solfataricus, Dpo1, is apparently more tolerant of sugar pucker variation during nucleotide insertion and extension steps than the translesion enzyme Dpo4. Ongoing structural and functional investigations will attempt to understand the mechanistic basis for increased tolerance of different sugar puckers by Dpo1.
Acknowledgments
FUNDING This work was supported in part by U.S.P.H. Service grants R00 GM084460 (R.L.E.) and R01 GM055237 (M.E.). Supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by grants from the National Center for Research Resources (5P41RR015301-10) and the National Institute of General Medical Sciences (8 P41 GM103403-10) from the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357.
Footnotes
ACCESSION NUMBERS The atomic coordinates and structure factors (codes 4GC6 and 4GC7 for Dpo4•North, Dpo4•South, respectively) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ, U.S.A. (http://www.rscb.org/).
REFERENCES
- 1.Geacintov N, Broyde S. The Chemical Biology of DNA Damage. Wiley-VCH; Weinheim: 2010. [Google Scholar]
- 2.Friedberg EC, Walker GC, Siede W, Wood RD, Shultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd ed. ASM Press; Washington, D.C.: 2006. [Google Scholar]
- 3.Guengerich FP. Interactions of carcinogen-bound DNA with individual DNA polymerases. Chem. Rev. 2006;106:420–452. doi: 10.1021/cr0404693. [DOI] [PubMed] [Google Scholar]
- 4.Kunkel TA. DNA replication fidelity. J. Biol. Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 5.Lazzaro F, Novarina D, Amara F, Watt DL, Stone JE, Costanzo V, Burgers PM, Kunkel TA, Plevani P, Muzi-Falconi M. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol. Cell. 2012;45:99–110. doi: 10.1016/j.molcel.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. (U. S. A.) 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Astatke M, Ng K, Grindley ND, Joyce CM. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc. Natl. Acad. Sci. (U. S. A.) 1998;95:3402–3407. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown JA, Fiala KA, Fowler JD, Sherrer SM, Newmister SA, Duym WW, Suo Z. A novel mechanism of sugar selection utilized by a human X-family DNA polymerase. J. Mol. Biol. 2009;395:282–290. doi: 10.1016/j.jmb.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown JA, Suo Z. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry. 2011;50:1135–1142. doi: 10.1021/bi101915z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLucia AM, Chaudhuri S, Potapova O, Grindley ND, Joyce CM. The properties of steric gate mutants reveal different constraints within the active sites of Y-family and A-family DNA polymerases. J. Biol. Chem. 2006;281:27286–27291. doi: 10.1074/jbc.M604393200. [DOI] [PubMed] [Google Scholar]
- 12.Joyce CM. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc. Natl. Acad. Sci. (U. S. A.) 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirouac KN, Suo Z, Ling H. Structural mechanism of ribonucleotide discrimination by a Y-family DNA polymerase. J. Mol. Biol. 2011;407:382–390. doi: 10.1016/j.jmb.2011.01.037. [DOI] [PubMed] [Google Scholar]
- 14.Sherrer SM, Beyer DC, Xia CX, Fowler JD, Suo Z. Kinetic basis of sugar selection by a Y-family DNA polymerase from Sulfolobus solfataricus P2. Biochemistry. 2010;49:10179–10186. doi: 10.1021/bi101465n. [DOI] [PubMed] [Google Scholar]
- 15.Saenger W. Principles of nucleic acid structure. Springer-Verlag; New York: 1984. [Google Scholar]
- 16.Olson WK, Sussman JL. How flexible is the furanose ring? 1. A comparison of experimental and theoretical studies. J. Am. Chem. Soc. 1982;104:270–278. [Google Scholar]
- 17.Nakamura T, Zhao Y, Yamagata Y, Hua YJ, Yang W. Watching DNA polymerase eta make a phosphodiester bond. Nature. 2012;487:196–201. doi: 10.1038/nature11181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaisman A, Kuban W, McDonald JP, Karata K, Yang W, Goodman MF, Woodgate R. Critical amino acids in Escherichia coli UmuC responsible for sugar discrimination and base-substitution fidelity. Nucleic Acids Res. 2012;40:6144–6157. doi: 10.1093/nar/gks233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. (U. S. A.) 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marquez VE. Chemical and biological consequences of locking the conformation of nucleosides at the two antipodal extremes of the pseudorotational cycle. Nucleic Acids Symp. Ser. (Oxf) 2004:11–12. doi: 10.1093/nass/48.1.11. [DOI] [PubMed] [Google Scholar]
- 21.Boyer PL, Julias JG, Marquez VE, Hughes SH. Fixed conformation nucleoside analogs effectively inhibit excision-proficient HIV-1 reverse transcriptases. J. Mol. Biol. 2005;345:441–450. doi: 10.1016/j.jmb.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 22.Eoff RL, McGrath CE, Maddukuri L, Salamanca-Pinzon SG, Marquez VE, Marnett LJ, Guengerich FP, Egli M. Selective modulation of DNA polymerase activity by fixed-conformation nucleoside analogues. Angew. Chem. Int. Ed. Engl. 2010;49:7481–7485. doi: 10.1002/anie.201003168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marquez VE, Ezzitouni A, Russ P, Siddiqui MA, Ford H, Jr., Feidman RJ, Mitsuya H, George C, Barchi JJ., Jr. HIV-1 Reverse Transcriptase can discriminate between two conformationally locked carbocyclic AZT triphosphate analogues. J. Am. Chem. Soc. 1998;120:2780–2789. [Google Scholar]
- 24.Marquez VE, Ben-Kasus T, Barchi JJ, Jr., Green KM, Nicklaus MC, Agbaria R. Experimental and structural evidence that herpes 1 kinase and cellular DNA polymerase(s) discriminate on the basis of sugar pucker. J. Am. Chem. Soc. 2004;126:543–549. doi: 10.1021/ja037929e. [DOI] [PubMed] [Google Scholar]
- 25.Zang H, Goodenough AK, Choi JY, Irimia A, Loukachevitch LV, Kozekov ID, Angel KC, Rizzo CJ, Egli M, Guengerich FP. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4: analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1,N2-ethenoguanine. J. Biol. Chem. 2005;280:29750–29764. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- 26.Choi JY, Eoff RL, Pence MG, Wang J, Martin MV, Kim EJ, Folkmann LM, Guengerich FP. Roles of the four DNA polymerases of the crenarchaeon Sulfolobus solfataricus and accessory proteins in DNA replication. J. Biol. Chem. 2011;286:31180–31193. doi: 10.1074/jbc.M111.258038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eoff RL, Irimia A, Egli M, Guengerich FP. Sulfolobus solfataricus DNA polymerase Dpo4 is partially inhibited by “wobble” pairing between O6-methylguanine and cytosine, but accurate bypass is preferred. J. Biol. Chem. 2007;282:1456–1467. doi: 10.1074/jbc.M609661200. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLano WL. DeLano Scientific. San Carlos, CA, USA: 2002. [Google Scholar]
- 32.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng G, Lu XJ, Olson WK. Web 3DNA--a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 2009;37:W240–246. doi: 10.1093/nar/gkp358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 35.Ferraro P, Franzolin E, Pontarin G, Reichard P, Bianchi V. Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res. 2010;38:e85. doi: 10.1093/nar/gkp1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pallan PS, Marquez VE, Egli M. The conformationally constrained N-methanocarba-dT analogue adopts an unexpected C4'-exo sugar pucker in the structure of a DNA hairpin. Biochemistry. 2012;51:2639–2641. doi: 10.1021/bi300215k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005;24:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ketkar A, Zafar MK, Banerjee S, Marquez VE, Egli M, Eoff RL. A Nucleotide-Analogue-Induced Gain of Function Corrects the Error-Prone Nature of Human DNA Polymerase iota. J. Am. Chem. Soc. 2012;134:10698–10705. doi: 10.1021/ja304176q. [DOI] [PMC free article] [PubMed] [Google Scholar]