Abstract
Multidrug efflux pumps are chromosomally encoded genetic elements capable of mediating resistance to toxic compounds in several life forms. In bacteria, these elements are involved in intrinsic and acquired resistance to antibiotics. Unlike other well-known horizontally acquired antibiotic resistance determinants, genes encoding for multidrug efflux pumps belong to the core of bacterial genomes and thus have evolved over millions of years. The selective pressure stemming from the use of antibiotics to treat bacterial infections is relatively recent in evolutionary terms. Therefore, it is unlikely that these elements have evolved in response to antibiotics. In the last years, several studies have identified numerous functions for efflux pumps that go beyond antibiotic extrusion. In this review we present some examples of these functions that range from bacterial interactions with plant or animal hosts, to the detoxification of metabolic intermediates or the maintenance of cellular homeostasis.
Keywords: multidrug efflux pumps, host/bacteria interactions, plant/bacteria interactions, quorum sensing, antibiotic resistance, bacterial homeostasis, bacterial virulence
INTRODUCTION
Multidrug resistance (MDR) efflux pumps are relevant elements that contribute to both intrinsic and acquired resistance to toxic compounds in diverse life forms, including humans where they have a role in resistance to anti-cancer drugs (Wu et al., 2011), to bacteria, where they are involved in resistance to antibiotics (Webber and Piddock, 2003; Li and Nikaido, 2004, 2009; Poole, 2005, 2007). Unlike well-known horizontally acquired antibiotic resistance determinants, MDR efflux pumps are usually chromosomally encoded and the structural components of different systems are highly conserved in all members of a given bacterial species (Saier et al., 1998; Paulsen et al., 2001; Saier and Paulsen, 2001; Paulsen, 2003; Baquero, 2004; Andam et al., 2011). MDR systems are ancient elements, present in bacterial genomes long before the use of antibiotics for the treatment of human infections (Martinez et al., 2009a). This, along with their ubiquity in different organisms, suggests that the main function of these elements goes beyond providing resistance to antibiotics. The fact that quinolones, a family of synthetic antibiotics, constitute a common substrate of MDR efflux pumps supports this notion (Alonso et al., 1999; Hernandez et al., 2011b). These observations also suggest that the recent selective pressure imposed by the use of antibiotics is not the main evolutionary driver for MDR efflux pumps (Alonso et al., 2001; Martinez et al., 2009b).
Bacterial MDR efflux pumps can be grouped into five different structural families: the adenosine triphosphate (ATP)-binding cassette (ABC) superfamily (Lubelski et al., 2007), the multidrug and toxic compound extrusion (MATE) family (Kuroda and Tsuchiya, 2009), the major facilitator superfamily (MFS) (Pao et al., 1998), the small multidrug resistance (SMR) family (Chung and Saier, 2001), and the resistance/nodulation/division (RND) superfamily (Murakami et al., 2006; Nikaido and Takatsuka, 2009; Nikaido, 2011). The activity of an efflux pump depends on the different types of energy source each system uses: ABC transporters are fueled by ATP hydrolysis; MFS, RND, and SMR use the proton-motive force and MATE transporters consist of Na+/H+ drug antiport systems (Piddock, 2006a).
The RND family includes several members that are relevant to antibiotic resistance in Gram-negative bacteria, whereas the MATE family has been mainly associated to resistance in Gram-positive microorganisms (Piddock, 2006a; Vila and Martinez, 2008). This review will focus exclusively on RND efflux systems.
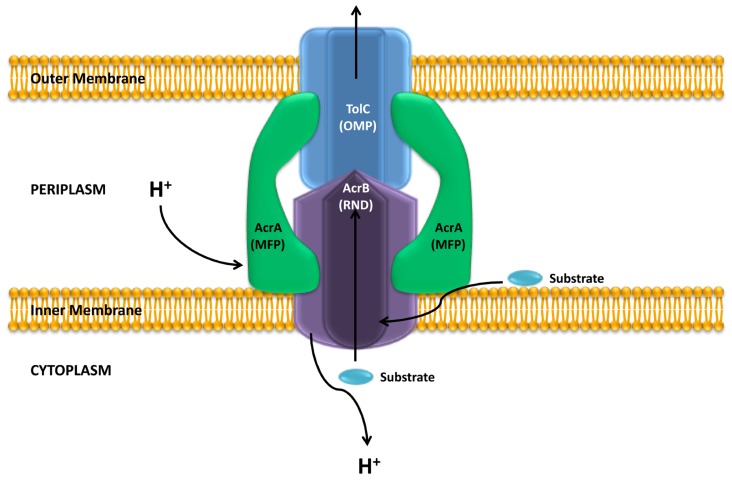
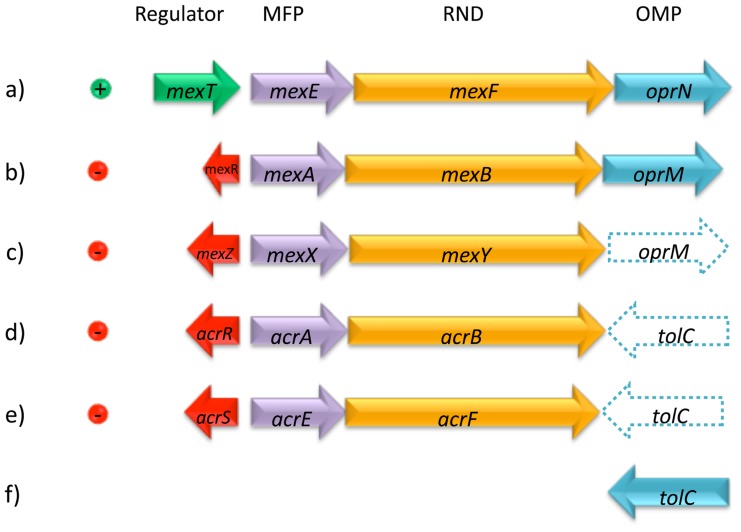
The crystallographic analysis of AcrB, a model member of the RND family, revealed that this protein forms a homotrimer (Murakami et al., 2002; Nakashima et al., 2011), that associates into a tripartite complex along with an outer membrane protein (OMP, TolC) and a periplasmic membrane-fusion protein (MFP, AcrA; Figure 1). Usually the genes encoding for these proteins are found in a single operon, however, the gene encoding for the OMP can also be found elsewhere in the chromosome, as it happens with TolC in Escherichia coli (Koronakis et al., 2000, 2004); or is part of an operon encoding for a different efflux pump (Figure 2). The Pseudomonas aeruginosa RND efflux pump MexXY is an example of the latter, where the system uses the OprM porin encoded in the mexAB-OprM operon (Figure 2; Mine et al., 1999).
FIGURE 1.
Structure of an RND efflux pump. The figure shows a scheme of the structure of the E. coli AcrAB-TolC system. As shown, the system is a tripartite complex formed by the inner membrane AcrB protein, the outer membrane protein TolC and the membrane fusion protein AcrA. The activity of the AcrB RND protein is coupled to the proton gradient. It has been shown that these efflux pumps can extrude different compounds form the bacterial cytoplasm and the periplasm. Adapted from Blair and Piddock (2009).
FIGURE 2.
Representative examples of transcriptional regulation and genetic organization of RND efflux systems. Local regulators can be either transcriptional activators, such as MexT (a) or transcriptional repressors, such as MexR, MexZ, AcrR, or AcrS (b, c, d, and e). The three structural components may be organized in a single operon, such as in the MexEF-OprN (a) or MexAB-OprM (b) systems; alternatively, a given system may use the OMP from another system, such as in the MexXY system using OprM (c). The OMP component may be located elsewhere in the chromosome, such as TolC (f) and can be used by one or more different systems as in the case of AcrAB (d) and AcrEF (e). MexEF-OprN, MexAB-OprM, and MexXY belong to P. aeruginosa; AcrAB and AcrEF belong to E. coli.
In this review we will address the different functional roles that RND efflux pumps may have in addition to mediating antibiotic resistance. Exhaustive information on structure, regulatory aspects, and antibiotic resistance can be found elsewhere (Saier et al., 1998; Paulsen et al., 2001; Saier and Paulsen, 2001; Paulsen, 2003; Webber and Piddock, 2003; Li and Nikaido, 2004, 2009; Poole, 2004, 2007; Piddock, 2006a; Blair and Piddock, 2009; Nikaido, 2009, 2011; Nikaido and Takatsuka, 2009).
Some of the most relevant roles so far identified include involvement in bacterial virulence (Piddock, 2006b), plant–bacteria interactions (Maggiorani Valecillos et al., 2006), trafficking of quorum sensing molecules (Evans et al., 1998; Kohler et al., 2001), and detoxification processes from metabolic intermediates, and toxic compounds such as heavy metals, solvents, or antimicrobials produced by other microorganisms (Aendekerk et al., 2002, 2005; Ramos et al., 2002; Nies, 2003; Sekiya et al., 2003; Burse et al., 2004a). A comprehensive review of all potential functions identified to date for all RND efflux pumps is beyond the scope of this review. Instead, we would like to discuss some selected examples of the ecological role that these systems may have in the absence of antibiotics. As stated above, we believe that the evolution of bacterial RND efflux pumps has been primarily driven by their physiological functions and not by the selective pressure imposed by the relatively recent human use of antibiotics. We consider the important role RND efflux pumps currently play in antibiotic resistance to be an evolutionary novelty stemming from the aforementioned use of antibiotics by humankind (Martinez, 2008; Baquero et al., 2009).
REGULATION OF RND EFFLUX SYSTEMS BY NATURAL EFFECTORS
The regulation of bacterial RND efflux systems is often mediated by global and local regulators, resulting in a multilayered control to optimize gene expression in response to specific cues. A number of positive and negative regulators along with their known mechanism of action have been reviewed elsewhere (Grkovic et al., 2002; Li and Nikaido, 2009).
In most cases a transcriptional regulator (typically a repressor) is encoded upstream the operon coding for the efflux pump (Figure 2). This local regulator usually keeps expression of the efflux pump at a very low-level. High-level expression can be achieved either through an effector-mediated release of the repressed state or through mutations in one or more regulators (Hernandez et al., 2009, 2011a). Activation may occur at different levels: (1) By inactivation of the local repressor that blocks the expression of the pump’s structural genes such as AcrR in E. coli (Ma et al., 1996), MexR in P. aeruginosa (Poole et al., 1996; Sanchez et al., 2002c), or SmeT in Stenotrophomonas maltophilia (Sanchez et al., 2002a); (2) By activation of a global transcriptional regulator like SoxS, RobA, or RamA in E. coli (Martin et al., 2008; Zhang et al., 2008; Perez et al., 2012); (3) By switching on–off one or more steps that interlink regulatory cascades such as MtrR of Neisseria gonorrhoeae (Johnson et al., 2011); and (4) Through the emergence and selection of mutations in key genes like mexT in P. aeruginosa (Kohler et al., 1999).
Multidrug efflux pumps extrude a wide range of substrates. However, the number of effectors regulating them is lower in comparison. Understanding the mechanisms of regulation may help in deciphering the function of RND efflux pumps, since it is expected that different effectors trigger expression only when a given pump is required. RND efflux systems whose expression is controlled by natural inducers normally encountered during the course of infective processes have been studied in detail. Induction of expression by bile salts and fatty acids in enteric bacteria are perhaps the best studied examples of substances capable of modulating expression of these systems.
Expression of the acrAB system in E. coli is induced by decanoate and unconjugated bile salts usually encountered by the organism in the intestinal tract (Ma et al., 1995). The mechanism involves binding of these effectors to the Rob transcriptional regulator (Rosenberg et al., 2003). Bile salts also induce expression of acrAB in Salmonella, however, in this case the effector binds the RamA transcriptional regulator (Nikaido et al., 2008). Interestingly, in both cases the inductor is also a substrate for the efflux system, thus allowing the cell to respond quickly to deleterious environmental substances. Additional examples of bile salts-mediated induction include the cmeABC system in Campylobacter jejuni, the vexD gene in Vibrio cholerae and various RND-type efflux system genes in Bacteroides fragilis (Lin et al., 2005b; Pumbwe et al., 2007). These examples strongly suggest that these systems are relevant to bacterial adaptation for surviving in the gut and that this may be their original function.
In this regard, it has been suggested that some efflux pumps from human commensals and pathogens have evolved to overcome the innate immunity of the host (Blair and Piddock, 2009). For instance, the susceptibility to vertebrate antibacterial peptides in N. gonorrhoeae depends on the activity of the MtrCDE RND efflux pump (Shafer et al., 1998). Notably, this efflux pump is required to achieve mutation-driven resistance to penicillin in N. gonorrhoeae (Veal et al., 2002) and overexpression mutants present reduced susceptibility to several antibiotics and show an increase in in vivo fitness (Warner et al., 2007). MtrCDE (Jerse et al., 2003), enhances experimental gonococcal genital tract infections in female mice, whereas the FarAB-MtrE efflux pump (Lee and Shafer, 1999) is not needed to colonize this environment. It has been suggested that FarAB-MtrE is important for the resistance of N. gonorrhoeae to certain long-chained fatty acids that are present in the rectum (Lee and Shafer, 1999). Altogether these studies indicate that N. gonorrhoeae harbors efflux pumps each one responding to different environmental cues that enable adaptation for survival in different ecosystems.
Metal cations are another example of natural compounds capable of inducing expression of RND efflux pumps. Metals are required as cofactors in several bacterial processes. However, they are toxic at high concentrations. Consequently, bacteria harbor systems to maintain the cellular metal homeostasis. In some cases, this regulation implies that the efflux pump is involved in the extrusion of these toxic effectors (Nies, 2003). However, in other cases the situation is more complex and the effector is simply an environmental cue that indicates the type of ecosystem surrounding the organism. The cusCBA system in E. coli and mtrCDE system in N. gonorrhoeae constitute two of the most studied examples of metal-induced regulation among pathogenic bacteria. The CusCBA system confers tolerance to copper and silver ions (Franke et al., 2001; Grass and Rensing, 2001). Both substrates serve as natural inducers for cusCBA expression (Franke et al., 2001; Yamamoto and Ishihama, 2005), suggesting that this RND efflux pump may have been first selected to overcome the toxicity of these metals. As stated above, the MtrCDE system is involved in resistance to host-derived antibacterial peptides (Shafer et al., 1998). It was recently reported that mtrCDE expression is indirectly regulated by free levels of iron through the regulation of its major transcriptional repressor, MtrR, by the MpeR transcriptional regulator (Mercante et al., 2012). Under the proposed model, expression of the efflux system would increase under iron-limited conditions, a situation that bacteria can encounter over the course of the infection process (Martinez et al., 1990). The P. aeruginosa CzcABC efflux system confers tolerance to zinc, cadmium, and cobalt and constitutes another example of metal-induced expression. The regulation occurs through the metal-inducible CzcRS two-component system that is activated in the presence of the system’s substrates or indirectly in the presence of copper (Caille et al., 2007; Dieppois et al., 2012).
THE ROLE OF EFFLUX PUMPS IN PLANT–BACTERIA INTERACTIONS
The rhizosphere is a complex ecosystem characterized by a high microbial activity that results in a bacterial population density that can be two orders of magnitude higher than in bulk soil (Matilla et al., 2007). The structure of the rhizosphere’s microbiota is governed by the release of nutrients through plant root exudates and by the ecological relationships of the microorganisms present in this ecosystem. A transcriptomic analysis of Pseudomonas putida grown in the rhizosphere of maize revealed that the expression of different efflux pumps is induced in this ecosystem (Matilla et al., 2007), thus suggesting a relevant function for the colonization of this environment. Plant exudates have been identified as good effectors of RND efflux pumps, and it has been shown that these secondary metabolites bind regulators of RND efflux pumps such as TtgR (Alguel et al., 2007), the local repressor of the TtgABC system in P. putida (Teran et al., 2003). Some compounds produced by plants have antibacterial effects and it has been described the RND efflux pumps are required from the first steps of bacterial plant colonization (Espinosa-Urgel et al., 2000) to survival in plant tissues (Barabote et al., 2003), possibly due to their involvement in protection against these compounds. This is the case of Erwinia amylovora, the cause of fire blight disease in rosaceous plants (Eastgate, 2000). The plantlet toxic metabolites naringenin and phloretin are good inducers of the efflux pump acrAB in this bacterial species, and E. amylovora acrAB mutants are much less virulent that their wild-type counterpart (Burse et al., 2004b). A similar situation occurs in Agrobacterium tumefaciens. Coumestrol, an antimicrobial root-exudated flavonoid, is both a substrate and an inducer of expression of the ifeABR efflux system (Palumbo et al., 1998). The fact that this system is needed for effective root colonization indicates that it plays an important role in A. tumefaciens resistance to plant-produced antimicrobials.
Comprehensive analyses on Erwinia chrysanthemi RND efflux pumps revealed that each system may differentially contribute to host specificity. Mutants defective in each of the pumps were differently affected in their virulence in diverse hosts and the susceptibility to plant-produced antimicrobials was specific for each pump (Maggiorani Valecillos et al., 2006). As discussed in the case of N. gonorrhoeae, this suggests that each of the several efflux pumps encoded in the genome of a given bacterial species may have a different function. This adaptation does not rely exclusively on the extrusion of toxic antimicrobial plant exudates. For instance, salicylic acid, an important signaling molecule produced by plants (Loake and Grant, 2007), induces the expression of the E. chrysanthemi efflux pumps acrAB and emrAB (Ravirala et al., 2007). This indicates that RND efflux pumps are relevant elements mediating bacteria/plant interactions at different levels that include the response to toxic compounds, host specificity and interspecies signal trafficking. This functional role is not confined to plant-infective bacteria. Mutants of the mutualistic symbiont Rhizobium etli lacking the RmrAB efflux pump form fewer nodules on its host Phaseolus vulgaris than the corresponding wild-type strain (Gonzalez-Pasayo and Martinez-Romero, 2000). Similarly, the SmeAB efflux pump plays an important role in the nodulation competitiveness in Sinorhizobium meliloti (Eda et al., 2011). The effect of efflux pumps on plant–bacteria interactions can be host-specific. For instance, BdeAB from Bradyrhizobium japonicum is needed for the symbiotic nitrogen-fixation activity on soybean, but not on other host plants such as mung bean and cowpea (Lindemann et al., 2010).
THE ROLE OF EFFLUX PUMPS IN BACTERIAL VIRULENCE
From a clinical point of view, antibiotic resistance could be considered as a colonization factor since only those organisms surviving within a treated patient will be able to cause an infection (Martinez and Baquero, 2002). However, in this section we would like to address the direct role that RND efflux pumps play in the virulence of different human pathogens. As mentioned in a previous section, the expression of different RND efflux pumps is triggered by human-produced compounds, and they contribute to the colonization of different environments in the human host. Although the role of efflux pumps on virulence has been studied for several organism (Piddock, 2006b), only in a few cases comprehensive studies including different systems from a single bacterial species have been performed. Below we discuss some of these examples
Vibrio cholerae RND EFFLUX PUMPS AND VIRULENCE
Vibrio cholerae possesses six different operons encoding for RND-type efflux systems: vexAB, vexCD (breAB), vexEF, vexGH, vexIJK, and vexLM (Kitaoka et al., 2011). While different RND efflux systems often share an OMP, it is rather common that operons encode for a cognate OMP for each system (Figure 2). In the case of V. cholerae, it seems that all six different RND efflux systems operate with the same OMP, encoded by the tolC gene (Bina et al., 2008; Cerda-Maira et al., 2008).
During the course of V. cholerae infections, bacteria colonize primarily the small intestine, where they penetrate the mucus lining coating the intestinal epithelium. In addition to factors produced by the innate immune system, the intestinal environment is rich in substances such as bile salts and organic acids that are capable of inhibiting bacterial growth (Reidl and Klose, 2002). Predictably, four V. cholerae RND efflux systems have been implicated in in vitro resistance to bile salts and detergents similar to detergent-like molecules the organism is likely to encounter during colonization of the intestinal epithelium.
Susceptibility studies with single and multiple mutant combinations revealed that VexB has broad substrate specificity and that it is the primary RND efflux system responsible for resistance to bile salts in vitro (Bina et al., 2008). VexD, VexK, and VexH have also been implied in resistance to bile salts, which denotes redundancy among the different RND efflux systems (Taylor et al., 2012). Moreover, the expression of vexD is induced in the presence of bile salts (Cerda-Maira et al., 2008). The VexK and VexH contribution to bile salts resistance is only evident in a ΔvexBD double mutant background, which suggests a supportive role for VexK and VexH. However, as Taylor et al. (2012) point out, this hierarchy might be limited to their in vitro experimental conditions. In fact, the increasing attenuation levels displayed by combination mutants in in vivo colonization experiments (ΔvexBDK ΔvexBDH ΔvexBDHK < ΔRND), suggest that VexH plays a more relevant role than VexK during the infection process.
RND efflux systems are also required for optimal expression of the genes encoding for two of the most important V. cholerae virulence factors: cholera toxin (CT) and the toxin-coregulated pilus (TCP). A ΔRND mutant exhibited decreased transcription of the tcpA and toxT genes, the latter encoding for a transcriptional activator responsible for transcription of the genes encoding for CT, and a concomitant decrease in CT and TCP production (Bina et al., 2008). While VexB is able to complement this phenotype, a vexBDHK still exhibits a decrease in CT and TCP, thus suggesting a role for VexM and VexF in virulence factor production (Bina et al., 2008; Taylor et al., 2012).
The mechanism through which the V. cholerae RND efflux systems modulate the production of virulence factors has not been elucidated. However, it has been proposed that deletion of systems with redundant functions could lead to the accumulation of a low molecular weight molecule that normally functions as a negative effector molecule involved in fine-tuning the expression of the affected virulence factors (Taylor et al., 2012). V. cholerae inhabits aquatic environments where it normally grows associated with zooplankton or egg masses (Reidl and Klose, 2002). It is possible that some of the RND efflux systems have dedicated functions specific to this portion of the organism’s life cycle. This may be particularly true for VexM and VexF, for which no function in resistance to bile salts and antimicrobials has been identified to date.
Mycobacterium tuberculosis RND EFFLUX PUMPS AND VIRULENCE
The M. tuberculosis genome possesses 13 different genes encoding for RND proteins (Cole et al., 1998). Several domains in these proteins are unique to mycobacteria and are thus designated as MmpL (Mycobacterial membrane protein Large). Four mmpL genes appear to be in operons also containing an mmpS gene. The latter are predicted to encode for proteins equivalent to the MFPs in other bacterial RND systems (Domenech et al., 2005).
In spite of the documented M. tuberculosis resistance against first and second line antimicrobial therapy, none of the RND systems have been associated with antibiotic efflux to date, the only possible exception being MmpL7, which is capable of conferring isoniazid resistance when overexpressed in Mycobacterium smegmatis (Pasca et al., 2005; De Rossi et al., 2006; da Silva et al., 2011). Moreover, deletion mutants created in 11 mmpL genes failed to exhibit significantly altered drug susceptibility in M. tuberculosis (Domenech et al., 2005).
The primary role of most MmpL proteins appears to be the transport of lipids to be incorporated on the cell envelope. The complex mycobacterial cell wall is composed of peptidoglycan, arabinogalactan, and mycolic acids, the surface of which is covered by non-covalently associated lipids that include trehalose monomycolate (TMM), trehalose dimycolate (TDM), sulfolipids, phenolic glycolipids, and phthiocerol dimycocerosates (PDIMs; Tahlan et al., 2012). These lipids play important roles in protection against host-derived toxic molecules, bear an immunomodulatory activity and contribute to M. tuberculosis pathogenicity (Neyrolles and Guilhot, 2011). Lipid transport functions have been ascribed to MmpL3, MmpL7, and MmpL8, and in some cases deletion mutants have demonstrated the contribution of additional MmpL proteins to host survival and pathogenicity.
The inability to create an mmpL3 deletion mutant combined with its absence in transposon mutant collections suggests that this gene is essential to M. tuberculosis (Domenech et al., 2005; Lamichhane et al., 2005). A recent study aimed at identifying the target of a novel M. tuberculosis antibiotic found data that suggests that MmpL3 transports TMM out of the cell and that its inhibition prevents the incorporation of de novo-synthesized mycolic acids into the cell envelope (Tahlan et al., 2012).
MmpL7 is required for PDIM transport to the cell surface and was the first MmpL protein implicated in lipid transport in M. tuberculosis (Cox et al., 1999). In addition, MmpL7 appears to function as a scaffold for the PpsE polyketide synthase required for the final step of phthiocerol synthesis, thus coupling transport and synthesis (Jain and Cox, 2005). At least two different studies have determined that mmpL7 mutants display an attenuation phenotype in murine virulence models (Cox et al., 1999; Domenech et al., 2005). MmpL8 has been implicated in the transport of the SL-N, a precursor of the SL-1 sulfolipid, with a similar mechanism to that of MmpL7 where synthesis and transport appear to be coupled (Converse et al., 2003; Domenech et al., 2004). mmpL8 mutants also display attenuated lethality in murine virulence models (Converse et al., 2003; Domenech et al., 2004, 2005).
Domenech et al. (2005) determined that an mmpL4 mutant has both impaired growth kinetics and impaired lethality in a virulence murine model. The same study determined that while an mmpL11 mutant shows a growth pattern similar to that of the wild-type during the active growth phase, the mutant is attenuated during the course of chronic infections in an in vivo model. No substrate has been identified for these transporters. A role in heme uptake has been recently proposed for MmpL11 and such a function would be in line with the attenuated virulence phenotype observed with an mmpL11 mutant (Tullius et al., 2011). Furthermore, a role in extrusion of host-derived antimicrobials similar to that observed for V. cholerae RND efflux systems cannot be ruled out for those MmpL proteins that appear to be involved in the M. tuberculosis infection process.
Helicobacter pylori RND EFFLUX PUMPS AND VIRULENCE
The gastric colonizer Helicobacter pylori possesses three different operons encoding for RND efflux systems (Tomb et al., 1997). Over the years the systems have received different nomenclatures that may often lead to confusion when revising the literature: hp0605–hp0607 is also referred to as hefABC; hp0969–hp0971 was originally denominated as hefFDE and is currently known as cznABC; finally, the system encoded by hp1329–hp1327 was originally named hefIHG and currently hp1329 and hp1328 are known as czcA and czcB, respectively, while hp1327 is known as crdB.
Bina et al. (2000) initially assessed in vitro and in vivo expression profiles of each system as well as the individual contribution to intrinsic antibiotic susceptibility. The study revealed that hp0607 (hefC) and hp0969 (hefF) are expressed both in in vivo and in vitro, while hp1329 (hefI) is only expressed in vivo. Knockouts in each system failed to identify a contribution to intrinsic antibiotic susceptibility with 19 different compounds. However, overexpression of selected components has been associated with antibiotic resistance and different studies revisiting the contribution of each system to antibiotic susceptibility determined that hp0607 (hefC) and hp0605 (hefA) are involved in intrinsic antibiotic resistance to diverse antibiotics (Kutschke and de Jonge, 2005; Liu et al., 2008; Hirata et al., 2010; Tsugawa et al., 2011).
H. pylori is exposed to bile salts resulting from reflux into the human stomach; bile salts have an inhibitory effect on H. pylori growth, yet the ability to thrive in the presence of a bile gradient suggests that this organism has bile resistance mechanisms in place (Worku et al., 2004; Shao et al., 2008). HefC was recently found to play a role in resistance to bile salts and ceragenins (synthetic bile salt derivatives with antimicrobial activity; Trainor et al., 2011). A hefC mutant exhibited increased susceptibility to deoxycholate, cholate, glycodeoxycholate, taurodeoxycholate, taurocholate and to ceragenin 11(CSA 11); while no changes in susceptibility were observed with mutants in the other two efflux systems. Moreover, HefC appears to have substrate specificity for bile salts, since no change in susceptibility was observed with detergents. Although direct efflux of bile salts through HefC has not been experimentally demonstrated yet, it is likely that this system contributes to H. pylori successful colonization of bile-containing environments.
During the course of gastric colonization, H. pylori is exposed to additional environmental stresses, including low pH gradients (4.0–6.0) and acid shock. Acidic environments impact the bioavailability of metals like iron and nickel, which play an essential role in bacterial metabolism. In addition, environmental metal fluctuations are expected to arise from damaged epithelium and diffusion from ingested food (Stoof et al., 2008). Maintaining a cytoplasmic metal homeostasis is crucial to bacteria, as excessive concentrations can lead to severe cellular damage. The other two H. pylori RND efflux systems are involved in metal efflux.
The system encoded by hp1327–1329 (crdB, czcB, and czcA) constitutes a novel copper efflux pump. Expression of hp1329 is induced in the presence of copper and growth of hp1327 and hp1328 mutants is inhibited in the presence of this metal (Waidner et al., 2002). The same study found that expression of hp1326 (renamed as crdA), encoding for a secreted protein, is strongly induced in the presence of copper and growth of an hp1326 mutant was also impaired in the presence of copper. hp1326 is transcribed as a monocistronic unit, but is believed to constitute a copper resistance system along with hp1327–1329. A follow up study revealed that copper-mediated expression of hp1326 requires the CrdRS two-component system (Waidner et al., 2005); the study did not address expression of hp1327–1229. Mutants lacking the CrdRS system are unable to colonize the stomach of mice (Panthel et al., 2003). This suggests that hp1326 and hp1327–1329 might play an important role during the infective process of H. pylori.
The RND efflux system encoded by hp0969–0971 (renamed as cznABC) has been implicated in cadmium, zinc, and nickel resistance (Stahler et al., 2006). Stahler et al. (2006) showed growth inhibition of individual mutants in the presence of these metals. The H. pylori urease, a nickel-containing enzyme, is an essential colonization factor that enables survival in acidic conditions. Urease activity and expression is regulated in response to nickel availability (van Vliet et al., 2001), accordingly, cznC and cznA mutants exhibited enhanced urease activity (Stahler et al., 2006). The authors propose that the cznABC system plays an important role in fine-tuning urease activity, as nickel efflux reduces activity, while cadmium and zinc efflux prevents inhibition of this enzyme. High urea concentrations are toxic at neutral pH, therefore, untimely activation of this enzyme resulting from perturbations in metals homeostasis can be detrimental to the cell (Meyer-Rosberg et al., 1996; Rektorschek et al., 1998). The inability of cznA, cznB, and cznC mutants to achieve gastric colonization in a gerbil animal model and the failure of a cznA mutant to survive in acidic conditions might be linked to urease activity (Bijlsma et al., 2000; Stahler et al., 2006).
EFFLUX PUMPS AND GLOBAL BACTERIAL PHYSIOLOGY
One of the putative functions of RND efflux pumps is detoxification from detrimental intermediates derived from bacterial metabolism (Neyfakh, 1997). Studies on this subject have been mainly performed using mutants that overproduce RND efflux pumps. It is conceivable that overexpression of these elements might cause a metabolic burden on bacterial populations (Martinez et al., 2007, 2011; Andersson and Hughes, 2010). Indeed, different publications have shown that overproduction of RND efflux pumps may impact bacterial physiology (Sanchez et al., 2002b; Ruiz-Diez et al., 2003; Alonso et al., 2004; Linares et al., 2005; Lertpiriyapong et al., 2012; Olivares et al., 2012). Moreover, the uncontrolled production of these elements can affect the ability of pathogenic bacteria to infect experimental animal models, seriously impairing their virulence (Cosson et al., 2002; Hirakata et al., 2002; Warner et al., 2007; Lertpiriyapong et al., 2012; Perez et al., 2012).
The energy expenditure required to constantly maintain the activity of an efflux pump could lead to a fitness reduction upon overproduction of these elements. However, our group has recently shown that overproduction of the P. aeruginosa MexEF-OprN efflux system does not produce a fitness cost as measured in classical competition tests, although it alters several physiological aspects, including elements relevant for P. aeruginosa virulence such as Type III and Type VI secretion (Tian et al., 2009a,b; Olivares et al., 2012). Notably, this effect is specific to each pump and might be associated to their functional role, as overexpression of either MexAB-OprM of MexXY does not produce the same effect (Linares et al., 2005).
As mentioned before, efflux pumps might be involved in the elimination of endogenous toxic compounds. The P. aeruginosa MexGHI-OpmD efflux system might be implicated in the extrusion of anthranilate, a toxic intermediate of the Pseudomonas quinolone signal (PQS) synthetic pathway (Aendekerk et al., 2002, 2005; Sekiya et al., 2003), whereas MexEF-OprN extrudes kynurenine, another intermediate in the same pathway (Olivares et al., 2012). A recent study has shown that kynurenine and its derivatives have relevant effects in different human diseases, including modulation of the activation of glutamate and nicotinic receptors, the modification of the immune response in situations of inflammation and infection, and the generation and removal of reactive oxygen species (Stone et al., 2012). Any potential impact that the constant extrusion of kynurenine by a MexEF-OprN overexpression mutant may have on the pathogenic behavior of P. aeruginosa remains to be established.
Pseudomonas quinolone signal is one of the quorum sensing (QS) signals produced by P. aeruginosa (Mcknight et al., 2000). Strains overexpressing MDR efflux pumps capable of extruding QS signals or their intermediates are likely to be impaired in the QS response. Indeed, overexpression of MexEF-OprN impairs the QS response of P. aeruginosa (Kohler et al., 2001; Olivares et al., 2012). Previous studies also showed that MexAB-OprM likely extrudes the 3O12-HSL QS signal (Evans et al., 1998; Pearson et al., 1999), and that overproduction of this efflux pump reduced the expression of selected QS-regulated genes. The P. aeruginosa QS regulon comprises approximately 5% of this organism’s genome (Schuster et al., 2003); including several genes involved in virulence. Expression of some of these genes might be energetically costly. However, once the QS signals reach a specific threshold, expression of the regulon is maintained. It has been suggested that being signal-blind can be a good adaptive strategy to avoid this energetic burden (Haas, 2006). Whether the efflux pump-mediated extrusion of QS signals may be beneficial to P. aeruginosa under specific conditions remains to be determined.
Efflux pumps may also compensate for the effects that other bacterial elements may have on the organism. This might be the case of C. jejuni, a leading cause of food-borne enterocolitis worldwide (Ruiz-Palacios, 2007). As an intestinal pathogen this bacterium must overcome the antimicrobial effects of the bile salts secreted into the intestinal tract (Hofmann and Eckmann, 2006). The RND-type efflux pump CmeABC confers resistance to a broad range of antibacterial substances including bile salts, fatty acids, and detergents (Lin et al., 2005a). On the other hand, it has been demonstrated that the type VI secretion system (T6SS) plays a key role in the colonization of the intestinal tract (Lertpiriyapong et al., 2012). The activation of the T6SS may enable bile salts to enter inside the bacterium through the open secretion channel (Bidlack and Silverman, 2004); and this can compromise bacterial viability and infective capability. Bile salts trigger the expression of the CmeABC efflux pump; which extrudes the bile salts immediately outside the cell thus alleviating the entrance through the T6SS (Lin et al., 2005b). The functional interaction between the T6SS and CmeABC might be crucial for intestinal colonization by C. jejuni, thus playing a key role in the virulence of this bacterial pathogen (Lertpiriyapong et al., 2012).
Given the integration of RND efflux systems in bacterial metabolic networks, it is not surprising that their regulation is also incorporated in global regulatory networks. Global regulators such as MarA, RamA, and SoxS can activate the expression of efflux pumps such as AcrAB-TolC in E. coli and in additional Enterobacteriaceae (Davin-Regli et al., 2008). Similarly, the pleiotropic regulator MgrA (Luong et al., 2006) controls autolysis, virulence, biofilm formation, and efflux pump activity in Staphylococcus aureus (Ingavale et al., 2003, 2005; Truong-Bolduc et al., 2003, 2005; Trotonda et al., 2008). The control of efflux pumps by this global regulator is specific for each pump. Increased expression of mgrA in vivo in a subcutaneous abscess model upregulates expression of the norB and tet38 efflux pumps, whereas expression of norA and norC is downregulated (Ding et al., 2008). The relevance of these pumps for the in vivo growth of S. aureus has been studied; norB and tet38 defective mutants present a growth defect in a mice abscess model and the phenotype was not attributable to a staphylococcal stress response (Deng et al., 2012).
MexT, the transcriptional activator of MexEF-OprN in P. aeruginosa (Figure 2), constitutes another example of global regulation. MexT regulates the expression of several P. aeruginosa genes (Tian et al., 2009a). A portion of this regulation is mediated by the activity of the pump through the extrusion of a precursor of the PQS QS signal, and the concomitant impairment of the QS response (Olivares et al., 2012). However, the expression of other genes is directly regulated by MexT (Tian et al., 2009a). A recent study demonstrated that MexT functions as a redox-responsive regulator (Fargier et al., 2012), indicating that it might be involved in controlling cellular redox homeostasis. The fact that a local transcriptional regulator of an efflux pump behaves as a global regulator further supports the involvement of these elements in general processes of bacterial physiology and not simply as a response to the presence of antibiotics in the environment.
CONCLUDING REMARKS
The emergence of antibiotic resistance in bacterial human pathogens is a very recent process in the evolutionary timescale. It is often assumed that resistance genes have been mainly originated in antibiotic producers where they play a detoxification role (Benveniste and Davies, 1973; Webb and Davies, 1993; Davies, 1997). However, in the few cases where the origin of resistance genes has been tracked, the original hosts are not antibiotic producers. The QnrA gene from Shewanella algae constitutes a prime example, as it confers resistance to quinolones, which are synthetic antibiotics (Poirel et al., 2005). This indicates that, at least in some cases, antibiotic resistance would be an emergent function that has been recently selected due to the use of antibiotics for treating infections (Martinez, 2008, 2009a,b; Baquero et al., 2009; Fajardo et al., 2009). As we have seen in this review, MDR efflux pumps also fall within this category, since they exhibit multiple functions relevant to bacterial physiology in addition to mediating antibiotic resistance. A complete understanding of these functions is important in order to define the networks that connect antibiotic resistance with other basic physiological processes (Linares et al., 2010; Martinez and Rojo, 2011), both during the course of infections and in natural, non-clinical ecosystems.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work in our laboratory is supported by grants BIO2011-25255 from the Spanish Ministry of Science and Innovation, PROMPT from CAM, and HEALTH-F3-2011-282004 (EVOTAR) and HEALTH-F3-2010-241476 (PAR) from the European Union. Jorge Olivares is the recipient of a fellowship from the Government of Chile.
REFERENCES
- Aendekerk S., Diggle S. P., Song Z., Hoiby N., Cornelis P., Williams P., et al. (2005). The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology 151 1113–1125 [DOI] [PubMed] [Google Scholar]
- Aendekerk S., Ghysels B., Cornelis P., Baysse C. (2002). Characterization of a new efflux pump, MexGHI-OpmD, from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiology 148 2371–2381 [DOI] [PubMed] [Google Scholar]
- Alguel Y., Meng C., Teran W., Krell T., Ramos J. L., Gallegos M. T., et al. (2007). Crystal structures of multidrug binding protein TtgR in complex with antibiotics and plant antimicrobials. J. Mol. Biol. 369 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A., Morales G., Escalante R., Campanario E., Sastre L., Martinez J. L. (2004). Overexpression of the multidrug efflux pump SmeDEF impairs Stenotrophomonas maltophilia physiology. J. Antimicrob. Chemother. 53 432–434 [DOI] [PubMed] [Google Scholar]
- Alonso A., Rojo F., Martinez J. L. (1999). Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environ. Microbiol. 1 421–430 [DOI] [PubMed] [Google Scholar]
- Alonso A., Sanchez P., Martinez J. L. (2001). Environmental selection of antibiotic resistance genes. Environ. Microbiol. 3 1–9 [DOI] [PubMed] [Google Scholar]
- Andam C. P., Fournier G. P., Gogarten J. P. (2011). Multilevel populations and the evolution of antibiotic resistance through horizontal gene transfer. FEMS Microbiol. Rev. 35 756–767 [DOI] [PubMed] [Google Scholar]
- Andersson D. I., Hughes D. (2010). Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8 260–271 [DOI] [PubMed] [Google Scholar]
- Baquero F. (2004). From pieces to patterns: evolutionary engineering in bacterial pathogens. Nat. Rev. Microbiol. 2 510–518 [DOI] [PubMed] [Google Scholar]
- Baquero F., Alvarez-Ortega C., Martinez J. L. (2009). Ecology and evolution of antibiotic resistance. Environ. Microbiol. Rep. 1 469–476 [DOI] [PubMed] [Google Scholar]
- Barabote R. D., Johnson O. L., Zetina E., San Francisco S. K., Fralick J. A, San Francisco M. J. (2003). Erwinia chrysanthemi tolC is involved in resistance to antimicrobial plant chemicals and is essential for phytopathogenesis. J. Bacteriol. 185 5772–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste R., Davies J. (1973). Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. U.S.A. 70 2276–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidlack J. E., Silverman P. M. (2004). An active type IV secretion system encoded by the F plasmid sensitizes Escherichia coli to bile salts. J. Bacteriol. 186 5202–5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma J. J., Lie A. L. M., Nootenboom I. C., Vandenbroucke-Grauls C. M., Kusters J. G. (2000). Identification of loci essential for the growth of Helicobacter pylori under acidic conditions. J. Infect. Dis. 182 1566–1569 [DOI] [PubMed] [Google Scholar]
- Bina J. E., Alm R. A., Uria-Nickelsen M., Thomas S. R., Trust T. J., Hancock R. E. (2000). Helicobacter pylori uptake and efflux: basis for intrinsic susceptibility to antibiotics in vitro. Antimicrob. Agents Chemother. 44 248–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina X. R., Provenzano D., Nguyen N., Bina J. E. (2008). Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect. Immun. 76 3595–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair J. M., Piddock L. J. (2009). Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr. Opin. Microbiol. 12 512–519 [DOI] [PubMed] [Google Scholar]
- Burse A., Weingart H., Ullrich M. S. (2004a). NorM, an Erwinia amylovora multidrug efflux pump involved in in vitro competition with other epiphytic bacteria. Appl. Environ. Microbiol. 70 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burse A., Weingart H., Ullrich M. S. (2004b). The phytoalexin-inducible multidrug efflux pump AcrAB contributes to virulence in the fire blight pathogen, Erwinia amylovora. Mol. Plant Microbe Interact. 17 43–54 [DOI] [PubMed] [Google Scholar]
- Caille O., Rossier C., Perron K. (2007). A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J. Bacteriol. 189 4561–4568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda-Maira F. A., Ringelberg C. S., Taylor R. K. (2008). The bile response repressor BreR regulates expression of the Vibrio cholerae breAB efflux system operon. J. Bacteriol. 190 7441–7452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y. J., Saier M. H. , Jr (2001). SMR-type multidrug resistance pumps. Curr. Opin. Drug Discov. Dev. 4 237–245 [PubMed] [Google Scholar]
- Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393 537–544 [DOI] [PubMed] [Google Scholar]
- Converse S. E., Mougous J. D., Leavell M. D., Leary J. A., Bertozzi C. R., Cox J. S. (2003). MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc. Natl. Acad. Sci. U.S.A. 100 6121–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P., Zulianello L., Join-Lambert O., Faurisson F., Gebbie L., Benghezal M., et al. (2002). Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J. Bacteriol. 184 3027–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. S., Chen B., Mcneil M., Jacobs W. R. , Jr (1999). Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402 79–83 [DOI] [PubMed] [Google Scholar]
- da Silva P. E., Von Groll A., Martin A., Palomino J. C. (2011). Efflux as a mechanism for drug resistance in Mycobacterium tuberculosis. FEMS Immunol. Med. Microbiol. 63 1–9 [DOI] [PubMed] [Google Scholar]
- Davies J. E. (1997). Origins, acquisition and dissemination of antibiotic resistance determinants. Ciba Found. Symp. 207 15–27 [PubMed] [Google Scholar]
- Davin-Regli A., Bolla J. M., James C. E., Lavigne J. P., Chevalier J., Garnotel E., et al. (2008). Membrane permeability and regulation of drug “influx and efflux” in enterobacterial pathogens. Curr. Drug Targets 9 750–759 [DOI] [PubMed] [Google Scholar]
- Deng X., Sun F., Ji Q., Liang H., Missiakas D., Lan L., et al. (2012). Expression of multidrug resistance efflux pump gene norA is iron responsive in Staphylococcus aureus. J. Bacteriol. 194 1753–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rossi E., Ainsa J. A., Riccardi G. (2006). Role of mycobacterial efflux transporters in drug resistance: an unresolved question. FEMS Microbiol. Rev. 30 36–52 [DOI] [PubMed] [Google Scholar]
- Dieppois G., Ducret V., Caille O., Perron K. (2012). The transcriptional regulator CzcR modulates antibiotic resistance and quorum sensing in Pseudomonas aeruginosa. PLoS ONE 7:e38148 10.1371/journal.pone.0038148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Onodera Y., Lee J. C., Hooper D. C. (2008). NorB, an efflux pump in Staphylococcus aureus MW2, contributes to bacterial fitness in abscesses. J. Bacteriol. 190 7123–7129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech P., Reed M. B, Barry C. E., III (2005). Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 73 3492–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech P., Reed M. B., Dowd C. S., Manca C., Kaplan G, Barry C. E., III (2004). The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J. Biol. Chem. 279 21257–21265 [DOI] [PubMed] [Google Scholar]
- Eastgate J. (2000). Erwinia amylovora: the molecular basis of fireblight disease. Mol. Plant Pathol. 1 325–329 [DOI] [PubMed] [Google Scholar]
- Eda S., Mitsui H., Minamisawa K. (2011). Involvement of the smeAB multidrug efflux pump in resistance to plant antimicrobials and contribution to nodulation competitiveness in Sinorhizobium meliloti. Appl. Environ. Microbiol. 77 2855–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Urgel M., Salido A., Ramos J. L. (2000). Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J. Bacteriol. 182 2363–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K., Passador L., Srikumar R., Tsang E., Nezezon J., Poole K. (1998). Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 180 5443–5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo A., Linares J. F., Martinez J. L. (2009). Towards an ecological approach to antibiotics and antibiotic resistance genes. Clin. Microbiol. Infect. 15(Suppl. 1) 14–16 [DOI] [PubMed] [Google Scholar]
- Fargier E., Mac Aogáin M., Mooij M. J., Woods D. F., Morrissey J. P., Dobson A. D. W., et al. (2012). MexT functions as a redox-responsive regulator modulating disulfide stress resistance in Pseudomonas aeruginosa. J. Bacteriol. 194 3502–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke S., Grass G., Nies D. H. (2001). The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology 147 965–972 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Pasayo R., Martinez-Romero E. (2000). Multiresistance genes of Rhizobium etli CFN42. Mol. Plant Microbe Interact. 13 572–577 [DOI] [PubMed] [Google Scholar]
- Grass G., Rensing C. (2001). Genes involved in copper homeostasis in Escherichia coli. J. Bacteriol. 183 2145–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grkovic S., Brown M. H., Skurray R. A. (2002). Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66 671–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas D. (2006). Cost of cell–cell signalling in Pseudomonas aeruginosa: why it can pay to be signal-blind. Nat. Rev. Microbiol. 4 562 [DOI] [PubMed] [Google Scholar]
- Hernandez A., Mate M. J., Sanchez-Diaz P. C., Romero A., Rojo F., Martinez J. L. (2009). Structural and functional analysis of SmeT, the repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. J. Biol. Chem. 284 14428–14438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez A., Ruiz F. M., Romero A., Martinez J. L. (2011a). The binding of triclosan to SmeT, the repressor of the multidrug efflux pump SmeDEF, induces antibiotic resistance in Stenotrophomonas maltophilia. PLoS Pathog. 7:e1002103 10.1371/journal.ppat.1002103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez A., Sanchez M. B., Martinez J. L. (2011b). Quinolone resistance: much more than predicted. Front. Microbiol. 2:22 10.3389/fmicb.2011.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakata Y., Srikumar R., Poole K., Gotoh N., Suematsu T., Kohno S., et al. (2002). Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J. Exp. Med. 196 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata K., Suzuki H., Nishizawa T., Tsugawa H., Muraoka H., Saito Y., et al. (2010). Contribution of efflux pumps to clarithromycin resistance in Helicobacter pylori. J. Gastroenterol. Hepatol. 25(Suppl. 1) S75–S79 [DOI] [PubMed] [Google Scholar]
- Hofmann A. F., Eckmann L. (2006). How bile acids confer gut mucosal protection against bacteria. Proc. Natl. Acad. Sci. U.S.A. 103 4333–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingavale S., Van Wamel W., Luong T. T., Lee C. Y., Cheung A. L. (2005). Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect. Immun. 73 1423–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingavale S. S., Van Wamel W., Cheung A. L. (2003). Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 48 1451–1466 [DOI] [PubMed] [Google Scholar]
- Jain M., Cox J. S. (2005). Interaction between polyketide synthase and transporter suggests coupled synthesis and export of virulence lipid in M. tuberculosis. PLoS Pathog. 1:e2 10.1371/journal.ppat.0010002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerse A. E., Sharma N. D., Simms A. N., Crow E. T., Snyder L. A., Shafer W. M. (2003). A gonococcal efflux pump system enhances bacterial survival in a female mouse model of genital tract infection. Infect. Immun. 71 5576–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. J., Stringer V. A., Shafer W. M. (2011). Off-target gene regulation mediated by transcriptional repressors of antimicrobial efflux pump genes in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 55 2559–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka M., Miyata S. T., Unterweger D., Pukatzki S. (2011). Antibiotic resistance mechanisms of Vibrio cholerae. J. Med. Microbiol. 60 397–407 [DOI] [PubMed] [Google Scholar]
- Kohler T., Epp S. F., Curty L. K., Pechere J. C. (1999). Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 181 6300–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler T., Van Delden C., Curty L. K., Hamzehpour M. M., Pechere J. C. (2001). Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183 5213–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis V., Eswaran J., Hughes C. (2004). Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 73 467–489 [DOI] [PubMed] [Google Scholar]
- Koronakis V., Sharff A., Koronakis E., Luisi B., Hughes C. (2000). Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405 914–919 [DOI] [PubMed] [Google Scholar]
- Kuroda T., Tsuchiya T. (2009). Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta 1794 763–768 [DOI] [PubMed] [Google Scholar]
- Kutschke A, de Jonge B. L. (2005). Compound efflux in Helicobacter pylori. Antimicrob. Agents Chemother. 49 3009–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane G., Tyagi S., Bishai W. R. (2005). Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs. Infect. Immun. 73 2533–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. H., Shafer W. M. (1999). The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol. Microbiol. 33 839–845 [DOI] [PubMed] [Google Scholar]
- Lertpiriyapong K., Gamazon E. R., Feng Y., Park D. S., Pang J., Botka G., et al. (2012). Campylobacter jejuni type VI secretion system: roles in adaptation to deoxycholic acid, host cell adherence, invasion, and in vivo colonization. PLoS ONE 7:e42842 10.1371/journal.pone.0042842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Z., Nikaido H. (2004). Efflux-mediated drug resistance in bacteria. Drugs 64 159–204 [DOI] [PubMed] [Google Scholar]
- Li X. Z., Nikaido H. (2009). Efflux-mediated drug resistance in bacteria: an update. Drugs 69 1555–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Akiba M., Sahin O., Zhang Q. (2005a). CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob. Agents Chemother. 49 1067–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Cagliero C., Guo B., Barton Y. W., Maurel M. C., Payot S., et al. (2005b). Bile salts modulate expression of the CmeABC multidrug efflux pump in Campylobacter jejuni. J. Bacteriol. 187 7417–7424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares J. F., Lopez J. A., Camafeita E., Albar J. P., Rojo F., Martinez J. L. (2005). Overexpression of the multidrug efflux pumps MexCD-OprJ and MexEF-OprN is associated with a reduction of type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187 1384–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares J. F., Moreno R., Fajardo A., Martinez-Solano L., Escalante R., Rojo F., et al. (2010). The global regulator Crc modulates metabolism, susceptibility to antibiotics and virulence in Pseudomonas aeruginosa. Environ. Microbiol. 12 3196–3212 [DOI] [PubMed] [Google Scholar]
- Lindemann A., Koch M., Pessi G., Muller A. J., Balsiger S., Hennecke H., et al. (2010). Host-specific symbiotic requirement of BdeAB, a RegR-controlled RND-type efflux system in Bradyrhizobium japonicum. FEMS Microbiol. Lett. 312 184–191 [DOI] [PubMed] [Google Scholar]
- Liu Z. Q., Zheng P. Y., Yang P. C. (2008). Efflux pump gene hefA of Helicobacter pylori plays an important role in multidrug resistance. World J. Gastroenterol. 14 5217–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loake G., Grant M. (2007). Salicylic acid in plant defence-the players and protagonists. Curr. Opin. Plant Biol. 10 466–472 [DOI] [PubMed] [Google Scholar]
- Lubelski J., Konings W. N., Driessen A. J. (2007). Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol. Mol. Biol. Rev. 71 463–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong T. T., Dunman P. M., Murphy E., Projan S. J., Lee C. Y. (2006). Transcription profiling of the mgrA regulon in Staphylococcus aureus. J. Bacteriol. 188 1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Alberti M., Lynch C., Nikaido H., Hearst J. E. (1996). The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19 101–112 [DOI] [PubMed] [Google Scholar]
- Ma D., Cook D. N., Alberti M., Pon N. G., Nikaido H., Hearst J. E. (1995). Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16 45–55 [DOI] [PubMed] [Google Scholar]
- Maggiorani Valecillos A., Rodriguez Palenzuela P., Lopez-Solanilla E. (2006). The role of several multidrug resistance systems in Erwinia chrysanthemi pathogenesis. Mol. Plant Microbe Interact. 19 607–613 [DOI] [PubMed] [Google Scholar]
- Martin R. G., Bartlett E. S., Rosner J. L., Wall M. E. (2008). Activation of the Escherichia coli marA/soxS/rob regulon in response to transcriptional activator concentration. J. Mol. Biol. 380 278–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. L. (2008). Antibiotics and antibiotic resistance genes in natural environments. Science 321 365–367 [DOI] [PubMed] [Google Scholar]
- Martinez J. L. (2009a). Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 157 2893–2902 [DOI] [PubMed] [Google Scholar]
- Martinez J. L. (2009b). The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. R. Soc. Lond. B Biol. Sci. 276 2521–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. L., Baquero F. (2002). Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clin. Microbiol. Rev. 15 647–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. L., Baquero F., Andersson D. I. (2007). Predicting antibiotic resistance. Nat. Rev. Microbiol. 5 958–965 [DOI] [PubMed] [Google Scholar]
- Martinez J. L., Baquero F., Andersson D. I. (2011). Beyond serial passages: new methods for predicting the emergence of resistance to novel antibiotics. Curr. Opin. Pharmacol. 11 439–445 [DOI] [PubMed] [Google Scholar]
- Martinez J. L., Delgado-Iribarren A., Baquero F. (1990). Mechanisms of iron acquisition and bacterial virulence. FEMS Microbiol. Rev. 6 45–56 [DOI] [PubMed] [Google Scholar]
- Martinez J. L., Fajardo A., Garmendia L., Hernandez A., Linares J. F., Martinez-Solano L., et al. (2009a). A global view of antibiotic resistance. FEMS Microbiol. Rev. 33 44–65 [DOI] [PubMed] [Google Scholar]
- Martinez J. L., Sanchez M. B., Martinez-Solano L., Hernandez A., Garmendia L., Fajardo A., et al. (2009b). Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 33 430–449 [DOI] [PubMed] [Google Scholar]
- Martinez J. L., Rojo F. (2011). Metabolic regulation of antibiotic resistance. FEMS Microbiol. Rev. 35 768–789 [DOI] [PubMed] [Google Scholar]
- Matilla, M. A., Espinosa-Urgel M., Rodriguez-Herva J. J., Ramos J. L., Ramos-Gonzalez M. I. (2007). Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol. 8 R179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcknight S. L., Iglewski B. H., Pesci E. C. (2000). The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182 2702–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercante A. D., Jackson L., Johnson P. J., Stringer V. A., Dyer D. W., Shafer W. M. (2012). MpeR regulates the mtr efflux locus in Neisseria gonorrhoeae and modulates antimicrobial resistance by an iron-responsive mechanism. Antimicrob. Agents Chemother. 56 1491–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Rosberg K., Scott D. R., Rex D., Melchers K., Sachs G. (1996). The effect of environmental pH on the proton motive force of Helicobacter pylori. Gastroenterology 111 886–900 [DOI] [PubMed] [Google Scholar]
- Mine T., Morita Y., Kataoka A., Mizushima T., Tsuchiya T. (1999). Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43 415–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S., Nakashima R., Yamashita E., Matsumoto T., Yamaguchi A. (2006). Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443 173–179 [DOI] [PubMed] [Google Scholar]
- Murakami S., Nakashima R., Yamashita E., Yamaguchi A. (2002). Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419 587–593 [DOI] [PubMed] [Google Scholar]
- Nakashima R., Sakurai K., Yamasaki S., Nishino K., Yamaguchi A. (2011). Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 480 565–569 [DOI] [PubMed] [Google Scholar]
- Neyfakh A. A. (1997). Natural functions of bacterial multidrug transporters. Trends Microbiol. 5 309–313 [DOI] [PubMed] [Google Scholar]
- Neyrolles O., Guilhot C. (2011). Recent advances in deciphering the contribution of Mycobacterium tuberculosis lipids to pathogenesis. Tuberculosis 91 187–195 [DOI] [PubMed] [Google Scholar]
- Nies D. H. (2003). Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27 313–339 [DOI] [PubMed] [Google Scholar]
- Nikaido E., Yamaguchi A., Nishino K. (2008). AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J. Biol. Chem. 283 24245–24253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. (2009). Multidrug resistance in bacteria. Annu. Rev. Biochem. 78 119–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. (2011). Structure and mechanism of RND-type multidrug efflux pumps. Adv. Enzymol. Relat. Areas Mol. Biol. 77 1–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H., Takatsuka Y. (2009). Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta 1794 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares J., Alvarez-Ortega C., Linares J. F., Rojo F., Kohler T., Martinez J. L. (2012). Overproduction of the multidrug efflux pump MexEF-OprN does not impair Pseudomonas aeruginosa fitness in competition tests, but produces specific changes in bacterial regulatory networks. Environ. Microbiol. 14 1968–1981 [DOI] [PubMed] [Google Scholar]
- Palumbo J. D., Kado C. I., Phillips D. A. (1998). An isoflavonoid-inducible efflux pump in Agrobacterium tumefaciens is involved in competitive colonization of roots. J. Bacteriol. 180 3107–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panthel K., Dietz P., Haas R., Beier D. (2003). Two-component systems of Helicobacter pylori contribute to virulence in a mouse infection model. Infect. Immun. 71 5381–5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao S. S., Paulsen I. T., Saier M. H. , Jr (1998). Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca M. R., Guglierame P., De Rossi E., Zara F., Riccardi G. (2005). mmpL7 gene of Mycobacterium tuberculosis is responsible for isoniazid efflux in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 49 4775–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen I. T. (2003). Multidrug efflux pumps and resistance: regulation and evolution. Curr. Opin. Microbiol. 6 446–451 [DOI] [PubMed] [Google Scholar]
- Paulsen I. T., Chen J., Nelson K. E., Saier M. H. , Jr (2001). Comparative genomics of microbial drug efflux systems. J. Mol. Microbiol. Biotechnol. 3 145–150 [PubMed] [Google Scholar]
- Pearson J. P., Van Delden C., Iglewski B. H. (1999). Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181 1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez A., Poza M., Aranda J., Latasa C., Medrano F. J., Tomas M., et al. (2012). Effect of the transcriptional activators SoxS, RobA and RamA on expression of the multidrug efflux pump AcrAB-TolC in Enterobacter cloacae. Antimicrob. Agents Chemother. 56 6256–6266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock L. J. (2006a). Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19 382–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock L. J. (2006b). Multidrug-resistance efflux pumps – not just for resistance. Nat. Rev. Microbiol. 4 629–636 [DOI] [PubMed] [Google Scholar]
- Poirel L., Rodriguez-Martinez J. M., Mammeri H., Liard A., Nordmann P. (2005). Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 49 3523–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. (2004). “Efflux pumps,” in Pseudomonas, ed Ramos J.-L. (New York: Kluwer Academic/Plenum Publisher: ) 636–641 [Google Scholar]
- Poole K. (2005). Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 56 20–51 [DOI] [PubMed] [Google Scholar]
- Poole K. (2007). Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 39 162–176 [DOI] [PubMed] [Google Scholar]
- Poole K., Gotoh N., Tsujimoto H., Zhao Q., Wada A., Yamasaki T., et al. (1996). Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21 713–724 [DOI] [PubMed] [Google Scholar]
- Pumbwe L., Skilbeck C. A., Nakano V., Avila-Campos M. J., Piazza R. M., Wexler H. M. (2007). Bile salts enhance bacterial co-aggregation, bacterial-intestinal epithelial cell adhesion, biofilm formation and antimicrobial resistance of Bacteroides fragilis. Microb. Pathog. 43 78–87 [DOI] [PubMed] [Google Scholar]
- Ramos J. L., Duque E., Gallegos M. T., Godoy P., Ramos-Gonzalez M. I., Rojas A., et al. (2002). Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56 743–768 [DOI] [PubMed] [Google Scholar]
- Ravirala R. S., Barabote R. D., Wheeler D. M., Reverchon S., Tatum O., Malouf J., et al. (2007). Efflux pump gene expression in Erwinia chrysanthemi is induced by exposure to phenolic acids. Mol. Plant Microbe Interact. 20 313–320 [DOI] [PubMed] [Google Scholar]
- Reidl J., Klose K. E. (2002). Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol. Rev. 26 125–139 [DOI] [PubMed] [Google Scholar]
- Rektorschek M., Weeks D., Sachs G., Melchers K. (1998). Influence of pH on metabolism and urease activity of Helicobacter pylori. Gastroenterology 115 628–641 [DOI] [PubMed] [Google Scholar]
- Rosenberg E. Y., Bertenthal D., Nilles M. L., Bertrand K. P., Nikaido H. (2003). Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48 1609–1619 [DOI] [PubMed] [Google Scholar]
- Ruiz-Diez B., Sanchez P., Baquero F., Martinez J. L., Navas A. (2003). Differential interactions within the Caenorhabditis elegans–Pseudomonas aeruginosa pathogenesis model. J. Theor. Biol. 225 469–476 [DOI] [PubMed] [Google Scholar]
- Ruiz-Palacios G. M. (2007). The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin. Infect. Dis. 44 701–703 [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr., Paulsen I. T. (2001). Phylogeny of multidrug transporters. Semin. Cell Dev. Biol. 12 205–213 [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr., Paulsen I. T., Sliwinski M. K., Pao S. S., Skurray R. A., Nikaido H. (1998). Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 12 265–274 [DOI] [PubMed] [Google Scholar]
- Sanchez P., Alonso A., Martinez J. L. (2002a). Cloning and characterization of SmeT, a repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. Antimicrob. Agents Chemother. 46 3386–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez P., Linares J. F., Ruiz-Diez B., Campanario E., Navas A., Baquero F., et al. (2002b). Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J. Antimicrob. Chemother. 50 657–664 [DOI] [PubMed] [Google Scholar]
- Sanchez P., Rojo F., Martinez J. L. (2002c). Transcriptional regulation of mexR, the repressor of Pseudomonas aeruginosa mexAB-oprM multidrug efflux pump. FEMS Microbiol. Lett. 207 63–68 [DOI] [PubMed] [Google Scholar]
- Schuster M., Lostroh C. P., Ogi T., Greenberg E. P. (2003). Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185 2066–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya H., Mima T., Morita Y., Kuroda T., Mizushima T., Tsuchiya T. (2003). Functional cloning and characterization of a multidrug efflux pump, mexHI-opmD, from a Pseudomonas aeruginosa mutant. Antimicrob. Agents Chemother. 47 2990–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer W. M., Qu X., Waring A. J., Lehrer R. I. (1998). Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. U.S.A. 95 1829–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C., Zhang Q., Sun Y., Liu Z., Zeng J., Zhou Y., et al. (2008). Helicobacter pylori protein response to human bile stress. J. Med. Microbiol. 57 151–158 [DOI] [PubMed] [Google Scholar]
- Stahler F. N., Odenbreit S., Haas R., Wilrich J., Van Vliet A. H., Kusters J. G., et al. (2006). The novel Helicobacter pylori CznABC metal efflux pump is required for cadmium, zinc, and nickel resistance, urease modulation, and gastric colonization. Infect. Immun. 74 3845–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone T. W., Forrest C. M., Stoy N., Darlington L. G. (2012). Involvement of kynurenines in Huntington’s disease and stroke-induced brain damage. J. Neural. Transm. 119 261–274 [DOI] [PubMed] [Google Scholar]
- Stoof J., Belzer C, Van Vliet A. H. M. (2008). “Metal metabolism and transport in Helicobacter pylori,” in Helicobacter pylori: Molecular Genetics and Cellular Biology, ed Yamaoka Y. (Norfolk: Caister Academic Press; ) 165–177 [Google Scholar]
- Tahlan K., Wilson R., Kastrinsky D. B., Arora K., Nair V., Fischer E., et al. (2012). SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 56 1797–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. L., Bina X. R., Bina J. E. (2012). Vibrio cholerae VexH encodes a multiple drug efflux pump that contributes to the production of cholera toxin and the toxin co-regulated pilus. PLoS ONE 7:e38208 10.1371/journal.pone.0038208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teran W., Felipe A., Segura A., Rojas A., Ramos J. L., Gallegos M. T. (2003). Antibiotic-dependent induction of Pseudomonas putida DOT-T1E TtgABC efflux pump is mediated by the drug binding repressor TtgR. Antimicrob. Agents Chemother. 47 3067–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z. X., Fargier E., Mac Aogain M., Adams C., Wang Y. P, O’Gara F. (2009a). Transcriptome profiling defines a novel regulon modulated by the LysR-type transcriptional regulator MexT in Pseudomonas aeruginosa. Nucleic Acids Res. 37 7546–7559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z. X., Mac Aogain M., O’Connor H. F., Fargier E., Mooij M. J., Adams C., et al. (2009b). MexT modulates virulence determinants in Pseudomonas aeruginosa independent of the MexEF-OprN efflux pump. Microb. Pathog. 47 237–241 [DOI] [PubMed] [Google Scholar]
- Tomb J. F., White O., Kerlavage A. R., Clayton R. A., Sutton G. G., Fleischmann R. D., et al. (1997). The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388 539–547 [DOI] [PubMed] [Google Scholar]
- Trainor E. A., Horton K. E., Savage P. B., Testerman T. L., Mcgee D. J. (2011). Role of the HefC efflux pump in Helicobacter pylori cholesterol-dependent resistance to ceragenins and bile salts. Infect. Immun. 79 88–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotonda M. P., Tamber S., Memmi G., Cheung A. L. (2008). MgrA represses biofilm formation in Staphylococcus aureus. Infect. Immun. 76 5645–5654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong-Bolduc Q. C., Dunman P. M., Strahilevitz J., Projan S. J., Hooper D. C. (2005). MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J. Bacteriol. 187 2395–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong-Bolduc Q. C., Zhang X., Hooper D. C. (2003). Characterization of NorR protein, a multifunctional regulator of norA expression in Staphylococcus aureus. J. Bacteriol. 185 3127–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa H., Suzuki H., Muraoka H., Ikeda F., Hirata K., Matsuzaki J., et al. (2011). Enhanced bacterial efflux system is the first step to the development of metronidazole resistance in Helicobacter pylori. Biochem. Biophys. Res. Commun. 404 656–660 [DOI] [PubMed] [Google Scholar]
- Tullius M. V., Harmston C. A., Owens C. P., Chim N., Morse R. P., Mcmath L. M., et al. (2011). Discovery and characterization of a unique mycobacterial heme acquisition system. Proc. Natl. Acad. Sci. U.S.A. 108 5051–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet A. H., Kuipers E. J., Waidner B., Davies B. J., De Vries N., Penn C. W., et al. (2001). Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect. Immun. 69 4891–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal W. L., Nicholas R. A., Shafer W. M. (2002). Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J. Bacteriol. 184 5619–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila J., Martinez J. L. (2008). Clinical impact of the over-expression of efflux pump in nonfermentative gram-negative bacilli, development of efflux pump inhibitors. Curr. Drug Targets 9 797–807 [DOI] [PubMed] [Google Scholar]
- Waidner B., Melchers K., Ivanov I., Loferer H., Bensch K. W., Kist M., et al. (2002). Identification by RNA profiling and mutational analysis of the novel copper resistance determinants CrdA (HP1326), CrdB (HP1327), and CzcB (HP1328) in Helicobacter pylori. J. Bacteriol. 184 6700–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waidner B., Melchers K., Stahler F. N., Kist M., Bereswill S. (2005). The Helicobacter pylori CrdRS two-component regulation system (HP1364/HP1365) is required for copper-mediated induction of the copper resistance determinant CrdA. J. Bacteriol. 187 4683–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner D. M., Folster J. P., Shafer W. M., Jerse A. E. (2007). Regulation of the MtrC-MtrD-MtrE efflux-pump system modulates the in vivo fitness of Neisseria gonorrhoeae. J. Infect. Dis. 196 1804–1812 [DOI] [PubMed] [Google Scholar]
- Webb V., Davies J. (1993). Antibiotic preparations contain DNA: a source of drug resistance genes? Antimicrob. Agents Chemother. 37 2379–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber M. A., Piddock L. J. (2003). The importance of efflux pumps in bacterial antibiotic resistance. J. Antimicrob. Chemother. 51 9–11 [DOI] [PubMed] [Google Scholar]
- Worku M. L., Karim Q. N., Spencer J., Sidebotham R. L. (2004). Chemotactic response of Helicobacter pylori to human plasma and bile. J. Med. Microbiol. 53 807–811 [DOI] [PubMed] [Google Scholar]
- Wu C. P., Hsieh C. H., Wu Y. S. (2011). The emergence of drug transporter-mediated multidrug resistance to cancer chemotherapy. Mol. Pharm. 8 1996–2011 [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Ishihama A. (2005). Transcriptional response of Escherichia coli to external copper. Mol. Microbiol. 56 215–227 [DOI] [PubMed] [Google Scholar]
- Zhang A., Rosner J. L., Martin R. G. (2008). Transcriptional activation by MarA, SoxS and Rob of two tolC promoters using one binding site: a complex promoter configuration for tolC in Escherichia coli. Mol. Microbiol. 69 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]