Abstract
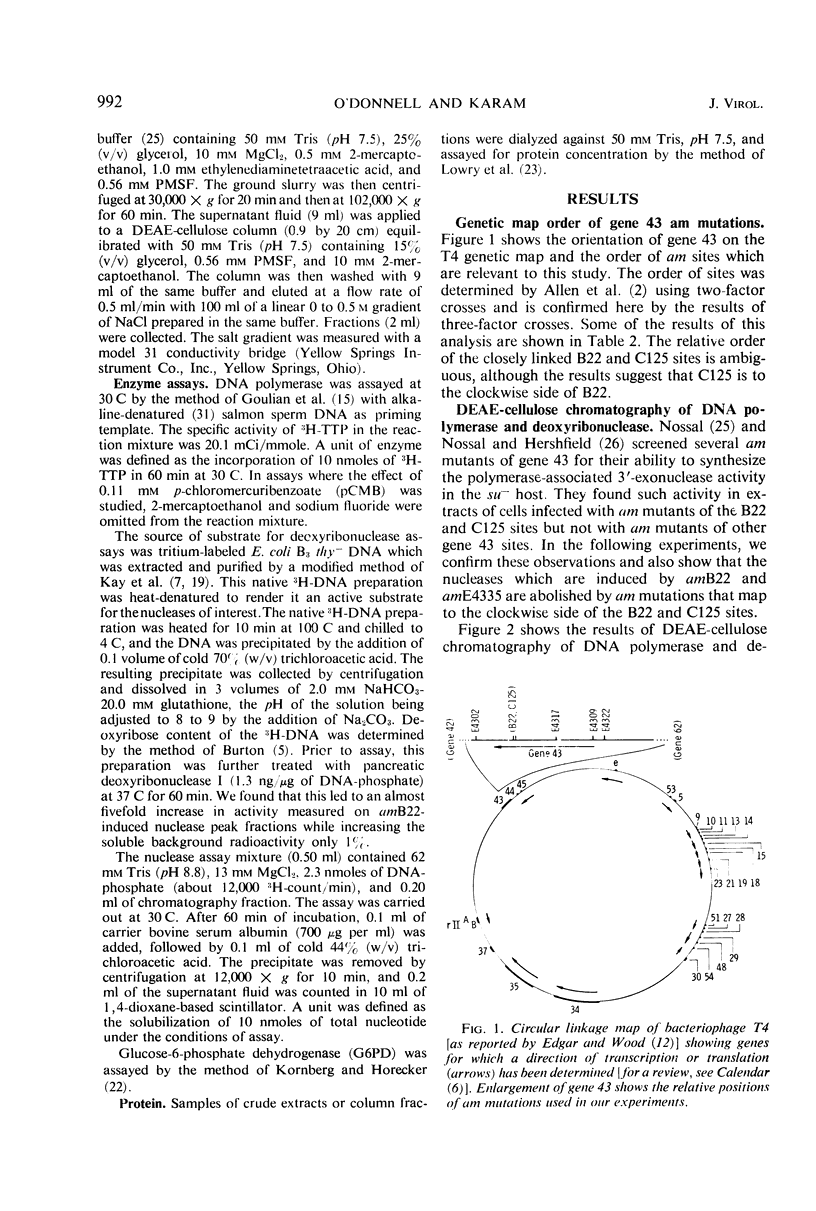
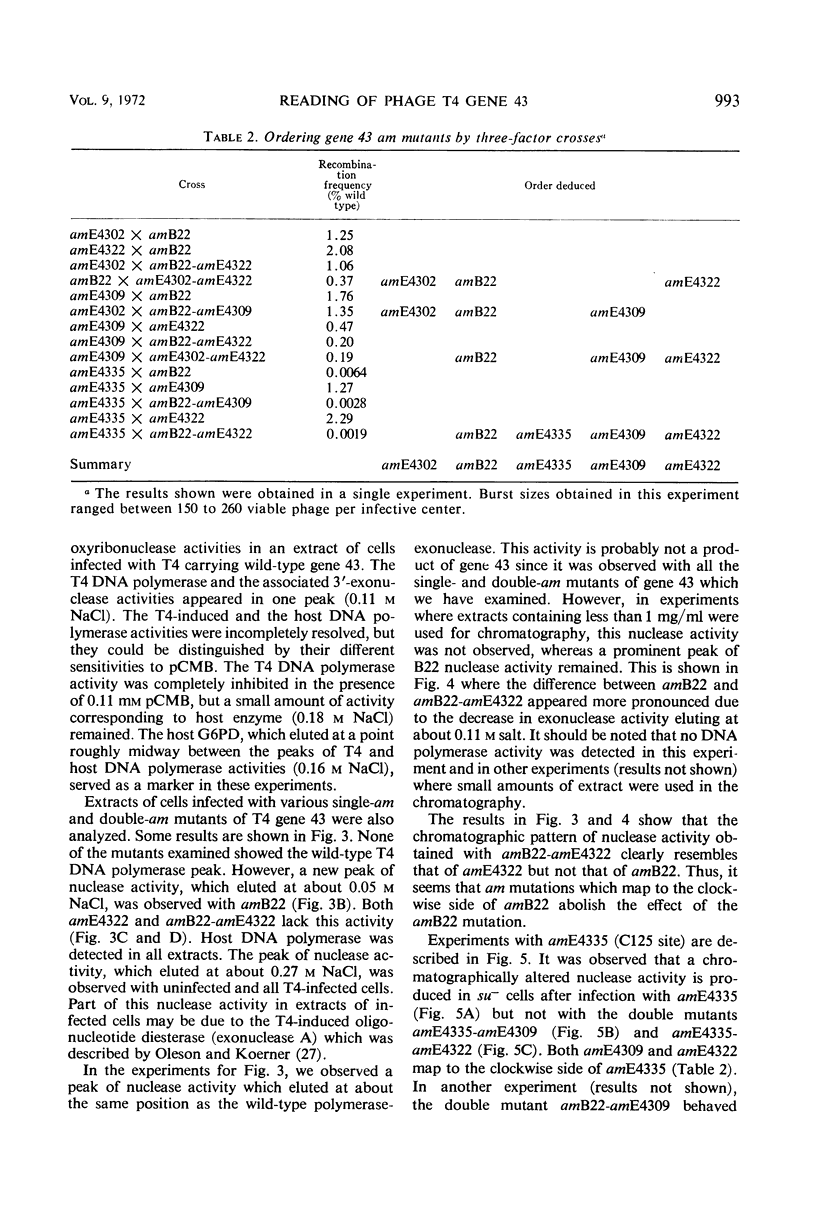
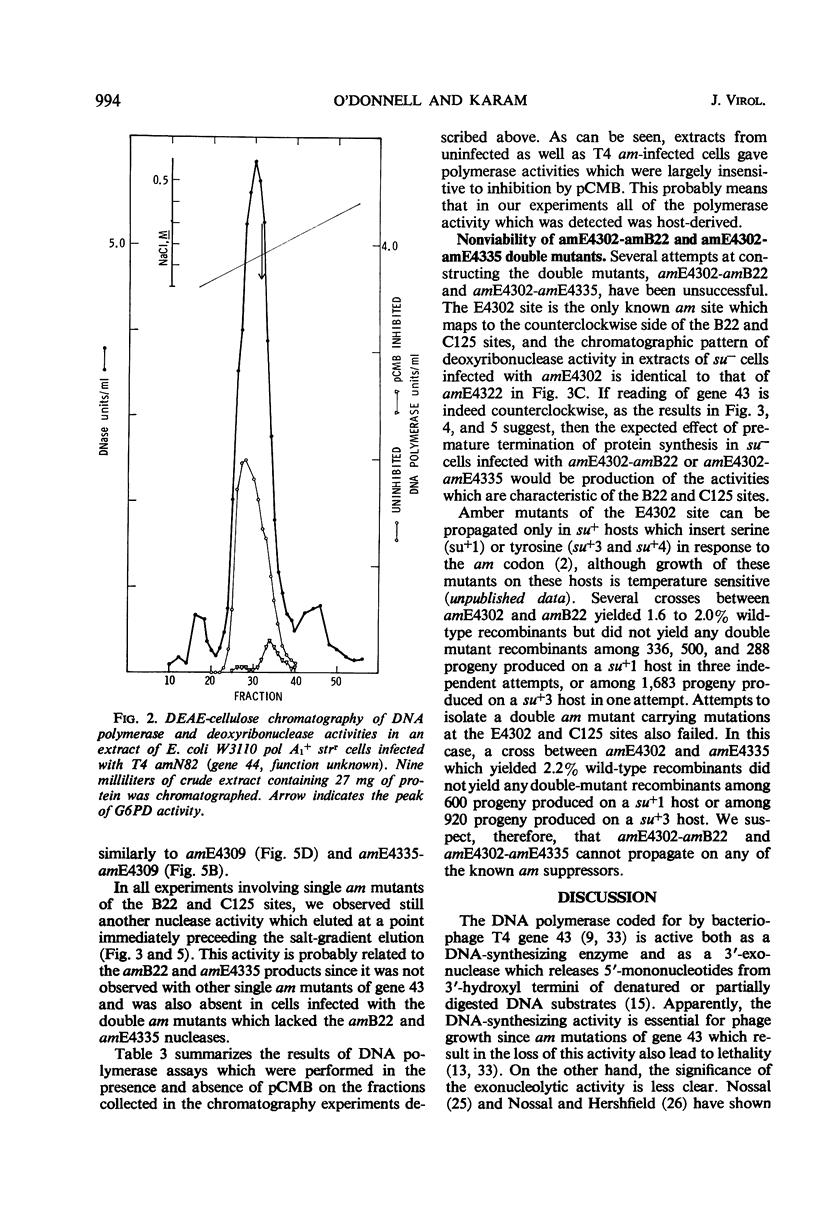
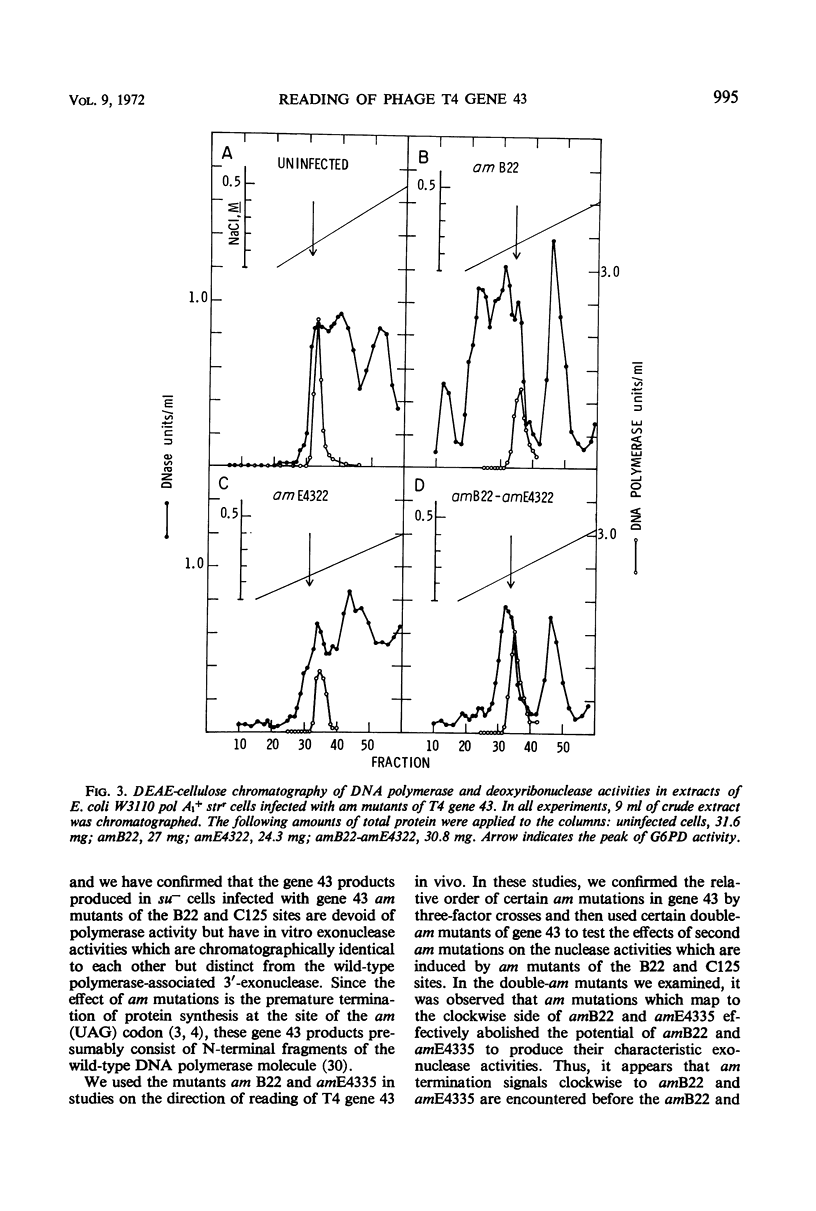
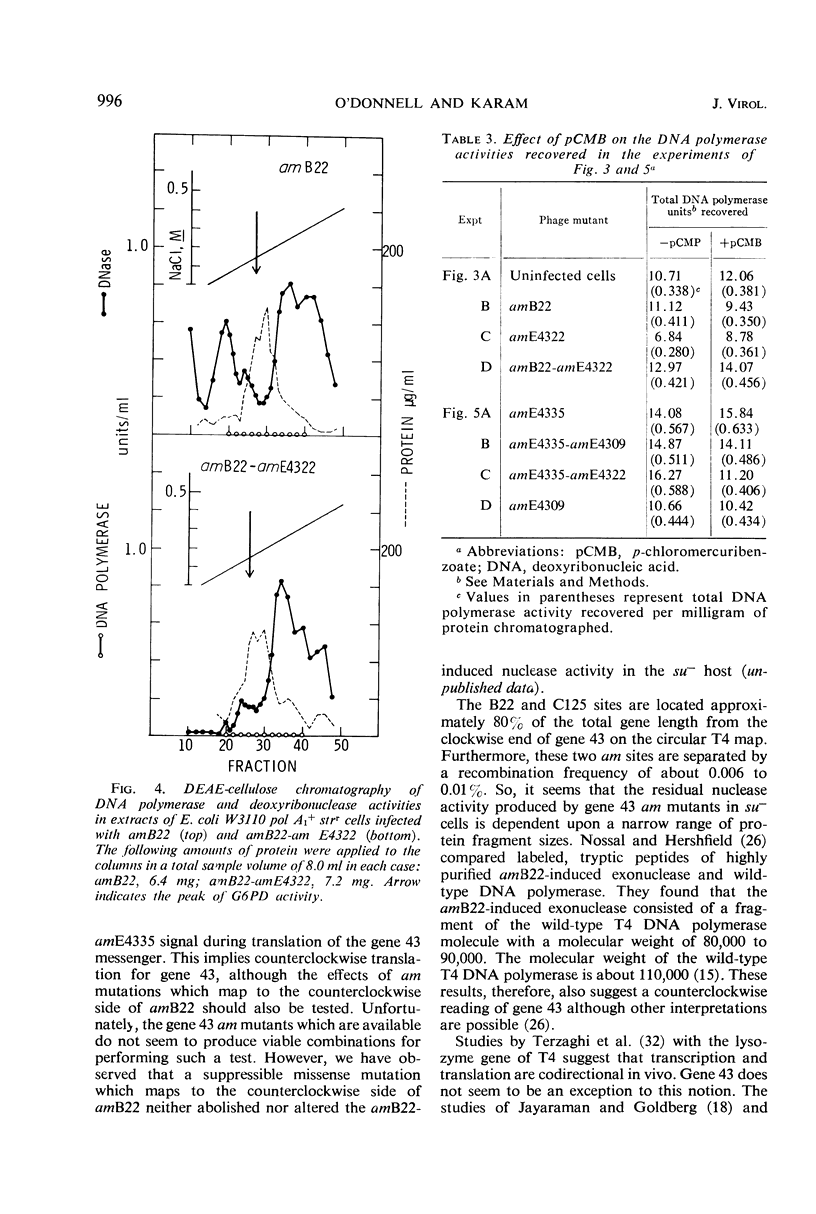
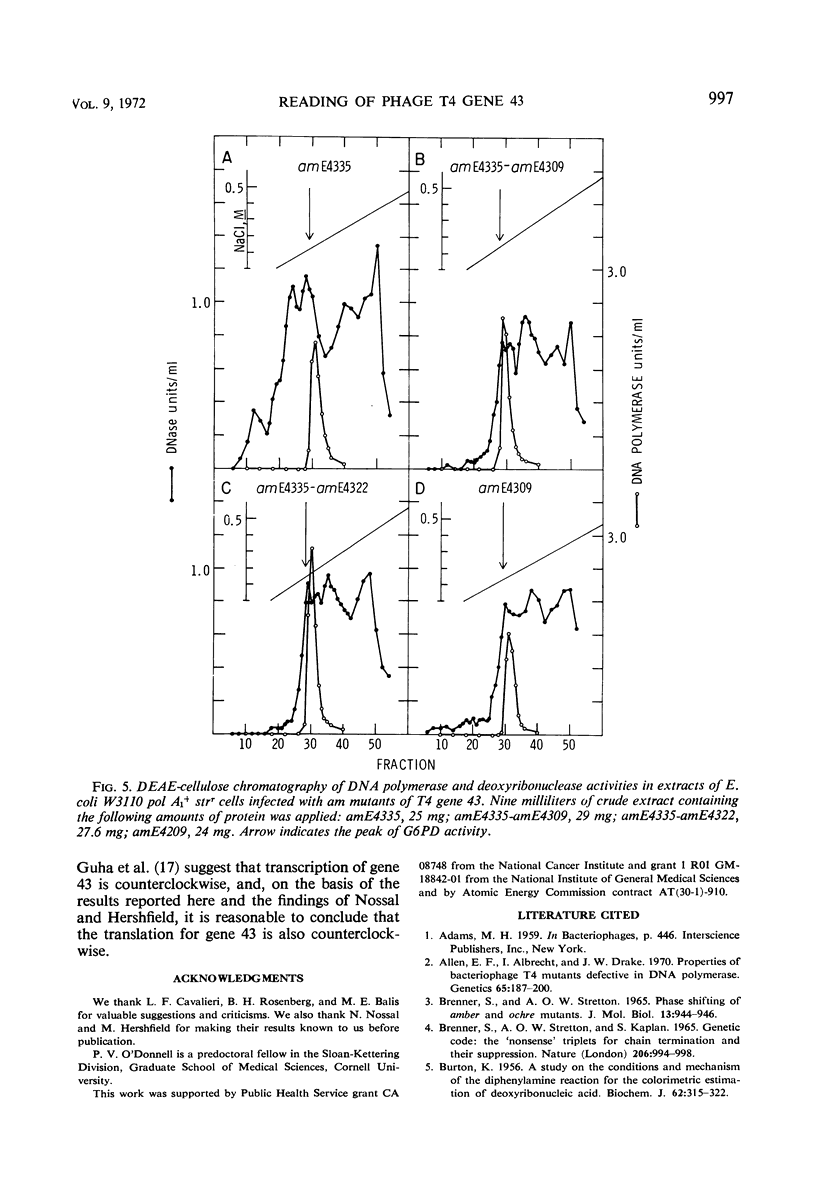
Amber (am) mutants of the two closely linked sites, B22 and C125, in bacteriophage T4 gene 43 [deoxyribonucleic acid (DNA) polymerase] synthesize in the nonpermissive (su−) Escherichia coli host gene 43 products which are devoid of DNA polymerase activity, but which retain a 3′-exonuclease activity. Diethylaminoethyl-cellulose chromatographic analysis of DNA polymerase and deoxyribonuclease activities from extracts of su− cells infected with single- and double-am mutants of T4 gene 43 showed that the exonuclease activity which is observed with amB22 is not seen with double mutants carrying, in addition to amB22, am mutations which map to the clockwise side of the B22 site on the circular genetic map of T4. Similarly, am mutations which map to the clockwise side of the C125 site abolish the exonuclease activity which is observed with an am mutant (amE4335) of this site. It was concluded that in these double mutants termination signals to the clockwise side of amB22 and amE4335 are encountered before the amB22 and amE4335 signals during translation of the messenger ribonucleic acid from T4 gene 43. Thus, it seems that the T4 DNA polymerase is synthesized in vivo in a direction which corresponds to a counterclockwise reading of gene 43.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen E. F., Albrecht I., Drake J. W. Properties of bacteriophage T4 mutants defective in DNA polymerase. Genetics. 1970 Jun;65(2):187–200. doi: 10.1093/genetics/65.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BURTON K. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochem J. 1956 Feb;62(2):315–323. doi: 10.1042/bj0620315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., Stretton A. O., Kaplan S. Genetic code: the 'nonsense' triplets for chain termination and their suppression. Nature. 1965 Jun 5;206(988):994–998. doi: 10.1038/206994a0. [DOI] [PubMed] [Google Scholar]
- Calendar R. The regulation of phage development. Annu Rev Microbiol. 1970;24:241–296. doi: 10.1146/annurev.mi.24.100170.001325. [DOI] [PubMed] [Google Scholar]
- De Lucia P., Cairns J. Isolation of an E. coli strain with a mutation affecting DNA polymerase. Nature. 1969 Dec 20;224(5225):1164–1166. doi: 10.1038/2241164a0. [DOI] [PubMed] [Google Scholar]
- De Waard A., Paul A. V., Lehman I. R. The structural gene for deoxyribonucleic acid polymerase in bacteriophages T4 and T5. Proc Natl Acad Sci U S A. 1965 Oct;54(4):1241–1248. doi: 10.1073/pnas.54.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doermann A H, Hill M B. Genetic Structure of Bacteriophage T4 as Described by Recombination Studies of Factors Influencing Plaque Morphology. Genetics. 1953 Jan;38(1):79–90. doi: 10.1093/genetics/38.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W. Some irregularities in the nomenclature of bacteriophage T4 DNA polymerase amber mutants. Genetics. 1971 Oct;69(2):273–273. doi: 10.1093/genetics/69.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. S., Wood W. B. Morphogenesis of bacteriophage T4 in extracts of mutant-infected cells. Proc Natl Acad Sci U S A. 1966 Mar;55(3):498–505. doi: 10.1073/pnas.55.3.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garen A., Garen S., Wilhelm R. C. Suppressor genes for nonsense mutations. I. The Su-1, Su-2 and Su-3 genes of Escherichia coli. J Mol Biol. 1965 Nov;14(1):167–178. doi: 10.1016/s0022-2836(65)80238-8. [DOI] [PubMed] [Google Scholar]
- Goulian M., Lucas Z. J., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XXV. Purification and properties of deoxyribonucleic acid polymerase induced by infection with phage T4. J Biol Chem. 1968 Feb 10;243(3):627–638. [PubMed] [Google Scholar]
- Gross J., Gross M. Genetic analysis of an E. coli strain with a mutation affecting DNA polymerase. Nature. 1969 Dec 20;224(5225):1166–1168. doi: 10.1038/2241166a0. [DOI] [PubMed] [Google Scholar]
- Guha A., Szybalski W., Salser W., Geiduschek E. P., Pulitzer J. F., Bolle A. Controls and polarity of transcription during bacteriophage T4 development. J Mol Biol. 1971 Jul 28;59(2):329–349. doi: 10.1016/0022-2836(71)90054-4. [DOI] [PubMed] [Google Scholar]
- Kelley W. S., Whitfield H. J. Purification of an altered DNA polymerase from an E. coli strain with a pol mutation. Nature. 1971 Mar 5;230(5288):33–36. doi: 10.1038/230033a0. [DOI] [PubMed] [Google Scholar]
- Klein A., Niebch U. Host cell reactivation in strains of E. coli lacking DNA polymerase activity in vitro. Nat New Biol. 1971 Jan 20;229(3):82–84. doi: 10.1038/newbio229082a0. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Monk M., Peacey M., Gross J. D. Repair of damage induced by ultraviolet light in DNA polymerase-defective Escherichia coli cells. J Mol Biol. 1971 Jun 14;58(2):623–630. doi: 10.1016/0022-2836(71)90376-7. [DOI] [PubMed] [Google Scholar]
- Nossal N. G. A T4 bacteriophage mutant which lacks deoxyribonucleic acid polymerase but retains the polymerase-associated nuclease. J Biol Chem. 1969 Jan 10;244(1):218–220. [PubMed] [Google Scholar]
- Nossal N. G., Hershfield M. S. Nuclease activity in a fragment of bacteriophage T4 deoxyribonucleic acid polymerase induced by the amber mutant am B22. J Biol Chem. 1971 Sep 10;246(17):5414–5426. [PubMed] [Google Scholar]
- OLESON A. E., KOERNER J. F. A DEOXYRIBONUCLEASE INDUCED BY INFECTION WITH BACTERIOPHAGE T2. J Biol Chem. 1964 Sep;239:2935–2943. [PubMed] [Google Scholar]
- STEINBERG C. M., EDGAR R. S. A critical test of a current theory of genetic recombination in bacteriophage. Genetics. 1962 Feb;47:187–208. doi: 10.1093/genetics/47.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STRETTON A. O., BRENNER S. MOLECULAR CONSEQUENCES OF THE AMBER MUTATION AND ITS SUPPRESSION. J Mol Biol. 1965 Jun;12:456–465. doi: 10.1016/s0022-2836(65)80268-6. [DOI] [PubMed] [Google Scholar]
- Smithsm, Ymonds N., White P. The Kornberg polymerase and the repair of irradiated T4 bacteriophage. J Mol Biol. 1970 Dec 14;54(2):391–393. doi: 10.1016/0022-2836(70)90438-9. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Conformational changes of single-stranded DNA. J Mol Biol. 1969 Apr;41(2):189–197. doi: 10.1016/0022-2836(69)90384-2. [DOI] [PubMed] [Google Scholar]
- Terzaghi E., Okada Y., Streisinger G., Emrich J., Inouye M., Tsugita A. Change of a sequence of amino acids in phage T4 lysozyme by acridine-induced mutations. Proc Natl Acad Sci U S A. 1966 Aug;56(2):500–507. doi: 10.1073/pnas.56.2.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H. R., Barnes J. E. Deoxyribonucleic acid synthesis in Escherichia coli infected with some deoxyribonucleic acid polymerase-less mutants of bacteriophage T4. Virology. 1966 Jan;28(1):100–107. doi: 10.1016/0042-6822(66)90310-2. [DOI] [PubMed] [Google Scholar]


