SUMMARY
Transcriptional pausing by multi-subunit RNA Polymerases (RNAPs) is a key mechanism for regulating gene expression in both prokaryotes and eukaryotes, and is a prerequisite for transcription termination. Pausing and termination states are thought to arise through a common, elemental pause state that is inhibitory for nucleotide addition. We report three crystal structures of Thermus RNAP elemental paused elongation complexes (ePECs). The structures reveal the same relaxed, open-clamp RNAP conformation in the ePEC that may arise by failure to reestablish DNA contacts during translocation. A kinked bridge-helix sterically blocks the RNAP active site, explaining how this conformation inhibits RNAP catalytic activity. Our results provide a framework for understanding how RNA hairpin formation stabilizes the paused state and how the ePEC intermediate facilitates termination.
INTRODUCTION
Transcription of DNA into RNA, a fundamental process in every living cell, is the first step in gene expression and a major point of regulation. All stages of the transcription cycle, from initiation to elongation to termination, are subject to regulation. During elongation, RNA extension by the DNA-dependent RNA polymerase (RNAP) is not a smooth, continuous process; instead, RNAP is prone to sequence-dependent pausing, where RNAP molecules delay extension of the RNA transcript. Transcriptional pausing is not simply the cause of inefficient elongation, but plays key regulatory roles in prokaryotes and eukaryotes (Core and Lis, 2008; Landick, 2006). In bacteria, pausing i) allows the coordination of transcription with translation (Landick et al., 1985; Richardson, 1991), ii) facilitates the proper folding of nascent RNA transcripts (Pan et al., 1999), iii) provides opportunities for regulatory factors to bind the elongation complex (Artsimovitch and Landick, 2002; Roberts et al., 1998), and iv) is thought to play an obligatory role in transcriptional termination (Farnham and Platt, 1981; Lau et al., 1983).
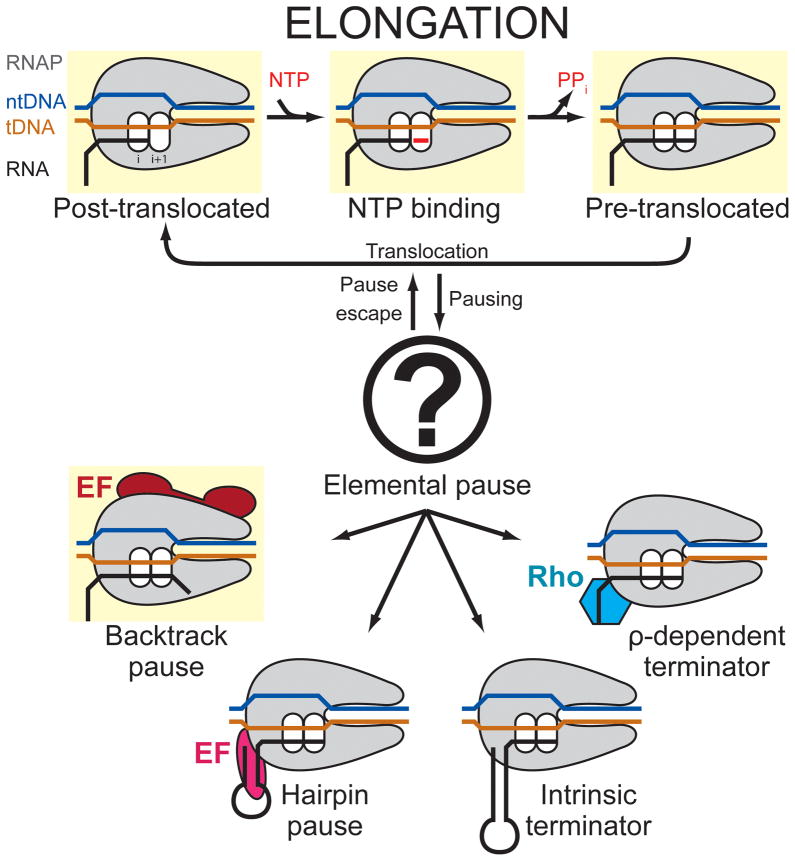
Pause mechanisms have been studied in greatest detail for bacterial RNAP. The current view of transcriptional pausing is that a common, unproductive elemental conformational intermediate of the RNAP active site underlies all transcriptional pauses (Herbert et al., 2006; Kireeva and Kashlev, 2009; Landick, 2006, 2009; Neuman et al., 2003). The elemental pause state is off-pathway from the normal elongation cycle; the efficiency of RNAP entering the elemental pause state at pause-prone sites along DNA is less than 100% and is encoded in the underlying nucleic acid sequence of the RNA/DNA hybrid and the duplex DNA just downstream, nucleic acid elements that are enclosed within, and in close contact with, the RNAP (Figure 1; Vassylyev et al., 2007a). Genetic, biochemical, and single-molecule studies have established that the pause signal is multipartite.
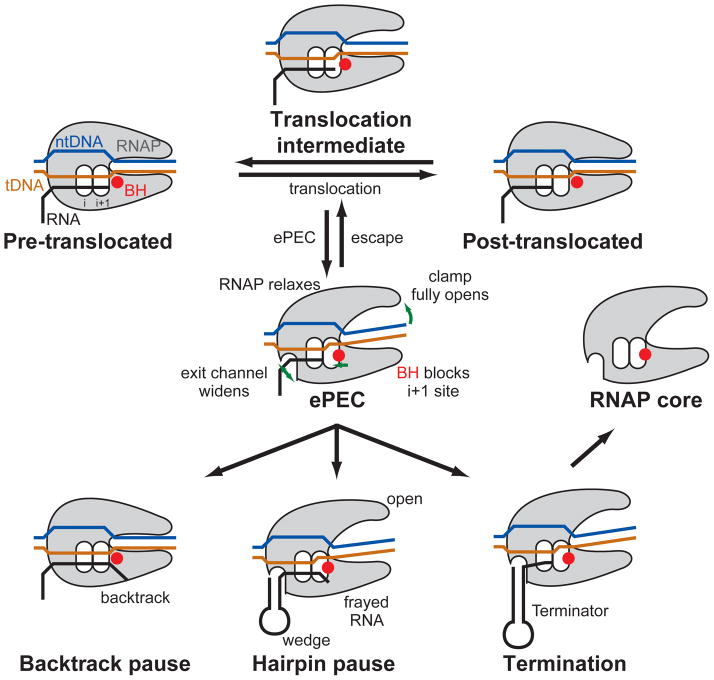
Figure 1. Schematic of the transcription elongation cycle.
States for which X-ray crystal structures are available are highlighted in yellow. Pausing occurs as an off-pathway event with the formation of an initial elemental paused state, which further isomerizes into transcriptional pausing (backtracking or RNA hairpin) or termination (instrinsic or ρ-dependent) states. The elemental pause state crystallized in the present study is symbolized by the question mark.
Once the elemental pause state is entered, the pause duration can be increased by subsequent events, such as backtracking or folding of a hairpin in the nascent RNA (Figure 1; Artsimovitch and Landick, 2000). Crystal structures of backtracked states of eukaryotic RNAP II provide a framework for a mechanistic understanding of backtrack pauses (Cheung and Cramer, 2011; Wang et al., 2009). On the other hand, the elemental pause state, as well as its stabilization by the nascent RNA hairpin, is poorly understood, due in part to the absence of structural information. Here, we report crystal structures of bacterial RNAP paused elongation complexes (PECs). The structures define the RNAP conformation corresponding to the elemental pause state (ePECs), explain how this conformation inhibits the RNAP active site, and provide a framework for understanding how RNA hairpin formation extends the pause.
RESULTS
Thermus RNAPs form PECs on nucleic acid scaffolds
Crystallographic studies of RNAP elongation states have been greatly facilitated by the use of nucleic acid scaffolds, which allow the formation of homogeneous complexes from short, pre-annealed synthetic DNA and RNA oligonucleotides (Daube and von Hippel, 1992; Komissarova et al., 2003; Sidorenkov et al., 1998). The biochemical and kinetic behavior of Escherichia coli (Eco) RNAP PECs assembled from nucleic acid scaffolds derived from the Eco his pause (Figure 2A) recapitulate the features of normal PECs, including the enhancement of pausing by NusA (Kyzer et al., 2007). To further facilitate structural studies, we tested whether scaffolds could similarly recapitulate pausing with Thermus thermophilus (Tth) or Thermus aquaticus (Taq) RNAPs, the source for all high-resolution bacterial RNAP structures to date (Murakami et al., 2002; Vassylyev et al., 2002; Vassylyev et al., 2007a; Zhang et al., 1999).
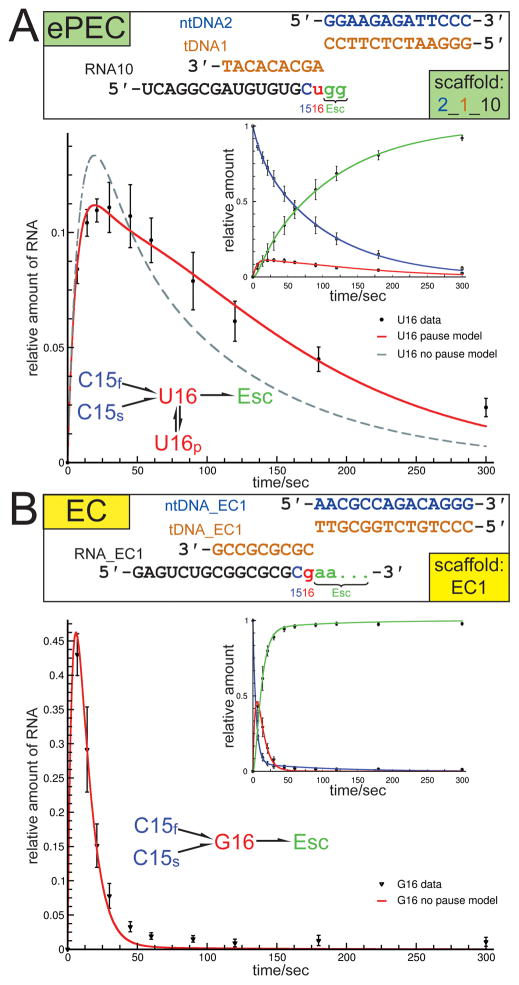
Figure 2. Tth RNAP forms ePECs on a minimal nucleic acid scaffold (see also Figure S1 and Tables S1–S3).
(A) (top) Schematic showing the scaffold variants used to assemble the elemental pause elongation complexes (ePECs) for pause kinetics and crystallization trials. The non-template DNA (ntDNA) is shown in blue, the template DNA (tDNA) in orange. Positions just before the target pause site (C15, blue), the target pause site (U16, red), and after pause escape (Esc, green) were compared. (bottom) Results from transcription kinetic assays: The inset shows the data points and model curves (using the branched kinetic scheme shown) for the C15, U16, and escaped transcripts (color-coded as in the scaffold schematic). The blow-up shows the U16 pause target site data. The red curve corresponds to the branched kinetic scheme containing the pause state, the dashed line is the best fit model that does not include the pause state. Typical for halted complexes, two rates of nucleotide addition were required to fit the C15 complexes (C15f and C15s). C15 complexes on the ePEC scaffold also exhibited pronounced pausing. Inclusion of an off-line pause state (U16p) was required to fit U16 on the PEC scaffold. Error bars depict standard deviations of 6 independent experiments.
(B) (top) Schematic showing the scaffold variants used to assemble the canonical elongation complexes (ECs) for kinetic analysis (color-coded as B); the same sequence was used in crystal structures of bacterial RNAP ECs (Vassylyev et al., 2007a; Vassylyev et al., 2007b). (bottom) Results from transcription kinetic assays: The inset shows the data points and model curves (using the unbranched kinetic scheme shown) for the C15, G16, and escaped transcripts (color-coded as in the scaffold schematic). The blow-up shows the G16 data (corresponds to the U16 target pause site of the ePEC scaffolds). The red line is the best fit model that does not include the pause state. A simple one-state model with a 6-fold faster elongation rate than U16p (0.121 s−1 vs. 0.017 s−1) accounted for all but a neglible fraction of G16 complexes on the EC scaffold. Error bars depict standard deviations of 6 independent experiments.
Transcription kinetics of Tth RNAP elongating through the expected target pause site of minimal Eco his pause nucleic acid scaffolds (Figure 2A, Table S1) were compared with a non-pausing scaffold that was used to generate the structure of a canonical Tth RNAP elongation complex (EC; Figure 2B; Vassylyev et al., 2007a; Vassylyev et al., 2007b). In these assays, Tth RNAP transcription complexes were generated using DNA scaffolds (Figure 2; orange and blue) annealed to 14-nt RNAs (black) with 3′-ends two nucleotides upstream of the target site (target site at position 16 of the RNA). The complexes were provided with [α-32P]-CTP, generating 3′-labeled C15 RNAs (Figure 2). Addition of the next two NTPs allowed formation of complexes at the target 16mer position (U16 for PECs, G16 for ECs), extension of the 16mer RNAs, and, for the ePEC scaffold, isomerization into the ePEC state (Figures 2, S1A). Although the occurrence of pausing also at the C15 position complicated kinetic analysis on the ePEC scaffold (see Supplemental Experimental Procedures), kinetic simulations required inclusion of an off-line pause state to account for the observed behavior of U16 complexes on the ePEC scaffold but not the behavior of G16 complexes on the EC scaffold (Figure 2).
When a hairpin was added to the ePEC scaffold (Table S1), Tth RNAP exhibited an increase in dwell time of the paused species, similar to the behavior of Eco RNAP (data not shown; Kyzer et al., 2007). Tth and Taq RNAP PECs also responded as expected to Tth and Taq NusA, respectively (Figure S1B; data not shown for Taq RNAP), an elongation factor universally conserved among bacteria and archaea (Ingham et al., 1999) that increases the duration of the his pause by Eco RNAP, and that requires interaction with the pause hairpin for full effect (Artsimovitch and Landick, 2000; Toulokhonov et al., 2001). These pause behaviors by Tth RNAP could also be demonstrated in standard transcription elongation assays after initiation at a promoter on linear duplex DNA templates containing the relevant ePEC, PEC-hairpin, or EC sequences within the transcription unit (Figure S1C–E). We conclude that Thermus RNAPs respond to the Eco his pause signal, forming the elemental pause state as well as the NusA-sensitive RNA hairpin-stabilized pause state, when assembled on the ePEC or PEC-hairpin scaffolds, respectively, validating the use of Tth or Taq RNAPs to investigate the ePEC structure.
Crystallization and structure determination of Thermus ePECs
Based on these results, complexes with Tth and Taq core RNAPs were assembled on minimal ePEC and PEC-hairpin scaffolds for crystallization trials (Table S1). Three new crystal forms were obtained. With Taq RNAP PECs, a crystal form yielding diffraction data that extended to ~7.8 Å-resolution was obtained (Table S2). Analysis of the diffraction data indicated the presence of partial hemihedral twinning (Table S2). With Tth RNAP PECs assembled on three different scaffold designs, two novel crystal forms yielding diffraction data extending to 3.6 and 4.5 Å-resolution were obtained (Table S3). We were unable to identify crystallization conditions compatible with the presence of the RNA pause hairpin in the nascent transcript. Although complexes assembled with PEC-hairpin scaffolds yielded crystals, subsequent analyses established that trace amounts of RNase present in the RNAP samples degraded the upstream RNA hairpin, allowing crystal growth. Crystallization trials with RNase-free RNAP samples assembled on PEC-hairpin and ePEC scaffolds confirmed that only ePEC scaffolds supported growth of the identified crystal forms (see Supplemental Experimental Procedures).
For each of the three crystal forms, clear molecular replacement solutions were obtained using a search model based on the Taq RNAP core (Taq crystal form) or Tth EC structure with the nucleic acids removed (Tth crystal forms; Figure 3A; PDB IDs 1HQM and 2O5I respectively; Vassylyev et al., 2007a; Zhang et al., 1999). In all three cases, unbiased Fourier difference analyses i) confirmed the presence of the RNA/DNA hybrid (Figures 3B, S2A) and the downstream duplex DNA (albeit in this case, poorly ordered due to high mobility; Figure S2B), indicating bound scaffold; and ii) indicated substantial conformational changes in the RNAP (Figures 3C, S2C). After rigid body refinement (Taq and Tth crystal forms), CNS DEN refinement (Schröder et al., 2010) and model building (Tth crystal forms), the three ePEC-scaffold-bound RNAPs all converged to the same conformation that differed significantly from the conformation of the canonical EC structures (Vassylyev et al., 2007a; Vassylyev et al., 2007b). Because of the low resolution and twinning problems of the Taq crystal form, we focused most of our structural analysis on the Tth ePEC crystal forms. Since the structures from the two Tth crystal forms were nearly identical, cross crystal averaging, as implemented in Phenix (Adams et al., 2010), was used to improve electron density maps for model building.
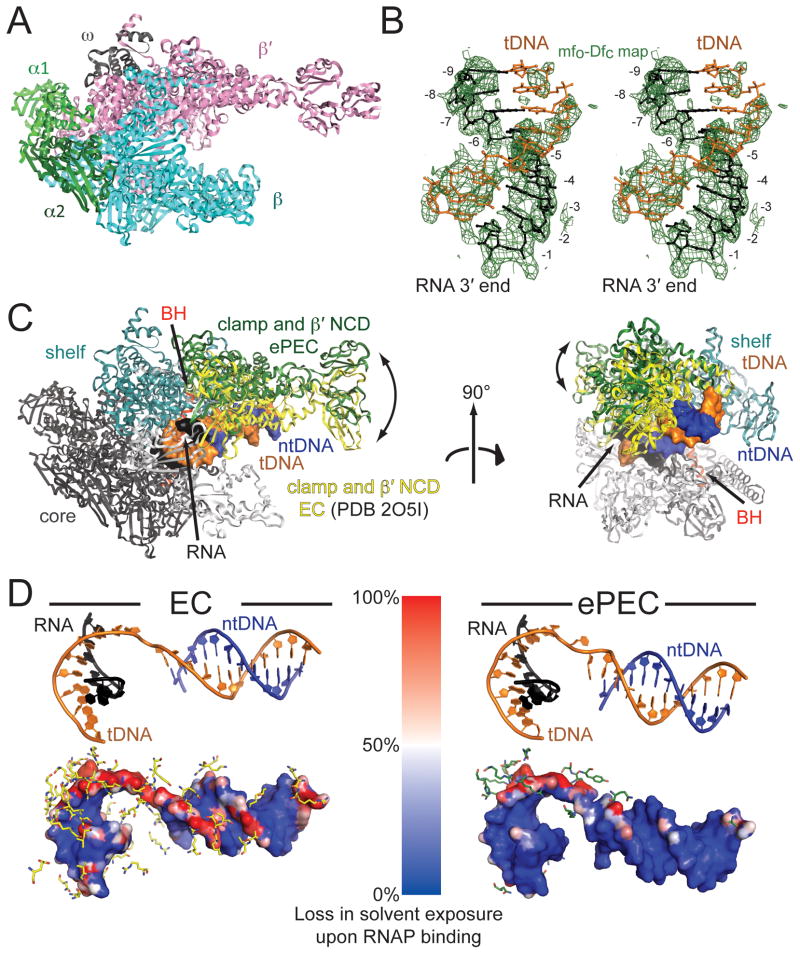
Figure 3. Overview of ePEC structure. (see also Figure S2).
(A) An overview of the bacterial RNAP structure is shown as an α-carbon backbone ribbon, with color-coded subunits.
(B) Stereo image showing the unbiased difference Fourier map (contoured at 1.5σ) for the RNA/DNA hybrid in the Tth ePEC structure.
(C) The ePEC and EC (PDB ID 2O5I; (Vassylyev et al., 2007a) structures were aligned via their ‘core’ modules (black). Differences in the orientation of the clamp domain are shown (ePEC clamp, green; EC clamp, yellow). Two orthogonal views are shown. In the ePEC the clamp opens and loosens its grip on the nucleic acids (the RNA/DNA hybrid and downstream DNA duplex are shown as a molecular surface, color-coded as in the schematic of Fig. 1B).
(D) Differential nucleic acid solvent accessibility (with vs. without the RNAP) for the EC (left) and ePEC (right). At the top, the same view of the nucleic acid scaffold (color-coded as in Figure 1) based on a structural superposition of the EC and ePEC core modules is shown. At the bottom, the molecular surface of the scaffold is colored by relative loss of solvent accessibility caused by the interactions with RNAP (blue no loss, red 100% loss). In an ePEC (right), the tDNA in particular is more solvent exposed, indicated by a reduced loss of solvent accessibility upon binding to RNAP (blue).
ePEC structures have an open-clamp conformation
The overall shape of RNAP resembles a crab claw, with one pincer formed primarily by the β subunit, and the other primarily by β′ (Figure 3A). Between the two pincers, the active-site channel accommodates the nucleic acids in the EC (Korzheva et al., 2000; Vassylyev et al., 2007a; Vassylyev et al., 2007b). The clamp (Figure 3C), a mobile structural module that makes up much of the β′ pincer, can undergo a swinging motion that opens the channel, or closes around the DNA and RNA/DNA hybrid to stabilize the nucleic acid-bound state, leading to the high processivity of the enzyme (Gnatt et al., 2001). All three RNAP structures exhibited an open-clamp conformation that was distinct from the closed-clamp conformation of the canonical ECs (Vassylyev et al., 2007a; Vassylyev et al., 2007b) but similar to the Taq core RNAP (Zhang et al., 1999) and to open-clamp structures described for Tth ECs bound to Gfh1 (Tagami et al., 2010). The root-mean-square differences (rmsd) for equivalent protein backbone atoms of the clamp module for various bacterial RNAP structures aligned with the ePEC structures based on the core module (Figure 3C) highlight the similarities with Taq core RNAP and the Gfh1-bound ECs on the one hand, and the differences with the canonical ECs on the other (Figures 3C, S2D, Table S4). While the rmsd differences for equivalent backbone atoms of the core module are ~1 Å for all of the structures, the differences for the clamp range from 3 – 4 Å for the core RNAP and Gfh1-ECs (reflecting small differences in the degrees of clamp opening), but 12 – 15 Å for the canonical ECs. In all cases, the fold of the clamp module itself is not altered (rmsd < 1.7 Å; Table S4). The open clamp conformation is unlikely due to crystal packing artifacts because most crystal contacts to the β′ pincer (comprised of clamp and β′ non-conserved-domain) are made via the non-conserved domain, which is flexible and has in fact a different orientation in the two Tth crystal forms. More importantly, the packing interactions around the clamp are different for each crystal form (Figure S2E).
The similarity of the ePEC structure with the structure of Taq core RNAP (Zhang et al., 1999) indicates that this open-clamp conformation is accessible to the free core RNAP, and that it may, in fact, represent the most stable conformation of the free core RNAP (see Discussion). The similar open-clamp structure in Gfh1-bound ECs is likely caused by Ghf1-binding (Tagami et al., 2010), although neither the contribution of scaffold sequence nor the presence of an RNA hairpin in the crystallized EC were explicitly tested (see Supplemental Information). As Tagami et al. (2010) point out, Gfh1 binding to the EC could not occur in the canonical closed-clamp EC conformation and requires the widened secondary channel of the open-clamp conformation, suggesting that Gfh1 binding likely stabilized the open-clamp conformation observed in the crystal. Moreover, our finding that Tth RNAP purified by standard procedures contains trace amounts of contaminating RNase that can degrade the RNA hairpin during crystallization trials suggests an explanation for the lack of hairpin electron density in the Tagami et al. (2010) structures since the presence of the hairpin was not confirmed biochemically.
In the canonical ECs, the clamp is closed and exerts a tight grip on the downstream DNA duplex (Figure 3C). In contrast, in the ePEC, because of the open clamp, most of these contacts are broken (Figures 3C, 3D). The loss of nucleic acid contacts can be visualized by increased solvent accessibility of the nucleic acids in the ePEC compared to a canonical EC (Figure 3D), and is consistent with the weak electron density for the downstream duplex DNA (Figure S2B), indicating its increased mobility. The orientation of the downstream duplex DNA relative to the DNA/RNA hybrid in the ePEC is also altered compared to the EC (Figure 3D).
A kinked bridge-helix sterically blocks the RNAP active site
During the elongation cycle, translocation and nucleotide incorporation cause the RNA 3′-end to alternatively occupy the i+1 (pre-translocated) and i (post-translocated) sites (register +1 and −1 or A- and P-site respectively; Figure 1). Unbiased difference Fourier maps revealed a 9-bp RNA/DNA hybrid in ePEC structures with scaffolds containing 9-bp hybrids (register −1 to −9; Figures 3B, S2A), defining the register with respect to the RNAP. Overall the RNA/DNA hybrid in the ePEC most closely resembles the hybrid in the α-amanitin-stalled yeast RNAP II EC (Brueckner and Cramer, 2008). The RNA 3′-end occupies the i-site, corresponding to the post-translocated state, but access of the template DNA (tDNA) to the i+1-site is sterically blocked by a kink in the bridge helix (BH; Figures 4A, 4B), a long, highly conserved α-helix that traverses from the β-pincer to the β′-pincer across the active site channel near the active site itself (Lane and Darst, 2010). In canonical EC structures, the BH is observed in a relatively straight conformation that leaves space for the tDNA +1 base, and for the incoming NTP substrate in the A-site (Figure 4C; Vassylyev et al., 2007b). Cycling of the BH between kinked and straight conformations has been proposed to be an integral part of the normal RNAP nucleotide addition cycle (Brueckner and Cramer, 2008; Gnatt et al., 2001; Temiakov et al., 2005; Tuske et al., 2005; Wang et al., 2006).
Figure 4. Comparison of RNAP active site region for ePEC and canonical EC structures. (see also Figure S3).
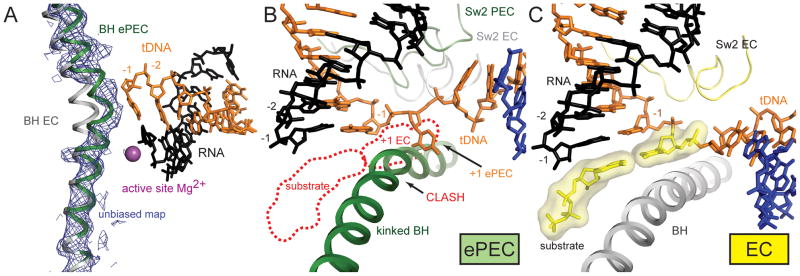
(A) An omit map, (blue mesh, contoured at 1.3σ) confirms the kinked BH in the ePEC (green) compared to an EC (grey).
(B) View of the ePEC kinked BH (green), along with the tDNA (orange) and RNA transcript (black) near the active site. A backbone trace of Sw2 is also shown (green). Superimposed are the base-paired +1 tDNA and nucleotide substrate from the EC (outlined as red dashed lines; (Vassylyev et al., 2007b), as well as the EC position of Sw2 (light grey). The kinked ePEC BH sterically clashes with the superimposed +1 tDNA and less severely with the nucleotide substrate. As a consequence, the +1 tDNA base sits in an alternate position at the side of the BH.
(C) View of the relatively straight BH of the EC, which leaves room for the +1 tDNA and incoming nucleotide substrate (yellow). Sw2 is also shown (yellow).
In the ePEC, the +1 base of the tDNA thus cannot be accommodated in the active site but is instead seen in an intermediate position to the side of the kinked BH (Figures 4B, S3). The incoming NTP substrate would also not interact productively with the A-site since i) it cannot base pair to the absent +1 base of the tDNA, and ii) modeling suggests the incoming NTP substrate would also clash (much less severely) with the kinked BH (Figure 4B). Supporting evidence for this was obtained by soaking the non-hydrolyzable NTP analogue GMPCPP (complementary to the +1 tDNA base) into the ePEC crystals (Table S3). The conformation of RNAP was identical to the other ePEC structures, with a kinked BH that blocked access for the +1 tDNA base to the active site. No density for the RNA 3′-end or the nucleotide analogue was apparent in the A-site. However, positive difference Fourier density was visible at the E-site (Figure S3B), identified previously as a binding site for mismatched incoming NTP substrates (Westover et al., 2004).
The trigger-loop (TL) is a structural element of the RNAP active site that undergoes a disorder/order transition in the presence of the matched nucleotide substrate in the A-site (Lane and Darst, 2010; Vassylyev et al., 2007b; Wang et al., 2006). The TL is disordered in all the ePEC structures, with or without the GMPCPP substrate analog, as it is in the Taq core RNAP and in the Gfh1-bound ECs (Tagami et al., 2010; Zhang et al., 1999). However, the BH, the TL, and the two αhelices flanking the TL are thought to function as a concerted structural unit, so conformational changes in one structural element likely influence the conformation of the others. Such an interaction could explain the strong effect of TL alterations on transcriptional pausing (Toulokhonov et al., 2007). We conclude that the kinked BH observed in all three ePEC structures sterically blocks both the +1 tDNA base and the incoming NTP substrate from accessing the A-site, providing a structural basis for the catalytic inactivity of the ePEC.
The RNAP Switch regions in the ePEC lose contact with the template DNA
In both pre- and post-translocated states of ECs, RNAP contacts to the RNA/DNA hybrid and the downstream duplex DNA are established almost exclusively through the sugar-phosphate backbone (Figure 5A; Vassylyev et al., 2007a; Vassylyev et al., 2007b). The Switch regions 1–3 (Sw1, Sw2, Sw3), and residues from the clamp, establish almost all of these contacts (Figure 5A). The Sw regions serve as hinges between mobile RNAP modules and couple nucleic acid interactions with clamp closure (Gnatt et al., 2001; Lane and Darst, 2010). The RNAP conformational changes in the ePEC move the Sw regions away from the nucleic acids, resulting in widening of the hybrid-binding site (Figure 5). In particular, contacts mainly to the tDNA are broken in the ePEC conformation and almost all the residues involved are part of the clamp, Sw1, Sw2, or Sw3 regions (Figure 5). In contrast, contacts to the RNA transcript, which occur primarily from the RNAP core module, are maintained, and some additional contacts are even formed (Figure 5). Consistent with these observations, the electron density for the template DNA is consistently weaker in all structures (Figure S4A), reflecting the loss in contacts to the Sw2, and Sw3 modules and confirming that it is more loosely bound.
Figure 5. Comparison of key nucleic acid contacts for ePEC and canonical EC structures. (also see Figure S4).
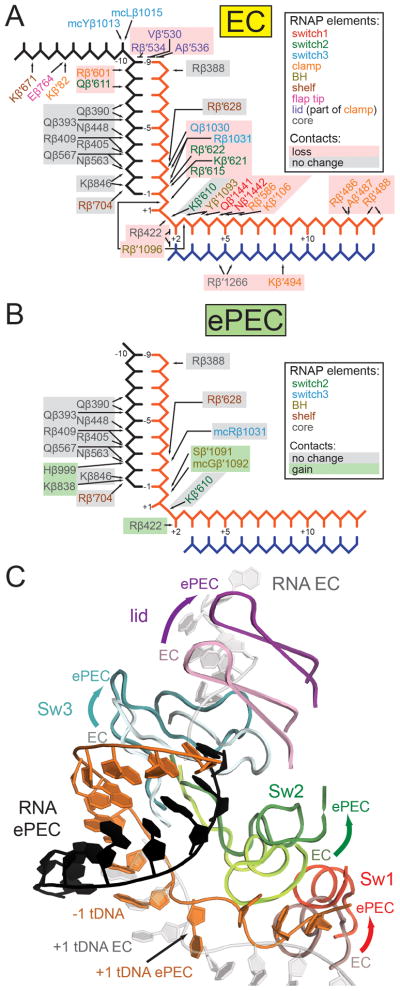
(A) Schematic representation of RNAP contacts to the nucleic acids in an EC (PDB ID 2O5I; (Vassylyev et al., 2007a). Text color corresponds to the RNAP protein element. Colored boxes indicate if a contact is maintained (grey), or lost/suboptimal (red) upon entering the elemental paused state.
(B) Same as (A) but for the ePEC. Newly formed contacts in the ePEC are highlighted by green boxes.
(C) Changes in contacts to the nucleic acids upon entering the elemental pause are exemplified by conformational changes in Sw1–3 and the lid (directions of movement from the EC to the ePEC structure are indicated by arrows).
The open clamp conformation of the ePEC likely allows formation of the RNA pause hairpin
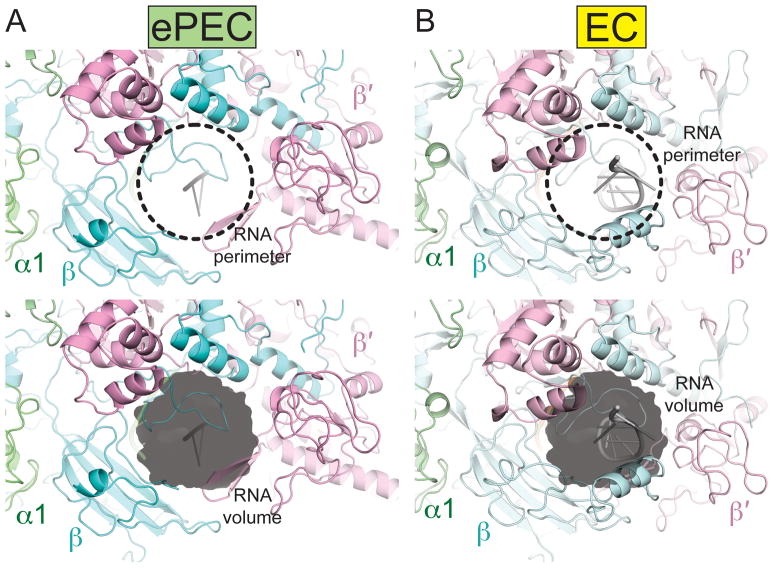
The opening of the clamp also significantly widens the RNA exit channel. Based on the location of Sw3 and the locations of the upstream RNA in the EC and ePEC structures, we modeled an RNA pause hairpin in the RNA exit channel. The hairpin is easily accommodated in the ePEC structure due to the opening of the clamp and widening of the RNA exit channel, but appears to clash with protein elements in the EC, with its closed-clamp and narrow RNA exit channel (Figure 6). This leads to the hypothesis that the RNA hairpin could serve as a steric block to stabilize the open-clamp conformation of the ePEC, explaining how formation of the RNA hairpin extends the duration of the paused state, but not the efficiency of the pause (Chan et al., 1997).
Figure 6. Comparison of the RNA exit channel in the ePEC and canonical EC structures.
In the ePEC (left), the RNA exit channel is wider compared to the EC (right). An RNA pause hairpin was modeled in the RNA exit channel. While it can be accommodated in the ePEC without major steric clashes, the exit channel seems to be too narrow in the EC.
DISCUSSION
Our results lead to many hypotheses regarding the entry of the elongating RNAP into the paused state. Below we explore these insights into the pausing mechanism.
Model for entry into the paused state
The observation that the ePEC is in an open-clamp state suggests the appealing model that pausing arises initially from a strained, closed-clamp EC by loss of nucleic-acid contacts during translocation. We note that bacterial core RNAP, in the absence of any bound factors or nucleic acids, crystallizes in an open-clamp state similar to the ePEC structures (Figure S2D, Table S4), suggesting that this state represents the low-energy, preferred conformation for the free enzyme (Zhang et al., 1999). In contrast, the EC of RNAP establishes numerous backbone contacts to the nucleic acids, stabilizing the closed-clamp conformation observed in crystal structures (Vassylyev et al., 2007a; Vassylyev et al., 2007b). FRET studies of bacterial RNAP in solution confirm these structural observations; an open clamp state is predominant for the core enzyme, whereas the EC is in a closed-clamp state (Chakraborty et al., 2012). We thus propose that the closed-clamp of the EC is analogous to a loaded spring, with a high-energy RNAP conformation stabilized by the backbone contacts to the nucleic acids. During translocation, these protein/nucleic acid contacts must be transiently broken and reestablished at the next position downstream as the enzyme moves along the DNA (Figure 7), but this process is unlikely to involve full clamp opening. The translocation intermediate between pre- and post-translocation provides a window of opportunity for RNAP to relax and enter the open-clamp elemental paused state if the nucleic acid contacts are not properly reestablished (Figure 7).
Figure 7. Schematic representation of our model for transcriptional pausing.
During translocation RNAP has an opportunity to relax to the open-clamp conformation if it fails to reform polar contacts to the nucleic acids, thus entering the elemental paused state. Key features of the ePEC are an open clamp, the kinked BH (red circle) blocking tDNA access to the active site, and a widened hybrid binding site and RNA exit channel. From the ePEC: i) the RNAP can backtrack (backtrack pause), ii) an RNA pause hairpin can form, wedging open the clamp in the pre-translocated hairpin pause, or iii) a terminator hairpin can form, resulting in dissociation of the RNAP from the nucleic acids (Termination).
Within bacterial RNAP and EC structures, there is a strict correlation between the open- or closed-clamp conformation, and whether or not the BH is kinked or straight, respectively. We thus propose that clamp opening (as in the core RNAP, Gfh1-EC, or ePEC structures, Table S4) is coupled with BH kinking (also see Tagami et al., 2010). Thus, according to our model, once RNAP has failed to reestablish the proper protein/nucleic acid contacts with the translocated nucleic acids and has relaxed into the elemental pause state, the open-clamp conformation couples with BH kinking, resulting in inactivation of the RNAP active site (Figure 4). The lifetime of this inactivated state, combined with the widened RNA exit channel (Figure 6) provides time and space to allow RNA hairpin formation, resulting in stabilization of the open-clamp/kinked-BH conformation and an increased lifetime of the paused state (Figure 7).
Implications for regulation of transcript elongation
Extrinsic regulators that modulate transcription elongation do so principally by suppressing or enhancing transcriptional pausing. Bacterial NusG and its archaeal and eukaryotic ortholog Spt5, the only universally conserved elongation regulators, are thought to suppress transcriptional pausing by binding to the RNAP clamp domain and preventing clamp opening by also contacting the opposite pincer across the RNAP active-site channel (Klein et al., 2011; Martinez-Rucobo et al., 2011; Sevostyanova et al., 2011). Our model for ePEC formation immediately suggests that these key elongation regulators work by stabilizing RNAP in the closed-clamp state, thereby decreasing the likelihood that RNAP would lose DNA contacts during translocation and preventing the fully open clamp state of the ePEC. Conversely, elongation regulators like bacterial NusA or NELF in metazoans (Chiba et al., 2010), which increase transcriptional pausing (Figure S1B), may increase the likelihood that DNA contacts are lost during translocation by increasing the likelihood of clamp opening.
Implications for the nature of the pause-inducing signal
Nucleic acid sequence elements, such as the downstream duplex DNA and RNA/DNA hybrid, are known to influence entry into the elemental pause state (Chan and Landick, 1993; Levin and Chamberlin, 1987). These effects can be understood from our model for entry into the ePEC state since weaker contacts to RNAP in the translocation intermediate or in the posttranslocated EC would increase the likelihood of clamp opening, thus increasing pause efficiency (Figure 7). DNA duplex and RNA/DNA hybrid conformations depend on the sequence context (Hunter, 1996; Shaw and Arya, 2008). Subtle differences in helix conformation could influence the likelihood of maintaining or reestablishing contacts between RNAP and the backbone of the DNA during translocation, thus influencing the kinetic partitioning between post-translocated and paused ECs (Figure 7).
A consensus pause-inducing signal, determined from analyses of ‘ubiquitous pausing’ observed in single-molecule traces of elongating bacterial RNAP (Herbert et al., 2006), includes +1G in the ntDNA just downstream of the RNA/DNA hybrid, consistent with the effects of the downstream DNA template sequence on pausing observed in other studies (Kireeva and Kashlev, 2009; Lee et al., 1990). In a bacterial initiation complex structure, it was noted that a guanosine at this position of the ntDNA can interact in a pocket formed by residues of the β subunit (Zhang et al., 2012), and that this could play a role in pausing. We do not observe electron density supporting this hypothesis in our ePEC structures. However, we cannot exclude that this is due to the use of the minimal ePEC scaffolds for crystallization, which lack the ntDNA strand upstream of position +1 (Figure 2A, Table S1).
Implications for the RNA-Hairpin-stabilized PEC
The ePEC is in the post-translocated state (Figure 4), but biochemical data indicate that RNAP in the RNA hairpin-stabilized pause is in the pre-translocated state with ‘fraying’ of the RNA 3′-nucleotide, but may be in equilibrium with the post-translocated state (Toulokhonov et al., 2007). Fraying of the RNA 3′-nucleotide must occur if formation of the RNA pause hairpin state causes the post-translocated ePEC to return to the pre-translocated state and the BH kink of the ePEC (Figure 4A) persists because the BH would sterically occlude base-pairing at the downstream end of the hybrid. Although the hairpin-stabilized PEC has been used to infer properties of the ePEC, it has been unclear if 3′-nt fraying was a cause or a consequence of pausing (Sydow et al., 2009; Toulokhonov et al., 2007). Thus, the hairpin-stabilized pause may arise from a post-translocated ePEC that reverts to a pre-translocated, paused state upon hairpin formation (Figure 7). More generally, paused ECs may equilibrate between pre- and post-translocated registers that remain catalytically inactive as long as the kinked BH persists (Kireeva and Kashlev, 2009).
Implications for termination
The elemental pause state is thought to be an obligate intermediate for both intrinsic and ρ-dependent termination (Figure 1; Landick, 2006). The same conformational determinants of the ePEC structure (open-clamp/kinked-BH/widened RNA exit channel) that provide time and space for the RNA pause hairpin to form would also allow a terminator hairpin to form. Termination arises from an initially paused state associated with U-rich RNA (Gusarov and Nudler, 1999); the rU-dA hybrid may be bound more weakly by RNAP and thus favor ePEC formation.
Changes near the ePEC RNA exit channel may also facilitate termination. The conformation of the RNA/DNA hybrid from base pairs -1 to -7 is virtually identical between the EC and the ePEC, but the base pairs at positions -8 and -9 of the ePEC are distorted from Watson-Crick geometry (Figure S4B). Movement of the β′-lid away from the upstream edge of the RNA/DNA hybrid in the ePEC, which stacks on the upstream end of the hybrid in the EC (Figure 5), likely accompanies the hybrid deformation. In addition, elements of the clamp, shelf, and Sw3 modules, which interact with the upstream RNA in EC structures (Figure 5), all move in the direction of the exiting RNA (Figure 5C). We infer that the conformational changes observed in the ePEC help pull the RNA transcript in the direction of exit, explaining the observed changes in the upstream end of the RNA/DNA hybrid and shift of RNA phosphate positions (Figure S4C). The distortion of the most upstream base pairs of the RNA/DNA hybrid in the ePEC (Figure S4) would facilitate disruption of the hybrid by the invading stem of the terminator hairpin. This, combined with the loss of contacts to the DNA and open-clamp conformation (that would facilitate the disassociation of the enclosed nucleic acids; Figures 3C, 3D, 5), suggests that the ePEC is primed for termination.
Conclusions
In summary, we have demonstrated that Thermus RNAPs assembled onto minimal nucleic acid scaffolds derived from the Eco his pause form the elemental pause and RNA hairpin-stabilized pause states (Figures 2, S1). Three independent crystal structures, using two RNAPs (Taq and Tth) in three unique crystal packing environments (Tables S2, S3, Figure S2E) establish that the ePEC comprises nucleic-acid bound RNAP (Figures 3B, S2A, S2B) in a relaxed, open clamp conformation, similar to the conformation of nucleic-acid free core RNAP observed in crystals (Figure S2D, Table S4) and in solution (Chakraborty et al., 2012). The elemental pause state may be entered when RNAP in an elongation translocation intermediate between pre- and post-translocated states fails to reestablish nucleic acid contacts (Figure 7). Compared to canonical (non-paused) EC structures, the ePEC RNAP conformation is characterized by coupled conformational changes that lead to an open-clamp (Figures 3C, Table S4), a kinked BH (Figure 4A), and a widened RNA exit channel (Figure 6). The open-clamp results in loss of contacts to the DNA template (Figures 3D, 5). The kinked BH sterically blocks the +1 tDNA base and, in turn, disfavors the substrate from entering the A-site (Figures 4, S3A), providing a structural basis for inhibition of the RNAP catalytic activity in the ePEC. The widened RNA exit channel provides space for the formation of the RNA pause hairpin, and suggests a role for the hairpin as a steric wedge that stabilizes the widened RNA exit channel/open-clamp conformation of the RNAP (Figures 6, 7), in agreement with the observation that the pause half-life and hairpin stability are positively correlated (Chan and Landick, 1993). The ePEC structures are consistent with the proposed role of the elemental pause state as an obligate intermediate in the termination process (Landick, 2006). These results provide many insights and hypotheses that lead to testable predictions, which can be addressed in future studies. We have summarized our results in a movie, which is available online (Supplemental Movie S1 available on Cell website).
EXPERIMENTAL PROCEDURES
Full details of Experimental Procedures are presented in the Supplemental Information.
Protein and Nucleic Acid Preparation
Tth core RNAP with C-terminally His10-tagged β′-subunit was purified from a Tth HB8rpoC::10H strain (Sevostyanova et al., 2007). The RNAP was initially purified by polyethyleneimine fractionation, ammonium sulfate precipitation, Ni2+-chelating chromatography, anion exchange chromatography, and gel filtration. Contaminating RNases were removed in subsequent purifications by a reverse-phase chromatography step before the gel filtration step. The purified RNAP was dialyzed into storage buffer (TGED: 10 mM Tris-HCl, pH 8.0, 5% glycerol, 0.1 mM EDTA, 1 mM DTT) + 0.15 M NaCl and 15% glycerol) and flash frozen. Aliquots were thawed, dialyzed into crystallization buffer (10 mM Tris-HCl pH 7.7, 0.15 M NaCl, 1% glycerol; (Tagami et al., 2010) and concentrated by centrifugal filtration to about 30 mg/ml for crystallization. Endogenous Taq RNAP was purified as described (Zhang et al., 1999), dialyzed into TGED + 0.15 M NaCl, and concentrated to about 30 mg/ml for crystallization. Tth and Taq NusA were sub-cloned into pET-based expression vectors and purified using standard methods.
DNA (TriLink BioTechnologies or IDT) and RNA (Dharmacon) oligonucleotides were chemically synthesized and gel purified by the manufacturer. RNA was deprotected following the protocols provided by the manufacturer. Both DNA and RNA were dissolved in RNase free water and stored at −80°C.
Pause assays
Pause assays were performed essentially as described (Kyzer et al., 2007; Landick et al., 1996). Kinetic modeling (Figure 2) was performed using Berkeley-Madonna (http://www.berkeleymadonna.com/).
Crystallization of complexes
Nucleic acid scaffolds for crystallization were reconstituted using equimolar amounts of each component and aliquots were stored at −80°C. Complexes were formed by mixing a 2.5-fold molar excess of scaffold with RNAP (10 mg/ml) in crystallization buffer (10 mM Tris-HCl pH 7.7, 0.15 M NaCl, 1% glycerol) and incubating at 37°C for 15 min. 1% DMSO and 5 mM Taurine were added to the Tth RNAP complexes and crystals were grown at 22°C via vapor diffusion by mixing equal volumes of the sample and a reservoir solution of: 3.5–6% PEG 8000, 0.1 M MES, pH 6.1–6.3, 0.3 M LiCl, 10 mM MgCl2 (Tth P3121 crystal form; derived from (Tagami et al., 2010), 5–7% PEG 3000, 0.1 M MES pH 6.1–6.3, 0.3 M LiCl, 10mM MgCl2 (Tth R3 crystal form), 2.7% PEG 8000, 0.1 M Na-cacodylate, pH 6.5, 26% MPD or 34–40% PEG 200, 0.1 M HEPES, pH 7–7.5 (Taq P41 crystal form). Tth crystals were cryo-protected using the reservoir solution containing 25% PEG 550MME or PEG 400, or a mixture of PEG 400 (12.5%) and MPD (12.5%) and flash-frozen in liquid nitrogen. Taq crystals were flash-frozen in liquid nitrogen directly from the mother liquor.
Data collection, refinement and model-building
X-ray diffraction data were collected at NSLS beamline X29 at Brookhaven National Laboratory and APS beamline 24ID-E at Argonne National Laboratory. The structures were solved by molecular replacement. Rigid body refinement (all crystal forms), CNS DEN refinement (Schröder et al., 2010) and limited model building (Tth crystal forms) yielded the final models (Tables S2, S3).
Supplementary Material
HIGHLIGHTS.
Three crystal structures of the bacterial RNAP elemental paused elongation complex
A conformational change blocks the RNAP active site, explaining catalytic inhibition
RNAP loosens its grip on nucleic acids
Structures provide insights into RNA hairpin pausing and transcriptional termination
Three independent crystal structures of bacterial RNA Polymerase shed light on the structural basis of transcriptional pausing, a key mechanism to regulate gene expression in all kingdoms of life and an obligate intermediate for termination of transcription. A conformational change in the paused state of RNA Polymerase is responsible for blocking the active site and inhibiting the catalytic activity. Contacts to the nucleic acids are loosened and the RNA exit channel is widened providing time and space for an RNA hairpin to form in the nascent transcript. The structures thus provide a framework for understanding termination of transcription.
Acknowledgments
We thank P. Hein for help with transcription assays; S. Leiman for help with Taq RNAP pausing assays; T. Terwilliger for help with Phenix cross crystal averaging; members of the Darst laboratory for helpful discussions; K.R. Rajashankar, S. Banerjee, F. Murphy, and K. Perry at APS NE-CAT beamline 24ID-E, W. Shi at NSLS beamline X29, and A. Heroux at NSLS beamline X25 for support with synchrotron data collection. A.W. was supported by an Anderson Cancer Center Postdoctoral Fellowship (The Rockefeller University) and a Human Frontier Science Program Long-term Postdoctoral Fellowship. This work was based, in part, on research conducted at the APS and the NSLS, supported by the US Department of Energy, Office of Basic Energy Sciences. The NE-CAT beamlines at the APS are supported by award RR-15301 from the NCRR at the NIH. This work was supported by NIH RO1 GM38660 to R.L. and NIH RO1 GM097458 to S.A.D.
Footnotes
ACCESSION NUMBERS
The structure factor files and X-ray crystallographic coordinates have been deposited in the Protein Data Bank with accession IDs 4GZY (Tth ePEC, P3121 crystal form) and 4GZZ (Tth ePEC, R3 crystal form).
Supplemental Data include four tables, four figures, and a movie, which can be found with this article online at http://www.cell.com/supplemental/***.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Wang D, Ebright YW, Korlann Y, Kortkhonjia E, Chowdhury S, Wisneshweraraj S, Irschik H, Jansen R, Nixon BT, et al. Opening and closing of the RNA polymerase clamp. Science. 2012;377:591. doi: 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CL, Landick R. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J Mol Biol. 1993;233:25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- Chan CL, Wang D, Landick R. Multiple interactions stabilize a single paused transcription intermediate in which hairpin to 3′ end spacing distinguishes pause and termination pathways. J Mol Biol. 1997;268:54–68. doi: 10.1006/jmbi.1997.0935. [DOI] [PubMed] [Google Scholar]
- Cheung ACM, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- Chiba K, Yamamoto J, Yamaguchi Y, Handa H. Promoter-proximal pausing and its release: molecular mechanisms and physiological functions. Exp Cell Res. 2010;316:2723–2730. doi: 10.1016/j.yexcr.2010.05.036. [DOI] [PubMed] [Google Scholar]
- Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daube SS, von Hippel PH. Functional transcription elongation complexes from synthetic RNA-DNA bubble duplexes. Science. 1992;258:1320–1324. doi: 10.1126/science.1280856. [DOI] [PubMed] [Google Scholar]
- Farnham PJ, Platt T. Rho-independent termination: dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro. Nucleic Acids Res. 1981;9:563–577. doi: 10.1093/nar/9.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, Block SM. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CA. Sequence-dependent DNA structure. Bioessays. 1996;18:157–162. doi: 10.1002/bies.950180212. [DOI] [PubMed] [Google Scholar]
- Ingham CJ, Dennis J, Furneaux PA. Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis. Mol Microbiol. 1999;31:651–663. doi: 10.1046/j.1365-2958.1999.01205.x. [DOI] [PubMed] [Google Scholar]
- Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc Nat Acad Sci USA. 2009;106:8900–8905. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BJ, Bose D, Baker KJ, Yusoff ZM, Zhang X, Murakami KS. RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Nat Acad Sci USA. 2011;108:546–550. doi: 10.1073/pnas.1013828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komissarova N, Kireeva ML, Becker J, Sidorenkov I, Kashlev M. Engineering of elongation complexes of bacterial and yeast RNA polymerases. Meth Enzymol. 2003;371:233–250. doi: 10.1016/S0076-6879(03)71017-9. [DOI] [PubMed] [Google Scholar]
- Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst SA. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- Kyzer S, Ha KS, Landick R, Palangat M. Direct versus limited-step reconstitution reveals key features of an RNA hairpin-stabilized paused transcription complex. J Biol Chem. 2007;282:19020–19028. doi: 10.1074/jbc.M701483200. [DOI] [PubMed] [Google Scholar]
- Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- Landick R. Transcriptional pausing without backtracking. Proc Nat Acad Sci USA. 2009;106:8797–8798. doi: 10.1073/pnas.0904373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R, Carey J, Yanofsky C. Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc Natl Acad Sci USA. 1985;82:4663–4667. doi: 10.1073/pnas.82.14.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R, Wang D, Chan CL. Quantitative analysis of transcriptional pausing by Escherichia coli RNA polymerase: his leader pause site as paradigm. Meth Enzymol. 1996;274:334–353. doi: 10.1016/s0076-6879(96)74029-6. [DOI] [PubMed] [Google Scholar]
- Lane WJ, Darst SA. Molecular evolution of multi-subunit RNA polymerases: structural analysis. J Mol Biol. 2010;395:686–704. doi: 10.1016/j.jmb.2009.10.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau LF, Roberts JW, Wu R. RNA polymerase pausing and transcript release at the lambda tR1 terminator in vitro. J Biol Chem. 1983;258:9391–9397. [PubMed] [Google Scholar]
- Lee DN, Phung L, Stewart J, Landick R. Transcription pausing by Escherichia coli RNA polymerase is modulated by downstream DNA sequences. J Biol Chem. 1990;265:15145–15153. [PubMed] [Google Scholar]
- Levin JR, Chamberlin MJ. Mapping and characterization of transcriptional pause sites in the early genetic region of bacteriophage T7. J Mol Biol. 1987;196:61–84. doi: 10.1016/0022-2836(87)90511-0. [DOI] [PubMed] [Google Scholar]
- Martinez-Rucobo FW, Sainsbury S, Cheung ACM, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011;30:1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc Natl Acad Sci U S A. 1999;96:9545–9550. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JP. Preventing the synthesis of unused transcripts by Rho factor. Cell. 1991;64:1047–1049. doi: 10.1016/0092-8674(91)90257-y. [DOI] [PubMed] [Google Scholar]
- Roberts JW, Yarnell W, Bartlett E, Guo J, Marr M, Ko DC, Sun H, Roberts CW. Antitermination by bacteriophage lambda Q protein. Cold Spring Harb Symp Quant Biol. 1998;63:319–325. doi: 10.1101/sqb.1998.63.319. [DOI] [PubMed] [Google Scholar]
- Schröder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevostyanova A, Belogurov GA, Mooney RA, Landick R, Artsimovich I. The b subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol Cell. 2011;43:253–262. doi: 10.1016/j.molcel.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevostyanova A, Djordjevic M, Kuznedelov K, Naryshkina T, Gelfand MS, Severinov K, Minakhin L. Temporal regulation of viral transcription during development of Thermus thermophilus bacteriophage phiYS40. J Mol Biol. 2007;366:420–435. doi: 10.1016/j.jmb.2006.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw NN, Arya DP. Recognition of the unique structure of DNA:RNA hybrids. Biochimie. 2008;90:1026–1039. doi: 10.1016/j.biochi.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Sidorenkov I, Komissarova N, Kashlev M. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- Sydow JF, Brueckner F, Cheung AC, Damsma GE, Dengl S, Lehmann E, Vassylyev D, Cramer P. Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Mol Cell. 2009;34:710–721. doi: 10.1016/j.molcel.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Tagami S, Sekine S, Kumarevel T, Hino N, Murayama Y, Kamegamori S, Yamamoto M, Sakamoto K, Yokoyama S. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468:978–982. doi: 10.1038/nature09573. [DOI] [PubMed] [Google Scholar]
- Temiakov D, Zenkin N, Vassylyeva MN, Perederina A, Tahirov TH, Kashkina E, Savkina M, Zorov S, Nikiforov V, Igarashi N, et al. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol Cell. 2005;19:655–666. doi: 10.1016/j.molcel.2005.07.020. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Tuske S, Sarafianos SG, Wang X, Hudson B, Sineva E, Mukhopadhyay J, Birktoft JJ, Leroy O, Ismail S, Clark ADJ, et al. Inhibition of bacterial RNA polymerase by streptolydigin: Stabilization of a straight-bridge-helix active-center conformation. Cell. 2005;122:541–552. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007a;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007b;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, Ebright RH. Structural basis of transcription initiation. Science. 2012 doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.