Abstract
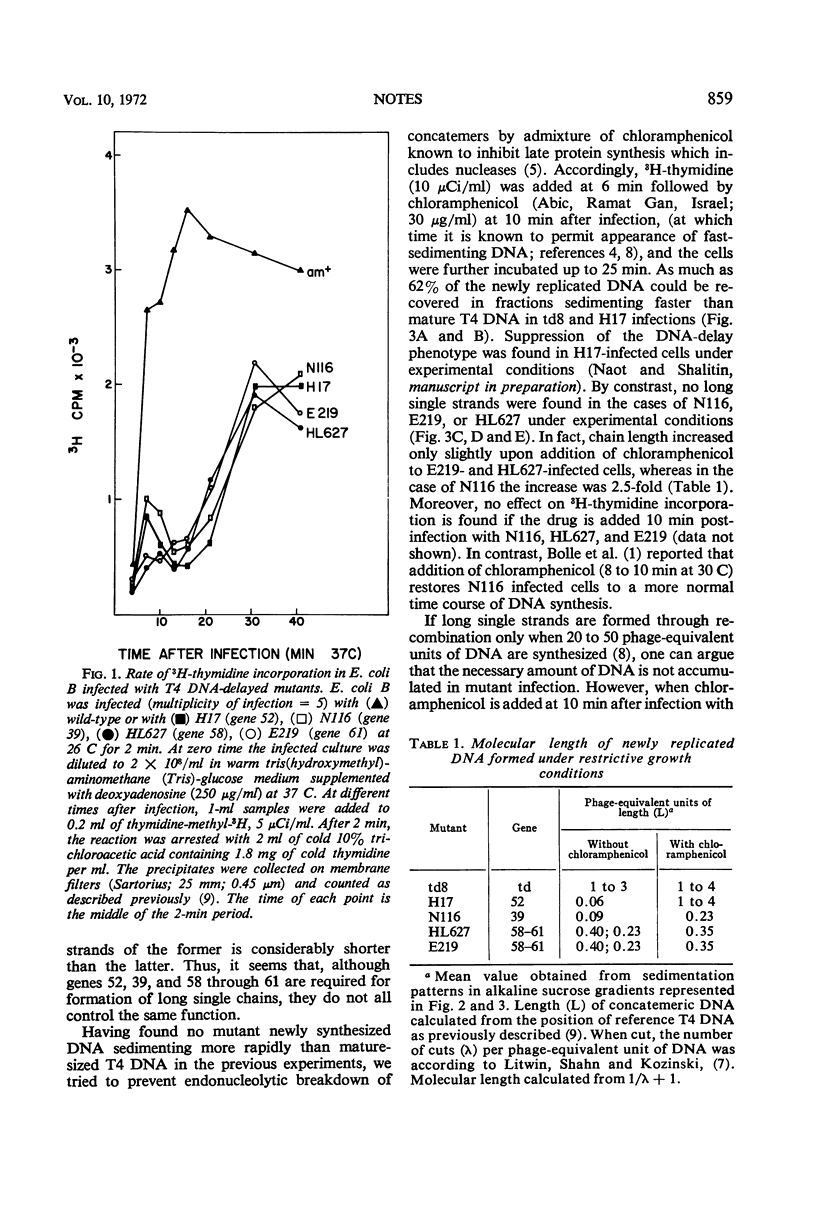
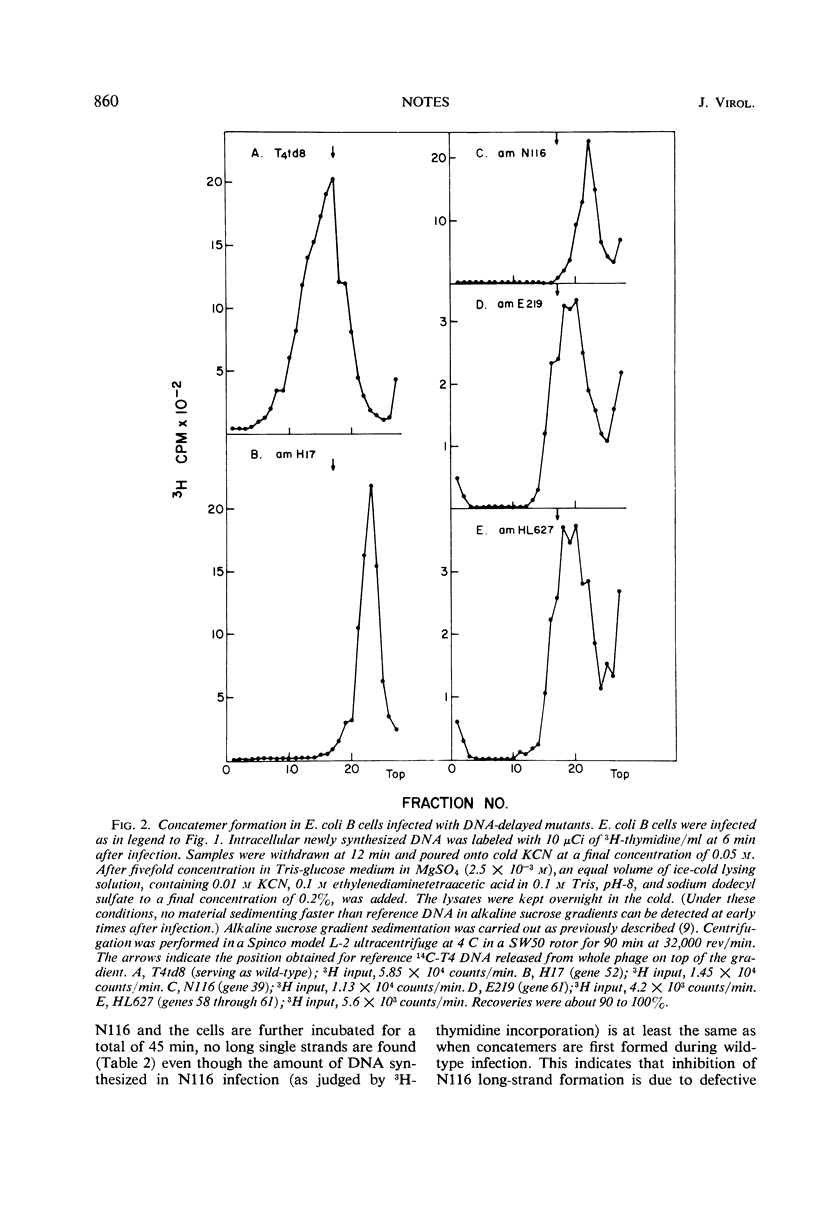
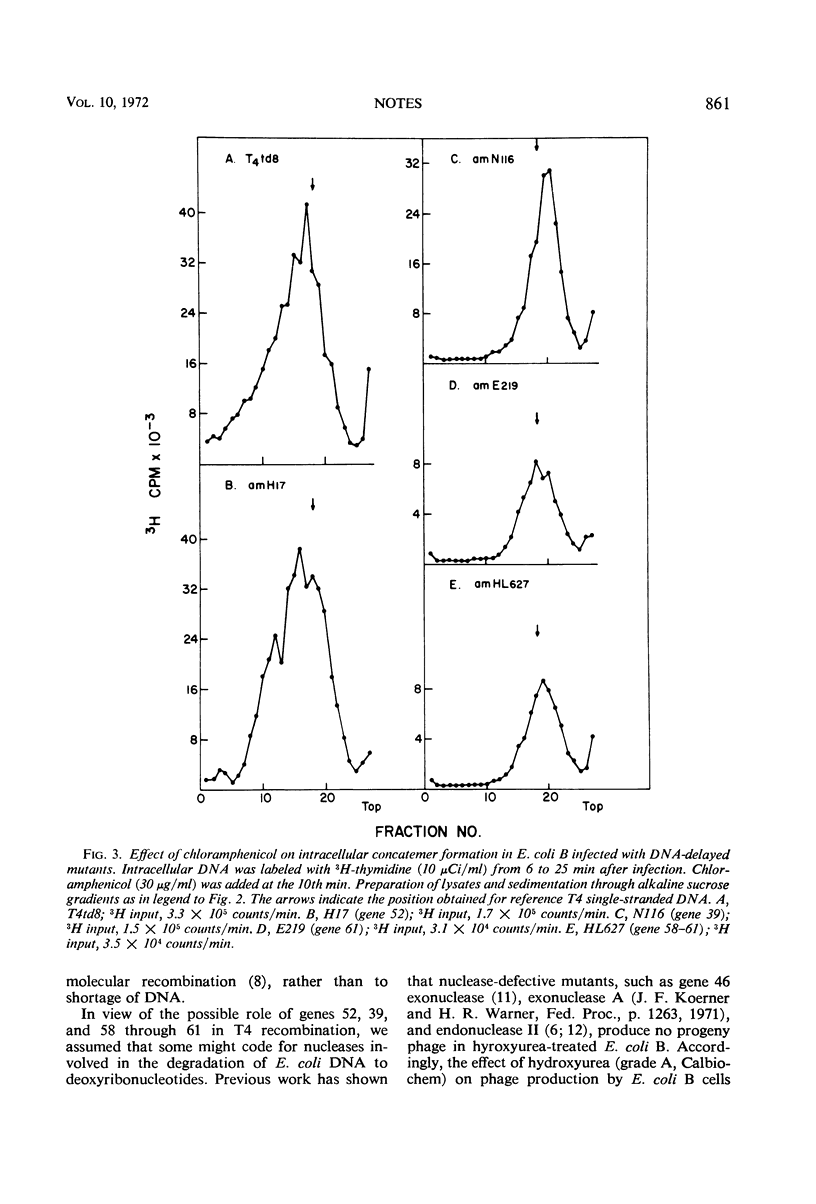
Nonpermissive host cells infected with phage T4 mutants in genes 52, 39, and 58 through 61 are shown to form short intracellular single-stranded deoxyribonucleic acid in contrast to wild-type infected cells, which form dimers and trimers of T4 genome size.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolle A., Epstein R. H., Salser W., Geiduschek E. P. Transcription during bacteriophage T4 development: requirements for late messenger synthesis. J Mol Biol. 1968 Apr 28;33(2):339–362. doi: 10.1016/0022-2836(68)90193-9. [DOI] [PubMed] [Google Scholar]
- Frankel F. R., Batcheler M. L., Clark C. K. The role of gene 49 in DNA replication and head morphogenesis in bacteriophage T4. J Mol Biol. 1971 Dec 28;62(3):439–463. doi: 10.1016/0022-2836(71)90147-1. [DOI] [PubMed] [Google Scholar]
- Frankel F. R. DNA replication after T4 infection. Cold Spring Harb Symp Quant Biol. 1968;33:485–493. doi: 10.1101/sqb.1968.033.01.056. [DOI] [PubMed] [Google Scholar]
- Frankel F. R. Evidence for long DNA strands in the replicating pool after T4 infection. Proc Natl Acad Sci U S A. 1968 Jan;59(1):131–138. doi: 10.1073/pnas.59.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercules K., Munro J. L., Mendelsohn S., Wiberg J. S. Mutants in a nonessential gene of bacteriophage T4 which are defective in the degradation of Escherichia coli deoxyribonucleic acid. J Virol. 1971 Jan;7(1):95–105. doi: 10.1128/jvi.7.1.95-105.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin S., Shahn E., Kozinski A. W. Interpretation of sucrose gradient sedimentation pattern of deoxyribonucleic acid fragments resulting from random breaks. J Virol. 1969 Jul;4(1):24–30. doi: 10.1128/jvi.4.1.24-30.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. C., Kozinski A. W., Litwin S. Molecular Recombination in T4 Bacteriophage Deoxyribonucleic Acid: III. Formation of Long Single Strands During Recombination. J Virol. 1970 Mar;5(3):368–380. doi: 10.1128/jvi.5.3.368-380.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin C., Naot Y. Role of gene 46 in bacteriophage T4 deoxyribonucleic acid synthesis. J Virol. 1971 Aug;8(2):142–153. doi: 10.1128/jvi.8.2.142-153.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H. R., Hobbs M. D. Effect of hydroxyurea on replication of bacteriophage T4 in Escherichia coli. J Virol. 1969 Mar;3(3):331–336. doi: 10.1128/jvi.3.3.331-336.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H. R., Hobbs M. D. Incorporation of uracil-14C into nucleic acids in Escherichia coli infected with bacteriophage T4 and T4 amber mutants. Virology. 1967 Nov;33(3):376–384. doi: 10.1016/0042-6822(67)90113-4. [DOI] [PubMed] [Google Scholar]
- Warner H. R., Snustad P., Jorgensen S. E., Koerner J. F. Isolation of bacteriophage T4 mutants defective in the ability to degrade host deoxyribonucleic acid. J Virol. 1970 Jun;5(6):700–708. doi: 10.1128/jvi.5.6.700-708.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegian C. D., Mueller M., Selzer G., Russo V., Stahl F. W. Properties of the DNA-delay mutants of bacteriophage T4. Virology. 1971 Dec;46(3):900–919. doi: 10.1016/0042-6822(71)90090-0. [DOI] [PubMed] [Google Scholar]


