Abstract
Intraspecific mate selectivity often is enforced by self-incompatibility (SI), a barrier to self-pollination that inhibits productive pollen-pistil interactions. In the Brassicaceae, SI specificity is determined by two highly-polymorphic proteins: the stigmatic S-locus receptor kinase (SRK) and its pollen coat-localized ligand, the S-locus cysteine-rich protein (SCR). Arabidopsis thaliana is self fertile, but several of its accessions can be made to express SI, albeit to various degrees, by transformation with functional SRK-SCR gene pairs isolated from its close self-incompatible relative, Arabidopsis lyrata. Here, we use a newly identified induced mutation that suppresses the SI phenotype in stigmas of SRK-SCR transformants of the Col-0 accession to investigate the regulation of SI and the SRK transgene. This mutation disrupts NRPD1a, a gene that encodes a plant-specific nuclear RNA polymerase required for genomic methylation and production of some types of silencing RNAs. We show that NRPD1a, along with the RNA-dependent RNA polymerase RDR2, is required for SI in some A. thaliana accessions. We also show that Col-0 nrpd1a mutants exhibit decreased accumulation of SRK transcripts in stigmas, which is not, however, responsible for loss of SI in these plants. Together, our analysis of the nrpd1a mutation and of SRK promoter activity in various accessions reveals that the SRK transgene is subject to several levels of regulation, which vary substantially by tissue type and by accession. This study thus helps explain the well-documented differences in expression of SI exhibited by SRK-SCR transformants of different A. thaliana accessions.
Keywords: self-incompatibility, S-locus receptor kinase, NRPD1a, Arabidopsis thaliana
In flowering plants, out-crossing often is enforced by genetic self-incompatibility (SI), a barrier to self-fertilization that prevents pollination of the pistil by pollen from genetically related plants while allowing pollination by pollen of dissimilar genotype. In the Brassicaceae family, SI is controlled by haplotypes of the S locus, and self-pollination is prevented when pollen and stigma are derived from plants that express the same S-locus variant. Two S-locus genes are known to be essential for specific recognition of “self” pollen in the SI response of this family: the S-locus receptor kinase (SRK) and the S-locus cysteine-rich protein (SCR) genes (reviewed in Tantikanjana et al. 2010). SRK is a receptor kinase localized in the plasma membrane of stigma epidermal cells, and its SCR ligand is located in the pollen coat. An SCR variant can bind and activate only the SRK variant encoded by the same S-locus haplotype. This “self” SRK-SCR interaction is thought to trigger a poorly understood signal transduction cascade within the stigma epidermal cell that prevents pollen hydration and germination as well as growth of the pollen tube into the stigma epidermal cell wall (Tantikanjana et al. 2010).
In recent years, Arabidopsis thaliana has emerged as a new model system for studies of SI in the Brassicaceae (Nasrallah et al. 2002, 2004; Boggs et al. 2009; Tantikanjana et al. 2009). Although A. thaliana is normally self-fertile, it can be made to express SI by transformation with SRK and SCR gene pairs isolated from its close self-incompatible relatives Arabidopsis lyrata and Capsella grandiflora (Nasrallah et al. 2002; Boggs et al. 2009). Importantly, the SI response exhibited by SRK-SCR transformants of several A. thaliana accessions, including C24, Cvi, Kas, Hodja, and Sha, is identical in strength and developmental regulation to the SI response observed in naturally self-incompatible A. lyrata and other members of the Brassicaceae: it is manifested by inhibition of “self” pollen grains at the stigma surface, it is first observed in stigmas prior to flower opening (a stage that corresponds to stage 13 of flower development in A. thaliana), and it is sustained throughout stigma development, resulting in very low, if any, seed set (Nasrallah et al. 2004; Boggs et al. 2009). In contrast, the stigmas of SRK-SCR transformants of some other accessions, such as Col-0 and Rld, express transient SI: they exhibit a robust SI response only during a narrow window of stigma development (stage 13 and early stage 14), after which the ability of their stigmas to inhibit “self” pollen is weakened and they set seed (Nasrallah et al. 2002).
As the determinant of SI in the stigma, SRK controls the strength of SI and its proper regulation during stigma development (Tantikanjana et al. 2010). For example, the transient SI phenotype characteristic of some A. thaliana accessions is determined primarily by a hypomorphic allele of the PLANT U-BOX8 (PUB8) gene, which regulates SRK expression: SRK-SCR plants homozygous for this hypomorphic PUB8 allele accumulate suboptimal levels of SRK transcripts at later stages of stigma development (Liu et al. 2007). However, a full understanding of the SI response in the transgenic A. thaliana SI model and its variable expression in different accessions requires an as-yet-unavailable detailed characterization of SRK transgene expression and the factors essential for its proper regulation. We therefore analyzed the regulation of the SRK transgene and the activity of its promoter in various A. thaliana accessions. This study was spurred by our identification of a newly identified recessive mutation that suppresses the SI phenotype of SRK-SCR transformants of the Col-0 accession. The mutation disrupts NRPD1a, a gene that encodes a plant-specific nuclear RNA polymerase required for genomic methylation and production of some types of silencing RNAs. Here we report on our analysis of this mutation in the context of SI. We show that NRPD1a plays a varied and complex role in the epigenetic regulation of SI in SRK-SCR transformants, possibly by acting on an accession-specific factor. By analyzing other genes known to be involved in gene silencing, we identify RDR2 as an additional requirement for the SI response in some accessions. We also show that differences among A. thaliana accessions for expression of SI are correlated with the activity of the SRK promoter.
Materials and Methods
Plant material and growth conditions
Transgenic plants belonging to the Col-0 accession and transformed with SRKb and SCRb, the SRK and SCR alleles derived from the Sb haplotype of A. lyrata, were previously described (Nasrallah et al. 2002; Tantikanjana et al. 2009). For plant growth, seeds were plated on MS media under sterile conditions and stratified for 3 d at 4°. Kanamycin at 25 μg/μL was used for selection of plants harboring the SRKb and SCRb transgenes. Seedlings were transplanted to soil once they had four true leaves, and plants were grown under long-day conditions (16-hr days/8-hr nights) at a temperature of 20° in a controlled-environment chamber.
Pollination assays
Pollination tests were performed on stigmas of floral buds at stage 13 of flower development (staging according to Smyth et al. 1990), which corresponds to 1 d prior to flower opening and is hereafter referred to as the −1 bud stage. To identify plants exhibiting a breakdown of SI, pollination tests were performed typically by pollinating three −1 bud-stage stigmas per individual plant with pollen from a plant known to express functional SCRb, as reflected by inhibition of its pollen on the stigmas of plants expressing SRKb. Pollinated buds were incubated at room temperature for 2 hr and subsequently processed for visualization with a ultraviolet-fluorescence microscope as described previously (Kho and Baer 1968). Under the aforementioned conditions, very few (<5) pollen tubes are observed on the stigmas of wild-type (WT) SRKb plants. Plants whose stigmas exhibited more than 20 pollen tubes that penetrated the epidermal cell wall were considered to exhibit a breakdown of the SI response and were scored as self-compatible.
Plant DNA gel blot analysis for identification of plants carrying single transgene integrations
Genomic DNA from leaf tissue was extracted using the CTAB method (Doyle and Doyle 1987) and digested overnight with EcoRI (New England Biolabs, Ipswich, MA). Capillary transfers were performed under alkaline conditions overnight onto Hybond N+ membrane (GE Healthcare, Bio-Sciences Corp., Piscataway, NJ). Membranes were probed with a 32P-labeled 1.6-kb probe derived from the 3′ UTR of the SRKb gene. Blots were exposed to phosphor screens (GE Healthcare Bio-Sciences Corp.) and scanned with a STORM 860 PhosphorImager (GE Healthcare, Bio-Sciences Corp.).
Map-based cloning and DNA sequencing
A mapping population of 1600 F2 plants was generated by crossing a plant homozygous for both the SRKb and SCRb transgenes and the mutation described in this paper as female parent to a WT plant of the Landsberg erecta (Ler-0) accession. Plants carrying the SRKb transgene were identified by polymerase chain reaction (PCR) amplification of SRKb using genomic DNA isolated from leaves with gene-specific primers as follows: (SRKbhvrF) 5′-TGGGTTGGGATGTCAAGAAAG-3′ and (SRKbhvrR) 5′-CAACTTCATCTTTCTCAGGCACAA-3′. F2 plants carrying the SRKb gene were analyzed by pollinating −1-stage stigmas with pollen from a plant known to express functional SCRb (hereafter SCRb pollen). Plants showing a consistent loss of SI in these stigmas were genotyped using simple-sequence length repeats and cleaved amplified polymorphism markers listed on The Arabidopsis Information Resource website (Supporting Information, Table S1). Additional simple-sequence length repeats and single-nucleotide polymorphism markers were generated using the Landsberg erecta random sequence database (www.tigr.org; Table S1). A total of 125 individuals informative for mapping were found. In addition, 28 F3 plants were screened both for mapping purposes and to confirm F2 phenotypes. In this manner, the mutation was mapped to a 165-kb region on chromosome 1. To identify the gene disrupted by the mutation within the mapping interval, gene-specific primers were designed for all 39 genic regions found in the interval. PCR products were sequenced at the Cornell Bioresource Center (Ithaca, NY) using big dye terminator chemistry and AmpliTaq-FS DNA polymerase and an Applied Biosystems 3730xl DNA analyzer.
Two Col-0 SALK lines (Alonso et al. 2003), SALK_143437 and SALK_128428, each containing a transfer DNA (T-DNA) insertion in the NRPD1a gene were obtained from the Arabidopsis Biological Resource Center (ABRC, Columbus, OH). Plants homozygous for each of the T-DNA insertions were crossed to a plant homozygous for the targeted mutation and for the SRKb and SCRb transgenes for complementation assays. Additionally, the following Col-0 strains carrying T-DNA insertions in several other genes involved in silencing pathways were obtained from the ABRC (mutant designation in parentheses): SALK_059661 (rdr2), SALK_071772 (ago4), and SALK_076129 (nrpd1b). The C24 nrpd1a ros1-1 double mutant was obtained from Craig Pikaard (Indiana University, Bloomington, IN). These lines were screened for homozygosity using gene-specific primers (Table S2). Plants homozygous for each of the T-DNA insertions were crossed to a Col-0 WT plant homozygous for the SRKb and SCRb transgenes, hereafter designated Col-0 WT[SRKb-SCRb] plant.
Analysis of genomic DNA methylation by chop PCR
The DNA methylation status of plants was assayed as follows. Genomic DNA isolated from leaf tissue was digested overnight with the methylation-sensitive enzyme, HaeIII, followed by amplification with specific primers. PCR primers for the methylated AtSN1 repeat and for the non-methylated At2g12990 gene were as described in a previous study (Hamilton et al. 2002).
RNA gel blot analysis
For RNA gel blot analysis, the following tissues were collected from 100 −1-stage flower buds: stigmas (collected by cutting pistils at the stigma-style boundary), styles (pistils lacking stigmas), petals, anthers, and sepals. PolyA RNA was extracted using the FastTrak RNA isolation kit (Invitrogen, Carlsbad, CA) and subjected to gel blot analysis as described previously (Kusaba et al. 2001). The blots were hybridized with a 32P-labeled probe generated from the first exon of SRKb and subsequently with a 32P-labeled actin probe. Visualization of hybridization signals was as described previously for DNA gel blot analysis. Signal intensities were quantified using the ImageQuant software package and normalized using actin hybridization signals.
Reverse transcription (RT)-PCR
For quantitative real-time RT-PCR of SRKb transcripts, RNA was isolated from 50 stigmas or styles using the Trizol reagent (Invitrogen). Three replicate RNA samples for each genotype were prepared. For each sample, the RNA was treated with DNase I and 1 μg was used for cDNA synthesis using the First Strand cDNA Synthesis Kit for Real-time PCR (United States Biochemical, Cleveland, OH) and oligo(dT) primers. Real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) on an ABI Prism 7900HT sequence detection system. The primers used for real-time PCR were as follows: for SRKb (rt-SRKb4), 5′-CTAAGCCTTGATTCTCATCTCTTTACA-3′ and 5′-GAAGTCCCCGAGCAATACCAT-3′; and for ubiquitin, gene-specific primers described in a previous study (Liu et al. 2007). To confirm that no genomic DNA remained in the RNA samples, both ubiquitin and SRKb primers were designed to span an intron. Results were analyzed using the Sequence Detection Systems software package (Applied Biosystems, Foster City, CA). The relative amounts of transcripts were calculated using the comparative ct method and normalized to ubiquitin (Livak and Schmittgen 2001). The mean values were calculated from three replicate samples collected on different days. The statistical significance of differences between genotypic groups was assessed using the Student’s t-test.
Analysis of SRKb promoter activity using the GUS reporter
An SRKbpr::uidA::nos gene was constructed as follows: a fragment corresponding to a 950-base pair region upstream of the SRKb initiating methionine codon was amplified by PCR from an SRKb-containing plasmid using the forward primer 5′- ATTGTTAGTTCTTTCATCAGTTCG-3′ and the reverse primer 5′- CACTTTCCCATGGCTCTCCTTC-3′. The reporter gene was introduced into plants of the Col-0, C24, Cvi-0, Hodja, Kas, Ler-0, Rld, and Sha accessions and into Col-0 nrpd1a[SRKb-SCRb] plants. In addition, the previously described chimeric Ats1pr::uidA::nos gene (Dwyer et al. 1994) was introduced into Col-0 and C24 as a control stigma-specific reporter. All transformations were performed using the Agrobacterium-mediated floral dip method (Clough and Bent 1998). Selection was on MS medium containing 40 μg/μL hygromycin for SRKbpr::uidA::nos transformants and 25 ug/ul kanamycin for Ats1pr::uidA::nos transformants. Homozygous plants carrying a single transgene integration were identified by DNA gel blot analysis and progeny screening on selective media. Histochemical assays for GUS activity were performed as described previously (Kusaba et al. 2001).
To quantitate GUS transcripts, real-time RT-PCR was performed using total RNA isolated from stigmas harvested from 30 floral buds. Real-time RT-PCR was performed as described previously using the GUS gene-specific primers 5′-TCCTACCGTACCTCGCATTACC-3′ and 5′-GACAGCAGCAGTTTCATCAATCAC-3′. Real-time RT-PCR of ubiquitin transcripts used primers described previously (Liu et al. 2007).
Results
Identification of NRPD1a as a modifier of SI in Col-0[SRKb] plants
Col-0 plants harboring the SRKb and SCRb genes, which are the SRK and SCR alleles isolated from the Sb haplotype of A. lyrata, express a robust SI response in −1-stage floral buds (Nasrallah et al. 2002). To identify mutations that cause loss of SI in these plants, we used a previously described Col-0 homozygous strain carrying SRKb and SCRb genes integrated at single but unlinked positions in the genome (Nasrallah et al. 2002). Seeds from this strain were mutagenized with ethyl methane sulfonate, and M2 plants derived from these mutagenized plants were screened by manual self-pollination of stigmas at the −1-bud stage as previously described (see Materials and Methods) (Tantikanjana et al. 2009). This screen identified a mutation, designated self-compatible 1 (sc1), which caused breakdown of SI in −1-stage floral buds of SRKb transformants. Pollination of sc1 mutants carrying the SRKb transgene (hereafter sc1[SRKb]) stigmas with pollen from WT plants harboring the SCRb transgene (hereafter WT[SCRb]) resulted in profuse pollen tube growth, whereas pollination of WT[SRKb] plants with sc1[SRKb] pollen did not result in pollen tube growth, indicating that the self-compatible phenotype of sc1[SRKb] plants was due to loss of stigma SI function. Crosses between an sc1[SRKb] mutant plant and WT[SRKb] restored SI in the −1-stage stigmas of F1 plants, demonstrating that the sc1 mutation is recessive. Additionally, when the sc1[SRKb] mutant was crossed to a Col-0 WT plant, the F1 plants were self-incompatible, demonstrating that the mutant phenotype is not the result of a loss-of-function mutation in the SRKb transgene.
Using the map-based cloning strategy described in Materials and Methods, we mapped the sc1 mutation to a 165.5-kb region between markers F16P17b and F9N12c on chromosome 1 (Table S1). Sequencing of the 39 genes within this region revealed a cytosine-to-thymine transition at position 3188 in the eighth exon of the NRPD1a gene, which results in a glutamine-to-asparagine substitution. The sc1[SRKb] mutant was then crossed to two Col-0 SALK lines (Alonso et al. 2003), SALK_143437 and SALK_128428, which contain a T-DNA insertion in the NRPD1a gene within exon 14 and exon 9, respectively (Swarbreck et al. 2007). The inability of both T-DNA lines to complement the phenotype of sc1[SRKb] plants demonstrates that a defect in NRPD1a is responsible for the breakdown of SI in this mutant. Because seven mutations (nrpd1a-1 to nrpd1a-7) had already been identified in NRPD1a, the sc1 mutation was named nrpd1a-8.
NRPD1a is a plant-specific nuclear RNA polymerase that functions in various silencing phenomena including the generation of small RNAs (Herr et al. 2005), the methylation of DNA (Onodera et al. 2005), and the spread and reception of the silencing signal (Palusa et al. 2007; Scascitelli et al. 2010). Alignments of the predicted amino-acid sequences of NRPD1a and RNA polymerase II from yeast had identified the putative structural domains of NRPD1a (Herr et al. 2005). Based on these alignments, the nrpd1a-8 mutation is located in the region homologous to the binding site of transcription factor IIb in yeast RNA polymerase II (Herr et al. 2005), specifically at amino acid 614 in the putative dock domain (Herr et al. 2005).
NRPD1a, DNA methylation, and the regulation of SRKb gene expression
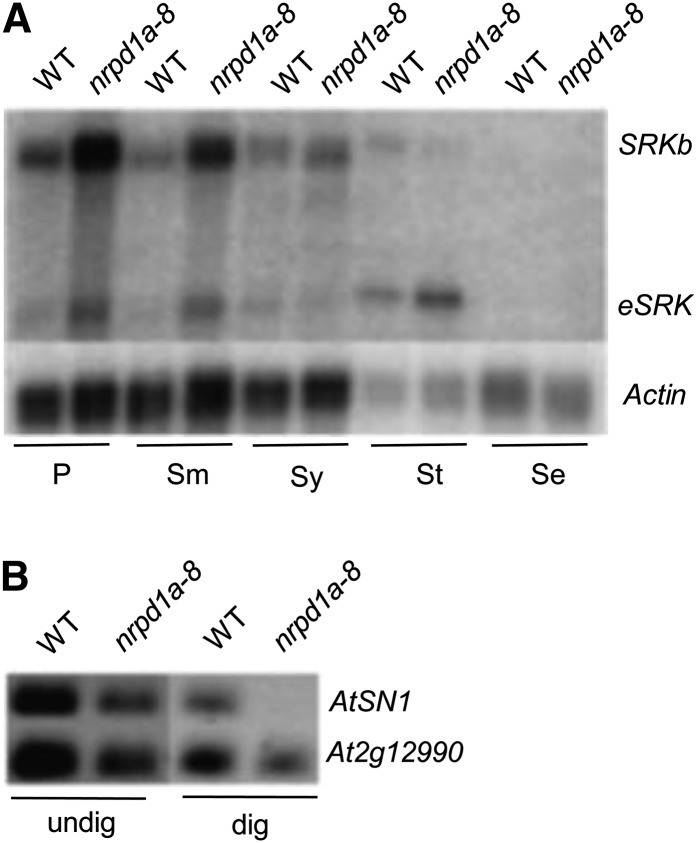
Suboptimal levels of SRK in stigmas are known to lead to weakening or loss of SI (Liu et al. 2007). Consequently, although nrpd1a mutations are expected to relieve the silencing of transgenes and therefore to increase the expression level of the SRKb transgene, we explored the possibility that the self-compatible phenotype of −1-stage stigmas in nrpd1a-8[SRKb] mutant plants might be due to reduced SRKb transcripts. Accordingly, we compared the relative amounts of SRKb transcripts in nrpd1a mutants harboring the SRKb transgene (designated nrpd1a-8[SRKb]) and WT[SRKb] plants. As previously described and shown by the gel blots in Figure 1A, the SRKb gene, like other SRK genes, produces two transcripts: a 3.0-kb transcript that encodes the full-length SRK protein and a 1.6-kb transcript, designated eSRK, which corresponds to the first exon of the gene and encodes a soluble form of the SRK extracellular domain whose function is not understood (Stein et al. 1991; Dixit et al. 2000; Kusaba et al. 2001). Furthermore, the SRKb gene is expressed in various floral tissues: in addition to stigmas, SRKb transcripts were detected in the styles, stamens, and petals of Col-0 WT[SRKb] plants (Figure 1A). This pattern of expression is comparable with the native expression pattern of the SRKb gene in A. lyrata (Figure S1), with the exception of expression in stamens, which might be due to position effects of the site of transgene integration in Col-0 WT[SRKb] plants.
Figure 1 .
Analysis of SRKb expression and genomic DNA methylation in nrpd1a-8 mutants. (A) Expression of SRKb and eSRKb in various floral tissues of nrpd1a-8 mutants and WT. P, petal; Sm, stamen; Sy, style; St, stigma; Se, sepal. (B) Loss of genomic methylation at the AtSN1 retroelement in nrpd1a-8 mutants. undig, undigested; dig, digested.
Comparison of nrpd1a-8[SRKb] and WT[SRKb] plants showed that the nrpd1a-8 mutation did affect the steady-state levels of SRKb transcripts, albeit in unexpected ways. In stigmas, the levels of the ~3-kb SRKb transcript were approximately 2.2-fold lower in nrpd1a-8[SRKb] plants than in WT[SRKb] plants, whereas the 1.6-kb SRKb transcripts were approximately 2-fold more abundant in nrpd1a-8[SRKb] plants than in WT[SRKb] plants (Figure 1A). In contrast, both SRKb and eSRKb transcripts were elevated in the styles, petals, and stamens of nrpd1a-8[SRKb] mutants relative to WT[SRKb] plants (Figure 1A). The nrpd1a-8 mutation did not, however, cause accumulation of SRKb transcripts in sepals, where the SRKb gene is not normally expressed (Figure 1A).
NRPD1a is known to function in the production of natural antisense (NAT) RNA and in DNA methylation (reviewed in Vaucheret 2006), both of which can have significant effects on gene expression. A previous study had shown that mutations in several genes of the NAT pathway did not exhibit loss of the SI response in Col-0[SRKb-SCRb] plants (Tantikanjana et al. 2009), indicating that NRPD1a does not exert its effect on SI via this pathway. We therefore assessed the possibility that NRPD1a regulates SI by regulating the methylation status, and therefore expression, of the SRKb gene or other genes that function in SI. We first examined nrpd1a-8 homozygotes to determine whether they exhibit a loss of DNA methylation similar to previously analyzed nrpd1a mutants. Toward this end, we used a standard method that assays methylation of the highly methylated AtSN1 retroelement. In this so-called “chop PCR” method (Onodera et al. 2005), genomic DNA is digested with the methylation-sensitive enzyme HaeIII before PCR amplification with AtSN1-specific primers, and an amplification product is obtained only if the DNA is methylated and therefore insensitive to HaeIII digestion. The At2g12990 gene, which contains no methylation sites, is used as a control. Figure 1B shows that AtSN1 fragments could not be amplified from HaeIII-digested genomic DNA of nrpd1a-8 homozygotes, indicating that the nrpd1a-8 mutation causes loss of genomic DNA methylation similar to other nrpd1a mutations.
To examine further the role of DNA methylation in the regulation of SI, strains containing T-DNA insertions in genes known to function in the DNA methylation pathway, including NRPD1a, NRPD1b, RDR2, and AGO4 (Matzke et al. 2009), were each crossed to a WT Col-0 plant carrying a single integration of a linked SRKb-SCRb gene pair (Nasrallah et al. 2004). NRPD1b is an alternative large subunit of Pol IV (Pontier et al. 2005), whereas RDR2 amplifies RNAs cleaved by AGO4 (Matzke et al. 2009). Based on the chop PCR assay, plants homozygous for the T-DNA insertion in each of these genes showed the expected loss of methylation of the AtSN1 retroelement (Figure S2). For each mutant line, F2 progenies that contained the SRKb-SCRb transgenes and were homozygous for the insertional mutation were analyzed by pollination assays of stigmas at the −1 bud stage. Of the mutants tested, only rdr2[SRKb-SCRb] plants exhibited a stigma-specific breakdown of SI similar to nrpd1a[SRKb-SCRb] plants. All other mutants exhibited an intense SI response in stigmas at the -1-bud stage.
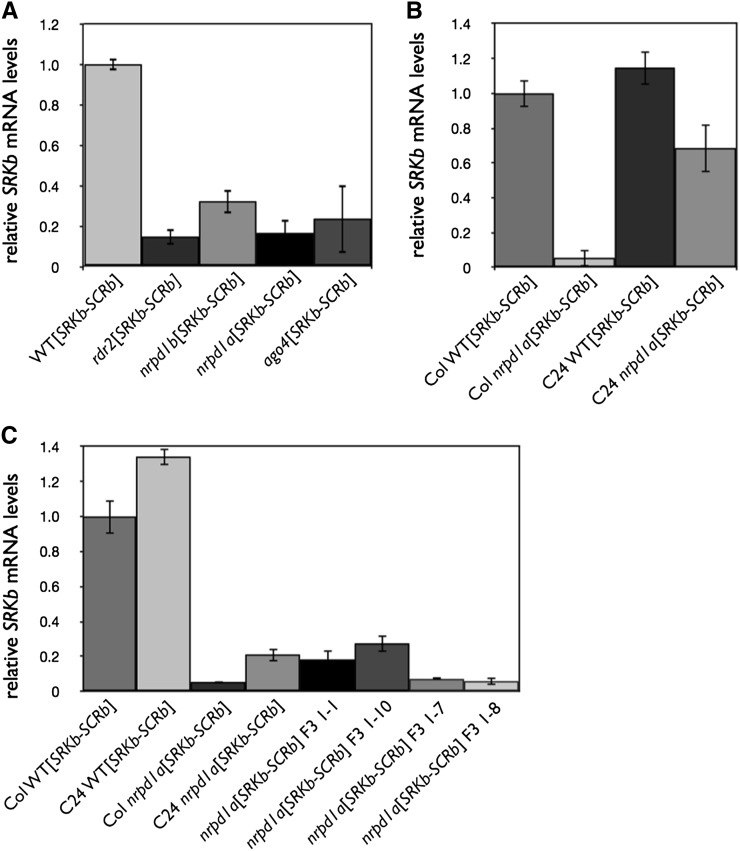
The different effects on SI of the nrpd1a and rdr2 mutations on the one hand and of mutations in genes of the NAT and DNA methylation pathways on the other hand might be due to differences in the effect of these mutations on SRKb transcript levels. To address this issue, we compared SRKb transcript levels in the self-compatible nrpd1a[SRKb-SCRb] and rdr2[SRKb-SCRb] plants and in the self-incompatible nrpd1b[SRKb-SCRb] and ago4[SRKb-SCRb] plants by real-time PCR using primers complementary to sequences in exons 4 and 5 and therefore specific for the full-length SRKb transcript. All methylation mutants tested were found to have a significant decrease in expression of full-length SRKb transcripts in the stigma (Figure 2A). The self-incompatible −1-bud stigmas of nrpd1b[SRKb-SCRb] and ago4[SRKb-SCRb] plants tended to have slightly greater expression than the self-compatible −1-bud stigmas of nrpd1a[SRKb-SCRb] and rdr2[SRKb-SCRb] plants. However, this difference was not significant in comparisons of ago4[SRKb-SCRb] with the self-compatible nrpd1a[SRKb-SCRb] (P = 0.8) and rdr2[SRKb-SCRb] plants (P = 0.4).
Figure 2 .
Effect of various mutations on the levels of SRKb transcripts in the stigmas of plants homozygous for the SRKb-SCRb trasngenes and for various genomic methylation mutations. Average fold-change values relative to WT[SRKb-SCRb] are shown for (A) rdr2, nrpd1b, nrpd1a, and ago4 mutants in the Col-0 background; (B) Col-0 nrpd1a [SRKb-SCRb] and C24 nrpd1a[SRKb-SCRb]; (C) Col-0 nrpd1a-8 × C24[SRKb-SCRb] F3 progeny. Among nrpd1a[SRKb-SCRb] F3 plants, plants 1−1 and 1−10 were self-incompatible whereas plants 1−7 and 1−8 were self-compatible.
Differential regulation of SRKb in different A. thaliana accessions
To determine whether disruption of the NRPD1a gene could cause breakdown of SI in SRKb-SCRb transformants of accessions that express a developmentally stable SI response, an nrpd1a mutant in the C24 background (Zheng et al. 2008) was obtained and crossed to a C24[SRKb-SCRb] plant. All 16 F2 plants that inherited the SRKb-SCRb transgenes were self-incompatible as demonstrated by little to no seed set. Among these plants, three nrpd1a[SRKb-SCRb] homozygotes showed a decrease in stigma SRKb transcript levels relative to C24 WT[SRKb-SCRb] (Figure 2B), despite having a self-incompatible phenotype. However, SRKb transcript levels in C24 nrpd1a[SRKb-SCRb] homozygotes were still ~12 fold greater than those observed in Col-0 nrpd1a[SRKb-SCRb] stigmas.
In parallel, a cross was made between a Col-0 nrpd1a-8[SRKb] homozygote and a C24 WT[SRKb-SCRb] homozygote, both of which contained a single integration of the SRKb or SRKb-SCRb transgenes. The F1 progeny of this cross were fully self-incompatible at all stages of stigma development. A total of 50 F2 plants from four different F1 plants were analyzed, and F2 plants were recovered whose stigmas did not inhibit SCRb pollen despite harboring the SRKb transgene. The ratios of “compatible” (C; stigmas failed to inhibit SCRb pollen) to “incompatible” (I; stigmas inhibited SCRb pollen) plants in these families were: 3 C:11 I in Family 1; 0 C:11 I in Family 2; 1 C:12 I in Family 3; and 1 C:11 I in Family 6. All of the “compatible” plants were homozygous for the nrpd1a-8 mutation, and their F3 progenies were uniformly “compatible”. However, some F2 nrpd1a-8 homozygotes were ”incompatible” (five of eight plants in Family 1; five of five plants in Family 2; two of three plants in Family 3; one of two plants in Family 6). All of the F2 SRKb plants that were homozygous or heterozygous for the NRPD1a WT allele were “incompatible.” Among F2 nrpd1a-8[SRKb] plants, the overall phenotypic ratio of “incompatible” to “compatible” plants was 2.6:1 (13 I:5 C). This ratio is not statistically different from the 3:1 ratio expected for segregation of a recessive allele at a locus other than NRPD1a that is required for SI (P = 0.23). Interestingly, all F2 nrpd1a-8[SRKb] plants tested, whether they were “incompatible” or “compatible,” were found to exhibit loss of AtSN1 methylation (Table S2). Additionally, based on DNA gel blot analysis, there was no correlation among F2 plants between SI phenotype and the source of the SRKb transgene (i.e., whether it was derived from the Col-0 or C24 parent) or the number of SRKb transgene integrations (i.e., whether the plant contained the Col-0-derived transgene, the C24-derived transgene, or both) (Table S2). Similar results were obtained in a cross between a Col-0 nrpd1a-8[SRKb] homozygote and an SRKb-SCRb transformant of Sha, an accession that expresses developmentally-stable SI like C24. In this cross, some but not all nrpd1a F2 plants exhibited a self-compatible phenotype with a ratio of self-incompatible to self-compatible plants of 7:2 (Table S3).
To determine whether variation in SRKb expression may be responsible for the observed phenotypes in these F2 populations, F3 nrpd1a homozygotes derived by selfing F2 individuals derived from the Col-0 nrpd1a-8[SRKb] × C24 WT[SRKb-SCRb] cross were generated and subjected to real-time PCR using stigma RNA. Among four F3 nrpd1a homozygotes tested (Figure 2C), two plants exhibited an incompatibility response toward SCRb pollen in pollination tests and expressed SRKb at levels comparable to C24 nrpd1a[SRKb-SCRb] plants as shown through real-time PCR analysis. In contrast, the remaining two plants exhibited a compatible response toward SCRb pollen and their SRKb transcript levels were on average threefold lower than those in C24 nrpd1a[SRKb-SCRb] plants.
At least two modifier loci have been shown to contribute to differences in the SI phenotype of SRKb-SCRb transformants of different accessions: PUB8 on chromosome 4 (Liu et al. 2007) and another as-yet-uncharacterized locus on chromosome 3 (Boggs et al. 2009). To determine whether either one of these modifier loci might be responsible for the accession-specific effect of nrpd1a-8 on SI, markers tightly linked to these modifiers were used to analyze F2 progenies of the Col-0 nrpd1a-8[SRKb] × C24 WT[SRKb-SCRb] cross. However, no association was found between SI phenotype and either of these markers. Additional markers located throughout the genome were therefore tested, and loose linkage of a putative modifier locus to marker NGA139 was detected on chromosome 5 (Table S4).
Differential activity of the SRKb promoter in A. thaliana
To understand the differential regulation of SRKb in the nrpd1a mutant and in various A. thaliana accessions, it is important to determine if differences in the steady-state levels of stigma SRKb transcripts result from differences in the transcription or stability of these transcripts. To address this issue, we assayed the activity of the SRKbpr::uidA:nos reporter gene in the pistils of the nrpd1a mutant and of several accessions that had previously been analyzed for expression of SI by transformation with the SRKb-SCRb gene pair (Nasrallah et al. 2004; Boggs et al. 2009): C24, Cvi, Hodja, Kas, and Sha, all of which express intense and developmentally-stable SI; and Col-0 and Rld as representatives of accessions that express PUB8-mediated transient SI (Liu et al. 2007).
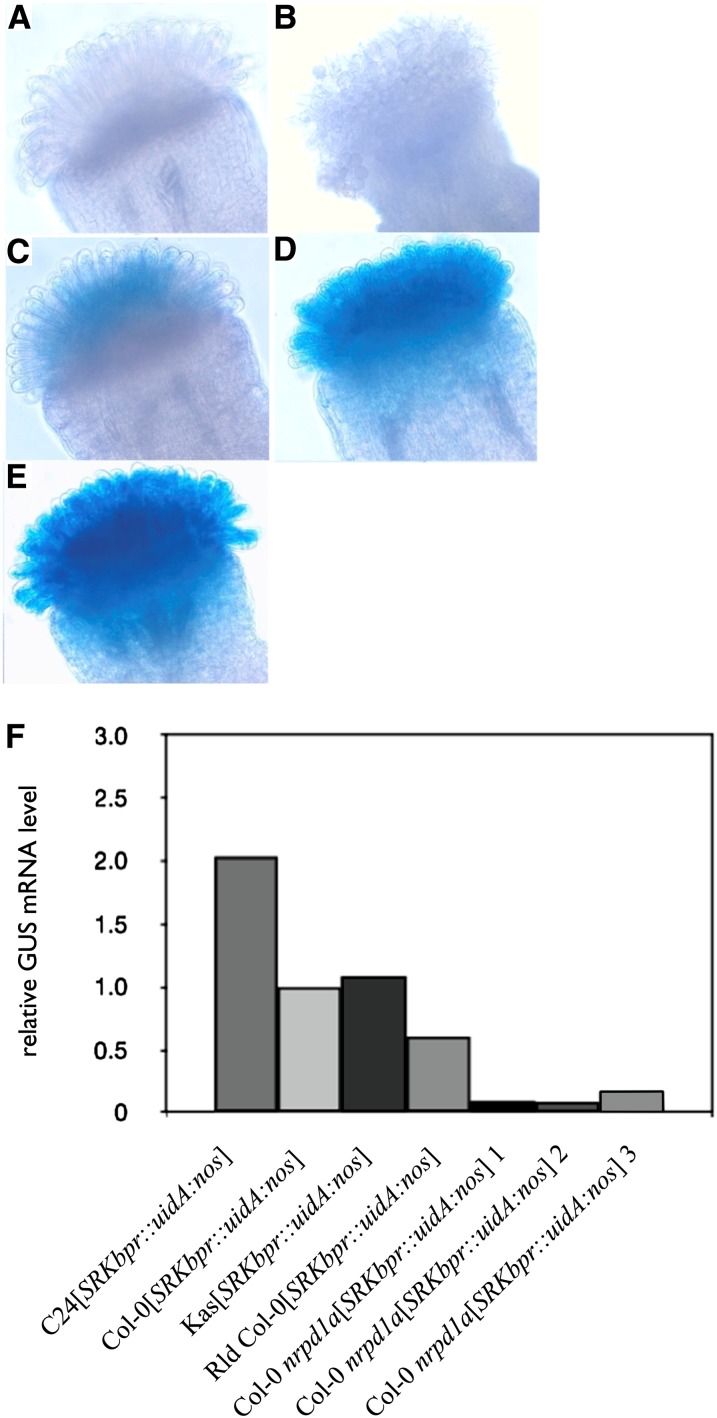
In histochemical analysis of GUS activity in Col-0 nrpd1a plants or Col-0 WT plants transformed with the SRKbpr::uidA:nos reporter, no staining was observed in stigma epidermal cells or other cells of the pistil (Figure 3, A and B; Table S1), and only little staining was observed in the stigma of Rld[SRKbpr::uidA:nos] transformants (Figure 3C; Table S1). In contrast, in all accessions that express developmentally-stable SI, SRKbpr::uidA:nos transformants were identified which exhibited GUS activity in stigma epidermal cells, and the majority of these transformants also exhibited GUS activity in the style (Figure 3D; Table S1). Interestingly, Col-0 plants transformed with an AtS1pr::uidA::nos construct exhibited strong GUS staining in stigma epidermal cells (Figure 3E), as expected for the stigma epidermal cell-specific AtS1 promoter (Dwyer et al. 1994). Thus, the lack of GUS staining in stigma epidermal cells of Col-0[SRKbpr::uidA:nos] plants is not due to general misregulation of promoters expressed in these cells, but was specific to the SRKb promoter.
Figure 3 .
Differential activity of the SRKb promoter in the Col-0 nrpd1a mutant and in various A. thaliana accessions. Activity of the SRKbpr::uidA:nos reporter was assayed in flowers at the +1-stage of development by histochemical analysis of GUS activity and by real-time RT-PCR of stigma tissue. (A−D) SRKbpr::uidA:nos transformants (A) Col-0 (B) Col-0 nrpd1a (C) Rld, and (D) C24. (E) Col-0[AtS1pr::uidA:nos] used as control. (F) Real-time RT-PCR of GUS transcripts in stigma tissue of SRKbpr::uidA:nos transformants of four A. thaliana accessions and the Col-0 nrpd1a mutant.
The lack of GUS staining in the pistils of Col-0[SRKbpr::uidA:nos] transformants and the weak GUS staining in Rld[SRKbpr::uidA:nos] transformants was surprising given the fact that SRKb-SCRb transformants of these accessions express SI in −1 bud-stage stigmas and that SRKb transcripts are detected in Col-0 [SRKb] stigmas by RNA gel blot analysis (Figures 1A and 2). To investigate this discrepancy, real-time PCR was used to compare the levels of stigma GUS transcripts in strains that showed no or little GUS staining on the one hand and in strains that exhibited strong GUS staining in stigma epidermal cells on the other hand. Figure 3F shows that GUS transcripts were detected in Col-0[SRKbpr::uidA:nos], Rld[SRKbpr::uidA:nos], and Col-0 nrpd1a [SRKbpr::uidA:nos] stigmas, albeit at lower levels than in the stigmas of C24 and Kas [SRKbpr::uidA:nos] transformants. GUS transcript levels in Col-0 and Rld transformants were, respectively, 2.2- and 1.4-fold lower than in C24 transformants and 1.7- and 1.1-fold lower than in Kas transformants. In Col-0 nrpd1a [SRKbpr::uidA:nos] plants, GUS transcript levels were 12- to ~24-fold lower than in C24[SRKbpr::uidA:nos] plants. Thus, the failure to observe GUS staining in the stigmas of [SRKbpr::uidA:nos] transformants of Col-0, Rld, and nrpd1a may be attributed to the relatively low sensitivity of histochemical GUS staining.
Discussion
The results described here reveal an unexpected complexity in the regulation of the SRKb transgene in A. thaliana. We found that NRPD1a is required for expression of SI in stigmas of Col-0[SRKb] plants. Because NRPD1a functions in transgene silencing at least partly by increasing DNA methylation, disruption of this gene is expected to cause reduced methylation and consequently increased expression of the SRKb transgene. In fact, SRKb plants homozygous for the nrpd1a-8 mutant allele identified here exhibited reduced overall genomic DNA methylation as determined by loss of methylation at the AtSN1 retroelement. They also accumulated increased levels of full-length SRKb and eSRKb transcripts relative to WT[SRKb] homozygotes in styles, stamens, and petals, in keeping with the abrogation of transgene silencing expected in eSRKb loss-of-function mutants. Unexpectedly, however, nrpd1a-8[SRKb] plants exhibited reduced steady-state levels of full-length SRKb transcripts relative to WT[SRKb] stigmas. In contrast, eSRKb transcripts levels were increased in nrpd1a[SRKb] stigmas relative to WT[SRKb] stigmas. A possible explanation for this difference in the effect of nrpd1a on the two SRKb transcripts is that the nrpd1a mutation causes different effects on the transcriptional machinery necessary for the production of these two transcripts.
The puzzling opposite effects of the nrpd1a-8 mutation on SRKb transcript levels in stigmas on the one hand and in styles, stamens, and petals on the other hand, suggest the existence of tissue-specific factors that differentially regulate expression of the SRKb transgene in different floral tissues. The existence of stigma-specific factors that control SRKb expression would not be surprising. Indeed, stigma-specific factors that regulate SRKb kinase activity have been previously invoked to explain the fact that the SRKb protein exhibits SCR-dependent activation in the stigma, where it functions in recognition of self pollen, but displays constitutive SCR-independent activity in the style, where it enhances cell division (Tantikanjana et al. 2009). Whether SRKb regulatory factors common to styles, stamens, and petals exist, and whether expression of SRK in non-pistil floral tissues has any biological significance remain to be determined.
There is a lack of a direct correlation between genomic DNA methylation status and SI phenotype in nrpd1a[SRKb-SCRb] plants. SRKb-SCRb plants homozygous for the nrpd1a, nrpd1b, ago4, and rdr2 mutations all showed a loss of genome methylation at the AtSN1 retroelement, but only nrpd1a[SRKb-SCRb] and rdr2[SRKb-SCRb] plants exhibited a breakdown of SI. Additionally, in the Col-0 nrpd1a-8[SRKb] × C24 WT[SRKb-SCRb] cross, all nrpd1a-8 mutant progenies exhibited a loss of genome methylation, but some retained SI. Silencing pathways in plants are known to be complex and various mechanisms exist by which DNA methylation may be initiated (Matzke et al. 2009; Jauvion et al. 2012). For example, impairment of RDR2 may reduce substrate competition between RDR2 and RDR6 and increase methylation in rdr2 mutants (Jauvion et al. 2012). The use of different pathways for SRKb transgene silencing and for genomic methylation would explain the increased silencing of SRKb in the stigma in the context of the overall reduced genomic DNA methylation observed in nrpd1a homozygotes.
Intriguingly, there was no absolute correlation between pollination phenotype and SRKb transcript levels. Although nrpd1a and rdr2 [SRKb-SCRb] plants that exhibited breakdown of SI often accumulated lower levels of SRKb transcripts than plants that retained SI, some plants, such as ago4[SRKb-SCRb] plants, retained SI despite expressing equally low SRKb transcript levels. Thus, the reduced accumulation of SRKb transcripts is not in itself sufficient to explain the complete breakdown of SI in nrpd1a or rdr2 homozygotes. Rather, our results suggest that at least one stigma factor found in the Col-0 genetic background is responsible for the self-fertile phenotype of Col-0 nrpd1a[SRKb] plants. In particular, the fact that disruption of the NRPD1a gene did not abrogate the SI phenotype of C24[SRKb-SCRb] plants, the 3:1 ratio of self-incompatible to self-compatible plants observed among nrpd1a-8 homozygous progenies of the Col-0 nrpd1a-8[SRKb] × C24 WT[SRKb-SCRb] cross, and preliminary evidence that implicates a locus on chromosome 5 in the breakdown of SI in Col-0 nrpd1a-8[SRKb] plants all suggest that NRPD1a and RDR2 act on a Col-0 locus that is involved in SI and whose phenotypic effect requires homozygosity for nrpd1a.
At present, we do not know what the relationship is, if any, between the factor encoded by this Col-0 locus and the accession-specific stigma factor that allows high-level expression of the SRKb::uidA::nos reporter in the stigmas of accessions that exhibit developmentally-stable SI but only little or no expression of the reporter in accessions that exhibit transient SI. It is clear however, that both factors act in trans on the SRKb promoter, as evidenced by the reduced GUS transcript levels observed in Col-0 nrpd1a[SRKbpr::uidA::nos] transformants.
Overall, our results are consistent with the conclusion that the SRKb transgene is subject to several levels of regulation that vary substantially by accession and by tissue, with involvement of epigenetic factors (as revealed by the misregulation of the SRKb gene in nrpd1a and other DNA methylation mutants) that might converge on the SRKb promoter. The nature of these factors and the mechanism(s) by which changes in DNA methylation effect changes in SRKb transcript levels remain to be determined. In any case, our analysis of the nrpd1a-8 mutation has identified one more SI modifier locus that exhibits natural variation among A. thaliana accessions, similar to PUB8, which also regulates SRK transcript levels (Liu et al. 2007), and to a previously identified SI modifier on chromosome 3 (Boggs et al. 2009). The polymorphic nature of these loci supports the conclusion that the switch to self-fertility in A. thaliana was accompanied, not only by inactivation of SRK or SCR genes, but also by mutations at SI modifier loci that arose stochastically in different accessions.
Supplementary Material
Acknowledgments
We thank Tiffany Crispell for plant transformation, GUS assays, and plant care; Craig Pikaard for C24 nrpd1a seed; and the Arabidopsis Biological Resource Center in Columbus, Ohio for various A. thaliana mutant seed. S.R.S. was supported in part by National Institutes of Health grant T32GM007617, a pre-doctoral training grant in Genetics and Development. This material is based upon work supported by National Science Foundation grants IOS-0744579 and IOS-1146725. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Communicating editor: Z. Yang
Literature Cited
- Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., et al. , 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Boggs N. A., Nasrallah J. B., Nasrallah M. E., 2009. Independent S-locus mutations caused self-fertility in Arabidopsis thaliana. PLoS Genet. 5: e1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F., 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dixit R., Nasrallah M. E., Nasrallah J. B., 2000. Post-transcriptional maturation of the S receptor kinase of Brassica correlates with co-expression of the S-locus glycoprotein in the stigmas of two Brassica strains and in transgenic tobacco plants. Plant Physiol. 124: 297–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19: 11–15 [Google Scholar]
- Dwyer K. G., Kandasamy M. K., Mahosky D. I., Acciai J., Kudish B. I., et al. , 1994. A superfamily of S locus-related sequences in Arabidopsis: diverse structures and expression patterns. Plant Cell 6: 1829–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A., Voinnet O., Chappell L., Baulcombe D., 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21: 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr A. J., Jensen M. B., Dalmay T., Baulcombe D. C., 2005. RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120 [DOI] [PubMed] [Google Scholar]
- Jauvion V., Rivard M., Bouteiller N., Elmayan T., Vaucheret H., 2012. RDR2 partially antagonizes the production of RDR6-dependent siRNA in sense transgene-mediated PTGS. PLoS ONE 7: e29785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho Y. O., Baer J., 1968. Observing pollen tubes by means of fluorescence. Euphytica 17: 298–302 [Google Scholar]
- Kusaba M., Dwyer K., Hendershot J., Vrebalov J., Nasrallah J. B., et al. , 2001. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13: 627–643 [PMC free article] [PubMed] [Google Scholar]
- Liu P., Sherman-Broyles S., Nasrallah M. E., Nasrallah J. B., 2007. A cryptic modifier causing transient self-incompatibility in Arabidopsis thaliana. Curr. Biol. 17: 734–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Matzke M., Kanno T., Daxinger L., Huettel B, Matzke A. J. M., 2009. RNA-mediated chromatin-based silencing in plants. Curr. Opin. Plant Biol. 21: 367–376 [DOI] [PubMed] [Google Scholar]
- Nasrallah M. E., Liu P., Nasrallah J. B., 2002. Generation of self- incompatible Arabidopsis thaliana by transfer of two S locus genes from Arabidopsis lyrata. Science 297: 247–249 [DOI] [PubMed] [Google Scholar]
- Nasrallah M. E., Liu P., Sherman-Broyles S., Boggs N. A., Nasrallah J. B., 2004. Natural variation in expression of self-incompatibility in Arabidopsis thaliana: Implications for the evolution of selfing. Proc. Natl. Acad. Sci. USA 101: 16070–16074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y., Haag J. R., Ream T., Nunes P. C., Pontes O., et al. , 2005. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622 [DOI] [PubMed] [Google Scholar]
- Palusa S. G., Ali G. S., Reddy A. S. N., 2007. Alternative splicing of pre- mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 49: 1091–1107 [DOI] [PubMed] [Google Scholar]
- Pontier D., Yahubyan G., Vega D., Bulski A., Saez-Vasquez J., et al. , 2005. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 19: 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scascitelli M., Cognet M., Adams K. L., 2010. An interspecific plant hybrid shows novel changes in parental splice forms of genes for splicing factors. Genetics 184: 975–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D. R., Bowman J. L., Meyerowitz E. M., 1990. Early flower development in Arabídopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein J. C., Howlett B., Boyes D. C., Nasrallah M. E., Nasrallah J. B., 1991. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl. Acad. Sci. USA 88: 8816–8820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D., Wilks C., Lamesch P., Berardini T. Z., Garcia-Hernandez M., et al. , 2007. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 36: D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantikanjana T., Rizvi N., Nasrallah M. E., Nasrallah J. B., 2009. A dual role for the S-locus receptor kinase in self-incompatibility and pistil development revealed by an Arabidopsis rdr6 mutation. Plant Cell 21: 2642–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantikanjana T., Nasrallah M. E., Nasrallah J. B., 2010. Complex networks of self-incompatibility signaling in the Brassicaceae. Curr. Opin. Plant Biol. 13: 520–526 [DOI] [PubMed] [Google Scholar]
- Vaucheret H., 2006. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20: 759–771 [DOI] [PubMed] [Google Scholar]
- Zheng X., Pontes O., Zhu J., Miki D., Zhang F., et al. , 2008. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature 455: 1259–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.