Abstract
Aquaporins (AQPs) are membrane channels belonging to the major intrinsic proteins family and are known for their ability to facilitate water movement. While in Populus trichocarpa, AQP proteins form a large family encompassing fifty-five genes, most of the experimental work focused on a few genes or subfamilies. The current work was undertaken to develop a comprehensive picture of the whole AQP gene family in Populus species by delineating gene expression domain and distinguishing responsiveness to developmental and environmental cues. Since duplication events amplified the poplar AQP family, we addressed the question of expression redundancy between gene duplicates. On these purposes, we carried a meta-analysis of all publicly available Affymetrix experiments. Our in-silico strategy controlled for previously identified biases in cross-species transcriptomics, a necessary step for any comparative transcriptomics based on multispecies design chips. Three poplar AQPs were not supported by any expression data, even in a large collection of situations (abiotic and biotic constraints, temporal oscillations and mutants). The expression of 11 AQPs was never or poorly regulated whatever the wideness of their expression domain and their expression level. Our work highlighted that PtTIP1;4 was the most responsive gene of the AQP family. A high functional divergence between gene duplicates was detected across species and in response to tested cues, except for the root-expressed PtTIP2;3/PtTIP2;4 pair exhibiting 80% convergent responses. Our meta-analysis assessed key features of aquaporin expression which had remained hidden in single experiments, such as expression wideness, response specificity and genotype and environment interactions. By consolidating expression profiles using independent experimental series, we showed that the large expansion of AQP family in poplar was accompanied with a strong divergence of gene expression, even if some cases of functional redundancy could be suspected.
Introduction
Aquaporins (AQPs) are found in every organism but are especially abundant in plants [1]. In higher plants, AQPs have been classified into five subfamilies: plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), nodulin-26 intrinsic proteins (NIPs), small basic intrinsic proteins (SIPs) and unrecognized X intrinsic proteins (XIPs). These intrinsic channel proteins facilitate and regulate the passive movement of water molecules and other small neutral molecules (e.g. urea, glycerol, ammonium, metalloids) across biological membranes [2], [3]. AQPs are involved in major physiological processes such as root and leaf hydraulic plasticity, stomatal aperture, cell expansion, or acclimation to drought or salinity [3]. Some isoforms play important roles in other processes such as gas or nutrient uptake and translocation, and nitrogen remobilisation [4], [5]. The increase of AQP isoforms in plants has been suggested to “offer adaptive advantages for growth in different environmental conditions, possibly as a result of divergent transport selectivities or regulatory mechanisms” [6]. Although regulation of AQP activities relies on a complex interplay of post-transcriptional, translational and post-translational processes [7], monitoring gene expression has been a valuable tool to dissect AQP roles in plant functioning [8]–[10].
Fifty-five AQP genes were identified in Populus trichocarpa genome [11]. One of the main rationale motivating analyses of gene expression in Populus comes from its status of model system characterised by woodiness and perennial habit and thus developing structures and behaviours which are not questionable in herbaceous and annual models [12], [13]. In addition, P. trichocarpa has become a model to study the evolution of duplicated genes, the Salicoid duplication event having significantly contributed to the amplification of multigene families [14]–[16]. In various Populus species, regulations of AQP expression were reported during adventitious root development in P. trichocarpa x deltoides [17], mycorrhizal symbiosis in P. tremula x tremuloides [18] and recovery from xylem embolism in P. trichocarpa [19]. Some AQP members were found responsive to environmental challenges and hormone treatments (in P. balsamifera, P. simonii x balsamifera, P. alba x tremula, P. trichocarpa x deltoides) [20]–[23] and to be differentially expressed depending on genotypes [24]. Most of these studies focused on a few AQP genes and/or subfamilies. In several analyses of whole transcriptome response, some AQP members were listed among the most responsive genes to various environmental constraints [25]–[28]. Meanwhile, the sole family-wide picture of AQP expression drawn to date has been a visualization of transcript accumulation across nine tissues from Populus balsamifera [11].
Our aim was to provide new insights for functional characterisation of the AQP gene family in Populus by delineating their expression domain and distinguishing their responsiveness to developmental and environmental cues. Taking advantage from the large expression data set obtained with the Affymetrix GeneChip Poplar Genome Array, several sources of diversity were simultaneously investigated, namely species/genotypes, tissues/organs and various cues. In-silico strategy was optimised to control for previously identified biases in cross-species transcriptomics [25], [29]. Key aspects of AQP expression profiles were cross-validated using previously-published data such as expressed sequence tag libraries (EST), expression data from qPCR or from another platform array (GPL7424, NCBI, Gene Expression Omnibus). Our meta-analysis reveals the specificities of AQP expression which cannot be fully addressed in single experiments, such as expression wideness, response specificity as well as genotype-dependent diversity. Through the simultaneous investigation of experimental series, we show that the large expansion of AQP family in poplar was accompanied with a strong divergence of gene expression, even if some cases of functional redundancy could be suspected.
Materials and Methods
Database Search
Full-length sequences of all AQP genes of Populus trichocarpa were downloaded from Phytozome v8.0 [30]. A total of 429,444 Populus expressed sequence tags (EST) were downloaded from the GenBank database [31]. AQP coding regions were used as queries to perform BLASTN alignment against all EST [32]. NCBI BLAST 2.2.25+ executable was used on a local platform. Command line “blastn” was executed with task argument set as “blastn” and default parameters (word size: 11, expect threshold: 10, match/mismatch scores: 2/−3, gap penalties: existence 5, extension 2). Matches above 96% identity and over an alignment of at least 100 bp were considered as corresponding sequences of AQPs. Reverse BLASTN strategy (using EST as queries against AQP transcripts) was performed to assign each EST to a single AQP ID. Metadata associated to each EST were manually inspected for their tissue origin.
All publicly available Affymetrix GeneChip Poplar Genome Array data were downloaded from the NCBI Gene Expression Omnibus [33] and ArrayExpress [34] at the end of January 2012. Collection gathered 632 arrays from distinct experiments. Within each experiment, arrays were normalised with the GcRMA package (GcRMA 2.0 [35]) available in Bioconductor [36], followed by Log2 transformation and calculation of the mean for each condition [16]. AQP expression was explored in a subset of 110 “control” arrays, excluding “treatment” and “transgenic line” data. We discriminated eight sample types, namely suspension cells, seedling, catkin, shoot apex, leaf, stem, root and xylem. To analyse regulation of AQP expression, mean signal intensities were pair-wise compared and expressed as Log2 ratio. To prevent introduction of noise, computation of Log2 ratio was constrained, i.e. set to null when signal intensities of the two compared conditions were below background level (cut-off set to 3.2). Treated plants or “transgenic lines” were compared to their respective control or wild type. In the analysis of temporal series, successive time points were compared to the initial one (ie t = 0 or predawn). The present meta-analysis comprises 167 comparisons.
The Affymetrix GeneChip Poplar Genome Array contains 61,251 probe sets representing over 56,000 transcripts and predicted genes, and was generated from several Populus species (including P. trichocarpa genome v1.1). Probe sets corresponding to AQPs were identified using Batch Query and Probe Match, tools available at the NetAffx Analysis Center (http://www.affymetrix.com). “Batch Query” was run either using Gene Symbol from previous releases of P. trichocarpa genome (v1.1 and v2.0) or NCBI RefSeq. “Probe Match” found probes that identically match AQP sequences. Due to the criterion used for the array design (minimal overlap between EST/mRNA-based UniGene clusters and predicted genes), some probe sets were lacking of a gene model correspondence. To strengthen our annotation, the 61,251 target sequences (i.e. one per probeset on the array) were confronted to P. trichocarpa genome. Target sequences of probe sets were used as queries against P. trichocarpa genome, using a local BLASTN with parameters mentioned above. Each probeset were re-annotated according to the best BLAST hits per query.
Extracting Gene-level Information from Probe Set-based Information
Extracting reliable gene-level information from probe set-based information is still under debate [25]. While the use of median value takes advantage from the presence of multi-probe sets for automatic consolidation, it relies on the assumption that distinct probe sets are equally suitable for the detection of a given transcript (at least having equivalent matching probabilities). This assumption stands as long as probe sets x sample matrices are homogeneous [37]. Our analysis being based on experiments carried on distinct poplar species hybridised on a multispecies-designed array, one could expect that probe sets designed on EST from different species would differentially match depending on species matrices. We tested this hypothesis by screening signal intensity and Log2 ratio of all probe sets targeting a given gene and comparing information retrieved from median and maximal values (Figure S1). The two methods were mostly consistent. Median provided lower estimates of expression and/or regulation than maximum since it took into account absence of signal. Median depended not only on the number of probe set per gene but also on the compatibility between probe set and hybridised matrix, which makes it unsuitable in a meta-analysis (illustrated for PtPIP2;4 - Figure S1). Expression and regulation for each gene were thus extracted from probe set data using maximal values (either maximal signal intensity or maximal absolute Log2 ratio). Data were visualised by heatmap and hierarchical clustering, which was performed with 'hclust' function using Euclidean distance (R2.14.1, http://www.R-project.org). Based on the Log2 ratio distribution, regulations of gene expression were categorized according to their intensity applying a fold change threshold of 1.5 (fold change = 2Log2ratio).
Sequence Analysis
Phylogenetic relationships of AQP family have been previously described [11]. Populus genome had undergone several rounds of genome-wide duplication followed by multiple segmental and tandem duplications [13], [15]. Among them, the Salicoid duplication event had significantly contributed to the amplification of multigene families. Three interfaces were interrogated to identify duplicate pairs in the poplar AQP family (Gramene release 34b, http://gramene.org; PGDD [38]; Plaza v2.5 [39]). The genetic distance between syntenic gene pairs was examined on the basis of the proportion of four-fold degenerate nucleotide sites that underwent transversions (4DTV values) [13]. The 4DTV values were downloaded from Plaza v2.5 and from a recent genome-wide analysis of gene pair in poplar [15]. Synonymous (dS) and nonsynonymous (dN) substitution rates were estimated from nucleotide sequences in a pair-wise manner with CodonSuite interface [40].
Results and Discussion
The Aquaporin Family in Populus trichocarpa Genome
Gupta and Sankararamakrishnan [11] studied the AQP family in the Populus trichocarpa genome v1.1. They discarded nine invalid sequences, confirmed 54 AQP genes and identified a new AQP sequence. In subsequent versions of P. trichocarpa genome (v2.0 and v2.2), the functional annotation “Aquaporin” has been consistently up-dated, except two remaining invalid sequences (POPTR_0007s07950 and POPTR_1606s00200 [11]). PtXIP1;1 has been recently invalidated (truncated sequence POPTR_0009s13100) [23]. We thus considered 54 predicted genes and used the nomenclature of Gupta and Sankararamakrishnan [11]. Using these genomic sequences, we detected 2961 expressed sequence tags (EST).
Closely related AQP pairs were identified in previous phylogenetic analysis [11], [24]. The expansion of the AQP family in P. trichocarpa genome resulted from both segmental and tandem duplications (Table 1). Only six AQP genes could be considered as single copy in P. trichocarpa genome (PtNIP1;3 - PtNIP1;4 - PtNIP1;5 - PtNIP3;5 - PtPIP1;3 - PtXIP2;1). A lack of congruency across distinct information sources were detected for two clusters (PtSIP1;3/PtSIP1;4 - PtTIP3;1/PtTIP3;2). Two AQP pairs were retained following tandem duplication processes (PtPIP2;9/PtPIP2;10 - PtPIP2;5/PtPIP2;6). The peculiar mapping of XIP1s among P. trichocarpa linkage groups (PtXIP1;3 is located on LGIV while the three others genes are arranged head-to-tail on LGIX), precluded inferring the evolutionary processes that shaped the PtXIP1 subfamily. The genetic distance between pairs, that was determined on the basis of 4DTV values [13], indicated that several rounds of segmental duplication have shaped the poplar AQP family. PtNIP2;1, PtPIP2;8 and PtTIP4;1 shared complex evolutionary relationships with other members of their subfamily, indicating ancient duplication events. The Salicoid whole-genome duplication event strongly amplified the AQP family (16 pairs, Table 1). To explore AQPs divergence, the rates of non-synonymous (dN) and synonymous (dS) nucleotide substitutions were calculated (Table 1). dN/dS ratios ranked from 0.05 (PtTIP1;3/PtTIP1;4) to 0.47 (PtSIP2;1/PtSIP2;2), and were close to those observed within the poplar HD-ZIP family [16]. While the highest dN/dS ratios indicated that some pairs may have evolved more rapidly than others following duplication events, a limited functional divergence occurred between AQP pairs, at least in the coding region.
Table 1. Non-synonymous/synonymous ratio for AQP pairs.
| Gene pairs | duplication | dN | dS | dN/dS |
| PtNIP1;1/PtNIP1;2 | S | 0.062 | 0.293 | 0.212 |
| PtNIP3;1/PtNIP3;2 | S | 0.031 | 0.271 | 0.115 |
| PtNIP3;3/PtNIP3;4 | S | 0.042 | 0.246 | 0.172 |
| PtPIP1;1/PtPIP1;2 | S | 0.027 | 0.273 | 0.098 |
| PtPIP1;4/PtPIP1;5 | S | 0.101 | 0.280 | 0.362 |
| PtPIP2;1/PtPIP2;2 | S | 0.053 | 0.297 | 0.177 |
| PtPIP2;3/PtPIP2;4 | S | 0.056 | 0.294 | 0.191 |
| PtPIP2;5/PtPIP2;7 | S | 0.036 | 0.368 | 0.098 |
| PtPIP2;5/PtPIP2;6 | T | 0.008 | 0.022 | 0.347 |
| PtPIP2;9/PtPIP2;10 | T | 0.166 | 0.430 | 0.386 |
| PtSIP1;1/PtSIP1;2 | S | 0.095 | 0.223 | 0.423 |
| PtSIP1;3/PtSIP1;4 | Nd | 0.044 | 0.202 | 0.217 |
| PtSIP2;1/PtSIP2;2 | S | 0.087 | 0.186 | 0.466 |
| PtTIP1;1/PtTIP1;2 | S | 0.037 | 0.369 | 0.099 |
| PtTIP1;3/PtTIP1;4 | S | 0.016 | 0.325 | 0.050 |
| PtTIP1;5/PtTIP1;6 | S | 0.031 | 0.345 | 0.090 |
| PtTIP1;7/PtTIP1;8 | S | 0.059 | 0.288 | 0.203 |
| PtTIP2;1/PtTIP2;2 | S | 0.032 | 0.217 | 0.150 |
| PtTIP2;3/PtTIP2;4 | S | 0.029 | 0.310 | 0.093 |
| PtTIP3;1/PtTIP3;2 | Nd | 0.046 | 0.288 | 0.160 |
| PtTIP5;1/PtTIP5;2. | S | 0.033 | 0.211 | 0.157 |
| PtXIP1;3/PtXIP1;4 | Nd | 0.110 | 0.531 | 0.208 |
| PtXIP1;3/PtXIP1;5 | Nd | 0.242 | 1.240 | 0.195 |
| PtXIP1;1/PtXIP1;2 * | T | 0.017 | 0.021 | 0.824 |
Gene pairs resulted from segmental (S) or tandem (T) duplication. For two AQP pairs no unambiguous inference about duplication events can be provided (Nd).
PtXIP1;1 is a pseudogen.
Extracting AQP Information from Affymetrix Data
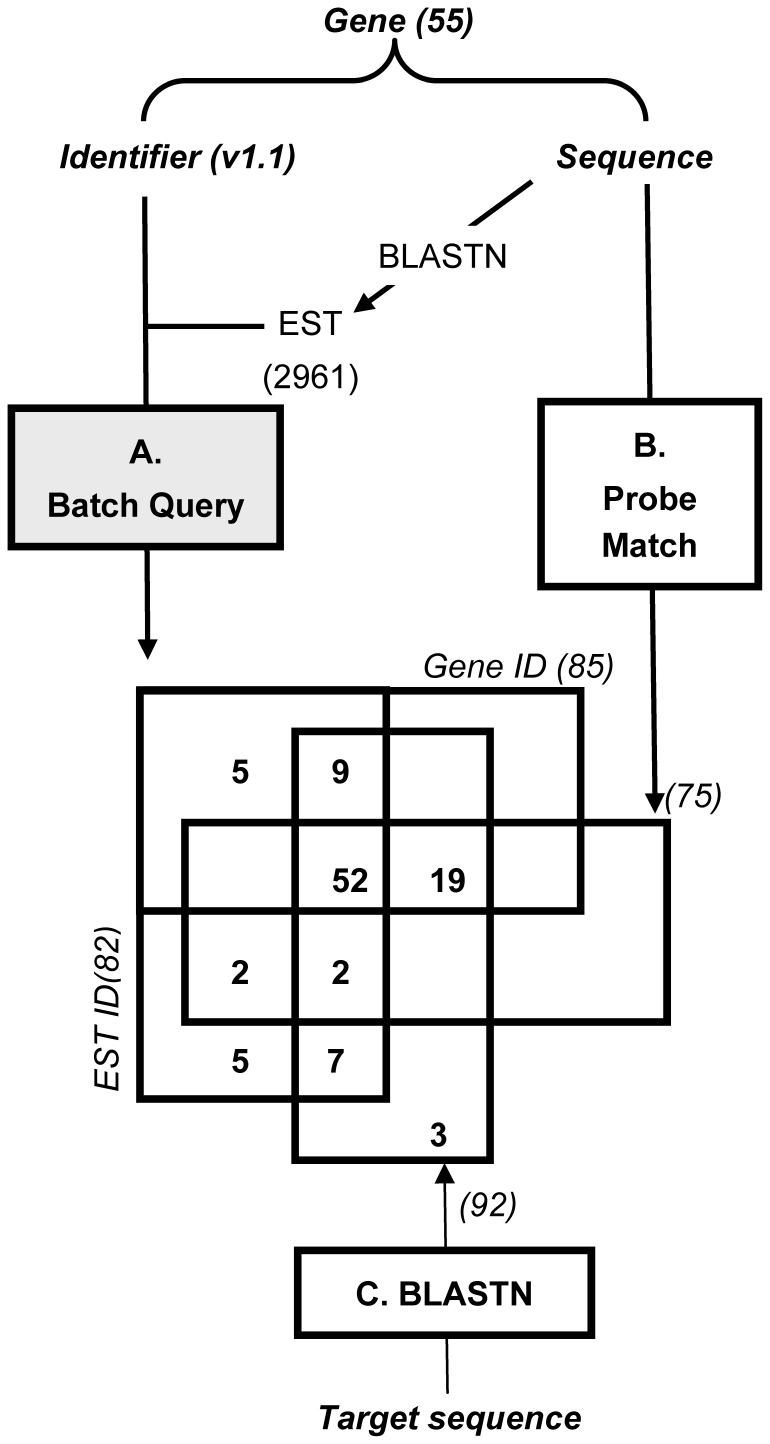
AQP-targeting probe sets were retrieved on the Affymetrix GeneChip Poplar Genome Array using several identifiers in a three-step strategy (Figure 1). Six probe sets matching invalid AQP sequences were filtered out (Table S1). As shown in Figure 1, 85 and 82 probe sets were identified using Gene ID or EST ID, respectively. This discrepancy reflects the multispecies design of the array and the incomplete gene prediction on genomic sequence (such as the lack of UTR prediction [11]). Running Probe Match with P. trichocarpa AQP sequences enabled the detection of 75 probe sets. Probe Match revealed only perfect identity – implying that hybridised sequence is known and/or was used in array design. To consider high similarity rather than identity, the 61,251 Affymetrix target sequences were confronted to P. trichocarpa genome (v1.1 and v2.0) using BLASTN. We confirmed 89 previously detected probe sets and revealed three new ones. As a final step, the AQP-targeting probe sets were in-silico evaluated. Three out of seven probe sets commonly detected by EST ID and BLASTN, and one out of the five probe sets found exclusively with EST ID were designed on minus strand and were discarded (Table S1). We also filtered out five probe sets for which gene assignation was ambiguous and one designed in an intronic region. Finally, 94 probe sets were deemed appropriate for targeting 53 AQPs, only PtNIP1;5 being missed. In details, these probe sets were designed on 12 Populus species, with a complex layout of species and redundancy as one-to-one probe set to gene relationships concerned only 31 AQPs (Table S2). Our analysis highlighted that retrieving information about a multigene family, or even a single gene, on multispecies-designed array cannot only rely on ID Query or Perfect Match but requires similarity-based screening (Figure 1). After proper filtering, the Affymetrix GeneChip Poplar Genome Array appeared to be a valuable tool for AQPs profiling. Besides, our optimised in-silico strategy is applicable to any multigene family and is suitable for any comparative transcriptomics based on multispecies-designed chips (such as most plant Affymetrix GeneChips).
Figure 1. Bio-informatics strategy for GeneChip screening.
AQP-targeting probe sets were identified using “Batch Query” and “Probe Match”, tools available as at the NetAffx Analysis Center. A. “Batch Query” was run either using Gene symbol or JGI transcript ID and NCBI RefSeq. B. “Probe Match” found probes that identically match AQP-coding sequences. C. AQP-targeting probe sets were identified through BLASTN alignment of target sequence and Populus trichocarpa genome sequences (v1.1 and v2.0). Venn diagram exhibits the number of probe sets retrieved from each procedure.
Profiling Reveals Tissue- or Organ-preferred Expression
First insight of AQP functions in poplar was provided by an analysis of transcript accumulation in distinct tissues and organs sampled under control conditions (Figure 2). Organs/tissues were not equally represented in the data set ranging from one for bark to 56 for leaf samples. In some cases, a confounding effect cannot be excluded since sample types were collected on a unique Populus species. Biological inferences focused on positive signals, absence call reporting either absence of transcription or unsuitable experimental data.
Figure 2. Expression profiles of AQP genes across tissues.
Expression domains were computed from 110 “control” arrays (i.e. without “treatment” and “transgenic line” data). Arrays were normalised with GcRMA within each experiment. Each row of the heatmap corresponds to an AQP member. Color scale depicts maximal Log2 expression level. White represents below background level. Columns correspond to the eight sample types, namely SC for suspension cells (2 arrays: GSE16773, GSE17804), catkin (2 arrays: GSE13990), seedling (3 arrays: GSE13990), root (15 arrays: E-MEXP-1874, E-MEXP-2234, GSE13109, GSE13990, GSE16888, GSE16785, GSE17223, GSE17225, GSE19297), leaf (56 arrays: E-MEXP-1928, GSE9673, GSE13109, GSE13990, GSE14515, GSE14893, GSE15242, GSE16417, GSE16783, GSE16785, GSE17226, GSE17230, GSE21171, GSE24349, GSE27693, GSE16417), shoot apex (2 arrays: GSE16495, GSE21061), bud (14 arrays: GSE29335, GSE29336, GSE30320, GSE24349) bark (1 array: GSE29303), stem (4 arrays: GSE21480, GSE12152, GSE19467) and xylem (11 arrays: E-MEXP-2031, GSE13990, GSE16459, GSE20061, GSE27063, GSE3232). The number of arrays per tissue and the series accession numbers are given into brackets.
Most AQP members were expressed in most vegetative tissues (Figure 2). Only PtNIP1;4, PtNIP3;5, and PtXIP2;1 were called absent in all analysed tissues. PIPs, SIPs and TIPs exhibited higher expression levels than XIPs and NIPs. As can be expected, AQP expression pattern in suspension cells differed from multicellular tissues. The reproductive tissue showed the expression pattern the closest to that of suspension cells, both accumulating PtNIP1;2 and PtNIP1;3 transcripts but no PtTIP1;7/PtTIP1;8 transcripts. PtTIP5;1/PtTIP5;2 were preferentially expressed in mature catkins but not detected in floral bud nor in any vegetative organ. These results are in accordance with the predominant expressions of AtTIP5;1, AtNIP4;1 and AtNIP4;2 in Arabidopsis flowers and pollen [5], [41]. AtTIP5;1 has been suggested to be an urea transporter for pollen mitochondria and involved in nitrogen recycling in pollen tubes. Seedlings and roots were characterised by the broadest AQP expression patterns, with however some specific features. PtTIP3 transcripts were strongly and preferentially accumulated in seedlings (Figure 2). In line, three out of the four PtTIP3;1 EST were isolated from imbibed seeds and TIP3 were reported as specific for maturating and dry seeds in several species [42]–[44]. In Arabidopsis, a high TIP3 protein abundance is maintained until completion of germination [45] and AtTIP3;1 and AtTIP3;2 are the only detectable TIPs in embryos during seed maturation and the early stages of seed germination [46]. Eight AQPs exhibited a root-preferred expression (Figure 2). Within three experiments (GSE17223/GSE17230, GSE13109 and GSE16783), transcript profiling was performed in both leaves and roots, enabling a straight comparison based on Log2ratio computation (Figure S2). Eleven AQPs were expressed at a higher level in roots than in leaves and only three AQPs exhibited a leaf-preferred expression. This analysis confirmed previously detected root-preferred expression of PtNIP3;4, PtPIP2;8, PtTIP1;1/PtTIP1;2, and PtTIP2;3/PtTIP2;4, and revealed new contrasts (PtPIP2;2, PtPIP2;5, PtPIP2;7, PtTIP1;4 and PtTIP4;1, Figure S2). On the opposite, PtPIP2;9, PtTIP1;8 and PtXIP1;5 appeared consistently more expressed in leaves than in roots (Figure 2, Figure S2). The same expression patterns were reported for PtPIP2;7, PtPIP2;8, PtTIP1;2, PtTIP1;8 and PtTIP2;4, based on P. trichocarpa samples analyzed on NimbleGen platform [12]. Similar expression patterns of PtPIP2;8, PtPIP2;9 and PtXIP1;5 were confirmed on P. trichocarpa using qPCR technology [47]. However, the cases of PtPIP1;4/PtPIP1;`5, PtPIP2;3, PtTIP1;5/PtTIP1;6, PtTIP2;2 and PtXIP1;2 highlighted that tissue-preferred expression may vary across genotypes (Figure S2).
Changes in plant AQP expression are known to occur during leaf development [8]. Although exhibiting growth-driven regulations, species and/or culture conditions affected the expression patterns of PtNIP3;3/PtNIP3;4, PtTIP1;8 and PtXIP1;5 (Figure S3). Meanwhile PtPIP1;2, PtPIP2;6 and PtTIP4;1 transcripts were accumulated in mature leaves while PtPIP1;5, PtPIP2;2, PtPIP2;9, PtTIP1;4 and PtTIP1;6 were preferentially-expressed in young leaves (Figure S3). Dealing with a woody species, the xylem transcriptome has been investigated. Except for the PtXIP subfamily, AQP expression patterns were relatively similar in xylem and aerial parts - leaf, shoot apex, bud, stem and bark (Figure 2). No AQP exhibited a xylem-preferred expression, consistently with the presence of this tissue in all organs (Figure 2). PtPIP2;3/PtPIP2;4, PtTIP2;2 and PtTIP4;1, highly expressed in wood tissue, were more expressed in ray cambial cells as in fusiform cambial cells [48]. Interestingly PtPIP2;3, PtTIP2;2 and PtTIP4;1 proteins were detected in the plasma membrane of differentiating secondary vascular tissue [49].
Comprehensive Analysis of Poplar AQP Expression Under Various Situations Reveals Heterogeneity in AQP Subfamilies Responsiveness and Co-regulations
Transcriptional regulations of plant AQPs are known to be isoform-specific [50]. Even within a subfamily, transcriptional responses clearly depend on the experimental procedures and vary across species as recently shown for PIPs in the case of drought stress [51]. In this context, a wide collection of experiments was analysed to highlight key features about poplar AQP responsiveness (Figure S4 to S8). To better address the questions of where and how consistent AQP expression was regulated, all transcriptional regulations - for each AQP gene under each of the 145 tested conditions- were compiled in Table 2. Given that some AQPs exhibited tissue- or organ-prefered expression (Figure 2), tissues and organs were considered separately. Based on a post-hoc grouping of common cues, 5 groups were delineated, namely abiotic stress, nutrional status, hormonal signalling, biotic interactions and temporal oscillation.
Table 2. Map of AQP responsiveness.
| Abiotic stress | Nutrition | Hormone | Biotic | Temporal oscillation | ||||||||||||||||||||||||
| Water deficit | Osmoticum | Salt | R-hypoxia | L- wounding | Aluminium | Embolism, infiltration | Sarvation | Gln | Glc | Gln+Glc | CIM | SIM | MeJa | GA-modified | L-pathogen | Mycorrhiza | Seasonal | Diurnal | Diurnal | |||||||||
| L | X | R | R | X | R | R | L | L | R | R | X | L | B | B | B | B | St | Ca | SC | R | L | R | Lb | Fb | St | X | ||
| 40 | 1 | 6 | 1 | 2 | 3 | 3 | 1 | 3 | 1 | 3 | 2 | 6 | 3 | 2 | 2 | 2 | 2 | 2 | 1 | 5 | 4 | 2 | 3 | 4 | 1 | 18 | 4 | |
| PtNIP1;1 | x | C | 2 | C | 1 | x | – | – | 2 | C | – | – | 2 | 1 | 1 | – | C | 1 | – | – | 1 | 2 | – | 1 | 3 | – | x | – |
| PtNIP1;2 | x | – | 3 | C | – | – | C | – | x | – | – | – | 4 | – | – | – | – | – | – | – | 3 | 1 | – | 2 | – | – | x | – |
| PtNIP1;3 | 1 | – | – | – | – | – | – | C | – | – | – | – | – | 1 | C | – | – | – | – | – | – | – | – | – | – | – | – | – |
| PtNIP1;4 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| PtNIP2;1 | x | – | 4 | – | – | 2 | x | – | 2 | – | 1 | – | x | – | – | – | – | – | – | – | – | 1 | – | – | – | – | x | – |
| PtNIP3;1 | x | – | – | – | – | 1 | – | – | 1 | – | – | – | – | x | 1 | – | C | C | – | – | – | – | – | – | – | – | – | – |
| PtNIP3;2 | x | – | 1 | – | – | 1 | – | – | – | C | 1 | – | x | 2 | C | – | C | C | – | – | 1 | – | – | C | – | – | 1 | – |
| PtNIP3;3 | x | C | x | C | C | 2 | 2 | – | x | – | – | 1 | C | C | – | – | – | C | – | C | 4 | 1 | 1 | C | – | – | x | 2 |
| PtNIP3;4 | x | – | x | – | 1 | C | 2 | – | – | C | 1 | 1 | 2 | C | – | – | 1 | 1 | C | – | – | – | 1 | C | 3 | – | 7 | 2 |
| PtNIP3;5 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| PtPIP1;1 | x | – | 3 | C | – | – | – | – | x | – | – | 1 | 2 | 1 | – | – | – | x | 1 | C | 1 | – | – | C | 3 | – | 7 | 1 |
| PtPIP1;2 | x | C | – | C | 1 | 1 | C | – | 2 | – | 1 | 1 | x | 1 | – | – | – | 1 | 1 | C | 4 | 1 | 1 | 2 | 1 | – | x | 1 |
| PtPIP1;3 | x | C | – | – | – | – | – | – | x | – | – | – | – | – | – | – | – | C | 1 | – | – | – | – | 1 | – | – | 5 | – |
| PtPIP1;4 | x | C | 3 | C | 1 | 2 | – | – | – | C | C | 1 | x | C | 1 | – | C | C | C | – | – | – | 1 | 2 | 3 | – | x | – |
| PtPIP1;5 | x | C | 4 | – | – | 2 | 2 | – | 1 | – | 1 | – | x | C | C | – | C | x | – | – | – | 1 | – | – | 2 | – | x | 1 |
| PtPIP2;1 | x | – | 1 | C | C | – | 2 | – | 1 | – | 1 | 1 | – | 2 | – | – | 1 | C | – | – | – | – | – | x | 3 | – | x | – |
| PtPIP2;2 | x | – | 3 | C | C | x | 1 | – | 1 | – | 1 | – | 3 | – | – | 1 | 1 | C | 1 | – | – | 3 | 1 | C | 2 | C | x | – |
| PtPIP2;3 | x | C | – | – | 1 | – | C | – | 1 | – | 1 | – | 3 | 2 | 1 | – | 1 | C | – | C | 1 | 1 | – | C | – | – | x | – |
| PtPIP2;4 | x | – | 2 | C | 1 | – | – | – | x | – | 2 | C | x | 2 | – | – | 1 | – | – | – | 4 | 1 | – | 2 | x | C | x | – |
| PtPIP2;5 | x | C | 4 | – | 1 | 2 | – | – | 2 | – | 1 | 1 | 2 | 2 | 1 | – | C | x | 1 | – | C | – | 1 | – | – | C | x | 1 |
| PtPIP2;6 | x | C | – | C | 1 | 1 | – | – | 2 | – | 1 | 1 | 4 | 1 | – | – | 1 | x | C | C | 3 | 2 | – | x | 2 | – | x | 1 |
| PtPIP2;7 | x | C | C | C | 1 | 2 | C | – | 1 | – | x | C | 3 | x | 1 | – | – | 1 | C | – | C | 1 | 1 | x | 1 | – | x | 1 |
| PtPIP2;8 | x | – | 5 | C | 1 | – | C | – | x | – | – | – | 2 | 2 | C | – | C | 1 | C | – | 4 | – | – | C | 1 | – | x | 1 |
| PtPIP2;9 | x | – | – | – | – | – | – | – | 1 | – | – | – | 3 | 1 | – | – | 1 | C | C | – | – | 2 | – | C | 3 | – | x | – |
| PtPIP2;10 | x | – | 5 | C | – | x | C | – | x | C | 1 | – | 2 | 2 | – | x | 1 | 1 | 1 | – | 2 | 1 | 1 | – | 1 | C | x | – |
| PtSIP1;1 | – | – | – | C | – | – | – | – | – | – | – | – | – | C | – | – | – | – | – | – | – | – | – | C | 3 | – | – | – |
| PtSIP1;2 | x | C | 3 | – | C | – | 2 | – | 2 | – | 1 | 1 | 1 | C | – | – | – | – | – | – | 2 | 1 | – | C | 1 | – | x | – |
| PtSIP1;3 | x | C | 2 | – | 1 | 1 | 2 | – | 1 | – | – | – | – | C | 1 | – | 1 | 1 | – | – | – | – | 1 | C | 1 | C | x | – |
| PtSIP1;4 | x | – | – | C | – | 1 | 1 | – | – | – | – | – | x | – | C | – | C | 1 | C | – | – | – | 1 | 1 | 1 | – | 3 | – |
| PtSIP2;1 | x | C | – | C | – | – | 1 | C | 2 | C | – | – | – | 1 | – | – | C | x | C | – | – | 2 | – | C | 1 | – | x | – |
| PtSIP2;2 | – | – | 2 | – | – | – | x | – | – | – | – | – | 1 | – | – | – | – | x | C | C | 2 | 1 | – | – | – | – | – | – |
| PtTIP1;1 | x | C | 4 | C | 1 | C | C | – | 2 | C | – | 1 | 3 | – | 1 | 1 | 1 | 1 | C | – | C | – | x | – | – | C | x | 2 |
| PtTIP1;2 | – | C | 5 | C | 1 | C | C | – | – | – | 1 | C | – | – | – | – | – | – | – | – | C | – | x | – | – | C | – | 2 |
| PtTIP1;3 | x | – | C | – | – | C | 2 | – | x | – | – | 1 | 4 | x | – | – | C | C | C | C | C | 1 | 1 | C | 2 | – | 14 | 2 |
| PtTIP1;4 | x | C | C | C | 1 | C | – | – | x | – | – | C | 5 | C | 1 | – | 1 | C | C | – | C | 1 | x | – | 3 | – | x | 2 |
| PtTIP1;5 | x | C | 1 | C | 1 | 2 | C | – | x | – | – | – | 3 | C | C | 1 | C | C | 1 | – | 1 | 2 | 1 | 2 | 1 | – | x | 1 |
| PtTIP1;6 | x | C | – | – | 1 | 1 | C | – | 2 | – | – | – | 4 | C | 1 | – | – | C | – | C | 2 | 3 | 1 | 1 | 1 | – | x | – |
| PtTIP1;7 | x | – | 1 | C | – | – | 2 | – | 2 | – | – | 1 | 3 | 1 | – | – | – | C | 1 | – | C | 1 | – | C | 1 | C | x | – |
| PtTIP1;8 | x | – | – | – | – | 2 | C | – | C | – | 2 | – | 5 | C | C | 1 | C | C | x | – | C | 1 | 1 | C | 1 | – | x | – |
| PtTIP2;1 | x | – | C | C | – | C | 1 | – | 1 | – | – | 1 | x | 2 | C | – | – | C | 1 | – | 1 | 3 | 1 | C | – | – | x | – |
| PtTIP2;2 | x | C | C | C | 1 | – | – | – | x | C | 1 | – | 1 | C | C | – | 1 | C | 1 | – | 1 | 1 | – | x | – | – | x | 2 |
| PtTIP2;3 | – | – | 4 | C | – | 1 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | C | – | – | – | – | – | – | – |
| PtTIP2;4 | – | – | 4 | C | – | C | 1 | – | – | – | 1 | – | – | – | – | – | – | – | – | – | C | – | 1 | – | – | – | – | – |
| PtTIP3;1 | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – | – | C | – | – | – | – | – | – | – | – |
| PtTIP3;2 | x | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – |
| PtTIP4;1 | x | C | 1 | – | 1 | C | C | – | x | – | – | 1 | 3 | C | – | – | – | C | x | – | – | 1 | – | C | 1 | – | x | – |
| PtTIP5;1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| PtTIP5;2 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| PtXIP1;2 | – | C | C | C | – | 2 | – | – | 1 | – | 1 | – | – | – | – | – | – | – | – | – | – | – | C | – | – | – | – | – |
| PtXIP1;3 | – | – | 4 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| PtXIP1;4 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | C | – | – |
| PtXIP1;5 | x | – | – | – | – | – | – | – | 2 | – | – | – | x | – | – | – | C | C | C | – | – | 2 | – | C | 1 | C | x | – |
| PtXIP2;1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
Regulations of AQP expression in response to distinct cues are depicted in distincft organs and tissues: leaf (L), root (R), xylem (X), bark (B), stem (St), floral bud (Fb), leaf bud (Lb) and suspension cell (SC). The number of experiments analysed is given in the heading. Responsiveness is described as follow: “−” denotes absence of regulation (FC<1.5), “C” denotes consistent regulations (FC≥1.5, 100% cases), a number denotes intermediary cases (FC≥1.5, number of cases). A cross indicates interaction: both down- and up-regulations are observed within a category (FC≥1.5).
Transcriptional Regulation of AQP Expression Accompanying Abiotic Challenges
As expected for a model plant of agronomic interest, water deficit was the most studied abiotic stress (Table 2). While about one third of AQPs were not responsive to water deficit, PtPIP1;2 and PtPIP2;7 expressions were consistently up-regulated in all organs (Table 2, Figure S4A). Identified as preferentially expressed in roots under control conditions (Figure 2), PtPIP2;8, PtTIP1;2 and PtTIP2;3/PtTIP2;4 were even more expressed in roots under water deficit. In addition, PtTIP2;2 expression was one of the strongest water deficit-induced up-regulations in roots while expressions of PtNIP2;1, PtPIP2;2 and PtPIP2;10 were down-regulated. Osmotic stress and soil water deficit induced similar patterns of AQP regulation in Soligo root apices except for PtNIP1;1 and PtXIP1;2. Drought-driven regulations occurring in leaves were found to be mostly inconsistent across the 40 comparisons, reflecting either wide genotype diversity or large number of experiments (Table 2, Figure S4A). Accordingly, drought-driven transcriptome response in leaf has been shown to be shaped by time of day, to be dependent on genotype x treatment interaction and on clone history [27], [29], [52]. However the strongest drought responses were down-regulations of PtPIP1;5, PtPIP2;9, PtTIP1;6, PtTIP1;8 and PtXIP1;5 expressions (Figure S4A). The highest up-regulations of expression were found for PtTIP1;1 and PtTIP1;4 in xylem and for early response in leaf of a drought-tolerant genotype (Figure S4A). Using qPCR approaches, similar drought responses have been reported in poplar leaves for PtPIP1;2, PtPIP2;7 and PtXIP1;5 [23], [24]. While some AQPs have been suggested as playing a role in regulation of leaf hydraulics with a possible link to stomatal conductance and drought tolerance, such as PIP2;5 orthologs [24], the above-cited AQPs could be considered as drought markers.
Salt and hypoxia-driven responses were similar in roots, both stresses repressing several AQP expressions (Figure S4B). However the accumulation of PtPIP2;10 transcripts seemed to be a key AQP signature of hypoxia in roots, but this result has to be confirmed in other Populus species. The impact of wounding on AQP patterns was strong but clearly dependent on both leaf plastochron index and time after treatment, thus precluding general conclusion (Figure S4C). In leaves collected on tree submitted to nitrogen limitation, expression of several AQPs (PtNIP3;3/PtNIP3;4, PtPIP2;3, PtPIP2;5/PtPIP2;6, PtPIP2;7, PtTIP1;1, PtTIP1;3/PtTIP1;4 and PtTIP4,1) tended to be up-regulated while the expression of some others (PtNIP2;1, PtPIP2;2, PtPIP2;9/PtPIP2;10, PtTIP1;5/PtTIP1;6, PtTIP1;7/PtTIP1;8 and PtXIP1;5) was down-regulated under prolonged starvation (Figure S4D). In lines, incubation in water of partially defoliated stem led to the accumulation of PtNIP3;3/PtNIP3;4, PtPIP2;5/PtPIP2;6 and PtTIP4;1 transcripts and to reduced expression of PtTIP1;5/PtTIP1;6 and PtTIP1;8 in bark. In addition, expressions of PtSIP1;1/PtSIP1;2 and PtSIP1;3 were up-regulated in bark of starved stem only (Table 2). In response to starvation, PtTIP1;4 showed a contrasting response in bark and leaf tissues (Figure S4D). Given that these transcriptional regulations were mostly reversed – or alleviated - when incubation media included glutamine (in combination or not with glucose) and that glucose feeding did not modify AQP expression (Table 2), these AQPs appeared to be responsive to nitrogen status.
Transcriptional Regulation of AQP Expression in Response to Other Cues
As previously observed in other species [53], poplar AQPs were also diversely responsive to modification of hormonal status (Table 2). Distinct phases of poplar micro-propagation induced large modifications of AQP expression (Figure S5A). Biotic interactions were also accompanied with transcriptional regulation of AQP expression (Table 2). In line with its root-preferred expression under control conditions (Figure 2), PtXIP1;2 expression was strongly induced by mycorrhization (Figure S6A). PtXIP1;2 being apparently devoid of water transport in Xenopus leavis oocyte [23], it suggests another role during mycorrhizal interactions, e.g urea or ammonium transport [3]. Meanwhile enhanced expression of root AQPs and increased root hydraulic conductivity was shown in ectomycorrhizal seedlings of P. tremula × P. tremuloides [18] and root hydraulic conductivity of balsam poplar (P. balsamifera) was differentially enhanced regarding mycorrhizal fungal species [54]. All in all, mycorhizal fungi appear to interfere with the aquaporin-mediated transport, whatever the transported substrate, as suggested for P. angustifolia [55]. As previously reported during infection by Melampsora larici-populina incompatible strain (PICME technology, [56]), foliar infection by another biotrophic fungi also repressed the expression of PtTIP1;5/PtTIP1;6, and PtPIP2.2 (Figure S6B). In soybean, 24 of 32 AQP genes were down-regulated in the hours following infection by avirulent Pseudomonas syringae [57]. These crosstalks suggest that controlling water movement could be a mechanism of pathogen inhibition during plant defense.
Seasonal variations of AQP expression were recorded in different organs of this perennial species (Table 2, Figure S7). The only regulation of the stem-preferred PtXIP1;4 was detected during winter hardening, opening new hypotheses. As in other species [58], [59], poplar AQP expression generally varied around the day (Figure S7B). Evidence of diurnal regulation was found, even if AQP expressions were often inconsistent across genotypes (Table 2). Nevertheless, PtPIP1;1, PtPIP2;3 and PtPIP2;7 were more expressed during dark period than during light period in leaf and xylem while PtPIP2;9 and PtXIP1;5 were clearly light induced in leaf (Figure S7B), indicating different physiological functions for these AQP members. Transcriptional regulations of AQPs co-occurring with the over-expression or silencing of several genes in distinct transgenic lines are not detailed here (see Figure S8). Result show that transgenic lines constitute a source of diversity, disturbing AQP expression.
Member-specific Expression Profiles
The expression of five AQPs was never regulated (Figure 3). Among those, our meta-analysis show no evidence of expression for PtNIP1;4, PtNIP3;5 and PtXIP2;1, suggesting very narrow expression patterns (Figures 2 and 3). The functionality of these genes may also be questioned since no corresponding EST has been reported to date in GenBank, and no clear evidence of their transcription was found in another genome-wide transcript profiling based on an independent platform [12]. While never regulated, PtTIP5;1/PtTIP5;2 expressions were restricted to mature catkins under control conditions (Figure 2, Figure S3) and their responsiveness within catkins could be suspected but not tested.
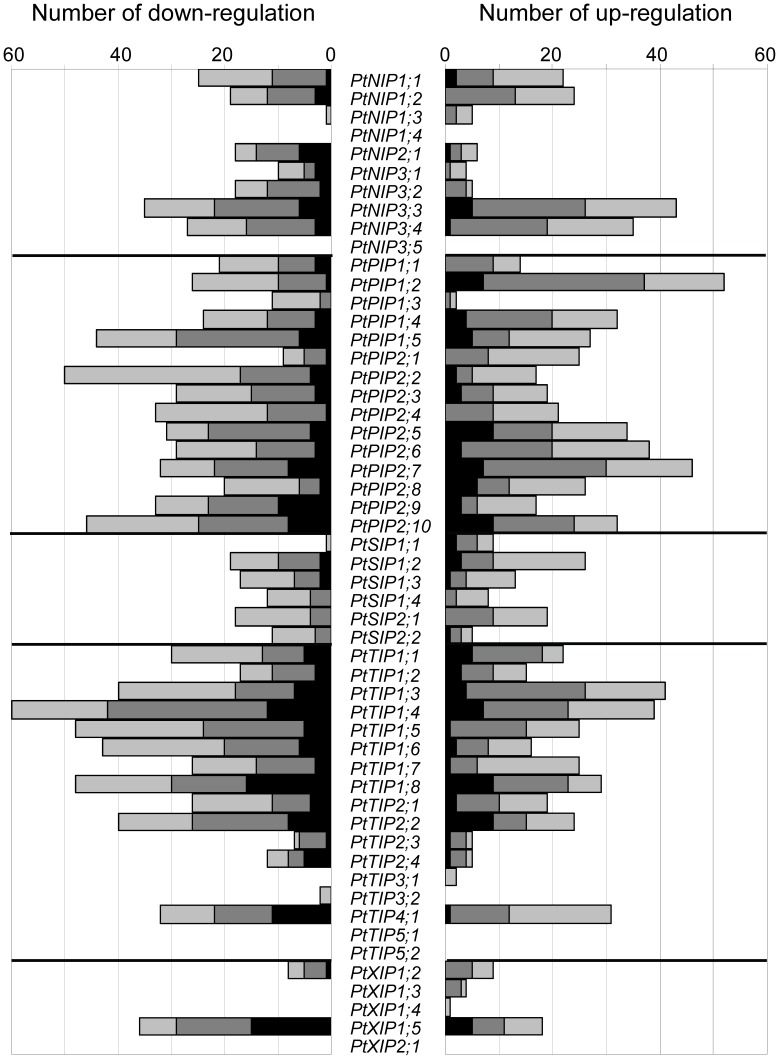
Figure 3. Distribution of regulations by class of fold-change.
For each AQP, up- and down-regulations were counted across 145 comparisons and classified according to the fold change (FC) level: weak regulation 1.5≤FC<2 (light grey); 2≤FC<4 moderate regulation (grey) and FC≥4 strong regulation (dark grey).
While expressed under control conditions, six AQPs were found to be hardly responsive to tested cues (Figure 3). PtNIP1;3 was mostly expressed in catkins and was only punctually regulated. PtXIP1;3 was found exclusively expressed in roots but could be expressed in other organs [12]. PtXIP1;4 was expressed in stem and to a lesser extent in xylem in accordance with literature [12], [15]. PtTIP3;1/PtTIP3;2 exhibited a seedling-preferred expression, but their transcripts have been previously detected in other organs [12]. Given that PtPIP1;3 was constitutively expressed at high level in all organs under control conditions but underwent only few and weak down-regulations, the absence of responsiveness was not linked to the wideness nor the intensity of expression under control conditions.
Concerning AQPs exhibiting numerous transcriptional regulations, PIP and TIP members were more frequently regulated than those of other subfamilies but regulations of expression were mainly of moderate intensity (fold-change ≤4, Figure 3). PtTIP1;4 was found to be the most responsive gene, regulated in almost 100 over 145 comparisons (Figure 3, Table 2). Expressions of PtTIP1;8 and PtXIP1;5 occurred preferentially in leaves, were strongly regulated (fold-change ≥4) and were responsive to many cues. XIP1;5 was recently found ubiquitously expressed [23], disagreeing with our results, i.e. absence of expression in roots as well as absence of root EST. More interestingly PtXIP1;5 was shown to function as water transporter in Xenopus leavis oocyte [23]. Beside, the strong regulation of PtXIP1;5 expression in the ProHSP:FT lines was not found under constitutive over-expression of FT1 and FT2 (Figure S8C), suggesting a potential response to heat induction, which is consistent with its demonstrated drought sensitivity [23].
Divergence and Redundancy of AQP Duplicates
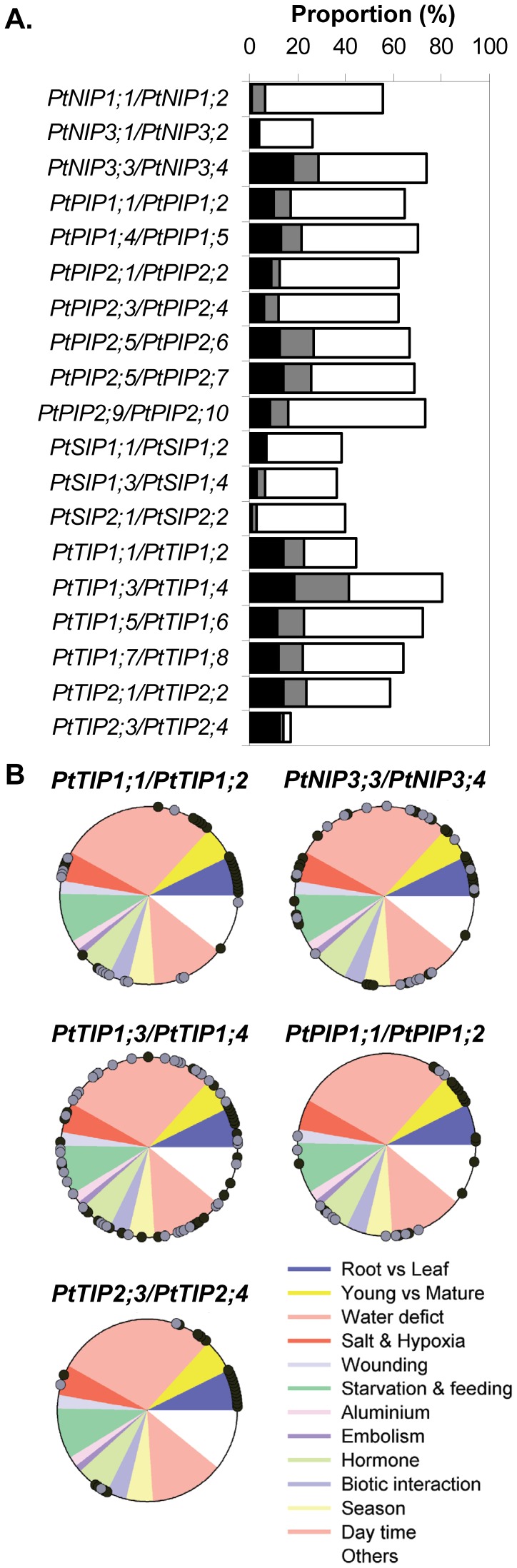
Our meta-analysis gave access to co-expression patterns of AQP members. To test whether duplicates were functionally redundant, their regulation patterns were pair-wise compared (Figure 4). For each AQP pair, we determined the percentage of comparisons for which none of the duplicates underwent transcriptional regulation. Varying from 20% (PtTIP1;3/PtTIP1;4) to 80% (PtTIP2;3/PtTIP2;4), this proportion reflects the wideness of gene pairs expression and the over-representation of studies carried on leaf (Figure 4A). Then, we determined the percentage of comparisons for which a divergence in response was observed, i.e. either only one member of the pair being regulated or both oppositely regulated. In Arabidopsis, AtPIP2;2/AtPIP2;3 shared a high structural similarity and were found to be functionally divergent on the basis of distinct expression properties [60]. Such divergent responses concerned more than 50% of regulation events for all gene pairs, except for PtTIP2;3/PtTIP2;4 (Figure 4A). The latter exhibited similar expression patterns under control conditions (being preferentially expressed in root and seedling, Figure 2), were responsive to few cues and shared convergent responses to modification of gibberellin status and to water deficit (Table 2, Figure 4). These results suggest functional redundancy of these paralogs. While exhibiting balanced proportion of convergent and divergent regulations, convergent regulations of PtTIP1;1/PtTIP1;2 were mainly observed in comparisons carried on root and xylem, suggesting a putative functional redundancy in these organs (Figure 4B). In contrast, convergent regulations of PtTIP1;3/PtTIP1;4 expression were observed in 51% cases but occurred over a large panel of cues, organs and species. This random distribution precluded concluding about functional redundancy but indicated that this gene pair encodes generic AQPs. For some gene pairs, convergent regulations were observed in response to specific cues. For instance, most convergent regulations of PtPIP1;1/PtPIP1;2 expression were observed in response to leaf maturity, hormonal treatment and day time (Figure 4B). Both genes were strongly expressed in leaves and their expressions were commonly enhanced during leaf aging. Concerted regulations of PtNIP3;3/PtNIP3;4 expression mainly occurred in response to abiotic stresses and day time. Co-regulation within five gene pairs was very scarce (less than 20% of regulation events) whatever the responsiveness of the pairs, indicating clear functional divergence between duplicates. In bream and salmon respectively, two and three functional AQP paralogs were differentially distributed and regulated in the intestinal epithelium [61], [62]. These results suggested a fine regulation of transcellular transport in regards to regulation of AQP paralogs. The divergence between duplicates could be partly due to the larger responsiveness of one duplicate as compared to its counterpart (for instance, see PtSIP1;1/PtSIP1;2, Figure 3). Globally, the convergence level was slightly higher for the PtTIP pairs than for the PtPIP pairs, especially if absence of regulation of the two pair members is considered as convergence too (Figure 4A). Then residual functional redundancy may have been conserved at a higher level in the PtTIP subfamilly than in the PtPIP subfamilly. In addition, most PtTIP pairs expression patterns showed higher tissue-specificity than those of PtPIPs (Figure 2), suggesting that TIPs could more contribute to cell identity than PIPs.
Figure 4. Occurrence of convergent regulation within gene pairs.
A. Proportion of convergent versus divergent regulations. Percent of comparison in which duplicates underwent convergent regulations is shown in grey (1.5≤FC<2) or in black (FC≥2). White bar indicates the proportion of comparison inducing divergent regulation of expression. B. Repartition of convergent regulation over experimental categories within five gene pairs. Each dot indicates that gene duplicates underwent convergent regulations under one comparison. Experimental categories are depicted by colour sectors. Dot colour denotes fold change level, black: FC≥2 and grey: 1.5≤FC<2.
Conclusions
While considered as molecular entry into plant water relations, diversity of AQP functions in plants together with family amplification make their characterisation challenging. As a step towards a better understanding of transcriptional regulation, this meta-analysis of all Affymetrix data publicly available has provided a comprehensive picture of poplar AQP expression and regulation at the whole family scale. Through a detailed confrontation with literature, our results were globally validated by previously published information on AQP expression and regulation. In the meantime, gathering usually un-compared cues (for instance biotic vs abiotic) provided novel information. The responsiveness of all genes to a given cue as well as the impact of many cues on the expression of each member were provided without a priori, revealing key features but also highlighting the strong functional divergence within the AQP family.
Supporting Information
Analysis of “sibling” probe sets reveals the sensitivity of median-based procedure for the automatic extraction of gene-centred information in the case of multi-experiment comparison.
(PDF)
Root- and leaf-preferred expression of AQPs.
(PDF)
Young leaf- and mature leaf-preferred expression of AQPs.
(PDF)
Transcriptional regulation of AQP expression under abiotic challenges.
(PDF)
Transcriptional regulation of AQP expression in response to hormonal signalling.
(PDF)
Transcriptional regulation of AQP expression under biotic interactions.
(PDF)
Seasonal or diurnal regulation of AQP expression.
(PDF)
Differential AQP expression in response to other cues.
(PDF)
Listing of Aquaporin-targeting probe sets. The table details for each Affymetrix probe set, its ID, the tools enabling its detection (see “materials and methods” and Figure 1), its target gene ID and its annotation.
(XLS)
Number of probe sets per AQP member.
(XLS)
Acknowledgments
Part of the computations was performed at the Vital-IT (http://www.vital-it.ch) Center for high-performance computing of the SIB Swiss Institute of Bioinformatics.
Funding Statement
This work was supported by the Laboratory of Excellence ARBRE [ANR-11-LABX-02-01], the French Research Agency [POPSEC-project number ANR-Genoplante GPLA06028G] and the Region Lorraine [grant number 12000158]. DC was supported by the European Regional Development Fund [project number 3709]. PEC is an Ambizione fellow of the Swiss National Science Foundation [PZ00P3_136651]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Danielson JA, Johanson U (2008) Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biology 8: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bertl A, Kaldenhoff R (2007) Function of a separate NH3-pore in Aquaporin TIP2;2 from wheat. FEBS Letters 581: 5413–5417. [DOI] [PubMed] [Google Scholar]
- 3. Maurel C, Verdoucq L, Luu DT, Santoni V (2008) Plant aquaporins: Membrane channels with multiple integrated functions. Annual Review of Plant Biology 59: 595–624. [DOI] [PubMed] [Google Scholar]
- 4.Bienert GP, Chaumont F (2011) Plant Aquaporins: Roles in Water Homeostasis, Nutrition, and Signaling Processes. In: Geisler M, Venema K, editors. Transporters and Pumps in Plant Signaling. 3–36.
- 5. Soto G, Fox R, Ayub N, Alleva K, Guaimas F, et al. (2010) TIP5;1 is an aquaporin specifically targeted to pollen mitochondria and is probably involved in nitrogen remobilization in Arabidopsis thaliana. Plant Journal 64: 1038–1047. [DOI] [PubMed] [Google Scholar]
- 6. Chaumont F, Moshelion M, Daniels MJ (2005) Regulation of plant aquaporin activity. Biology of the Cell 97: 749–764. [DOI] [PubMed] [Google Scholar]
- 7. Zardoya R (2005) Phylogeny and evolution of the major intrinsic protein family. Biology of the Cell 97: 397–414. [DOI] [PubMed] [Google Scholar]
- 8. Besse M, Knipfer T, Miller AJ, Verdeil J-L, Jahn TP, et al. (2011) Developmental pattern of aquaporin expression in barley (Hordeum vulgare L.) leaves. Journal of Experimental Botany 62: 4127–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maurel C, Chrispeels MJ (2001) Aquaporins. A molecular entry into plant water relations. Plant Physiology 125: 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sakurai J, Ishikawa F, Yamaguchi T, Uemura M, Maeshima M (2005) Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant and Cell Physiology 46: 1568–1577. [DOI] [PubMed] [Google Scholar]
- 11.Gupta AB, Sankararamakrishnan R (2009) Genome-wide analysis of major intrinsic proteins in the tree plant Populus trichocarpa: Characterization of XIP subfamily of aquaporins from evolutionary perspective. BMC Plant Biology 9. [DOI] [PMC free article] [PubMed]
- 12. Quesada T, Li Z, Dervinis C, Li Y, Bocock PN, et al. (2008) Comparative analysis of the transcriptomes of Populus trichocarpa and Arabidopsis thaliana suggests extensive evolution of gene expression regulation in angiosperms. New Phytologist 180: 408–420. [DOI] [PubMed] [Google Scholar]
- 13. Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604. [DOI] [PubMed] [Google Scholar]
- 14. Barakat A, Bagniewska-Zadworna A, Choi A, Plakkat U, DiLoreto DS, et al. (2009) The cinnamyl alcohol dehydrogenase gene family in Populus: phylogeny, organization, and expression. BMC Plant Biology 9: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rodgers-Melnick E, Mane SP, Dharmawardhana P, Slavov GT, Crasta OR, et al. (2012) Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Research 22: 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu R, Chi X, Chai G, Kong Y, He G, et al. (2012) Genome-wide identification, evolutionary expansion, and expression profile of homeodomain-leucine zipper gene family in poplar (Populus trichocarpa). PLoS One 7: e31149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kohler A, Delaruelle C, Martin D, Encelot N, Martin F (2003) The poplar root transcriptome: analysis of 7000 expressed sequence tags. FEBS Letters 542: 37–41. [DOI] [PubMed] [Google Scholar]
- 18. Marjanovic Z, Uehlein N, Kaldenhoff R, Zwiazek JJ, Weiss M, et al. (2005) Aquaporins in poplar: What a difference a symbiont makes! Planta. 222: 258–268. [DOI] [PubMed] [Google Scholar]
- 19. Secchi F, Zwieniecki MA (2010) Patterns of PIP gene expression in Populus trichocarpa during recovery from xylem embolism suggest a major role for the PIP1 aquaporin subfamily as moderators of refilling process. Plant Cell and Environment 33: 1285–1297. [DOI] [PubMed] [Google Scholar]
- 20. Almeida-Rodriguez AM, Hacke UG, Laur J (2011) Influence of evaporative demand on aquaporin expression and root hydraulics of hybrid poplar. Plant Cell and Environment 34: 1318–1331. [DOI] [PubMed] [Google Scholar]
- 21. Bae EK, Lee H, Lee JS, Noh EW (2011) Drought, salt and wounding stress induce the expression of the plasma membrane intrinsic protein 1 gene in poplar (Populus alba x P. tremula var. glandulosa). Gene 483: 43–48. [DOI] [PubMed] [Google Scholar]
- 22. Hacke UG, Plavcova L, Almeida-Rodriguez A, King-Jones S, Zhou WC, et al. (2010) Influence of nitrogen fertilization on xylem traits and aquaporin expression in stems of hybrid poplar. Tree Physiology 30: 1016–1025. [DOI] [PubMed] [Google Scholar]
- 23. Lopez D, Bronner G, Brunel N, Auguin D, Bourgerie S, et al. (2012) Insights into Populus XIP aquaporins: evolutionary expansion, protein functionality, and environmental regulation. Journal of Experimental Botany 63: 2217–2230. [DOI] [PubMed] [Google Scholar]
- 24. Almeida-Rodriguez AM, Cooke JEK, Yeh F, Zwiazek JJ (2010) Functional characterization of drought-responsive aquaporins in Populus balsamifera and Populus simonii x balsamifera clones with different drought resistance strategies. Physiologia Plantarum 140: 321–333. [DOI] [PubMed] [Google Scholar]
- 25.Janz D, Behnke K, Schnitzler JP, Kanawati B, Schmitt-Kopplin P, et al.. (2010) Pathway analysis of the transcriptome and metabolome of salt sensitive and tolerant poplar species reveals evolutionary adaption of stress tolerance mechanisms. BMC Plant Biology 10. [DOI] [PMC free article] [PubMed]
- 26. Kreuzwieser J, Hauberg J, Howell KA, Carroll A, Rennenberg H, et al. (2009) Differential Response of Gray Poplar Leaves and Roots Underpins Stress Adaptation during Hypoxia. Plant Physiology 149: 461–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wilkins O, Waldron L, Nahal H, Provart NJ, Campbell MM (2009) Genotype and time of day shape the Populus drought response. Plant Journal 60: 703–715. [DOI] [PubMed] [Google Scholar]
- 28. Street NR, James TM, James T, Mikael B, Jaakko K, et al. (2011) The physiological, transcriptional and genetic responses of an ozone-sensitive and an ozone tolerant poplar and selected extremes of their F2 progeny. Environmental Pollution 159: 45–54. [DOI] [PubMed] [Google Scholar]
- 29. Cohen D, Bogeat-Triboulot MB, Tisserant E, Balzergue S, Martin-Magniette ML, et al. (2010) Comparative transcriptomics of drought responses in Populus: a meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genomics 11: 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goodstein DM, Shu SQ, Howson R, Neupane R, Hayes RD, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40: D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2011) GenBank. Nucleic Acids Research 39: D32–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 33. Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, et al. (2011) ArrayExpress update–an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Research 39: D1002–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- 36. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biology 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stalteri MA, Harrison AP (2007) Interpretation of multiple probe sets mapping to the same gene in Affymetrix GeneChips. BMC Bioinformatics 8: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tang H, Bowers JE, Wang X, Ming R, Alam M, et al. (2008) Synteny and collinearity in plant genomes. Science 320: 486–488. [DOI] [PubMed] [Google Scholar]
- 39. Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, et al. (2009) PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell 21: 3718–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schneider A, Cannarozzi GM, Gonnet GH (2005) Empirical codon substitution matrix. BMC Bioinformatics 6: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alexandersson E, Fraysse L, Sjovall-Larsen S, Gustavsson S, Fellert M, et al. (2005) Whole gene family expression and drought stress regulation of aquaporins. Plant Molecular Biology 59: 469–484. [DOI] [PubMed] [Google Scholar]
- 42. Johnson KD, Herman EM, Chrispeels MJ (1989) An abundant, highly conserved tonoplast protein in seeds. Plant Physiology 91: 1006–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maeshima M, Haranishimura I, Takeuchi Y, Nishimura M (1994) Accumulation of vacuolar H+-Pyrophosphatase and H+-ATPaseduring reformation of the central vacuole in germinating pumpkin seeds. Plant Physiology 106: 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oliviusson P, Salaj J, Hakman I (2001) Expression pattern of transcripts encoding water channel-like proteins in Norway spruce (Picea abies). Plant Molecular Biology 46: 289–299. [DOI] [PubMed] [Google Scholar]
- 45. Vander Willigen C, Postaire O, Tournaire-Roux C, Boursiac Y, Maurel C (2006) Expression and inhibition of aquaporins in germinating Arabidopsis seeds. Plant and Cell Physiology 47: 1241–1250. [DOI] [PubMed] [Google Scholar]
- 46. Gattolin S, Sorieul M, Frigerio L (2011) Mapping of Tonoplast Intrinsic Proteins in Maturing and Germinating Arabidopsis Seeds Reveals Dual Localization of Embryonic TIPs to the Tonoplast and Plasma Membrane. Molecular Plant 4: 180–189. [DOI] [PubMed] [Google Scholar]
- 47. Secchi F, Maciver B, Zeidel ML, Zwieniecki MA (2009) Functional analysis of putative genes encoding the PIP2 water channel subfamily in Populus trichocarpa. Tree Physiology 29: 1467–1477. [DOI] [PubMed] [Google Scholar]
- 48. Goué N, Lesage-Descauses MC, Mellerowicz EJ, Magel E, Label P, et al. (2008) Microgenomic analysis reveals cell type-specific gene expression patterns between ray and fusiform initials within the cambial meristem of Populus. New Phytologist 180: 45–56. [DOI] [PubMed] [Google Scholar]
- 49. Song D, Xi W, Shen J, Bi T, Li L (2011) Characterization of the plasma membrane proteins and receptor-like kinases associated with secondary vascular differentiation in poplar. Plant Molecular Biology 76: 97–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Luu DT, Maurel C (2005) Aquaporins in a challenging environment: molecular gears for adjusting plant water status. Plant Cell and Environment 28: 85–96. [Google Scholar]
- 51. Aroca R, Porcel R, Ruiz-Lozano JM (2012) Regulation of root water uptake under abiotic stress conditions. Journal of Experimental Botany 63: 43–57. [DOI] [PubMed] [Google Scholar]
- 52. Raj S, Brautigam K, Hamanishi ET, Wilkins O, Thomas BR, et al. (2011) Clone history shapes Populus drought responses. Proceedings of the National Academy of Sciences of the United States of America 108: 12521–12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Parent B, Hachez C, Redondo E, Simonneau T, Chaumont F, et al. (2009) Drought and Abscisic Acid Effects on Aquaporin Content Translate into Changes in Hydraulic Conductivity and Leaf Growth Rate: A Trans-Scale Approach. Plant Physiology 149: 2000–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Siemens JA, Zwiazek JJ (2008) Root hydraulic properties and growth of balsam poplar (Populus balsamifera) mycorrhizal with Hebeloma crustuliniforme and Wilcoxina mikolae var. mikolae. Mycorrhiza 18: 393–401. [DOI] [PubMed] [Google Scholar]
- 55. Gehring CA, Mueller RC, Whitham TG (2006) Environmental and genetic effects on the formation of ectomycorrhizal and arbuscular mycorrhizal associations in cottonwoods. Oecologia 149: 158–164. [DOI] [PubMed] [Google Scholar]
- 56. Rinaldi C, Kohler A, Frey P, Duchaussoy F, Ningre N, et al. (2007) Transcript profiling of poplar leaves upon infection with compatible and incompatible strains of the foliar rust Melampsora larici-populina. Plant Physiology 144: 347–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zou J, Rodriguez-Zas S, Aldea M, Li M, Zhu J, et al. (2005) Expression profiling soybean response to Pseudomonas syringae reveals new defense-related genes and rapid HR-specific downregulation of photosynthesis. Mol Plant Microbe Interact 18: 1161–1174. [DOI] [PubMed] [Google Scholar]
- 58. Vandeleur RK, Mayo G, Shelden MC, Gilliham M, Kaiser BN, et al. (2009) The Role of Plasma Membrane Intrinsic Protein Aquaporins in Water Transport through Roots: Diurnal and Drought Stress Responses Reveal Different Strategies between Isohydric and Anisohydric Cultivars of Grapevine. Plant Physiology 149: 445–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Voicu MC, Zwiazek JJ (2011) Diurnal and seasonal changes of leaf lamina hydraulic conductance in bur oak (Quercus macrocarpa) and trembling aspen (Populus tremuloides). Trees-Structure and Function 25: 485–495. [Google Scholar]
- 60. Javot H, Lauvergeat V, Santoni V, Martin-Laurent F, Guclu J, et al. (2003) Role of a single aquaporin isoform in root water uptake. Plant Cell 15: 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Madsen SS, Olesen JH, Bedal K, Engelund MB, Velasco-Santamaria YM, et al. (2011) Functional characterization of water transport and cellular localization of three aquaporin paralogs in the salmonid intestine. Front Physiol 2: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Raldua D, Otero D, Fabra M, Cerda J (2008) Differential localization and regulation of two aquaporin-1 homologs in the intestinal epithelia of the marine teleost Sparus aurata. Am J Physiol Regul Integr Comp Physiol 294: R993–1003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of “sibling” probe sets reveals the sensitivity of median-based procedure for the automatic extraction of gene-centred information in the case of multi-experiment comparison.
(PDF)
Root- and leaf-preferred expression of AQPs.
(PDF)
Young leaf- and mature leaf-preferred expression of AQPs.
(PDF)
Transcriptional regulation of AQP expression under abiotic challenges.
(PDF)
Transcriptional regulation of AQP expression in response to hormonal signalling.
(PDF)
Transcriptional regulation of AQP expression under biotic interactions.
(PDF)
Seasonal or diurnal regulation of AQP expression.
(PDF)
Differential AQP expression in response to other cues.
(PDF)
Listing of Aquaporin-targeting probe sets. The table details for each Affymetrix probe set, its ID, the tools enabling its detection (see “materials and methods” and Figure 1), its target gene ID and its annotation.
(XLS)
Number of probe sets per AQP member.
(XLS)