Abstract
To date, it remains impossible to guarantee that short-term treatment given to a patient suffering from a major depressive episode (MDE) will improve long-term efficacy. Objective biological measurements and biomarkers that could help in predicting the clinical evolution of MDE are still warranted. To better understand the reason nearly half of MDE patients respond poorly to current antidepressive treatments, we examined the gene expression profile of peripheral blood samples collected from 16 severe MDE patients and 13 matched controls. Using a naturalistic and longitudinal design, we ascertained mRNA and microRNA (miRNA) expression at baseline, 2 and 8 weeks later. On a genome-wide scale, we detected transcripts with roles in various biological processes as significantly dysregulated between MDE patients and controls, notably those involved in nucleotide binding and chromatin assembly. We also established putative interactions between dysregulated mRNAs and miRNAs that may contribute to MDE physiopathology. We selected a set of mRNA candidates for quantitative reverse transcriptase PCR (RT-qPCR) to validate that the transcriptional signatures observed in responders is different from nonresponders. Furthermore, we identified a combination of four mRNAs (PPT1, TNF, IL1B and HIST1H1E) that could be predictive of treatment response. Altogether, these results highlight the importance of studies investigating the tight relationship between peripheral transcriptional changes and the dynamic clinical progression of MDE patients to provide biomarkers of MDE evolution and prognosis.
Keywords: antidepressant, biomarker, miRNA, mood disorder, PBMC, transcriptome
Introduction
Responses to current treatments of a major depressive episode (MDE) are often inconsistent and unpredictable. More than one-third of patients will not respond to the first antidepressant prescribed. Less than half of treated patients demonstrate complete remission.1, 2 Practice guidelines recommend waiting from 2 to 8 weeks with the same treatment to evaluate initial clinical response3, 4, 5 (that is, a 50% reduction in depressive symptoms as measured by depression rating scales such as the Hamilton rating scale for depression (HDRS) or the Montgomery–Asberg depression rating scale (MADRS)).6 Moreover, studies suggest that clinical response generally occurs after about a month,7 and could occur after 10 weeks of treatment initiation.8 On the other hand, waiting for such a long time without efficacy is associated with extended suffering, disability and risk of suicide.9 Without clinical parameters clearly predictive of treatment outcome, it is therefore challenging to make a valid prognosis for MDE. In addition, there has not been any established biological marker at present that can predict treatment response for the individual patient before the initiation or during the early course of antidepressant treatment.10
Epidemiological studies suggest that environmental factors, especially exposure to stressful life events, have a significant role in triggering major depression.11 It has been proposed that the combination of certain environmental factors with genetic predispositions would result in an epigenetic and persistent dysregulation of cerebral gene programs, leading to phenotypic manifestations of psychiatric disorders, including depression.12 To study such gene program alterations in a central nervous system disorder, we and others underlined the potential interest of using a peripheral tissue sample, such as blood, as these tissues share numerous biological pathways with the central nervous system.13, 14, 15 Indeed, significant gene expression similarities were found between blood and brain,16, 17 including numerous candidate genes in mood disorders.18, 19 Moreover, compared with post-mortem tissues or cell lines cultured with long-term serial passage, freshly collected blood cells offer the possibility to more precisely correlate the stage of clinical evolution and treatment with biological observations. Several publications have shown transcriptional variations in peripheral blood that discriminate MDE patients from controls14, 20, 21, 22, 23, 24, 25 and also patients during several weeks of treatment.13, 19, 26, 27, 28, 29, 30, 31
Of note, most gene expression studies investigating MDE have been conducted with a candidate gene design that is biased toward pre-existing theory. On the other hand, pangenomic studies offer insights from novel molecular signatures and possible underlying biological mechanisms, but have the drawback of multiple testing and PCR validation. In addition, when validating potential biomarkers with or without hypothesis-driven investigations of transcriptional signatures, the physiological and stochastic variability of gene expression of samples within the same patient, between different patients, and more importantly within and between healthy controls, remain largely underestimated, raising the possibility of biases and false-positive gene candidates. Thus, although some studies suggest that the majority of gene expression is stable within individuals and have low dependence on physiological parameters,32 repetitive measurements of gene expression in both patients and controls may help discriminate real variations triggered by disease evolution and treatment pharmacodynamics from other causes of gene expression variability.
We and others have shown that genes dysregulated during MDE belong to various molecular families.13, 33 One possible explanation for this convergent regulation of structurally distant markers might be due to common master regulator(s) yet to be characterized. MicroRNAs (miRNAs) are a class of non-coding RNAs that regulate gene expression by binding to their target mRNAs. Recently, it has been hypothesized that miRNAs have a role in brain disorders and emerging evidence suggests that they regulate neuropathology associated processes, such as brain development, dendritic spine morphology and neurite outgrowth.34 The estimated number of human miRNAs exceeds 1500 (miRBase release 18) with >60% of all human mRNA predicted to contain conserved miRNA targets. Thus, miRNAs represent candidates for psychiatric diseases-associated genetic and epigenetic factors and constitute promising targets for novel drugs.35 So far, although several studies investigated altered levels of miRNAs in schizophrenia, only one work comprehensively examined miRNA expression in the prefrontal cortex of depressed suicide subjects.36
In this study, we undertook a longitudinal follow-up of severe MDE patients and their matched controls, recruited in naturalistic conditions, to further characterize transcriptional biomarkers of MDE evolution. After conducting genome-wide screens and quantitative reverse transcriptase PCR (RT-qPCR) validation, we evaluated whether specific RNA signatures (both mRNA and miRNA) from blood collected at 0, 2 and 8 weeks after study inclusion can distinguish MDE patients from controls and may be predictive of the treatment response.
Materials and methods
Design setting and subjects
The study design was a naturalistic, prospective, longitudinal and comparative study with assessments of MDE patients and healthy controls at baseline (week 0), 2 and 8 weeks after study inclusion. Sixteen patients who met the Diagnostic and Statistical Manual of Mental Disorders, fourth edition, text revision (DSM-IV-TR) criteria for major depressive disorder participated to the study.37 Inclusion criteria were: (i) treated or untreated MDE (ii) 17-item HDRS score ⩾20 corresponding to severe or very severe MDE.38
At the end of the 8-week clinical follow-up, patients were classified as responders and nonresponders according to the consensual definition of clinical response corresponding to a minimal reduction of 50% of the HDRS score from the initial evaluation.6
For the control group, age- and sex-matched subjects were evaluated to exclude any psychiatric disorder history using the French version of standardized interview validated for health control subjects (SCID-NP).
Blood mRNA extraction
Peripheral blood mononuclear cells (PBMCs) were isolated from the blood by Ficoll density centrifugation. Total RNAs were extracted from the PBMCs with the mirVana miRNA isolation kit (Ambion, Austin, TX, USA) according to the manufacturer's protocol. RNA concentration was determined using a nanodrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). RNA integrity was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
Preparation of samples and microarray assay
Sample amplification, labeling and hybridization onto Agilent whole human genome oligo microarrays containing 50 599 different oligonucleotide probes (SurePrint G3 Human GE 8 × 60 K, Agilent Technologies) essentially followed the one-color microarray-based gene expression analysis (low RNA input linear amplification PLUS kit) recommended by Agilent Technologies (details in Supplementary Information). The scanned images were analyzed with Agilent feature extraction software 10.5.1.1 to obtain background subtracted and spatially detrended processed signal intensities. All data were normalized by quantile normalization, and only 39 784 oligonucleotide probes with signal intensities detectable (that is, above background according to the ‘gIsWellAboveBg' Agilent feature extraction value) in ⩾70% of samples (from either MDE patients or controls and at either baseline or 8 weeks later) were subsequently analyzed. The microarray data are available from the gene expression omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) under the series accession number GSE38206.
Gene and chromosome band enrichment analyses
Lists of significant genes were uploaded on DAVID (database for annotation, visualization and integrated discovery) for identifying statistically relevant signaling pathways,39 with high classification stringency, P-value (p)<0.05 and false discovery rate (FDR) ⩽5%. To determine the chromosomal cytoband enrichment in our lists of significant genes, we used the ToppFun algorithm and restricted the search to P<0.05.40
miRNA quantification
Four hundred nanogram of total RNA were reverse transcribed using the TaqMan MicroRNA RT kit (Applied Biosystems, Foster City, CA, USA) in combination with the stem-loop Megaplex primer pools (A and B v3.0, Applied Biosystems) without preamplification and according to manufacturer's recommendation. For each RT pool, the equivalent of 320 ng of total RNA converted into cDNA was mixed with TaqMan Universal PCR Master Mix II No AmpErase UNG (Applied Biosystems). Hundred microliter of mix (A or B) was loaded into each port of the corresponding 384 wells Human miRNA TaqMan low density array (that is, A or B) and run for 40 cycles on a ABI PRISM 7900 HT according to manufacturer's protocol with SDS v2.4 software. Normalized expression level of each miRNA was quantified as 2−ΔΔCt with the DataAssist software (Applied Biosystems, v3.0), relatively to the normalized expression level of the same miRNA in a calibrator sample (details in Supplementary Information).
Target prediction
For each list of differentially expressed miRNAs the Ingenuity pathway analysis software (Ingenuity systems), which relies on three popular algorithms (TargetSan, TarBase and miRecords), was queried to identify targets within lists of differentially expressed genes in our microarray analysis. Only highly predicted and experimentally validated targets were considered for further analysis.
Real-time RT-PCR for candidate gene validation
RNA was reverse transcribed with the High Capacity cDNA archive kit (Applied Biosystems). The resulting cDNA was combined with a TaqMan universal PCR Master Mix (Applied Biosystems) and 48 PCR reactions were simultaneously performed in triplicate on an ABI PRISM 7900HT thermocycler using tLDA technology according to manufacturer recommendations (Applied Biosystems). For each tested candidate gene, primer sets and probes were selected using the web portal of the manufacturer (Applied Biosystems). The expression level of each candidate gene was calculated as 2−ΔΔCt with the DataAssist software (Applied Biosystems, v3.0), in which each candidate gene is quantified relative to the expression of one reference gene, or a combination of two reference genes exhibiting proximal level of expression compared with the target gene, and also with a calibrated sample (details in Supplementary Information).
Statistical analysis
Demographic and clinical data
Demographic and clinical variables were compared between patients and matched-controls for cohort A and between responder and nonresponder patients with a Fisher exact test for qualitative variables and a t-test for quantitative variables.
Microarray data
Differential gene expression were obtained using the MultiExperiment Viewer 4 (MeV4, TM4 software suite) and SAM (significant analysis of the microarrays program) were measured with a FDR threshold set at 5%.41, 42 Student's t-tests were also applied to determine P-values. The data were analyzed using either a two-class unpaired (for patients versus controls comparison) or two-class paired (for internal comparisons within patients or within controls) response type. Details are provided in Supplementary Information and include the Supplementary Figure 1 for the calculation of the number of samples required.
miRNA study
To select differentially expressed miRNAs (at 0 and 8-week visits), non-parametric unpaired Mann–Whitney tests were used to compare the fold change (FC) between patients and matched-controls with a threshold P-value of 0.05. Non-parametric paired Wilcoxon tests were used to compare the FC between the two evaluations within the MDE group with a threshold P-value of 0.05.
Candidate gene validation
For exploratory purposes and to compare microarray data with RT-qPCR data, parametric unpaired t-tests were used to compare the FC between patients and matched controls at first evaluation and 8 weeks later within either the whole cohort (FCall), the initial sub-cohort A (FCA), or the subsequent sub-cohort B (FCB).
To explore differences between responders and nonresponders, we calculated and compared marginal estimated mean of each group in a mixed linear model.
To explore the association between mRNA expression of best candidates and clinical evolution, we calculated Spearman's correlations between HDRS evolution (0 to 8- and 2 to 8-week intervals) and FC at inclusion or 2 weeks later, respectively. A discriminant function analysis was used with the goal of establishing a predictive score to classify patients in the responder versus nonresponder groups. Sensitivity, specificity, positive/negative predictive value and the confidence intervals of either each selected mRNA candidates or a combination of the best classifiers were computed for the combination of selected variables. Receiver-operating characteristic (ROC) curves analysis was used to determine the area under the curve.
Results
Demographic and clinical variables
We recruited a group of 16 severe MDE patients (MDE group) and 13 healthy sex- and age-matched controls (control group). A summary of clinical and therapeutic data for each patient is reported in Table 1. Patients and controls were all of Caucasian origin and the analysis of socio-demographic parameters (Table 1 and Supplementary Tables 1 and 2) showed no significant difference among patients and controls (Supplementary Table 3).
Table 1. Major depression sub-cohorts and the paired control cohorts.
| Cohort | Pairs |
Major depression group
|
Control group
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Inclusion
|
2 weeks
|
8 weeks
|
||||||||||
| Sex | Age at first visit | Episode number | HDRS | Medications | HDRS | Medications | HDRS | Medications | Sex | Age at first visit | ||
| A | 1 | F | 61 | 2 | 40 | Venl, Ola, Arip, Bzd | 36 | Mirt, Arip, Bzd | 14 | ECT, Mirt, Arip, Hydrox | F | 59 |
| 2 | F | 58 | 4 | 27 | Dul, Mirt, Li | 24 | Dul, Mirt, Li | 2 | Dul, Li | F | 56 | |
| 3 | F | 46 | 1 | 25 | N | 18 | Esc | 3 | Esc | F | 48 | |
| 4 | M | 42 | 3 | 26 | Dul, Mirt, Bzd | 13 | Dul, Mirt, Bzd | 8 | Dul, Mirt, Bzd | M | 37 | |
| 5 | F | 63 | 5 | 28 | Dul, Mirt, Bzd | 9 | Dul, Mirt, Li, Arip, Bzd | 7 | Dul, Mirt, Li, Arip, Bzd | F | 65 | |
| 6 | M | 57 | 1 | 22 | Miln, Mirt | 9 | Miln, Mirt | 8 | Miln, Mirt, Flx | M | 44 | |
| 7 | F | 70 | 4 | 26 | Clom, Flx, Venl, Amlp, Bzd | 16 | Venl, Amlp, Bzd | 4 | Venl, Amlp, Bzd | F | 64 | |
| 8 | M | 59 | 4 | 28 | Venl, Mirt, Arip | 23 | Venl, Mirt | 7 | ECT, Venl, Mirt | M | 55 | |
| 9 | M | 59 | 3 | 29 | Flv, Mirt, Bzd | 17 | Dul, Li, Bzd | 14 | Dul, Clom, Li, Bzd | M | 56 | |
| Mean | 5F/4M | 57.2 | 3 | 27.9 | 18.3 | 7.44 | 5F/4M | 53.8 | ||||
| s.e.m. | 2.83 | 0.47 | 1.66 | 2.83 | 1.43 | 3.07 | ||||||
| B | 10 | M | 56 | 1 | 30 | Ami | 16 | Ami, Bzd | 16 | Venl, Ami, Li, Bzd | M | 53 |
| 11 | M | 74 | 2 | 37 | Prx, Ola, Bzd | 29 | Prx, Ola, Bzd, Hydro | 32 | Sert, Mirt, Li, Ola, Bzd | M | 72 | |
| 12 | F | 47 | 4 | 21 | Dul | 9 | Dul | 8 | Dul | F | 53 | |
| 13 | F | 51 | 1 | 28 | Sert, Bzd | 25 | Sert, Bzd | — | F | 60 | ||
| 14 | M | 65 | 5 | 21 | Venl, Ola | 17 | Moclo, Ola | 20 | Moclo, Ola | — | — | |
| 15 | M | 41 | 2 | 24 | Venl, Bzd | 8 | Venl, Bzd | 20 | Venl | — | — | |
| 16 | M | 35 | 2 | 22 | Venl, Mirt, Esc, Ola | 23 | ECT, Esc, Venl, Ola | 25 | ECT, Esc, Venl, Mirt | — | — | |
| Mean | 2F/5M | 52.7 | 2.43 | 26.1 | 18.1 | 20.0 | 2F/2M | 59.5 | ||||
| s.e.m. | 5.12 | 0.57 | 2.24 | 3.01 | 3.36 | 4.48 | ||||||
| Total | Mean | 7F/9M | 55.3 | 2.75 | 27.1 | 18.3 | 12.5 | 7F/6M | 55.5 | |||
| s.e.m. | 2.71 | 0.36 | 1.33 | 2.00 | 2.24 | 2.55 | ||||||
Abbreviations: Amlp, amisulpride; Ami, amitryptiline; Arip, aripiprazole; Bzd, benzodiazepine; Clom, clomipranime; Dul, duloxetine; ECT, electro-convulsivo-therapy; Esc, escitalopram; Flv, Fluvoxamine; Flx, fluoxetine; HDRS, Hamilton rating scale for depression; Hydrox, hydroxyzine; Ipro, iproniazide; Li, lithium; Miln, milnacipram; Mirt, mirtazapine; Moclo, moclobémide; N, no; Ola, olanzapine; Prx, paroxetine; Sert, Sertraline; Venl, venlafaxine.
MDE patients demonstrated different patterns of clinical evolution. Ten patients showed a clinical response (responders), whereas six patients did not experience a clinical response at 8 weeks of follow-up (nonresponders). To increase the chances of identifying transcriptional variations between MDE and control subjects, we decided to use only samples obtained from responder MDE patients and their matched controls for genome-wide screening. For simplification, we separated our study cohort into sub-cohort A (including nine MDE responders and nine matched-controls) and sub-cohort B, corresponding to the 11 other individuals (seven MDE patients (one responder and six nonresponder) and four controls) whose samples were not screened by microarray. In addition, we did not find any significant differences between responders and nonresponders based on variables reported on Table 1 and the Supplementary Table 1, such as HDRS score at inclusion, episode duration before inclusion, total number of MDE, history of suicide attempts, and familial history of MDE or other severe psychiatric disorders (Supplementary Table 4).
Microarray analysis of altered mRNA expression
The 36 samples obtained after collecting PBMC total RNAs from responder patients and matched controls of cohort A were used to characterize MDE genome-wide microarray transcriptional signatures. After conducting multiple SAM analysis with FDR set at 5%, we identified lists of transcripts differentially expressed between MDE patients at week 0 (HDRS⩾20) and controls, as well as between MDE patients at week 8 (with minimal reduction of ⩾50% of HDRS) and controls. To challenge the statistical significance of the microarray results, we also performed t-test analysis, using an arbitrary chosen cutoff ⩾1.50 FC in expression and P-value of ⩽0.001. About 200 transcripts were dysregulated among MDE patients at baseline compared with controls (Supplementary Tables 5 and 6), while nearly 130 were altered in MDE patients at 8 weeks (Supplementary Tables 7 and 8), with a core of 70 transcripts remaining dysregulated in the same way at both inclusion and 8 weeks later in MDE compared with controls (Supplementary Tables 9 and 10). We found similar numbers of overexpressed and underexpressed transcripts when comparing MDE patients to controls. The strong decrease in the number of dysregulated transcripts in MDE patients at 8 week is suggestive of a state-dependent signature of MDE in relation to clinical improvement. Thus, we also performed paired statistical analysis to identify transcripts whose level of expression would change with either treatment response or clinical evolution in patients (baseline versus 8-week samples) but did not identify such a signature at a pangenomic level at an FDR of 5%.
Nevertheless, because we tested each control twice in the microarray analysis, we also determined whether certain candidate transcripts showing variations between patients and controls may be false positives. Indeed, a SAM analysis of paired control samples at baseline and at 8 weeks showed that false positive transcripts were not present in our lists of dysregulated transcripts between MDE and control subjects with FDR set at 5%. Furthermore, we determined that variations for 53 transcripts reached statistical significance using a t-test (FC⩾1.50; P<0.001; Supplementary Tables 11 and 12). Importantly, among transcripts exhibiting variations compared with controls, we were able to find a few previously identified candidate genes for MDE physiopathology, such as NPY and GRIK5, highlighting the importance of multiple measurements in control samples to exclude transcriptional variations not specifically associated to disease pathophysiology.
As presented in Table 2 and Supplementary Table 13, using gene ontology analysis of dysregulated MDE transcripts, the most significant biological processes associated with MDE signatures were related to the nucleosome and chromatin. This result suggests a trait-dependent signature because it was observed for both baseline and 8-week samples. For overexpressed transcripts, processes identified included cell death and vacuole or lysosome organization. In addition, the chromosomal localization of dysregulated transcripts revealed significant enrichment of certain cytobands (FDR⩽1%), such as the 6p22 region containing histone genes, and the 2q21, 9p21 and 22q13 cytobands (Supplementary Table 14).
Table 2. Gene ontology analysis of dysregulated genes (FDR⩽5%).
| Category | ID | Term | Genes | Count | % | P-value |
|---|---|---|---|---|---|---|
| MDE 0w vs C (FC⩾1.50) | ||||||
| Biological process | GO:0008219 | Cell death | API5, FIG4, GSPT1, ARHGEF6, APAF1, ELMO2, HIPK3, PPT1, PDCD6IP, PKM2, SGPP1, TPP1, KRAS | 13 | 12.4 | 1.6E-3 |
| Biological process | GO:0007033 | Vacuole organization | FIG4, HEXB, PPT1, TPP1 | 4 | 3.8 | 1.9E-3 |
| Cellular component | GO:0016023 | Cytoplasmic membrane-bounded vesicle | AGFG1, DENND1A, CHIC2, HEXB, HGF, MAPKAP1, PPT1, PDCD6IP, SNAP23, TPP1 | 10 | 9.5 | 3.3E-3 |
| Molecular function | GO:0000166 | Nucleotide binding | ACTBL2, ACSL4, APAF1, ARL8B, ATP2A2, CMPK1, CTBP1, CELF2, GNAQ, GSPT1, HNRNPF, HIPK3, KRAS, MAT2A, MAPK6, MYH9, PKM2, POTEE, POTEF, POTEKP, PTBP1, RAB18, RAP2C, UBE2E2, UBE2H | 25 | 23.8 | 4.2E-3 |
| MDE 0w vs C (FC⩽−1.50) | ||||||
| Cellular component | GO:0000786 | Nucleosome | HIST1H1A, HIST1H1D, HIST1H1E, HIST1H3F/HIST2H3A/HIST1H3D, HIST1H4E/HIST1H4A/HIST1H4K/HIST1H4L | 5 | 8.9 | 8.6E-6 |
| Cellular component | GO:0044427 | Chromosomal part | CENPP, HIST1H1A, HIST1H1D, HIST1H1E, HIST1H3F/HIST2H3A/HIST1H3D, HIST1H4E/HIST1H4A/HIST1H4K/HIST1H4L | 6 | 10.7 | 1.1E-3 |
| Biological process | GO:0006325 | Chromatin organization | HIST1H1A, HIST1H1D, HIST1H1E, HIST1H3F/HIST2H3A/HIST1H3D, HIST1H4E/HIST1H4A/HIST1H4K/HIST1H4L, TLK2 | 6 | 10.7 | 1.1E-3 |
| MDE 8w (responders) vs C (FC⩾1.50) | ||||||
| Biological process | GO:0007040 | Lysosome organization | HEXB, PPT1, TPP1 | 3 | 5.5 | 2.6E-3 |
| MDE 8w (responders) vs C (FC⩽−1.50) | ||||||
| Biological process | GO:0006323 | DNA packaging | ASH1L, HIST1H1C, HIST1H1D, HIST1H1E, HIST1H3F/HIST1H3D, HIST1H4J/HIST1H4K/HIST1H4L/HIST1H4E/HIST1H4A | 6 | 12.5 | 7.4E-6 |
| Biological process | GO:0006334 | Nucleosome assembly | HIST1H1C, HIST1H1D, HIST1H1E, HIST1H3F/HIST1H3D, HIST1H4J/HIST1H4K/HIST1H4L/HIST1H4E/HIST1H4A | 5 | 10.4 | 4.4E-5 |
| Biological process | GO:0051276 | Chromosome organization | ASH1L, HIST1H1C, HIST1H1D, HIST1H1E, HIST1H3F/HIST1H3D, HIST1H4J/HIST1H4K/HIST1H4L/HIST1H4E/HIST1H4A, SMC1A, TLK2 | 8 | 16.7 | 1.1E-4 |
| Biological process | GO:0006325 | Chromatin organization | ASH1L, HIST1H1C, HIST1H1D, HIST1H1E, HIST1H3F/HIST1H3D, HIST1H4J/HIST1H4K/HIST1H4L/HIST1H4E/HIST1H4A, TLK2 | 7 | 14.6 | 2.2E-4 |
| Biological process | GO:0034622 | Cellular macromolecular complex assembly | HIST1H1C, HIST1H1D, HIST1H1E, HIST1H3F/HIST1H3D, HIST1H4J/HIST1H4K/HIST1H4L/HIST1H4E/HIST1H4A, RPL24 | 6 | 12.5 | 8.3E-4 |
| Biological process | GO:0048562 | Embryonic organ morphogenesis | CLIC5, EDN1, HOXA3, HOXA7 | 4 | 8.3 | 3.7E-3 |
Abbreviations: FC, fold change; FDR, false discovery rate; GO, gene ontology; PPT1, palmitoyl-protein thioesterase 1; RT-qPCR, quantitative reverse transcriptase PCR. Bold entries refer to candidate transcripts tested for RT-qPCR validation.
Differentially expressed miRNAs in MDE
In addition to microarray screening, the same RNA samples were used to search for miRNAs that could be differentially expressed between MDE and control subjects. Out of the 762 miRNAs assayed in parallel by multiplex RT-qPCR, approximately one-third revealed mean expression values above the background level for detectability (243 miRNAs were amplified with a Ct<33) and were further analyzed (Supplementary Table 15). We compared miRNAs expression in MDE and controls and found significantly up- and also downregulated miRNAs (FC>1.20 or<−1.20; P<0.05), as shown in Table 3. Comparison of miRNA expression between MDE patients and controls at baseline and at 8 weeks showed a similar number of dysregulated RNAs (14 miRNAs, with nine miRNAs upregulated and five downregulated). Only two miRNAs showed stable overexpression in MDE patients during the 8-week follow-up compared with controls, miR-941 and miR-589. We also identified miRNAs exhibiting significant variations of expression among patients with clinical improvement (seven upregulated and one downregulated). Thus, our results confirm the potential utility of miRNA signatures as markers of MDE evolution.
Table 3. Dysregulated miRNAs.
| miRNA | Cytoband | FC | P-value | miRNA | Cytoband | FC | P-value |
|---|---|---|---|---|---|---|---|
| MDE 0w vs C | |||||||
| hsa-miR-589 | 7p22.1 | 4.05 | 2.99E-3 | hsa-miR-517b | 19q13.42 | −2.60 | 1.01E-2 |
| hsa-miR-579 | 5p13.3 | 2.49 | 4.34E-2 | hsa-miR-636 | 17q25.1 | −2.22 | 5.67E-3 |
| hsa-miR-941 | 20q13.33 | 2.10 | 1.33E-2 | hsa-miR-1243 | 4p25 | −2.01 | 2.79E-2 |
| hsa-miR-133a | 18q11.2 | 2.04 | 3.50E-2 | hsa-miR-381 | 14q32.31 | −1.57 | 3.50E-2 |
| hsa-miR-494 | 14q32.31 | 1.75 | 2.20E-2 | hsa-miR-200c | 12p13.31 | −1.47 | 3.50E-2 |
| hsa-miR-107 | 10q23.31 | 1.68 | 2.20E-2 | ||||
| hsa-miR-148a | 7p15.2 | 1.48 | 2.79E-2 | ||||
| hsa-miR-652 | Xq23 | 1.35 | 4.35E-2 | ||||
| hsa-miR-425-3p | 3p21.31 | 1.21 | 2.79E-2 | ||||
| MDE 8w vs C | |||||||
| hsa-miR-941 | 20q13.33 | 3.49 | 1.55E-2 | hsa-miR-376a-5p | 14q32.31 | −10.84 | 2.05E-2 |
| hsa-miR-589 | 7p22.1 | 3.00 | 2.50E-2 | hsa-miR-1267 | 13q33.3 | −4.38 | 4.39E-3 |
| hsa-miR-331-5p | 12q22 | 2.31 | 2.79E-2 | hsa-miR-100-3p | 11q24.1 | −2.29 | 4.34E-2 |
| hsa-miR-342-5p | 14q32.2 | 1.77 | 2.79E-2 | hsa-miR-571 | 4p16.3 | −1.78 | 1.17E-2 |
| hsa-let-7b | 22q13.31 | 1.67 | 4.35E-2 | hsa-miR-454 | 17q22 | −1.25 | 1.72E-2 |
| hsa-miR-345 | 14q32.2 | 1.50 | 7.62E-3 | ||||
| hsa-miR-33a-3p | 22q13.2 | 1.49 | 1.60E-2 | ||||
| hsa-miR-363 | Xq26.2 | 1.29 | 2.05E-2 | ||||
| hsa-miR-331-3p | 12q22 | 1.23 | 1.33E-2 | ||||
| MDE 0w.8w vs C | |||||||
| hsa-miR-941 | 20q13.33 | 3.33 | 8.69E-4 | ||||
| hsa-miR-589 | 7p22.1 | 3.12 | 2.28E-4 | ||||
| MDE 0w vs MDE 8w | |||||||
| hsa-miR-20b-3p | Xq26.2 | 3.77 | 2.34E-2 | hsa-miR-331-5p | 12q22 | −4.42 | 1.95E-2 |
| hsa-miR-433 | 14q32.2 | 2.34 | 3.91E-3 | ||||
| hsa-miR-409-3p | 14q32.31 | 2.32 | 2.34E-2 | ||||
| hsa-miR-410 | 14q32.31 | 2.00 | 3.91E-2 | ||||
| hsa-miR-485-3p | 14q32.31 | 1.86 | 2.73E-2 | ||||
| hsa-miR-133a | 18q11.2 | 1.43 | 3.91E-3 | ||||
| hsa-miR-145 | 5q32 | 1.25 | 3.91E-2 | ||||
Abbreviations : FC, fold change; miRNA, microRNA.
MiRNA target prediction
We next identified and analyzed predicted targets of miRNAs differentially expressed in MDE using the Ingenuity pathway analysis software. We performed an inverse correlation analysis at the probe level between the expression of a specific miRNA and the expression levels of all its predicted mRNA targets. Only differentially expressed targets from our own microarray screen were conserved. As shown in Supplementary Table 16, fourteen dysregulated miRNAs have putative mRNA targets that are differentially expressed in MDE, suggesting that a common RNA regulatory network functions in MDE.
qPCR validation of microarray results
To validate our microarray findings (Table 4), we selected fourteen mRNAs that are significantly dysregulated in MDE responders based on their function highlighted by the gene ontology analysis (Table 2), and because they have also been described in previous studies of MDE.13, 43, 44 Table 4 reports the values obtained after RT-qPCR of sub-cohort A (MDE responders only), of sub-cohort B (mostly MDE nonresponders) and of the entire cohort, in comparison with the FC obtained from the microarray screen. Importantly, these comparisons showed that the microarray and RT-qPCR data from cohort A are the most closely matched, with significant alterations of expression in MDE validated for IL1B, IRF2, PPT1 (palmitoyl-protein thioesterase 1), SORT1 and TNF. For many genes, the results obtained in cohort B were quite divergent from those obtained with cohort A, especially at baseline, including the histone genes, IL1B and TNF, suggesting that the transcriptional signature of MDE is closely related to the responder/nonresponder status of the patients.
Table 4. qPCR validation of candidate gene expression.
| Gene | Probe | 0-week | 8-week | Mixed linear model | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Microarray | qPCR | Micro-array |
qPCR
|
Micro-array |
qPCR
|
P-value | Group hierarchy | |||||
| |
|
|
FC
|
FC
A
|
FC
B
|
FC
all
|
FC
|
FC
A
|
FC
B
|
FC
all
|
|
|
| ACTBL2 | A_24_P6903 | Hs01101944_s1 | 1.69a | −1.32 | 1.52 | 1.06 | 1.76b | −1.03 | 1.17 | 1.05 | 0.025 | Responder<non responder |
| ATP2A2 | A_24_P141786 | Hs00155939_m1 | 1.72a | 1.60 | 1.00 | 1.36 | 1.55a | 1.56 | 1.41 | 1.50 | 0.904 | No |
| CELF2 | A_23_P115645 | Hs00990166_m1 | 1.54b | 1.05 | 1.08 | 1.06 | 1.48 | 1.14 | 1.01 | 1.09 | 0.985 | No |
| HIST1H1A | A_23_P70448 | Hs00271225_s1 | −1.93b | −1.20 | 1.71 | 1.18 | −1.76 | −1.05 | 1.24 | 1.07 | 0.019 | Responder<non responder |
| HIST1H1E | A_23_P7976 | Hs00271195_s1 | −1.94a | −1.22 | 1.15 | −1.05 | −1.66b | 1.10 | 1.16 | 1.13 | 0.005 | Responder<non responder |
| HIST1H4E | A_23_P415411 | Hs00374346_s1 | −2.23a | −1.18 | 1.44 | 1.08 | −2.18a | −1.15 | 1.14 | −1.03 | 0.010 | Responder<non responder |
| IL1B | A_23_P79518 | Hs00174097_m1 | 3.31b | 4.12## | −1.22 | 2.80 | 3.17 | 6.56 | −1.17 | 4.28 | 0.001 | Responder>non responder |
| IRF2 | A_23_P125082 | Hs01082884_m1 | 2.00a | 1.25# | 1.10 | 1.20 | 1.63b | 1.11 | 1.03 | 1.08 | 0.194 | No |
| NRG1 | A_23_P315815 | Hs00247624_m1 | 2.01b | 1.74 | 1.19 | 1.52 | 1.87 | 1.79 | 2.10 | 1.92 | 0.327 | No |
| POTEKP | A_32_P155776 | Hs02598440_g1 | 1.66a | −1.16 | 1.76 | 1.22 | 1.61b | 1.01 | 1.40 | 1.16 | 0.011 | Responder<non responder |
| PPT1 | A_24_P276628 | Hs00165579_m1 | 2.30a | 1.21## | −1.09 | 1.10 | 2.22b | 1.28# | 1.01 | 1.17 | 0.00005 | Responder>non responder |
| SORT1 | A_24_P325520 | Hs00361760_m1 | 1.85b | 1.68# | 1.31 | 1.53 | 1.47 | 1.25 | 1.22 | 1.23 | 0.775 | No |
| TNF | A_23_P376488 | Hs00174128_m1 | 1.68 | 1.54# | −1.03 | 1.31 | 2.61b | 2.81 | 1.11 | 2.13 | 0.00047 | Responder>non responder |
| TPP1 | A_24_P38815 | Hs00166099_m1 | 1.55a | 1.04 | −1.04## | 1.01 | 1.51b | 1.04 | −1.04 | 1.01 | 0.051 | Responder>non responder |
Abbreviations: FC, fold change; FDR, false discovery rate; PPT1, palmitoyl-protein thioesterase 1; qPCR, quantitative PCR.
#P<0.05; ##P<0.01.
The last 2 columns display the P-value and hierarchy of significance in differences between the responder versus non responder groups, using a statistical mixed linear model, taking into account two groups (responder and non responder), the three first visits (0, 2, 8-week), and corrected by age and sex parameters.
FDR<1%.
FDR<5%.
To analyze the transcriptional profile of responder versus nonresponder patients in more detail, we had also collected RT-qPCR samples 2 weeks after study inclusion. We applied a mixed linear statistical model to compare the two groups of patients at the three visits with correction for age and sex (Table 3). Results of the analysis demonstrated that an mRNA signature is closely associated with the degree of clinical response. Responders underexpress ACTBL2, histone genes and POTEKP, and conversely overexpress IL1B, TNF, PPT1 and TPP. As we noted that significantly more mirtazapine treatment was prescribed to responders at the second visit (Table 1, Fisher Exact Test P=0.026), we included mirtazapine treatment as a co-variable, adjusted by age and sex, in the mixed linear model. For all the genes except HIST1H1E, the statistical analyses remained significant (Supplementary Table 18). In addition, we evaluated other potential confounding factors with the same method, that is, tobacco consumption, MDE recurrence, age of onset, episode duration before inclusion, history of suicide attempts, treatment initiated before baseline and treatment types (monotherapy versus polytherapy, selective serotonin reuptake inhibitors, serotonin-norepinephrine reuptake inhibitors, other antidepressants, lithium, atypical antipsychotics and electro-convulsivo-therapy). For six transcripts assayed (HIST1H1A, HIST1H4E, IL1B, POTEKP, PPT1 and TNF), none of co-variables analyzed altered the statistical significance determined by the mixed linear model (Supplementary Table 18).
Correlation and predictive analysis
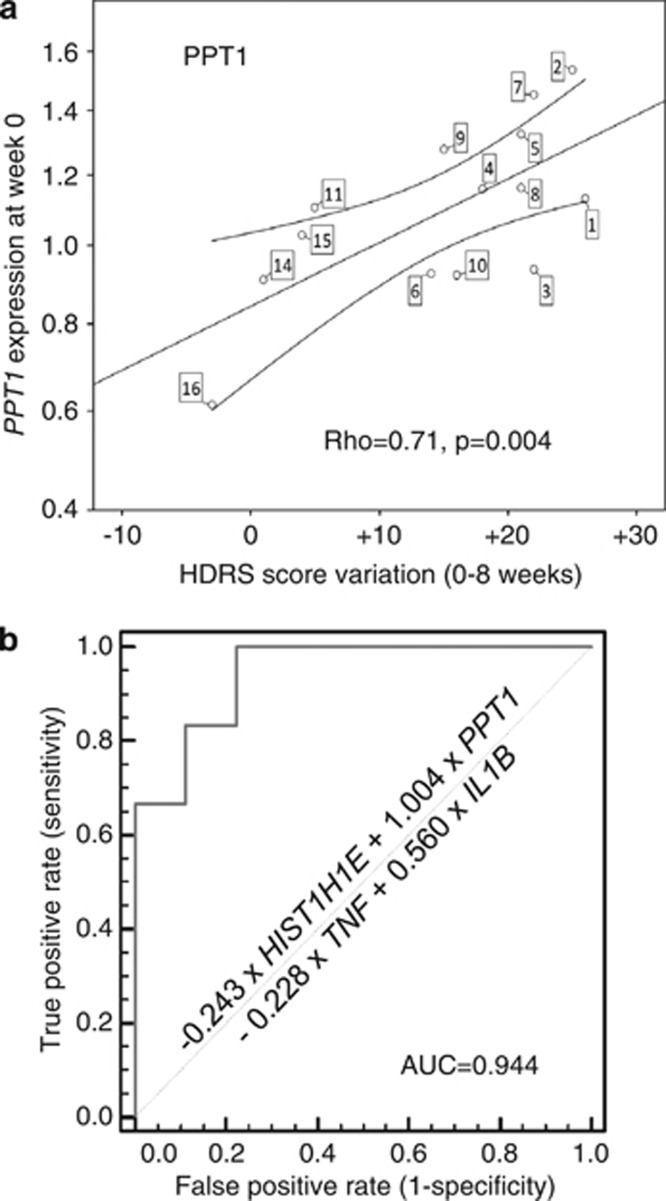
Significantly, PPT1 gene expression at baseline correlated with HDRS score evolution over 8 weeks, as shown in Figure 1a (ρ=0.71, P=0.004). We found a similar correlation between expression at week 2 and HDRS score evolution between weeks 2 and 8 for PPT1 (ρ=0.66, P=0.008, Supplementary Figure 2a) and CELF2 (ρ=−0.52, P=0.046, Supplementary Figure 2b). The other mRNAs tested in the qPCR validation did not show significant correlation with HDRS score evolution.
Figure 1.
Genes correlating to clinical evolution and predictive of outcome. (a) Spearman's correlation between HDRS score evolution and mRNA expression level for PPT1 (Spearman's correlation factor=0.67, P=0.009). Each circle indicates, for a specified MDE patient (the label attached to the circle refers to the case number in Table 1), the difference of HDRS scores at inclusion and 8 weeks after inclusion as a function of the PPT1 FC for MDE0w sample, calculated with the 2−ΔΔCt method. The calibrator was a mathematical pool of control sample Ct. Normalization was performed using GAPDH and PAFAH1B1. (b) ROC curve for the combined expression of four transcripts (that is, HIST1H1E, IL1B, PPT1 and TNF) to predict treatment response after 8 weeks. With the highest Youden index, the combination score has a sensitivity value of 100% (54.1–100), specificity value of 77.78% (40–97.2), a positive predictive value of 75% and a negative predictive value of 100%.
Moreover, to assess the potential predictive value of candidate mRNA expression at baseline in a categorical fashion, we performed a ROC analysis with area under the curve calculation for mRNAs that are differentially expressed between responders and nonresponders. Four mRNA candidates, HIST1H1E, IL1B, PPT1 and TNF, individually demonstrated significant area under the curve increase (P<0.05, Supplementary Figure 3). In order to reveal their capacity in discriminating responders from nonresponders, we combined these four candidates in the ROC analysis. From the discriminant model, (Wilk's λ=0.482; χ2=40.863; P<0.0001), the following equation was obtained: classification score=(−0.243 × HIST1H1E)+(1.004 × PPT1)−(0.228 × TNF)+(0.560 × IL1B). Finally, the area under the curve for the combination of best mRNA candidates reached 0.944 (confidence interval (0.696–0.999), P<0.0001) (Figure 1b).
Discussion
Whereas many microarray studies were previously conducted to identify mRNAs differentially expressed in MDE patients compared with controls,19, 21, 22, 43, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62 only one study had screened miRNAs.36 To our knowledge, we are the first to report a convergent analysis of genome-wide profiling of both mRNAs and miRNAs in MDE. In addition, only one previous study has evaluated variations of gene expression in association to treatment response over time.19 In this study, we aimed to correlate biological measure with clinical evaluations over an 8-week window after treatment in order to assess response to treatment. Moreover, our study is the first to account for gene expression variations in healthy controls to reduce false positive findings by testing freshly collected tissue samples rather than post-mortem samples for repeated measurements. Although PBMCs may not fully recapitulate the gene expression changes in the brain during MDE, increasing compelling evidence indicate that depression is not just limited to the nervous system, but components of the immune system can also be overactive in depressed patients, which raises the question of whether depression may be partly an inflammatory disorder.63
Our analysis revealed ∼200 transcripts that exhibit robust (FDR⩽5% and P⩽0.001) dysregulated expression at baseline in MDE patients compared with controls (Supplementary Tables 3 and 4). These transcripts encode proteins involved in a broad range of functions, including not only cell adhesion, cell cycle, cell junction, cellular ion homeostasis, cytoskeleton, transcription regulation, but also neuron differentiation and synapse (BAIAP2, CLIC5, DENND1A, FIG4, GNAQ, MACF1, MYH9, NPTN, PPT1, SNAP23 and STAT3). This confirms that molecules important for the function of the nervous system may also be identified in blood. Indeed, MDE candidate genes previously identified using brain tissues were also identified as dysregulated in our MDE PBMC screen, including RGS7BP, EDN1, PAQR6,46 PPM1K,55 and ELK3.56 Of the biological processes overrepresented within our lists of dysregulated transcripts, cell death, vacuole/lysosome organization, nucleotide binding, nucleosome/chromatin assembly were significantly affected in MDE at baseline and also after treatment response (Table 2). Importantly, all these processes were also found associated to MDE in previous investigations relying on both brain54, 55, 56 and blood materials.19, 62
Despite a growing interest within the field, miRNAs role in pathophysiology of psychiatric disorders is still not widely documented. Previous studies explored miRNA dysregulation in bipolar disorder and schizophrenia, both with peripheral tissues (PBMCs, serum)64, 65, 66 or with post-mortem brain tissues.65, 67, 68, 69, 70, 71 Overall, these studies reported conflicting data on the identities of altered miRNAs, as well as the magnitude of change in expression. These discrepancies are likely due to tissue-specific variations in expression levels, and heterogeneity in quantification and normalization procedures. Probably for the same reasons, our results did not overlap significantly with what was reported in Smalheiser et al.,36 a study conducted with brain cortical samples, which identified only two conserved dysregulated miRNA. Although miR-376a-5p was underexpressed in both our study and the Smalheiser et al.'s study, miR-494 was overexpressed in our PBMC samples while underexpressed in the cortex.36 In addition, out of the several studies conducted with schizophrenia samples, only one relied on PBMCs.66 Again, we shared only one dysregulated miRNA (miR-200c), while three miRNAs (miR-107, miR-342-5p, and let-7b) were dysregulated in opposing degrees. Nevertheless, as the present study reports the first MDE gene expression analysis with PBMC samples, the data are preliminary and open to further validation from other investigators.
miRNA dysregulation in MDE highlights the importance of gene expression and epigenetic regulation in mood disorders. As shown on Supplementary Table 16, putative interactions between dysregulated mRNAs and miRNAs can be identified without an a priori hypothesis. However, these types of interactions remain to be demonstrated in cellular or animal models. In addition, the nature and sequence of the miRNA determines its mRNA target. It was recently demonstrated that common human polymorphisms in the miRNA target element may regulate gene expression with a concomitant change in phenotype. For example, recent studies suggested that polymorphisms in miR-30e and pre-miR-182 might have a role in major depressive disorder susceptibility.34 Of note, as described in Supplementary Tables 5–10, other non-coding RNAs, such as long intergenic non-coding RNAs represent a significant portion of dysregulated RNAs in our MDE patients (21 out of 195 at baseline). Although, to our knowledge, this is the first study reporting the mis-expression of long intergenic non-coding RNAs in an affective disorder, functions for each of these molecules remain elusive and are likely additional mechanisms to regulate gene expression.72 As long intergenic non-coding RNAs are numerous and not yet fully described, their implications in biological pathways are still unclear, underscoring the importance of performing conservative ontology analysis. These complexities remind us that multiple layers of regulation are at play to fine-tune gene expression in both normal physiology and MDE.
After microarray profiling both mRNAs and miRNAs from PBMCs, a set of 14 mRNAs that discriminate responder patients from control subjects were selected for RT-qPCR validation. Of these, 10 candidates (ACTBL2, ATP2A2, CELF2, HIST1H1A, HIST1H1E, HIST1H4E, IRF2, POTEKP, PPT1 and TPP1) met the stringent statistical significance (FDR<1%) and/or the membership to a significantly dysregulated biological process. We also decided to test other genes for significance with lower stringency (FDR<5%), and found candidate genes (IL1B, NRG1, SORT1 and TNF) that had been identified for affective disorders in previous studies.13, 43, 44 We expanded the cohort of patient and healthy subjects for analysis and repeated measurements in three different time points (that is, 0, 2 and 8 weeks). A limitation in trying to reproduce microarray results with a different technology is the difficulty in generating amplicons covering the same portion of a specific transcript as the the microarray oligonucleotide probe. Despite this limitation, we obtained an almost complete overlap for ATP2A2, IL1B, NRG1, SORT1 and TNF in expression levels between the microarray and qPCR data for responder patients, while similar variations were also observed for HIST1H1A, HIST1H4E and IRF2 (Table 4). Only two genes (ACTBL2 and POTEKP) out of 14 exhibited conflicting results between the microarray and the qPCR data. Therefore, considering the limitation expressed above, these results indicate that our microarray procedures, which are exclusively based on responder patients, detected reliable variations in gene expression.
To further examine our hypothesis that responder and nonresponder MDE patients express different transcriptional signatures, the candidate genes were tested in a statistical model evaluating the contribution of potential confounding factors (Supplementary Table 18). It allowed us to define a smaller core of six markers, which resist the test of all co-variables and distinguish responders from nonresponders: HIST1H1A, HIST1H4E, IL1B, POTEKP, PPT1 and TNF.
Next, we sought to determine a candidate gene that is predictive of clinical evolution. Our analysis showed that PPT1 expression was correlated with the change in depression severity (Figure 1a). In addition, in a preliminary attempt to identify a transcriptional biomarker for MDE treatment response, we submitted the results obtained by RT-qPCR to ROC analysis. This led us to identify four genes that are individually predictive of clinical evolution in our sample: HIST1H1E, IL1B, PPT1 and TNF. Of note, the combination of these markers provided even better predictive value for treatment response. Although this is not the first attempt to build a classifier gene set for major depressive disorder based on blood gene expression, the seven diagnostic genes previously defined by Spijker et al.22 were identified after ex vivo stimulation of the blood cells with lipopolysaccharide.
Looking more specifically at our best predictive gene candidates, we are not surprised to find TNF and IL1B. These genes encode cytokines that are major players of disease behavior associated with pro-inflammatory responses.73 Inflammatory response is consistently cited as a key process in dysregulated pathway analysis from affective disorders.19, 21, 43, 44, 51, 54, 62, 74, 75, 76, 77 Many studies have validated dysregulation of IL1B and TNF mRNA/protein in major depression.78 Genetic studies have also underscored potential polymorphisms of these cytokines linked to MDE and/or treatment response.79 In this study, we observed that TNF and IL1B upregulation correlates to responder status, suggesting that a pro-inflammatory response may be associated with a better prognosis. However, this conclusion should be considered with caution as mRNAs are not necessarily correlated with protein expression and serum/plasma levels. Nevertheless, our results suggest that TNF and IL1β cytokines could be associated with treatment response in a naturalistic point of view. Although potential predictive values of TNF or IL1β expression in MDE patients for treatment response have been controversial, some recent studies have shown evidence in support of this hypothesis in animal and human studies.80
In addition to cytokines, we also observed dysregulated expression of several histone genes that contribute to chromatin organization. Several pangenomic studies have already implied a role of epigenetics in affective disorders,54, 62, 75 and chromatin remodeling is described as an important process in pathophysiology of major depression and its treatment.12 Moreover, key players of epigenetic control of chromatin structure such as histone deacetylases and DNA methyltransferases have been described to be dysregulated in MDE.13, 30, 81 In this study, we described a decreased expression of histone H1 family members (Table 4 and Supplementary Table 18) in a responder MDE patient, and a potential predictive value of treatment response for HIST1H1E (Supplementary Figure 2). Because linker histones H1 contribute to the so-called ‘histone code',82 we believe that our findings adds to the growing evidence of complex epigenetics regulating MDE pathophysiology and its treatment.
Finally, we discovered PPT1 expression was as one of the most consistently dysregulated genes associated with MDE response, making it an excellent predictive marker for clinical evolution (Table 4, Figure 1 and Supplementary Figure 2). PPT1 encodes a small glycoprotein involved in the catabolism of lipid-modified proteins during lysosomal degradation and synaptic vesicle endocytosis/recycling in brain.83 Mutations in PPT1 lead to infantile-onset neuronal ceroid lipofuscinosis, a recessive neurodegenerative disorder affecting the retina, cortex and cerebellum. To our knowledge, no previous genetic or proteomic study had implicated PPT1 in a psychiatric disorder, but its function could be relevant in the context of a neuroplasticity/neurodegeneration hypothesis. Future work will be required to explore the implication of PPT1 in MDE and its treatment.
Despite our exciting results, we acknowledge several general limitations to our experimental design. We chose to focus our study on the 8-week treatment response period, which is the first stage of recovery. However, a more extended follow-up is required to explore whether the same response and remission biomarker can be used in the long term. Second, although our sample size was determined to ensure enough statistical power for microarray screening and we performed repeated measures in both the MDE and control groups, our results are limited by the small size of the cohort and would require validation in larger cohort of MDE patients to better evaluate suitability and specificity of the proposed biomarkers. Third, as a consequence of the limited size of our cohort, we could not explore biomarkers for remission that are as important, if not more warranted than biomarkers for treatment response. Therefore a possible design for a more ambitious, future study, is the enrollment, still in naturalistic conditions, of a much larger cohort of patients. It will then be possible to stratify patients according to treatment regimen and to implement an additional time point (6 months) to the protocol during the follow-up of each patient, which would hopefully lead to the identification of remission markers in addition to the validation of treatment-response markers. Fourth, there is an open debate on whether the validation of a biomarker would require treatment-naive patients at baseline and whether it should be limited to a single treatment with a single targeted phenotype. Therefore, heterogeneities in MDE history and in the naturalistic treatments for all the patients in this study may present difficulties for drawing clear conclusions. Indeed, previous promising results for treatment response or clinical evolution in MDE obtained with biomarker analysis, and relying on biological, electrophysiological or brain imaging, have highlighted the limitation of MDE heterogeneity.84 Ideally, a biomarker assay for MDE needs to be easy to produce, cost effective and robust, regardless of all potential confounding factors as well as previous treatments received by the patients. Lastly, our study identified a short list of 14 gene candidates based on mRNA profiling that were validated by RT-qPCR according to arbitrary rules. Therefore, we cannot rule out whether among our list of ∼200 RNA candidates dysregulated in MDE at baseline, several other candidates would also show robust predictive value for clinical evolution. With this in mind, our next aim will be to test the biomarker predictive value of the most promising mRNA candidates that we validated by RT-qPCR, as well as other mRNA and miRNA candidates significantly dysregulated in our study, in a larger cohort.
In conclusion, we have profiled mRNA and miRNA expression in PBMCs from MDE patients compared with healthy controls in a prospective and naturalistic design. We report new insights into the biological description of MDE and identified PPT1 as a possible biomarker of MDE clinical evolution. Taken together, we propose that using RNAs from PBMCs as biomarkers may constitute an exciting new method to improve MDE prognosis and to develop personalized medicine in psychiatry.
Acknowledgments
This study was supported by a grant from Assistance Publique—Hôpitaux de Marseille, France (AORC, No. 2009/15 to RB) and by a national hospital clinical research program (PHRC, No. 2010–19 to JN). We are grateful to the participants and the nurse teams from VEGA and HDS for their technical and psychological supports. We thank Professor Frank Bellivier and Philippe Courtet for their suggestions and support. We are grateful to Jessica Fernandez for her advices with ROC analysis. We also thank Jeanne Hsu for her critical reading of the paper.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Fekadu A, Wooderson SC, Markopoulo K, Donaldson C, Papadopoulos A, Cleare AJ. What happens to patients with treatment-resistant depression? A systematic review of medium to long term outcome studies. J Affect Disord. 2009;116:4–11. doi: 10.1016/j.jad.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Kupfer DJ, Frank E, Phillips ML. Major depressive disorder: new clinical, neurobiological, and treatment perspectives. Lancet. 2012;379:1045–1055. doi: 10.1016/S0140-6736(11)60602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association Practice Guideline for the Treatment of Patients with Major Depressive Disorder3rd edn.American Psychiatric Association (APA): Arlington, VA; 2010152 [Google Scholar]
- Kennedy SH, Lam RW, Parikh SV, Patten SB, Ravindran AV. Canadian Network for Mood and Anxiety Treatments (CANMAT) clinical guidelines for the management of major depressive disorder in adults. Introduction. J Affect Disord. 2009;117 (Suppl 1:S1–S2. doi: 10.1016/j.jad.2009.06.043. [DOI] [PubMed] [Google Scholar]
- Bauer M, Whybrow PC, Angst J, Versiani M, Moller HJ. World Federation of Societies of Biological Psychiatry (WFSBP) Guidelines for Biological Treatment of Unipolar Depressive Disorders, Part 1: Acute and continuation treatment of major depressive disorder. World J Biol Psychiatry. 2002;3:5–43. doi: 10.3109/15622970209150599. [DOI] [PubMed] [Google Scholar]
- Frank E, Prien RF, Jarrett RB, Keller MB, Kupfer DJ, Lavori PW, et al. Conceptualization and rationale for consensus definitions of terms in major depressive disorder. Remission, recovery, relapse, and recurrence. Arch Gen Psychiatry. 1991;48:851–855. doi: 10.1001/archpsyc.1991.01810330075011. [DOI] [PubMed] [Google Scholar]
- Seemuller F, Riedel M, Obermeier M, Bauer M, Adli M, Kronmuller K, et al. Outcomes of 1014 naturalistically treated inpatients with major depressive episode. Eur Neuropsychopharmacol. 2010;20:346–355. doi: 10.1016/j.euroneuro.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Trivedi MH, Fava M, Wisniewski SR, Thase ME, Quitkin F, Warden D, et al. Medication augmentation after the failure of SSRIs for depression. N Engl J Med. 2006;354:1243–1252. doi: 10.1056/NEJMoa052964. [DOI] [PubMed] [Google Scholar]
- Uher R, Mors O, Rietschel M, Rajewska-Rager A, Petrovic A, Zobel A, et al. Early and delayed onset of response to antidepressants in individual trajectories of change during treatment of major depression: a secondary analysis of data from the Genome-Based Therapeutic Drugs for Depression (GENDEP) study. J Clin Psychiatry. 2011;72:1478–1484. doi: 10.4088/JCP.10m06419. [DOI] [PubMed] [Google Scholar]
- Tadic A, Wagner S, Gorbulev S, Dahmen N, Hiemke C, Braus DF, et al. Peripheral blood and neuropsychological markers for the onset of action of antidepressant drugs in patients with Major Depressive Disorder. BMC Psychiatry. 2011;11:16. doi: 10.1186/1471-244X-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim C, Binder EB. Current research trends in early life stress and depression: review of human studies on sensitive periods, gene-environment interactions, and epigenetics. Exp Neurol. 2012;233:102–111. doi: 10.1016/j.expneurol.2011.10.032. [DOI] [PubMed] [Google Scholar]
- Sun H, Kennedy PJ, Nestler EJ.Epigenetics of the Depressed Brain: Role of Histone Acetylation and Methylation Neuropsychopharmacology 2012. e pub ahead of print 14 June 2012.doi: 10.1038/npp.2012.73 [DOI] [PMC free article] [PubMed]
- Belzeaux R, Formisano-Treziny C, Loundou A, Boyer L, Gabert J, Samuelian JC, et al. Clinical variations modulate patterns of gene expression and define blood biomarkers in major depression. J Psychiatr Res. 2010;44:1205–1213. doi: 10.1016/j.jpsychires.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Mehta D, Menke A, Binder EB. Gene expression studies in major depression. Curr Psychiatry Rep. 2010;12:135–144. doi: 10.1007/s11920-010-0100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelk CH, Singhania A, Perez-Santiago J, Glatt SJ, Tsuang MT. The utility of gene expression in blood cells for diagnosing neuropsychiatric disorders. Int Rev Neurobiol. 2011;101:41–63. doi: 10.1016/B978-0-12-387718-5.00003-1. [DOI] [PubMed] [Google Scholar]
- Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med. 2006;147:126–132. doi: 10.1016/j.lab.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Jasinska AJ, Service S, Choi OW, DeYoung J, Grujic O, Kong SY, et al. Identification of brain transcriptional variation reproduced in peripheral blood: an approach for mapping brain expression traits. Hum Mol Genet. 2009;18:4415–4427. doi: 10.1093/hmg/ddp397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, Kurian SM, Yehyawi N, Dike C, Patel SD, Edenberg HJ, et al. Identifying blood biomarkers for mood disorders using convergent functional genomics. Mol Psychiatry. 2009;14:156–174. doi: 10.1038/mp.2008.11. [DOI] [PubMed] [Google Scholar]
- Mamdani F, Berlim MT, Beaulieu M-M, Labbe A, Merette C, Turecki G. Gene expression biomarkers of response to citalopram treatment in major depressive disorder. Transl Psychiatry. 2011;1:e13. doi: 10.1038/tp.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Kim YK. BDNF mRNA expression of peripheral blood mononuclear cells was decreased in depressive patients who had or had not recently attempted suicide. J Affect Disord. 2010;125:369–373. doi: 10.1016/j.jad.2010.01.074. [DOI] [PubMed] [Google Scholar]
- Yi Z, Li Z, Yu S, Yuan C, Hong W, Wang Z, et al. Blood-based gene expression profiles models for classification of subsyndromal symptomatic depression and major depressive disorder. PLoS One. 2012;7:e31283. doi: 10.1371/journal.pone.0031283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spijker S, Van Zanten JS, De Jong S, Penninx BW, van Dyck R, Zitman FG, et al. Stimulated gene expression profiles as a blood marker of major depressive disorder. Biol Psychiatry. 2010;68:179–186. doi: 10.1016/j.biopsych.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Pajer K, Andrus BM, Gardner W, Lourie A, Strange B, Campo J, et al. Discovery of blood transcriptomics markers for depression in animal models and pilot validation in subjects with early-onset major depression. Transl Psychiatry. 2012;2:e101. doi: 10.1038/tp.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galecki P, Galecka E, Maes M, Chamielec M, Orzechowska A, Bobinska K, et al. The expression of genes encoding for COX-2, MPO, iNOS, and sPLA2-IIA in patients with recurrent depressive disorder. J Affect Disord. 2012;138:360–366. doi: 10.1016/j.jad.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Zhang J, Chen Y, Zhang K, Yang H, Sun Y, Fang Y, et al. A cis-phase interaction study of genetic variants within the MAOA gene in major depressive disorder. Biol Psychiatry. 2010;68:795–800. doi: 10.1016/j.biopsych.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Numata S, Iga J, Nakataki M, Tayoshi S, Taniguchi K, Sumitani S, et al. Gene expression and association analyses of the phosphodiesterase 4B (PDE4B) gene in major depressive disorder in the Japanese population. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:527–534. doi: 10.1002/ajmg.b.30852. [DOI] [PubMed] [Google Scholar]
- Cattaneo A, Bocchio-Chiavetto L, Zanardini R, Milanesi E, Placentino A, Gennarelli M. Reduced peripheral brain-derived neurotrophic factor mRNA levels are normalized by antidepressant treatment. Int J Neuropsychopharmacol. 2010;13:103–108. doi: 10.1017/S1461145709990812. [DOI] [PubMed] [Google Scholar]
- Cattaneo A, Sesta A, Calabrese F, Nielsen G, Riva MA, Gennarelli M. The expression of VGF is reduced in leukocytes of depressed patients and it is restored by effective antidepressant treatment. Neuropsychopharmacology. 2010;35:1423–1428. doi: 10.1038/npp.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe N, Uchida S, Otsuki K, Hobara T, Yamagata H, Higuchi F, et al. Altered sirtuin deacetylase gene expression in patients with a mood disorder. J Psychiatr Res. 2011;45:1106–1112. doi: 10.1016/j.jpsychires.2011.01.016. [DOI] [PubMed] [Google Scholar]
- Higuchi F, Uchida S, Yamagata H, Otsuki K, Hobara T, Abe N, et al. State-dependent changes in the expression of DNA methyltransferases in mood disorder patients. J Psychiatr Res. 2011;45:1295–1300. doi: 10.1016/j.jpsychires.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Nakataki M, Iga J, Numata S, Yoshimoto E, Kodera K, Watanabe SY, et al. Gene expression and association analysis of the epithelial membrane protein 1 gene in major depressive disorder in the Japanese population. Neurosci Lett. 2011;489:126–130. doi: 10.1016/j.neulet.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Eady JJ, Wortley GM, Wormstone YM, Hughes JC, Astley SB, Foxall RJ, et al. Variation in gene expression profiles of peripheral blood mononuclear cells from healthy volunteers. Physiol Genomics. 2005;22:402–411. doi: 10.1152/physiolgenomics.00080.2005. [DOI] [PubMed] [Google Scholar]
- Fiori LM, Turecki G. Broadening our horizons: gene expression profiling to help better understand the neurobiology of suicide and depression. Neurobiol Dis. 2012;45:14–22. doi: 10.1016/j.nbd.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor RM, Dinan TG, Cryan JF. Little things on which happiness depends: microRNAs as novel therapeutic targets for the treatment of anxiety and depression. Mol Psychiatry. 2012;17:359–376. doi: 10.1038/mp.2011.162. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Rizavi HS, Torvik VI, Turecki G, Dwivedi Y. MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS One. 2012;7:e33201. doi: 10.1371/journal.pone.0033201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Text Revision (DSM-IV-TR) American Psychiatric Association: Arlington, VA; 2000. [Google Scholar]
- American Psychiatric Association Handbook of Psychiatric Measures2nd edn.American Psychiatric Association (APA): Washington, DC; 2008864 [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Shelton RC, Claiborne J, Sidoryk-Wegrzynowicz M, Reddy R, Aschner M, Lewis DA, et al. Altered expression of genes involved in inflammation and apoptosis in frontal cortex in major depression. Mol Psychiatry. 2011;16:751–762. doi: 10.1038/mp.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexhage RC, van der Heul-Nieuwenhuijsen L, Padmos RC, van Beveren N, Cohen D, Versnel MA, et al. Inflammatory gene expression in monocytes of patients with schizophrenia: overlap and difference with bipolar disorder. A study in naturalistically treated patients. Int J Neuropsychopharmacol. 2010;13:1369–1381. doi: 10.1017/S1461145710000799. [DOI] [PubMed] [Google Scholar]
- Duric V, Banasr M, Stockmeier CA, Simen AA, Newton SS, Overholser JC, et al. Altered expression of synapse and glutamate related genes in post-mortem hippocampus of depressed subjects Int J Neuropsychopharmacol 2012. e-pub ahead of print 22 February 2012;doi: 10.1017/S1461145712000016 [DOI] [PMC free article] [PubMed]
- Guilloux JP, Douillard-Guilloux G, Kota R, Wang X, Gardier AM, Martinowich K, et al. Molecular evidence for BDNF- and GABA-related dysfunctions in the amygdala of female subjects with major depression Mol Psychiatry 2011. e-pub ahead of print 14 September 2011;doi: 10.1038/mp.2011.113 [DOI] [PMC free article] [PubMed]
- Duric V, Banasr M, Licznerski P, Schmidt HD, Stockmeier CA, Simen AA, et al. A negative regulator of MAP kinase causes depressive behavior. Nat Med. 2010;16:1328–1332. doi: 10.1038/nm.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard R, Kerman IA, Thompson RC, Jones EG, Bunney WE, Barchas JD, et al. Altered expression of glutamate signaling, growth factor, and glia genes in the locus coeruleus of patients with major depression. Mol Psychiatry. 2011;16:634–646. doi: 10.1038/mp.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibille E, Wang Y, Joeyen-Waldorf J, Gaiteri C, Surget A, Oh S, et al. A molecular signature of depression in the amygdala. Am J Psychiatry. 2009;166:1011–1024. doi: 10.1176/appi.ajp.2009.08121760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira A, Mamdani F, Ernst C, Vawter MP, Bunney WE, Lebel V, et al. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PLoS One. 2009;4:e6585. doi: 10.1371/journal.pone.0006585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempan TA, Sequeira A, Canetti L, Lalovic A, Ernst C, ffrench-Mullen J, et al. Altered expression of genes involved in ATP biosynthesis and GABAergic neurotransmission in the ventral prefrontal cortex of suicides with and without major depression. Mol Psychiatry. 2009;14:175–189. doi: 10.1038/sj.mp.4002110. [DOI] [PubMed] [Google Scholar]
- Chu TT, Liu Y, Kemether E. Thalamic transcriptome screening in three psychiatric states. J Hum Genet. 2009;54:665–675. doi: 10.1038/jhg.2009.93. [DOI] [PubMed] [Google Scholar]
- Tochigi M, Iwamoto K, Bundo M, Sasaki T, Kato N, Kato T. Gene expression profiling of major depression and suicide in the prefrontal cortex of postmortem brains. Neurosci Res. 2008;60:184–191. doi: 10.1016/j.neures.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Adams DH, Simen A, Simen BB, Rajkowska G, Stockmeier CA, et al. Gene expression profiling in postmortem prefrontal cortex of major depressive disorder. J Neurosci. 2007;27:13329–13340. doi: 10.1523/JNEUROSCI.4083-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira A, Klempan T, Canetti L, ffrench-Mullen J, Benkelfat C, Rouleau GA, et al. Patterns of gene expression in the limbic system of suicides with and without major depression. Mol Psychiatry. 2007;12:640–655. doi: 10.1038/sj.mp.4001969. [DOI] [PubMed] [Google Scholar]
- Sequeira A, Gwadry FG, Ffrench-Mullen JM, Canetti L, Gingras Y, Casero RA, et al. Implication of SSAT by gene expression and genetic variation in suicide and major depression. Arch Gen Psychiatry. 2006;63:35–48. doi: 10.1001/archpsyc.63.1.35. [DOI] [PubMed] [Google Scholar]
- Choudary PV, Molnar M, Evans SJ, Tomita H, Li JZ, Vawter MP, et al. Altered cortical glutamatergic and GABAergic signal transmission with glial involvement in depression. Proc Natl Acad Sci USA. 2005;102:15653–15658. doi: 10.1073/pnas.0507901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston C, Jiang L, Sokolov BP. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Mol Psychiatry. 2005;10:309–322. doi: 10.1038/sj.mp.4001565. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Kakiuchi C, Bundo M, Ikeda K, Kato T. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Mol Psychiatry. 2004;9:406–416. doi: 10.1038/sj.mp.4001437. [DOI] [PubMed] [Google Scholar]
- Evans SJ, Choudary PV, Neal CR, Li JZ, Vawter MP, Tomita H, et al. Dysregulation of the fibroblast growth factor system in major depression. Proc Natl Acad Sci USA. 2004;101:15506–15511. doi: 10.1073/pnas.0406788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibille E, Arango V, Galfalvy HC, Pavlidis P, Erraji-Benchekroun L, Ellis SP, et al. Gene expression profiling of depression and suicide in human prefrontal cortex. Neuropsychopharmacology. 2004;29:351–361. doi: 10.1038/sj.npp.1300335. [DOI] [PubMed] [Google Scholar]
- Segman RH, Goltser-Dubner T, Weiner I, Canetti L, Galili-Weisstub E, Milwidsky A, et al. Blood mononuclear cell gene expression signature of postpartum depression. Mol Psychiatry. 2010;15:93–100. doi: 10.1038/mp.2009.65. [DOI] [PubMed] [Google Scholar]
- Raison CL, Miller AH. Is depression an inflammatory disorder. Curr Psychiatry Rep. 2011;13:467–475. doi: 10.1007/s11920-011-0232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CY, Yu SL, Hsieh MH, Chen CH, Chen HY, Wen CC, et al. MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS One. 2011;6:e21635. doi: 10.1371/journal.pone.0021635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Du J, Qi Y, Liang G, Wang T, Li S, et al. Aberrant expression of serum miRNAs in schizophrenia. J Psychiatr Res. 2012;46:198–204. doi: 10.1016/j.jpsychires.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Gardiner E, Beveridge NJ, Wu JQ, Carr V, Scott RJ, Tooney PA, et al. Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol Psychiatry. 17:827–840. doi: 10.1038/mp.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol Psychiatry. 2011;69:180–187. doi: 10.1016/j.biopsych.2010.09.030. [DOI] [PubMed] [Google Scholar]
- Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci USA. 2012;109:3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Reimers M, Maher B, Williamson V, McMichael O, McClay JL, et al. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res. 2010;124:183–191. doi: 10.1016/j.schres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. 2010;15:1176–1189. doi: 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer R, O'Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci. 2008;9:46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmos RC, Hillegers MH, Knijff EM, Vonk R, Bouvy A, Staal FJ, et al. A discriminating messenger RNA signature for bipolar disorder formed by an aberrant expression of inflammatory genes in monocytes. Arch Gen Psychiatry. 2008;65:395–407. doi: 10.1001/archpsyc.65.4.395. [DOI] [PubMed] [Google Scholar]
- Shao L, Vawter MP. Shared gene expression alterations in schizophrenia and bipolar disorder. Biol Psychiatry. 2008;64:89–97. doi: 10.1016/j.biopsych.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MM, Lockstone HE, Huffaker SJ, Wayland MT, Webster MJ, Bahn S. Gene expression analysis of bipolar disorder reveals downregulation of the ubiquitin cycle and alterations in synaptic genes. Mol Psychiatry. 2006;11:965–978. doi: 10.1038/sj.mp.4001875. [DOI] [PubMed] [Google Scholar]
- Kao CF, Jia P, Zhao Z, Kuo PH. Enriched pathways for major depressive disorder identified from a genome-wide association study. Int J Neuropsychopharmacol. 2012;15:1401–1411. doi: 10.1017/S1461145711001891. [DOI] [PubMed] [Google Scholar]
- Janssen DG, Caniato RN, Verster JC, Baune BT. A psychoneuroimmunological review on cytokines involved in antidepressant treatment response. Hum Psychopharmacol. 2010;25:201–215. doi: 10.1002/hup.1103. [DOI] [PubMed] [Google Scholar]
- Bufalino C, Hepgul N, Aguglia E, Pariante CM.The role of immune genes in the association between depression and inflammation: A review of recent clinical studies Brain Behav Immun 2012. e-pub ahead of print 15 May 2012;doi: 10.1016/j.bbi.2012.04.009 [DOI] [PubMed]
- Warner-Schmidt JL, Vanover KE, Chen EY, Marshall JJ, Greengard P. Antidepressant effects of selective serotonin reuptake inhibitors (SSRIs) are attenuated by antiinflammatory drugs in mice and humans. Proc Natl Acad Sci USA. 2011;108:9262–9267. doi: 10.1073/pnas.1104836108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobara T, Uchida S, Otsuki K, Matsubara T, Funato H, Matsuo K, et al. Altered gene expression of histone deacetylases in mood disorder patients. J Psychiatr Res. 2010;44:263–270. doi: 10.1016/j.jpsychires.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431:1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Saja S, Buff H, Smith AC, Williams TS, Korey CA. Identifying cellular pathways modulated by Drosophila palmitoyl-protein thioesterase 1 function. Neurobiol Dis. 2010;40:135–145. doi: 10.1016/j.nbd.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuchter AF, Cook IA, Hamilton SP, Narr KL, Toga A, Hunter AM, et al. Biomarkers to predict antidepressant response. Curr Psychiatry Rep. 2010;12:553–562. doi: 10.1007/s11920-010-0160-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.