Abstract
Transcriptome sequences are becoming more broadly available for multiple individuals of the same species, providing opportunities to derive population genomic information from these datasets. Using the 454 Life Science Genome Sequencer FLX and FLX-Titanium next-generation platforms, we generated 11−430 Mbp of sequence for normalized cDNA for 40 wild genotypes of the invasive plant Centaurea solstitialis, yellow starthistle, from across its worldwide distribution. We examined the impact of sequencing effort on transcriptome recovery and overlap among individuals. To do this, we developed two novel publicly available software pipelines: SnoWhite for read cleaning before assembly, and AllelePipe for clustering of loci and allele identification in assembled datasets with or without a reference genome. AllelePipe is designed specifically for cases in which read depth information is not appropriate or available to assist with disentangling closely related paralogs from allelic variation, as in transcriptome or previously assembled libraries. We find that modest applications of sequencing effort recover most of the novel sequences present in the transcriptome of this species, including single-copy loci and a representative distribution of functional groups. In contrast, the coverage of variable sites, observation of heterozygosity, and overlap among different libraries are all highly dependent on sequencing effort. Nevertheless, the information gained from overlapping regions was informative regarding coarse population structure and variation across our small number of population samples, providing the first genetic evidence in support of hypothesized invasion scenarios.
Keywords: normalized ESTs, allele clustering, 454 GS FLX Titanium, yellow starthistle, invasive species
Almost five decades of molecular genetic research has revealed that an abundance of genetic variation resides in the natural populations of most living organisms (e.g., Lewontin and Hubby 1966; Hamrick and Godt 1989; Morin et al. 2004; Ossowski et al. 2008; Durbin et al. 2010). This variation is necessarily shaped by the history of mating, demography, dispersal, and adaptation playing out within species and as a result allele frequency distributions in wild populations can provide unique insights into evolution and ecology (Avise 2004; Wakeley 2008). Most recently, genome-wide studies of allelic variation in natural populations are proving to be particularly powerful for detecting subtle and/or complex aspects of population structure, selection on specific regions of the genome, and associations between allelic and phenotypic variation (e.g., Drosophila 12 Genomes Consortium 2007; McCarthy et al. 2008; Zayed and Whitfield 2008; Emerson et al. 2010; Hancock et al. 2010). Genomic approaches were first accessible to population geneticists via targeted amplification of genome-wide markers (Vos et al. 1995), but the declining cost and increasing length of next-generation sequencing reads are now making bulk sequencing of the genome practical for allele discovery in nonmodel and outbred study subjects (Li et al. 2008; Davey et al. 2011; Nielsen et al. 2011).
Sequencing of transcribed genes, the ‘transcriptome,’ is an especially attractive strategy for surveying genomic variation at a relatively low cost because it provides a reduced fraction of the genome that is also rich in information (Bouck and Vision 2007). Coding sequences can be placed into reading frame, aspects of protein variation studied, and their general function inferred from homology to proteins in model organisms (Wheat 2010). Coding regions also may underlie phenotypic variation of interest and the associated protein variants identified as the direct targets of selection (e.g., Elmer et al. 2010; Renaut et al. 2010; Barreto et al. 2011). Simultaneously, a genome-wide panel of transcriptome-derived SNPs should also be largely suitable for standard population genetic analyses that assume neutrality (e.g., Barbazuk et al. 2007; Seeb et al. 2011; Ueno et al. 2010; Zakas et al. 2012).
Here, we explore the potential for de novo whole transcriptome sequences from many individuals to generate useful genetic polymorphism data across their genomes. We focus on a species of economic and ecological concern, yellow starthistle (Centaurea solstitialis L., Asteraceae). Yellow starthistle (hereafter YST) is an annual herb, native to Eastern Europe and the Caucasus, and hypothesized to be naturalized in Mediterranean Western Europe (Maddox et al. 1985). Historical records indicate that this species was introduced to South America as a crop contaminant via Spain in the mid-1600s and then to North America (primarily via Chile) in the 1800s (Gerlach 1997). Introduced YST have successfully invaded a wide variety of grass- and shrublands in western North America and temperate South America. In North America, this species has spread to 23 U.S. states and five Canadian provinces (Maddox et al. 1985), with the largest infestations occurring in California (>15 million acres) and the Pacific Northwest (>3.3 million acres) United States (Wilson et al. 2003).
For transcriptome sequencing, we generated normalized cDNA libraries from actively growing tissues in an effort to sequence as many of the coding regions of the genome as possible. In contrast to direct sequencing of cDNA for quantification of gene expression (i.e., ‘RNASeq’), normalization reduces the representation of common transcripts for better sequence coverage of both common and rare transcripts (Zhulidov et al. 2004; Christodoulou et al. 2011). We report on sequences of 40 YST plants: 19 from its invaded range (11 from North America and 8 from South America), 4 from its putative ancient naturalized range (Spain), and 17 from its native range (Eastern Europe/Caucasus). Plants are diploid (2N = 16) throughout these regions (Heiser and Whitaker 1948; Widmer et al. 2007; Ozturk et al. 2009), with a genome size of 1N = 851 Mbp (Brancheva and Greilhuber 2006).
We address two key issues using our dataset: (1) the accurate identification of allelic variation in these outbred individuals, and (2) the extent of sequence overlap among transcriptome libraries. YST propagates exclusively by seed and is self-incompatible (Sun and Ritland 1998), and so we expect allelic variation and observed heterozygosity to be relatively high in our populations (Sun 1997). Accurate identification of allelic variants of the same locus is not straightforward, however, because allele sequences vary widely in their divergence (e.g., Lawlor et al. 1988; Li and Sadler 1991; Moriyama and Powell 1996; Bergelson et al. 2001), overlapping the divergence of closely related but separate loci (paralogs) (Lynch and Conery 2000; Sebat et al. 2004; Hahn et al. 2005; Demuth et al. 2006; Hahn et al. 2007). This means that it is impossible to discriminate among alleles and paralogs by sequence divergence alone. We present a conservative strategy to filter for unique loci by using the putative allelic variation across all individuals under study to identify the minimum number of haplotypes within individuals. This strategy relies on phasing of multi-locus SNPs within individual genes, and so we use a long-read 454 pyrosequencing approach to increase our ability to discriminate accurately among haplotypes. We find that approximately one-half of our putative loci are suitable for population genetic analyses, that allele recovery is far more dependent on sequencing depth than is gene recovery, but that modest transcriptome sampling nevertheless generates thousands of informative markers observed across many individuals. In our study system, these markers indicate a history of admixture and the presence of high genetic variation in introduced populations of YST.
Materials and Methods
Library preparation
We prepared cDNA libraries from leaves or whole plants of 8-wk-old seedlings (n = 40), representing the native and introduced range of YST (Table 1). Seeds from these populations were reared in a greenhouse at 25° and 16-hr day/8-hr night conditions in a 1:1 mixture of sand and potting soil. RNA extraction followed Lai et al. (2012).
Table 1. Sequencing effort and assembly information for C. solstitialis normalized Roche 454 transcriptome libraries.
| Raw | Cleaned |
Assembled |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SampleTissue [Latitude, Longitude] | Platform | Plates | Mb | Mb | Read No. | Median bp | Mb | Unigene No. | Median bp | Contig No. | UCO No. | |||
| North America (introduced) | ||||||||||||||
| CA-1-1S [N 41° 59’, W 122° 36’] | FLX | 0.25 | 17.2 | 13.0 | 69939 | 211 | 2.4 | 9783 | 242 | 8769 | 10 | |||
| CA-1-2S [N 41° 59’, W 122° 36’] | FLX | 0.25 | 25.1 | 19.0 | 101468 | 213 | 3.2 | 12810 | 242 | 11237 | 18 | |||
| CA-2-2S [N 40° 25’, W 122° 16’] | FLX | 0.25 | 20.1 | 15.9 | 82739 | 216 | 2.8 | 10596 | 255 | 9236 | 25 | |||
| CA-2-4S [N 40° 25’, W 122° 16’] | FLX | 0.25 | 19.9 | 14.3 | 78770 | 210 | 2.4 | 9853 | 238 | 8551 | 14 | |||
| CA-3-1S [N 39° 12’, W 121° 06’] | FLX | 0.5 | 58.0 | 45.6 | 229635 | 224 | 7.1 | 23671 | 268 | 19905 | 77 | |||
| CA-3-2S [N 39° 12’, W 121° 06’] | FLX | 0.5 | 58.1 | 39.9 | 212773 | 219 | 5.8 | 21578 | 254 | 18377 | 28 | |||
| CA-4-1S [N 38° 31’, W 121° 45’] | FLX | 0.5 | 80.1 | 66.8 | 321955 | 226 | 10.1 | 32206 | 270 | 27121 | 109 | |||
| CA-4-3S [N 38° 31’, W 121° 45’] | FLX | 0.25 | 20.0 | 13.2 | 72154 | 214 | 2.3 | 9524 | 238 | 8655 | 11 | |||
| CA-4-4S [N 38° 31’, W 121° 45’] | Ti | 0.75 | 206.3 | 193.2 | 579859 | 366 | 24.8 | 42042 | 537 | 32786 | 252 | |||
| CA-5-3S [N 38° 16’, W 121° 49’] | FLX | 0.5 | 80.2 | 65.5 | 325569 | 223 | 9.9 | 32743 | 266 | 27222 | 111 | |||
| CA-5-4S [N 38° 16’, W 121° 49’] | FLX | 0.25 | 21.1 | 14.7 | 79481 | 215 | 2.3 | 9172 | 249 | 8112 | 13 | |||
| South America (introduced) | ||||||||||||||
| AR-1-24L [S 36° 26’, W 64° 17’] | Ti | 0.5 | 234.7 | 225.4 | 542881 | 462 | 33.4 | 46538 | 632 | 38346 | 299 | |||
| AR-1-25CL [S 36° 26’, W 64° 17’] | Ti | 0.56 | 153.7 | 132 | 369437 | 412 | 21.1 | 39579 | 532 | 29944 | 177 | |||
| AR-6-13L [S 37° 39’, W 64° 08’] | Ti | 0.5 | 256.4 | 247.1 | 596060 | 473 | 28.5 | 38572 | 630 | 31082 | 287 | |||
| AR-6-26L [S 37° 39’, W 64° 08’] | Ti | 0.5 | 181.7 | 169.2 | 485202 | 390 | 17.3 | 30641 | 537 | 23376 | 185 | |||
| AR-8-15L [S 38° 11’, W 64° 04’] | Ti | 0.5 | 166.1 | 154.6 | 443044 | 391 | 20.4 | 35628 | 549 | 26419 | 230 | |||
| AR-8-19L [S 38° 11’, W 64° 04’] | Ti | 0.5 | 259.3 | 249.9 | 600878 | 458 | 28.7 | 37199 | 673 | 30305 | 303 | |||
| AR-13-24L [S 36° 18’, W 65° 40’] | Ti | 0.5 | 274.6 | 264.7 | 622982 | 491 | 28.4 | 37408 | 679 | 29348 | 311 | |||
| AR-13-28L [S 36° 18’, W 65° 40’] | Ti | 0.63 | 183.5 | 171.5 | 488746 | 405 | 21.1 | 38160 | 540 | 28579 | 206 | |||
| Western Europe (putative ancient expansion) | ||||||||||||||
| SP-1-5L [N 39° 50’, W 2° 30’] | Ti | 0.5 | 152.5 | 135.6 | 393803 | 398 | 19.5 | 38699 | 519 | 27841 | 172 | |||
| SP-1-10L [N 39° 50’, W 2° 30’] | Ti | 0.5 | 269.7 | 258.7 | 604697 | 483 | 29.5 | 43236 | 637 | 32884 | 292 | |||
| SP-2-2L [N 41° 45’, W 4° 5′] | Ti | 0.63 | 181.9 | 162.1 | 522160 | 341 | 22.8 | 46071 | 474 | 34707 | 184 | |||
| SP-2-6L [N 41° 45’, W 4° 5′] | Ti | 1 | 332.6 | 315.9 | 815538 | 459 | 41.6 | 62444 | 583 | 48230 | 311 | |||
| Eastern Europe (native) | ||||||||||||||
| GA-1-1S [N 41° 56’, E 45° 27’] | FLX | 0.25 | 19.1 | 12.5 | 71584 | 209 | 2.0 | 8901 | 233 | 7823 | 8 | |||
| GA-2-1S [N 41° 56’, E 45° 35’] | FLX | 0.25 | 18.9 | 12.9 | 73656 | 210 | 2.0 | 8921 | 233 | 7659 | 11 | |||
| GA-3-4S [N 41° 44’, E 45° 12’] | FLX | 0.25 | 11.5 | 7.5 | 42595 | 212 | 1.3 | 5257 | 240 | 4704 | 3 | |||
| GA-4-3S [N 41° 43’, E 45° 16’] | FLX | 0.25 | 13.0 | 7.7 | 43567 | 215 | 1.3 | 5299 | 240 | 4811 | 11 | |||
| GA-5-4S [N 41° 38’, E 45° 38’] | FLX | 0.25 | 12.8 | 8.6 | 48417 | 214 | 1.5 | 6073 | 243 | 5407 | 4 | |||
| GA-5-24L [N 41° 38’, E 45° 38’] | Ti | 0.63 | 153.5 | 126.5 | 367110 | 390 | 17.4 | 33086 | 522 | 25382 | 165 | |||
| HU-1-8L [N 46° 58’, E 18° 41’] | Ti | 0.5 | 185.3 | 172.4 | 465081 | 421 | 22.3 | 39661 | 559 | 28967 | 213 | |||
| HU-2-10L [N 47° 19’, E 21° 02’] | Ti | 0.5 | 297.9 | 286.6 | 661490 | 489 | 32.5 | 40448 | 699 | 32643 | 311 | |||
| RO-1-6L [N 47° 42’, E 26° 2’] | Ti | 0.75 | 189.6 | 170.8 | 518480 | 376 | 22.6 | 43834 | 510 | 32820 | 182 | |||
| RO-5-10L [N 46° 15’, E 27° 39’] | Ti | 0.5 | 292.5 | 279.1 | 663787 | 479 | 33.7 | 52108 | 602 | 39456 | 293 | |||
| TK-1-3S [N 39° 45’, E 29° 06’] | FLX | 0.5 | 24.7 | 17.3 | 89230 | 229 | 3.2 | 11053 | 268 | 9841 | 26 | |||
| TK-1-5L [N 39° 45’, E 29° 06’] | Ti | 0.63 | 159.5 | 145.6 | 494855 | 327 | 20.2 | 41229 | 462 | 30361 | 207 | |||
| TK-2-3S [N 37° 02’, E 29° 47’] | FLX | 0.25 | 17.1 | 10.6 | 63668 | 201 | 1.8 | 8085 | 223 | 7106 | 4 | |||
| TK-2-4S [N 37° 02’, E 29° 47’] | FLX | 0.25 | 15.7 | 10.0 | 59734 | 204 | 1.5 | 7087 | 225 | 6191 | 6 | |||
| TK-3-2S [N 37° 50’, E 27° 51’] | FLX | 0.25 | 11.4 | 6.2 | 38205 | 198 | 1.0 | 4873 | 210 | 4373 | 2 | |||
| TK-5-4S [N 37° 01’, E 30° 22’] | Ti/FLX | 2.25 | 430.7 | 389.4 | 1258319 | 352 | 35.1 | 71045 | 418 | 48608 | 278 | |||
| TK-5-9L [N 37° 01’, E 30° 22’] | Ti | 0.5 | 252.1 | 241.7 | 554227 | 481 | 29.8 | 40147 | 671 | 32773 | 283 | |||
| Pseudo-reference | 39.6 | 43717 | 811 | 260 | ||||||||||
| Pseudo-reference filtered for polymorphic single loci | 21.2 | 22687 | 840 | 154 | ||||||||||
Tissues included are whole seedlings (S) or leaves (L). FLX, 454 Life Science Genome Sequencer FLX; Ti, FLX-Titanium.
To generate the full-length complementary DNA (cDNA) for the transcriptome analysis, we used a protocol from the Clontech Creator SMART cDNA library construction kit (Clontech Laboratories, Mountain View, CA). This requires an oligo-dT primer that anchors the polyA tail of mRNA to primer the cDNA synthesis process. However, mononucleotide runs reduce sequence quality and quantity due to excessive light production and crosstalk between neighboring cells (Margulies et al. 2005). To counteract this problem, we used two different approaches to synthesize cDNA. The first approach was to use a “broken chain” short oligo-dT primer (primer sequence: 5′- AAGCAGTGGTATCAACGCAGAGTCGCAGTCGGTACTTTTTTCTTTTTTV-3′, V = A, G, or C) to prime the poly(A) tail of mRNA during first-strand cDNA synthesis (Meyer et al. 2009). In the second approach we used two different modified oligo-dT primers: one (5′-AAGCAGTGGTATCAACGCAGAGT(T)4G(T)9C(T)10VN-3′) to prime the poly(A) tail of mRNA during first strand cDNA synthesis and another (5′-AAGCAGTGGTATCAACGCAGAGT(T)4GTC(T)4GTTCTG(T)3C(T)4VN-3′) to further break down the stretches of poly(A) sequence during second strand cDNA synthesis (Beldade et al. 2006). Approximately 1.5 µg of total RNA was reverse-transcribed to first-strand cDNA using these methods.
Double-stranded (ds) cDNA synthesis was performed using Phusion polymerase (New England Biolabs, Ipswich, MA) with a hot start of 98° for 30 sec, followed by 18 cycles of 98° for 7 sec, 66° for 20 sec, and 72° for 4 min. The ds-cDNA polymerase chain reaction product was purified using a QIAquick PCR Purification column (QIAGEN). Normalization was performed using a TRIMMER-DIRECT cDNA normalization kit (Evrogen, Moscow, Russia). Approximately 600−1200 ng of purified ds-cDNA was used as the starting amount for normalization. A mixture of 0.25 µL and 0.5 µL of DSN normalization tubes was used for the first and second amplifications.
After normalization, cDNA was fragmented to 500- to 800-bp fragments by sonication or nebulization and size-selected to remove small fragments using AMPure SPRI beads. The fragmented ends were polished and ligated with adaptors (Meyer et al. 2009). The optimal ligation products were selectively amplified and subjected to two rounds of size selection including gel electrophoresis and AMPure SPRI bead purification (Lai et al. 2012).
Sequencing and assembly
All cDNA libraries were sequenced on the 454 Life Science Genome Sequencer (Roche Applied Science, Branford, CT), using either FLX or FLX-Titanium chemistry (Table 1). Sequencing of each sample was performed by the 454 Life Science Sequencing Center at Roche Applied Science, the Center for Genomics and Bioinformatics at Indiana University, or the Genome Quebec Innovation Centre at McGill University. Primer/adapter and polyA/T sequences were trimmed from the reads using custom Perl trimming scripts and SeqClean (http://www.tigr.org/tdb/tgi/software/). Sequences composed of primer multimers were removed using TagDust (Lassmann et al. 2009), with a false discovery rate of 0.01. These cleaning steps were combined into a flexible automatic sequence cleaning pipeline ‘SnoWhite’ (v1.1.4), which we have made publicly available at http://evopipes.net (Barker et al. 2010).
Cleaned reads were assembled into contigs de novo using the assembler MIRA v3.0 (Chevreux et al. 2004). MIRA produced identical duplicate contigs in areas with high read depth, and these were merged using additional iterations of both MIRA and CAP3 (Huang and Madan 1999) at 97% minimum similarity, using the pipeline iAssembler v1.3 (Zheng et al. 2011). Successful resolution of highly similar alleles and/or paralogs into unique contigs was verified by examining synonymous site divergence among gene family members using the program DupPipe (Barker et al. 2008, 2010). Resolution of highly similar sequences within eukaryotic individuals should yield gene family phylogenetic trees with a characteristic L-shaped distribution of many recent nodes and diminishing numbers of older nodes (Lynch and Conery 2000).
Allele clustering with AllelePipe
Allele and single-nucleotide polymorphism (SNP) identification typically rely on mapping reads to a reference genome that represents a haploid set of genes (Bentley 2006; Charlesworth 2010). YST lacks a reference genome, and allelic variation in our samples prevented simple de novo creation of an accurate haploid reference sequence library. To circumvent this problem, we developed a novel software pipeline to cluster putative alleles within and among individuals, available as ‘AllelePipe’ (v1.0.25; Figure 1) at http://evopipes.net. Using the AllelePipe, we assessed similarity among all sequences from all 40 individuals with SSAHA2 (Ning et al. 2001), with 95% minimum similarity and 300-bp minimum alignment length between sequences. We also included sequences from a published assembly of a Sanger EST library for a YST genotype from the invasion in central California (Genbank #EH750647-EH791053) (Barker et al. 2008). AllelePipe was used to verify that similar sequences aligned throughout their region of overlap (expected for true alleles), and to cluster groups of similar sequences via single-linkage clustering. Single-linkage clustering generates maximal aggregation of sequences, and will bring together both closely related paralogs and their alleles (Dlugosch and Bonin 2012). Multiple alignments were created for sequences within each cluster and their consensus sequence generated using CAP3. Clusters were discarded from the analysis if they aggregated increasingly dissimilar sequences, preventing a single CAP3 consensus. From the resulting consensus sequences, a genomic ‘pseudo-reference’ FASTA file was generated for the entire dataset, suitable for anchoring contig alignments across individuals. We evaluated the quality of our overall clustering strategy by aligning our consensus sequences with known highly conserved eukaryotic single copy loci, including 357 ultra-conserved orthologs (UCOs, available at http://compgenomics.ucdavis.edu/compositae_reference.php; Kozik et al. 2008), using tblastx comparisons with maximum expectation (e-value) of 0.1 and minimum 30 protein residue alignments (Blast v2.2.24) (Altschul et al. 1997). Only one-to-one matches between UCOs and clusters are expected for properly clustered loci.
Figure 1 .
AllelePipe workflow for identifying alleles without a reference genome. Unigenes from all individuals are pooled and clustered by similarity. Clustered sequences are aligned and consensus sequences are generated, providing a pseudo-reference genome. Unigenes from the same and/or different individuals are aligned to the reference, and SNPs are identified. Multilocus SNP information is used to construct a minimum set of haplotypes for each individual, and clusters in which individuals are represented by an excess number of putative alleles are flagged as potential multigene clusters.
Finally, we filtered for (putatively) valid loci by removing those clusters with evidence of more than two alleles (multi-SNP haplotypes across the entire gene region) from any individual in the dataset. Excess alleles suggest that the cluster is not a single locus and instead a group of paralogous loci. This approach leverages the information gained from multiple individuals to infer paralogy, where it is not possible to do so from genomic DNA sequencing depth or alignment to a complete reference genome. Again using the AllelePipe, we identified SNPs for each sequence against the pseudo-reference by using ssahaSNP (Ning et al. 2001) with minimum 90% similarity for alignment to the pseudo-reference. Singleton SNPs (those seen only once across the dataset) were removed as potential errors, and the number of unique haplotypes for each individual in the cluster were evaluated for evidence of paralogous clustering. Those clusters with no more than two haplotypes per individual were retained, and their SNP variation (from ssahaSNP) retained for further analyses.
Gene annotation
Gene Ontology (GO) classifications for our pseudo-reference sequences and individual transcriptome assemblies were obtained through BLASTx searches against the Arabidopsis thaliana protein database from The Arabidopsis Information Resource (release TAIR10_pep_20101214; http://www.arabidopsis.org/), using an e-value cut off 1*10−10. To evaluate the impact of sequencing effort on the gene content of assemblies, we compared the representation of GO categories across libraries using a Chi-squared Contingency test in R (R Development Core Team 2010).
Population genetic analyses
We examined the impact of both sequencing effort and source region (native vs. invading) on assembled contig numbers, observed SNP positions, observed heterozygosity, and SNP overlap. SNP frequencies and overlap among individuals were quantified using custom Perl scripts. Significance of relationships was tested with linear model fits in R, using the lm function (R Development Core Team 2010). Geographic partitioning of genetic variation in the native range was assessed with STRUCTURE v2.3.2.1 for 1-4 subpopulations (K), using 10,000 burnin steps and 10,000 iterations of the models (Pritchard et al. 2000; Falush et al. 2003). Runs were repeated three to five times at each K to verify convergence. Naturalized and invading samples were then assigned to native sub-populations using STRUCTURE, and results were plotted using Distruct v 1.1 (Rosenberg 2004).
Results and Discussion
Transcriptome recovery
We obtained between 11.4 and 430.7 Mbp of raw sequence per individual as a result of a range of sequencing efforts across our samples (Table 1; NCBI Short Read Archive accession #SRA059334; assembly for AR-13-24 previously published in Lai et al. 2012 and available at doi:10.5061/dryad.cm7td/4). Our assemblies generated up to 71,054 unigenes (contigs and singleton reads combined), including up to 48,230 contigs (Figure 2). Gene number in the YST genome is not yet known, but our largest assemblies are at the top of the range of current annotation counts among complete angiosperm genomes (Barker et al. 2012), and somewhat higher than the ~35,000 loci predicted in the genome of the related asterid tomato (The Tomato Genome Consortium 2012). Given that our accessions are outbred individuals and allelic variation is expected, these numbers are consistent with a relatively complete view of expressed genes in this species. Indeed we find patterns of saturating gains in unique sequences and coverage of known conserved loci: Both unigene and contig numbers were positively related to sequencing effort, and these relationships were fit closely by logarithmic curves (regressions: unigenes, R2 = 0.93, P < 0.0001; contigs, R2 = 0.91, P < 0.0001, Figure 2), indicating that recovery of additional unique sequences was associated with exponential increases in sequencing effort. Although the longer-read GSFLX-Titanium chemistry resulted in longer contigs (Table 1), our contig numbers generated from those libraries followed the same logarithmic relationship with sequencing effort predicted by the shorter-read GS-FLX chemistry (nonsignificant interaction of chemistry type and sequencing effort in linear model: P = 0.43; Supporting Information, Figure S1). The proportion of UCOs recovered reached 87% (311 loci) among the largest libraries (Table 1). In general, gains from additional sequencing were relatively modest above approximately 100 Mb of usable sequence, which is likely to be less than 5x coverage of the YST transcriptome, given our assembly size. This result suggests that representative information can be gained from low coverage datasets, even while additional sequencing depth continues to yield further gene discovery (Lai et al. 2012).
Figure 2 .
Contig numbers in transcriptome libraries of native (triangles), naturalized (dashes), and invading (circles) genotypes, as a function of total sequence effort after cleaning by SnoWhite. A logarithmic fit is shown to variation across all individuals.
To evaluate our ability to resolve highly similar sequences in our assemblies, we examined frequency distributions of the synonymous site divergence at nodes in gene family trees for each of our datasets. Distributions of divergence times showed expected L-shaped patterns of abundant recent ancestry in each case, consistent with successful resolution of closely-related paralogs and alleles in our assemblies (Figure S2). The distributions also accurately reveal an additional small peak at ~Ks = 0.65, which has been shown previously to correspond to an ancient genome duplication event near the base of the family, based upon Sanger sequence data (Barker et al. 2008). Consistent with those previous analyses, there was no evidence of more recent genome duplication events in our YST individuals.
Allele clustering and library overlap
Single-linkage clustering of unigenes across all individuals using AlellePipe generated 43,717 total putative loci with a median consensus length of 811 bp. These aligned to 260 (73%) of the UCOs, and the majority of these UCOs aligned with only one cluster as expected (Figure S3). Alignment of the remaining UCOs to multiple clusters could reflect sequence divergence (low similarity with UCOs), duplication of the locus, or failure of the reads to cluster together. Divergence or duplication is possible but relatively unlikely in such highly conserved “single-copy” loci (Duarte et al. 2010); it is more likely that splitting of these clusters resulted from alternate splicing (Barbazuk et al. 2008) or insufficient overlap for observing similarity among sets of sequences. These latter processes will introduce clusters of sequences that are incomplete and/or redundant but nevertheless accurate variations on a gene region or splice form. Gene annotation of the consensus cluster sequences by similarity to A. thaliana proteins yielded 26,728 matches, 11,268 of which (25.8% of all clusters) were identified as unique, nonredundant annotations. Similar rates of homology to A. thaliana were observed in other transcriptome surveys of asterid plants, using both Sanger and next-generation sequencing platforms (Barker et al. 2008; Dempewolf et al. 2010; Angeloni et al. 2011; Lai et al. 2012).
After clustering, AllelePipe identified 22,687 polymorphic unigenes that conformed to expectations of no more than two alleles per individual. These loci annotated to 23,896 A. thaliana genes, indicating that most of the inferred multilocus clusters were not those that had successfully annotated, perhaps due to poor consensus formation. For annotated loci, GO representation differed among inferred single loci and all clusters (P < 0.001 for all three categories; Figure S4). Interestingly, single loci included a lower representation of transcriptome factors, and a greater representation of intracellular and plastid-associated components. These patterns contrast with patterns of duplicate retention from paleo genome duplication events in the Compositae family, consistent with the hypothesis that certain complete pathways are retained in duplicate from whole genome duplications due to dosage constraints, while other functions are free to vary in copy number [and the latter would generate our multilocus clusters (Barker et al. 2008, 2012) and references therein].
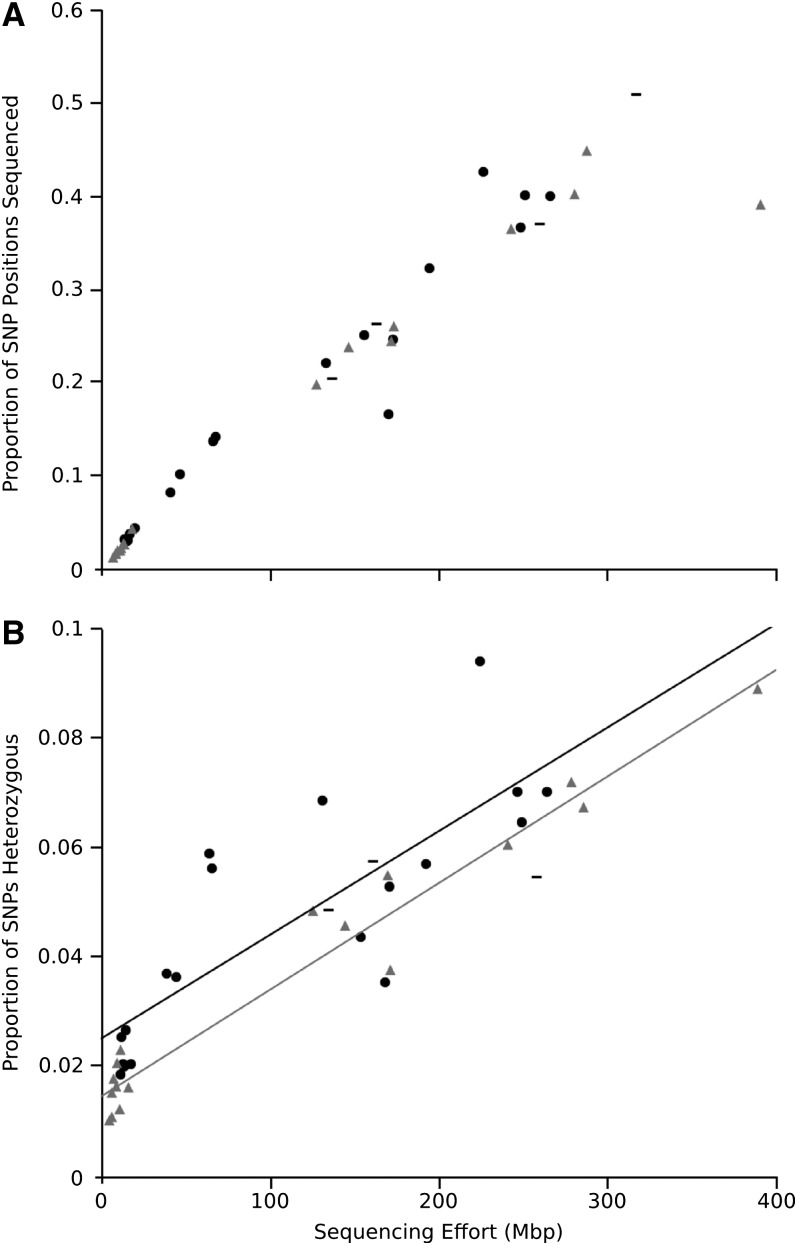
Aligning sequences from the 41 YST datasets against this filtered set of 22,687 pseudo-reference sequences revealed 237,034 polymorphic sites over the total sequence length of 21.2 Mbp, where each SNP variant was observed at least twice across all haplotypes (1 SNP per 89.6 bp across the sample). This low SNP density underscores the need for long-read approaches to accurately recover haplotypes within and among conspecific individuals (Dlugosch and Bonin 2012; Lai et al. 2012). The proportion of these sites sequenced in each individual increased sharply with sequencing effort (Figure 3A), with no indication of saturation; the largest libraries covered less than 60% of SNP positions. The number of observed heterozygous positions also showed a strong and linear increase with sequencing effort in both native and invading samples (Figure 3B). The accurate observation of heterozygosity is likely to be particularly important both for revealing population genetic information, and for validating the SNPs themselves (Seeb et al. 2011). Thus, despite saturating returns of additional unique sequences in the larger libraries, our ability to observe allelic variation at these loci did not plateau, and analyses of this type of variation must take sequencing effort into account.
Figure 3 .
Coverage of SNP positions identified across the dataset. (A) Proportion of SNPs that were sequenced in each individual, and (B) frequency of observed heterozygous loci among sequenced SNPs within native (triangles), naturalized (dashes), and invading (circles) individuals, relative to total sequencing effort after read cleaning. Linear fits are shown to native (gray line) and invading (black line) individuals.
Sequence overlap among samples also was improved by increased sequencing effort. In pairwise comparisons, the proportion of all polymorphic positions shared between individuals increased from <1% to almost 30% in the largest libraries (which shared nearly 70% of the SNP positions observed in any individual library; Figure 4). The 10 largest libraries included four native, four invading, and two naturalized individuals with more than 200 Mbp of sequencing effort each. Surveys of SNP sites—as identified from across the dataset—within just these greatest coverage libraries revealed that most SNP sites were sequenced in only a few individuals, though 6883 loci were present in all 10 libraries (Figure S5). The number of overlapping SNP positions was significantly greater than expected by chance, given the number of positions observed in each library (K-S Goodness of Fit test: P < 0.01, Figure S5). Overlapping positions are almost certainly biased toward the most commonly expressed loci, however our GO categorizations indicate that this does not represent a particularly biased subset of genome function. Functional categorization and distribution of the annotations within the three GO categories (biological process, cellular component, and molecular function), did not vary more than a few percent among the individual assemblies for any classification, though this modest variation was statistically significant (P < 0.0001 among libraries within each of the three categories, Figure 5). There was a consistent trend toward greater representation of loci of unknown function (although still similar to A. thaliana proteins) in larger libraries within each category (Figure 5), indicating greater recovery of less well-studied and presumably more rarely-expressed loci.
Figure 4 .
Pairwise overlap in observed SNPs. Isoclines reflect the proportion of all SNP positions that are observed in pairwise comparisons of 40 transcriptome libraries, as a function of sequencing effort in both samples.
Figure 5 .
The distribution of GO annotations to A. thaliana as a function of transcriptome sequencing effort.
Novel insights into YST population genetics
For introduced and invasive species, identifying the sources of their genetic variation can provide important insights into the circumstances that made these introductions so successful (Dlugosch and Parker 2008; Prentis et al. 2008; Lai et al. 2012). Native individuals assembled slightly higher numbers of both unigenes and contigs (Figure 2) than invading individuals, potentially indicating reduced sequence variation in introduced genotypes, but these differences were not significant in either case (analysis of covariance on Ln-transformed data: unigenes, P = 0.06; contigs, P = 0.23). Instead, the identification of allelic variation among our samples suggested different population genetic inferences in our YST transcriptome dataset: Across all inferred SNP positions, invaders had significantly greater heterozygosity than natives (analysis of covariance : F2,33 = 82.21, P = 0.005, Figure 4B) and a greater variance around predicted values based on sequencing effort (linear regressions: invader R2 = 0.67, native R2 = 0.96). These patterns are consistent with a known history of multiple, large introductions of YST (Gerlach 1997), which should have established substantial genetic diversity in the invaded range.
With the use of 2568 SNPs that were genotyped in at least five individuals per continent, STRUCTURE modeling was able to detect coarse genetic structure in the native range. Estimates of support for population number (ln Pr[X|K]) peaked at K = 2, and these two subpopulations were partitioned geographically, with one region including individuals from Hungary and Romania in eastern Europe and the other region including individuals from Turkey and the Republic of Georgia in the Caucasus (Figure 6A). Including invading and naturalized individuals together with natives in a single model obscured all genetic structure and supported no subpopulations, even when geographic groups were provided as prior information in the model, a pattern indicative of a large number of admixed individuals in the dataset (Pritchard et al. 2000). When the two native subpopulations were instead provided as a priori fixed groups, invading and naturalized individuals were both assigned as admixtures of the native subpopulations (Figure 6B). The evidence for admixture suggested by our coarse population sampling is consistent with a history of multiple introductions, and similar patterns in the Spanish collections provide some of the first support for the hypothesis that western European populations are not native and are themselves naturalized products of past introductions (Maddox et al. 1985).
Figure 6 .

STRUCTURE populations inferred from SNP variation. Vertical bars show the population assignment (color) for each individual by region. (A) Two major genetic groups (blue and orange) are supported for the native range, based upon genotypes from Hungary (HU), Romania (RO), Turkey (TK) and the Republic of Georgia (GA). (B) Admixture of these sources is suggested for both putatively naturalized genotypes in Spain (SP) and invading genotypes from California (CA) and Argentina (AR), when the two native genetic groups are fixed as potential source populations. SNP frequencies were based upon 2568 positions observed in at least five individuals per continent.
Inferences regarding introduction scenarios are only as robust as the sampling of the potential source populations (Dlugosch and Parker 2008), however, and our dataset is not extensive enough to rule out that unsampled source populations—rather than admixture of our observed native populations—have produced the current invasions. Thorough geographic sampling of the native range is essential for correctly identifying the most likely source genotypes. Moreover, dozens of individuals should be sampled from within each population to accurately estimate allele frequencies, and recover well-supported patterns of population structure (Pritchard et al. 2000; Avise 2004). Transcriptome studies generally have been considered too expensive to be used for this kind of broad population sampling, but as they becomes increasingly cost effective, investment in deeper sampling of individuals promises to be fruitful for robust population genomic studies.
Conclusions
By analyzing the realized outcome of a wide range of sequencing efforts, our dataset reveals that allele recovery is far more dependent on sequencing depth than is gene recovery, although both are enhanced by additional sequencing depth. These relationships are of particular concern for diversity comparisons among outbred individuals, which are increasingly taking center stage as genomic sequencing moves outside of its historical focus on inbred lines of traditional model organisms. Modest transcriptome sampling nevertheless generates thousands of informative markers observable across many individuals. This is promising news for further studies of coding variation among individuals, and our ability to combine datasets generated with different methods over time – one of the inherent strengths of sequence-based population genetics. Using our novel bioinformatic pipeline for allele identification, we were able to recover previously hypothesized population features using 40 individuals dispersed across a worldwide distribution, demonstrating the information-rich nature of transcriptome populations datasets.
Supplementary Material
Acknowledgments
We thank Ö. Eren, L. Khetsuriani, A. Diaconu, K. Török, and D. Montesinos for sharing seed collections; B. Boese and the 454 Sequencing Center for discussions about GS FLX sequencing techniques; M.S. Barker for discussions about sequence assembly; M. King for computing support; and two anonymous reviewers for helpful comments on the manuscript. Funding for this study was provided by a Roche Applied Science/454 Life Sciences sequencing prize to Z.L. and L.H.R., and a Natural Sciences and Engineering Research Council of Canada (NSERC) grant #353026 to L.H.R.
Footnotes
Communicating editor: J. Dekker
Literature Cited
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., et al. , 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeloni F., Wagemaker C. A. M., Jetten M. S. M., Op den Camp H. J. M., Jassen-Megens E. M., et al. , 2011. De novo transcriptome characterization and development of genomic tools for Scabiosa columbaria L. using next-generation sequencing techniques. Mol. Ecol. Res. 11: 662–674 [DOI] [PubMed] [Google Scholar]
- Avise J. C., 2004. Molecular Markers, Natural History, and Evolution. Sinauer Associates, Sunderland, MA [Google Scholar]
- Barbazuk W. B., Emrich S. J., Chen H. D., Li L., Schnable P. S., 2007. SNP discovery via 454 transcriptome sequencing. Plant J. 51: 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk W. B., Fu Y., McGinnis K. M., 2008. Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res. 18: 1381–1392 [DOI] [PubMed] [Google Scholar]
- Barker M. S., Kane N. C., Matvienko M., Kozik A., Michelmore W., et al. , 2008. Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Mol. Biol. Evol. 25: 2445–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker M. S., Dlugosch K. M., Dinh L., Challa R. S., Kane N. C., et al. , 2010. EvoPipes.net: bioinformatic tools for ecological and evolutionary genomics. Evol. Bioinf. 6: 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker M. S., Baute G. J., Liu S.-L., 2012. Duplication and turnover in plant genomes, pp. 155–169 in Plant Genome Diversity, edited by Wendel J. F. Springer, Vienna [Google Scholar]
- Barreto F. S., Moy G. W., Burton R. S., 2011. Interpopulation patterns of divergence and selection across the transcriptome of the copepod Tigriopus californicus. Mol. Ecol. 20: 560–572 [DOI] [PubMed] [Google Scholar]
- Beldade P., Rudd S., Gruber J. D., Long A. D., 2006. A wing expressed sequence tag resource for Bicyclus anynana butterflies, an evo-devo model. BMC Genomics 7: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. R., 2006. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 16: 545–552 [DOI] [PubMed] [Google Scholar]
- Bergelson J., Kreitman M., Stahl E. A., Tian D., 2001. Evolutionary dynamics of plant R-genes. Science 292: 2281–2285 [DOI] [PubMed] [Google Scholar]
- Bouck A., Vision T., 2007. The molecular ecologist’s guide to expressed sequence tags. Mol. Ecol. 16: 907–924 [DOI] [PubMed] [Google Scholar]
- Brancheva S., Greilhuber J., 2006. Genome size in Bulgarian Centaurea s.l. (Asteraceae). Plant Syst. Evol. 257: 95–117 [Google Scholar]
- Charlesworth B., 2010. Molecular population genomics: a short history. Genet. Res. 92: 397–411 [DOI] [PubMed] [Google Scholar]
- Chevreux B., Pfisterer T., Drescher B., Driesel A. J., Muller W. E. G., et al. , 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14: 1147–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou D. C., Gorham J. M., Herman D. S., Seidman J. G., 2011. Construction of normalized RNA-seq libraries for next-generation sequencing using the crab duplex-specific nuclease. Curr. Prot. Mol. Biol. 94: 4.12.1–4.12.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey J. W., Hohenlohe P. A., Etter P. D., Boone J. Q., Catchen J. M., et al. , 2011. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 12: 499–510 [DOI] [PubMed] [Google Scholar]
- Dempewolf H., Kane N. C., Ostevik K. L., Geleta M., Barker M. S., et al. , 2010. Establishing genomic tools and resources for Guizotia abyssinica (L.f.) Cass.—the development of a library of expressed sequence tags, microsatellite loci, and the sequencing of its chloroplast genome. Mol. Ecol. Res. 10: 1048–1058 [DOI] [PubMed] [Google Scholar]
- Demuth J. P., De Bie T., Stajich J. E., Cristianini N., Hahn M. W., 2006. The evolution of mammalian gene families. PLoS ONE 1: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlugosch K. M., Bonin A., 2012. Allele identification in assembled genomic sequence datasets, pp. 197–211 in Methods in Molecular Biology Series: Data Production and Analysis in Population Genomics, edited by Bonin A., Pompanon F. Springer, New York: [DOI] [PubMed] [Google Scholar]
- Dlugosch K. M., Parker I. M., 2008. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 17: 431–449 [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium , 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218 [DOI] [PubMed] [Google Scholar]
- Duarte J. M., Wall P. K., Edger P. P., Landherr L. L., Ma H., et al. , 2010. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol. Biol. 10: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R. M., Altshuler D., Abecasis G. R., Bentley D. R., Chakravarti A., et al. , 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer K. R., Fan S., Gunter H. M., Jones J. C., Boekhoff S., et al. , 2010. Rapid evolution and selection inferred from the transcriptomes of sympatric crater lake cichlid fishes. Mol. Ecol. 19: 197–211 [DOI] [PubMed] [Google Scholar]
- Emerson K. J., Merz C. R., Catchen J. M., Hohenlohe P. A., Cresko W. A., et al. , 2010. Resolving postglacial phylogeography using high-throughput sequencing. Proc. Natl. Acad. Sci. USA 107: 16196–16200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J. K., 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach J., 1997. How the west was lost: reconstructing the invasion dynamics of yellow starthistle and other plant invaders of western rangelands and natural areas. California Exotic Pest Plant Council Symp. Proc. 3: 67–72 [Google Scholar]
- Hahn M. W., De Bie T., Stajich J. E., Nguyen C., Cristianini N., 2005. Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome Res. 15: 1153–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. W., Han M. V., Han S.-G., 2007. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 3: e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamrick J. L., Godt M. J., 1989. Allozyme diversity in plant species, pp. 43–63 in Plant Population Genetics, Breeding and Germplasm Resources, edited byA. H. D ., Brown M. T., Clegg A. L., Kahler, Weir B. S. Sinauer, Sunderland [Google Scholar]
- Hancock A. M., Brachi B., Faure N., Horton M. W., Jarymowycz L. B., et al. , 2011. Adaptation to climate across the Arabidopsis thaliana genome. Science 334: 83–86 [DOI] [PubMed] [Google Scholar]
- Heiser C. B., Whitaker T. W., 1948. Chromosome number, polyploidy, and growth habit in California weeds. Am. J. Bot. 35: 179–186 [PubMed] [Google Scholar]
- Huang X., Madan A., 1999. CAP3: a DNA sequence assembly program. Genome Res. 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozik A., Matvienko M., Kozik I., Van Leeuwen H., Van Deynze A., et al. , 2008. Eukaryotic ultra conserved orthologs and estimation of gene capture in EST libraries. Plant and Animal Genome Conference XVI: P6 [Google Scholar]
- Lai Z., Kane N. C., Kozik A., Hodgins K., Dlugosch K. M., et al. , 2012. Genomics of Compositae weeds: EST libraries, microarrays, and evidence of introgression. Am. J. Bot. 99: 209–218 [DOI] [PubMed] [Google Scholar]
- Lassmann T., Hayashizaki Y., Daub C. O., 2009. TagDust—a program to eliminate artifacts from next generation sequencing data. Bioinfomatics 25: 2839–2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor D. A., Ward F. E., Ennis P. D., Jackson A. P., Parham P., 1988. HLA-A and B polymorphisms predate the divergence of humans and chimpanzees. Nature 335: 268–271 [DOI] [PubMed] [Google Scholar]
- Lewontin R. C., Hubby J. L., 1966. A molecular approach to the study of genic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics 54: 595–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ruan J., Durbin R., 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. H., Sadler L. A., 1991. Low nucleotide diversity in man. Genetics 129: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J. S., 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Maddox D. M., Mayfield A., Poritz N. H., 1985. Distribution of yellow starthistle (Centaurea solstitialis) and Russian knapweed (Centaurea repens). Weed Sci. 33: 315–327 [Google Scholar]
- Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., et al. , 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M. I., Abecasis G. R., Cardon L. R., Goldstein D. B., Little J., et al. , 2008. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 9: 356–369 [DOI] [PubMed] [Google Scholar]
- Meyer E., Aglyamova G. V., Wang S., Buchanan-Carter J., Abrego D., et al. , 2009. Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. BMC Genomics 10: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin P. A., Luikart G., Wayne R. K., SNP Wokshop Group , 2004. SNPs in ecology, evolution and conservation. Trends Ecol. Evol. 19: 208–216 [Google Scholar]
- Moriyama E. N., Powell J. R., 1996. Intraspecific nuclear DNA variation in Drosophila. Mol. Biol. Evol. 13: 261–277 [DOI] [PubMed] [Google Scholar]
- Nielsen R., Paul J. S., Albrechtsen A., Song Y. S., 2011. Genotype and SNP calling from next-generation sequencing data. Nat. Rev. Genet. 12: 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Z., Cox A. J., Mullikin J. C., 2001. SSAHA: a fast search method for large DNA databases. Genome Res. 11: 1725–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Clark R. M., Lanz C., Warthmann N., et al. , 2008. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 18: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk M., Martin E., Dinc M., Duran A., Ozdemir A., et al. , 2009. A cytogenetical study on some plant taxa in Nizip region (Aksaray, Turkey). Turk. J. Biol. 33: 35–44 [Google Scholar]
- Prentis P. J., Wilson J. R. U., Dormontt E. E., Richardson D. M., Lowe A. J., 2008. Adaptive evolution in invasive species. Trends Plant Sci. 13: 288–294 [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnell P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team , 2010. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Renaut S., Nolte A. W., Bernatchez L., 2010. Mining transcriptome sequences towards identifying adaptive single nucleotide polymorphisms in lake whitefish species pairs (Coregonus spp. Salmonidae). Mol. Ecol. 19: 115–131 [DOI] [PubMed] [Google Scholar]
- Rosenberg N. A., 2004. Distruct: a program for the graphical display of population structure. Mol. Ecol. Notes 4: 137–138 [Google Scholar]
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., et al. , 2004. Large-scale copy number polymorphism in the human genome. Science 305: 525–528 [DOI] [PubMed] [Google Scholar]
- Seeb J. E., Pascal C. E., Grau E. D., Seeb L. W., Templin W. D., et al. , 2011. Transcriptome sequencing and high-resolution melt analysis advance single nucleotide polymorphism discovery in duplicated salmonids. Mol. Ecol. Res. 11: 335–348 [DOI] [PubMed] [Google Scholar]
- Sun M., 1997. Population genetic structure of yellow starthistle (Centaurea solstitialis), a colonizing weed in the western United States. Can. J. Bot. 75: 1470–1478 [Google Scholar]
- Sun M., Ritland K., 1998. Mating system of yellow starthistle (Centaurea solstitialis), a successful colonizer in North America. Heredity 80: 225–232 [Google Scholar]
- The Tomato Genome Consortium , 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno S., Le Provost G., Léger V., Klopp C., Noirot C., et al. , 2010. Bioinformatic analysis of ESTs collected by Sanger and pyrosequencing methods for a keystone forest tree species: oak. BMC Genomics 11: 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P., Hogers R., Bleeker M., Reijans M., van de Lee T., et al. , 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23: 4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J., 2008. Coalescent Theory: An introduction. Roberts & Company Publishers, Greenwood Village. CO [Google Scholar]
- Wheat C. W., 2010. Rapidly developing functional genomics in ecological model systems via 454 transcriptome sequencing. Genetica 138: 433–451 [DOI] [PubMed] [Google Scholar]
- Widmer T., Guermache F., Dolgovskaia M., Reznick S., 2007. Enhanced growth and seed properties in introduced vs. native populations of yellow starthistle (Centaurea solstitialis). Weed Sci. 55: 465–473 [Google Scholar]
- Wilson L. M., Jette C., Connett J., McCaffrey J., 2003. Biology and Biological Control of Starthistle. USDA Forest Service / University of Nebraska Lincoln Faculty Publications, Lincoln, NB [Google Scholar]
- Zakas C., Schult N., McHugh D., Jones K. L., Wares J. P., 2012. Transcriptome analysis and SNP development can resolve population differentiation of Streblospio benedicti, a developmentally dimorphic marine annelid. PLoS ONE 7: e3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed A., Whitfield C. W., 2008. A genome-wide signature of positive selection in ancient and recent invasive expansions of the honey bee Apis mellifera. Proc. Natl. Acad. Sci. USA 105: 3421–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Zhao L., Gao J., Fei Z., 2011. iAssembler: a package for de novo assembly of Roche-454/Sanger transcriptome sequences. BMC Bioinf. 12: 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhulidov P. A., Bogdanova E. A., Shcheglov A. S., Vagner L. L., Khaspekov G. L., 2004. Simple cDNA normalization using kamchatka crab duplex specific nuclease. Nucleic Acids Res. 32: e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.