Abstract
Population-level measurements of phenotypic behaviour in biological systems may not necessarily reflect individual cell behaviour. To assess qualitative changes in the behaviour of a single cell, when alone and when part of a community, we developed an agent-based model describing the metabolic states of a population of quorum-coupled cells. The modelling is motivated by published experimental work of a synthetic genetic regulatory network (GRN) used in Escherichia coli cells that exhibit oscillatory behaviour across the population. To decipher the mechanisms underlying oscillations in the system, we investigate the behaviour of the model via numerical simulation and bifurcation analysis. In particular, we study the effect of an increase in population size as well as the spatio-temporal behaviour of the model. Our results demonstrate that oscillations are possible only in the presence of a high concentration of the coupling chemical and are due to a time scale separation in key regulatory components of the system. The model suggests that the population establishes oscillatory behaviour as the system's preferred stable state. This is achieved via an effective increase in coupling across the population. We conclude that population effects in GRN design need to be taken into consideration and be part of the design process. This is important in planning intervention strategies or designing specific cell behaviours.
Keywords: synthetic biology, genetic regulatory networks, oscillations, quorum-sensing
1. Introduction
Quantification of biological phenomena in cell populations is frequently based on average measurements. Any measurements, and thus any properties described by such measurements, may not necessarily reflect the properties of a single cell and, vice versa, properties of single cells may not be representative of population behaviour. This becomes even more important in synthetic biology where system design is based on previously quantified molecular components at the single-cell level, but in some cases analysis of the behaviour is assessed on the level of a population. A recent example that highlights this is Milias-Argeitis et al. [1], where a single-cell model is used to develop control principles that are tested over a population of cells. Knowledge of whether specific cellular states are possible only at single-cell and/or population-level is an essential step in designing effective intervention strategies. Examples include modification of genetic regulatory networks (GRNs) for maintaining health or preventing disease in many biological settings. This is equally important from a modelling perspective. It is essential to understand whether the predictive power of single-cell models can scale up accordingly to capture population-wide behaviour.
For instance, as was shown recently [2], a population of synthetically engineered bacteria communicating via quorum-sensing can produce synchronized oscillations or be entrained to an external periodic input. An outstanding question is to understand whether the oscillatory behaviour observed at the macroscopic, population level is a result of oscillatory behaviour at the single-cell level or not. Indeed, it is possible that oscillations emerge owing to the spatio-temporal dynamics of the entire population rather than as a result of synchronization of individual oscillating cells.
To shed light on these questions, whose answers are difficult to obtain experimentally, we developed an agent-based model of the experimental set-up of Danino et al. [2]. The model is studied over a range of scales to uncover the nature of oscillations as stated earlier. A full spatially explicit model describing a population of cells entrapped in a microfluidic chamber (illustrated in figure 1) was also studied to bridge the gap between the theoretical approach and the physical setting of the experiment [2]. We focus on whether population-level behaviour is carried over from a single cell by first asking whether the oscillatory behaviour is an intrinsic stable state at the single-cell level and then studying whether it persists when clusters of cells are considered.
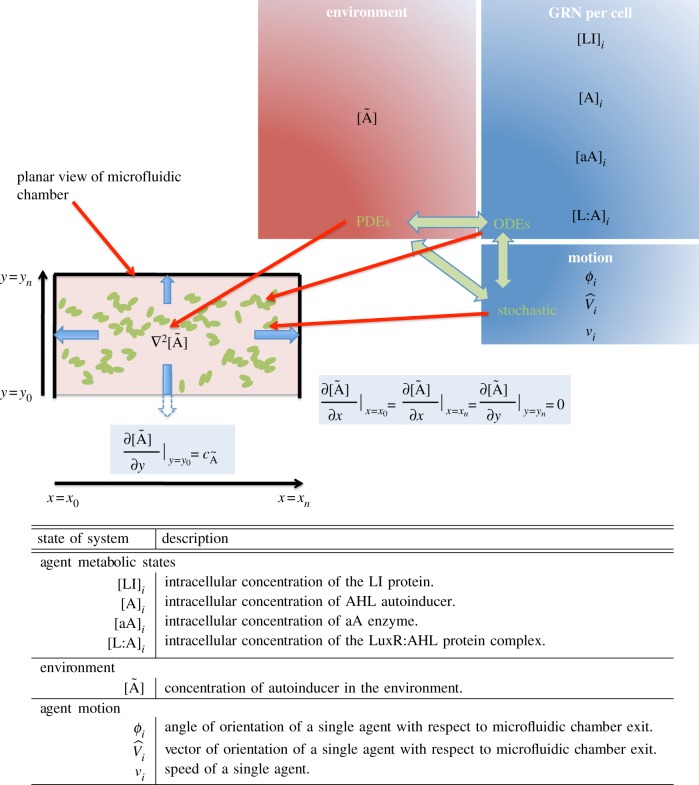
Figure 1.
The model with spatio-temporal dynamics. In the spatially explicit model, external AHL is modelled with a PDE, each cell's GRN with ODEs and motion is modelled stochastically. Mixed boundaries are implemented. There is no chemical flux in the xz and yz-planes. Non-zero flux is defined with a constant gradient at the y0 boundary only of the xy-plane, 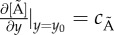 , where
, where 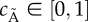 . This non-zero flux represents the chamber's exit.
. This non-zero flux represents the chamber's exit.
The system design in Danino et al. [2] is based on a synthetic version of the naturally occurring quorum-sensing mechanism found in many prokaryotes, including some strains of Escherichia coli [3]. The GRN is implemented in a laboratory strain of E. coli cells that, prior to transformation, lack this mechanism. Bacteria use quorum-sensing to communicate between themselves and sense the size of the colony [3–5]. Quorum-sensing can be implemented using a variety of proteins and molecular components [6]. However, the basic mechanism remains the same across bacterial species. Every bacterium in the quorum-sensing population is able to produce a small hormone molecule, termed an autoinducer, and secrete it into the environment. Once a threshold concentration of the autoinducer is exceeded, bacteria respond by binding the autoinducer to a receptor. This receptor is termed a sensor. The autoinducer–receptor complex is a transcription factor that regulates a GRN at the level of the promoter, allowing bacteria to perform tasks at the colony level.
As shown in figure 2a, the GRN in Danino et al. [2] is composed of three genes (luxI, aiiA and yemGFP) under the influence of the same promoter, li-P. Genes have a C-terminal degradation tag sequence that shortens the half-life of their proteins considerably [7] and are introduced into bacteria on separate plasmids. The luxI gene encodes for the LuxI protein (LI), which produces acyl homoserine lactone (abbreviated AHL in main text and as A in equations). AHL can interact with the constitutively expressed protein LuxR (LR), which is modelled as a constant in the GRN, to form the LuxR : AHL complex (L : A) and activate the promoter li-P, allowing for the transcription of all three genes. The AHL molecule is removed from the system by interacting with the acyl homoserine lactonase enzyme (aA), which degrades AHL. AHL can also freely diffuse across the cell membrane, allowing for communication and hence coupling between all cells in the population.
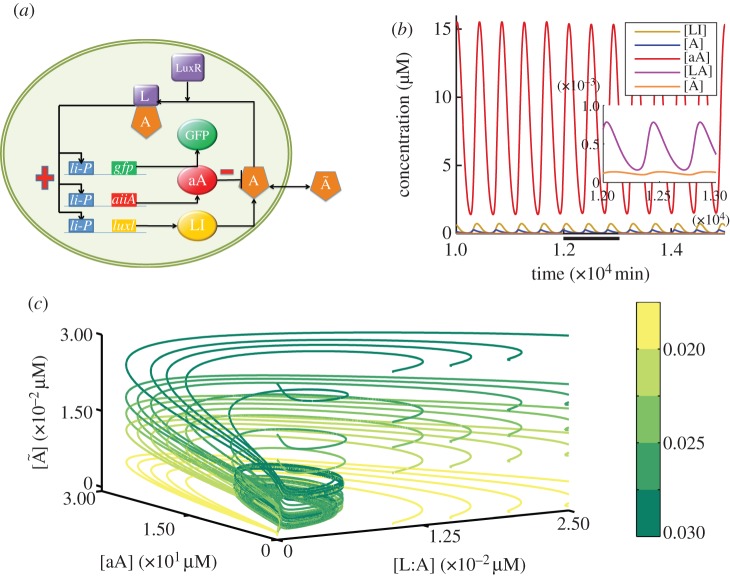
Figure 2.
Single-cell oscillating behaviour. (a) The GRN of a single cell in experiments presented in Danino et al. [2]. Th luxI gene encodes for the LuxI enzyme (LI), which produces the molecule acyl homoserine lactone (A). AHL combines with the constitutively expressed LuxR that is modelled as a constant, to form a complex, L:A, that can activate the promoter li-P. This drives the expression of all three genes: gfp, aiiA and luxI. AHL is removed from the system by enzymatic degradation catalysed by the aA enzyme (aA). AHL can also freely diffuse through the cell membrane. Positive and negative feedbacks steps within this network are indicated with a red plus/minus sign, respectively. (b) In the oscillating regime, oscillations are characterized by a period of approximately 410–420 min. The inset shows a magnification of the area that is highlighted with a bold line on the x-axis to illustrate the oscillations of the lower concentration components. (c) A projection of trajectories in three-dimensional space defined by the concentrations of aA, LA and Ã. Trajectories with sufficient external AHL concentration tend to the limit cycle. The gradient key illustrates the value of the increasing concentration of external AHL ([Ã]) in initial conditions of model simulations.
In order to systematically investigate the nature of the oscillatory behaviour observed in Danino et al. [2], we derive a mathematical model parametrized with values found in the literature to describe the metabolic states of cells as variables in a dynamical system. We use the mathematical formalism of ordinary differential equations (ODEs) that is extensively applied in modelling GRNs [8–11]. ODEs are amenable to analysis using mathematical tools from dynamical systems theory, such as bifurcation analysis [12], which enables the qualitative behaviour of the system (such as oscillations) to be studied as a function of key model parameter(s). Hence, the model can be used not only to replicate experimental observations but also to identify how parameter variation affects system behaviour.
To understand the behaviour at all levels, we use a hierarchical, bottom-up modelling approach to describe the biological process at different scales. We first analyse ODE-only models of a single cell as well as a population of cells, using a mean field and an agent-based approach. We then assess the impact of including explicit spatial components in the model system by performing large-scale simulations using BSim [13,14], a novel three-dimensional framework developed to study bacterial populations.
We find that oscillations are dependent on exceeding a threshold concentration of AHL. The concentration of external AHL varies as a function of cell density. An increase in cell density can be achieved either by decreasing the microfluidic device volume or by increasing the population size. This also increases the coupling strength across population members allowing oscillations to be the only stable behaviour in the system. The increase in coupling strength also allows for synchronized in-phase oscillatory behaviour. Our results highlight the importance of taking into account population as well as spatial effects in the process of GRN design and model development.
2. Model equations
The GRN, which is the basis of the model equations, is illustrated in figure 2a. We model the system's protein/molecule dynamics using quasi-steady-state assumptions on the dynamics of mRNA and remain consistent with the GRN description given in the introductory section. Proteins that are produced directly from the GRN (LI and aA) are described with two types of production rates, a basal rate a0X (where X is any protein), and Hill functions with production rates kpX and Hill coefficient equal to two. The concentration of free LuxR is obtained by subtracting the concentration of the LuxR : AHL (L : A) complex from the total concentration of LuxR, LTOT. Molecules not produced directly from the GRN are involved in linear reactions proportional to their concentration with rates kr−/+. With the exception of AHL (A), they also follow first-order degradation kinetics with the degradation rate constant τX. Degradation of AHL is enzymatic with rate kcataA represented by a Michaelis–Menten function. AHL diffuses in and out of the cell, depending on the intracellular (A) and extracellular (Ã) concentration difference with a rate ηcell/env. The parameter ηcell/env adjusts the concentration of AHL, depending on the volume of the environment where the molecule is found and thus also represents the cell density. The system of equations is as follows:
 |
2.1 |
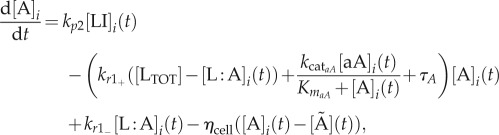 |
2.2 |
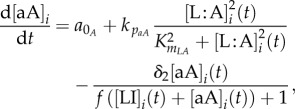 |
2.3 |
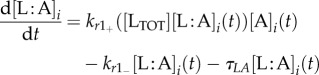 |
2.4 |
and
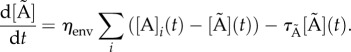 |
2.5 |
The terms at the end of equations (2.1) and (2.3) describe the enzymatic degradation of LI and aA, respectively, by the protease ClpX/P [15]. The parameters (δ1 and δ2) incorporate the maximum catalytic rate (Vmax) multiplied by the total ClpX/P concentration. The entire term is normalized by the Michaelis–Menten constant of enzyme ClpX/P (KMclx) for LI and aA, respectively. The parameter f is the inverse of the Michaelis–Menten constant of ClpX/P. Note that in the mean field approximation of the model and under quasi-steady-state assumptions on equation (2.5), an explicit expression can be derived that is proportional to the number of cells [16]. Detailed derivation of the model along with justification of the assumptions involved is presented in the supplementary material. The supplementary material also contains a list of abbreviations used throughout the text in table S2.
The ranges of possible values for each parameter were identified via an extensive search of the literature. We identified lower and upper bound values for each parameter and used particle swarm optimization to find optimal values that allow oscillatory behaviour in the system. Detailed explanation of the optimization process is given in the supplementary material, section S1.1.
A model describing the dynamics of this GRN has already being introduced in Danino et al. [2]. However, the model presented here differs in several aspects. The most notable difference is that in Danino et al. [2] the Hill function involves a delay term. Hence, delay differential equations (DDEs) are used to describe the dynamics of molecules produced directly from the GRN (here presented as aA and LI). The delay dynamics separate temporally the transcription and translation processes but in prokaryotes, there is no substantial delay associated with these processes [17–19]; therefore, we adopt the ODE modelling framework in this study. In Danino et al. [2], the values of the production rates for LI and aA are very different and adjusted with respect to cell density. In this study, we assume that LI and aA have similar production rates as they are under the influence of the same promoter. We also introduce an extra degree of freedom to describe the dynamics of the transcription factor, L : A, while in Danino et al. [2], AHL is assumed to be the transcription factor. We assume that production of AHL is linear and proportional to the concentration of LI, while in the model presented at [2], AHL production is nonlinear.
3. Single cell behaviour
We begin our study by investigation of the behaviour of the single-cell model (equations (2.1)–(2.5), i = 1). Specifically, we seek to determine whether the oscillations observed at the population level in Danino et al. [2] could be accounted for by this model. Direct numerical simulations illustrated in figure 2b indicate that there are parameter regimes where stable oscillatory behaviour is possible. Interestingly, we also find that the convergence to this oscillating state depends on the choice of initial conditions for the external AHL, whose dynamics are described by equation (2.5). This dependence is depicted in figure 2c, where we present a three-dimensional projection of the full phase space onto three of the model variables, namely the concentrations of aA, L:A and external AHL ([aA], [L : A],[Ã]). Figure 2c demonstrates that trajectories that start at concentrations of external AHL that are below the required threshold for oscillations converge to a fixed point characterized by zero amounts of the rest of the system's variables/proteins (depicted in yellow). Trajectories that exceed the threshold concentration of external AHL tend to a limit cycle attractor (depicted in green). These results indicate that exceeding the threshold concentration of external AHL is a sufficient condition for convergence to a stable oscillatory state in our model.
In the oscillatory regime, the model is characterized by a period in the order of hundreds of minutes (approx. 410 min), a value close to the experimental observations reported in Danino et al. [2]. A cycle of oscillation starts with the accumulation of the activated form of LuxR, L : A. This leads to production of LI and aA. aA quickly reaches maximum levels of expression, leading to enzymatic degradation of AHL. Removal of AHL stops further production of LI and aA; hence the dominating process becomes enzymatic degradation of both LI and aA by the ClpX/P protease. This completes a cycle of an oscillation. The maximum concentration of aA within a cycle of oscillation is higher than the maximum concentration of LI. We find the same qualitative behaviour after testing 10 parameter sets, with oscillatory period in the order of hours (simulations not shown). The parameters that we vary by taking evenly distributed values across each parameter range in the investigated sets (kpaA, kpli, KMaA, f,KcataA, δ1, δ2) are within the minimum and maximum ranges reported in the literature. Further details are given in the supplementary material, table S1. We observe similar oscillatory behaviour when the range of values a parameter can assume is increased in both minimum and maximum by an order of magnitude. This suggests that the model robustly supports stable oscillatory behaviour across a wide range of system parameters.
In order to identify the mechanisms leading to the onset of oscillations and also systematically investigate the behaviour of the system as a function of key parameters, we performed a bifurcation analysis of the single cell model (equations (2.1)– (2.5), i = 1). A projection of the bifurcation diagram constructed by varying the parameter kcataA is presented in the ([LI], kcataA) plane (figure 3a). We find that the system is bistable with respect to variation in kcataA; a stable steady state with nearly zero levels of expression (‘switched off’ cell) coexists with a stable limit cycle attractor for a significant range of control parameter values. In particular, for high values of the concentration of LI, we find a stable family (branch) of fixed points, shown in black in figure 3a, that loses stability in a supercritical Hopf bifurcation (HB1) as the control parameter (kcataA) increases. This family of fixed points regains stability in another supercritical Hopf bifurcation (HB2) as kcataA continues to increase. Eventually, the branch of stable fixed points turns around (to the left; not shown in figure 3a) as kcataA starts to decrease, whereas the concentration of LI has already dropped to nearly zero. At near-zero values of the concentration of LI, the system is characterized by a family of stable fixed points shown as a black solid line in figure 3a. Furthermore, for a range of control parameter values (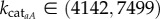 ), this stable family of (nearly zero) fixed point solutions coexists with the stable family of periodic orbits that are born and die in the pair of supercritical Hopf bifurcation points, HB1 and HB2, respectively. In biological terms, this means that in order for oscillations to occur, the cell must overcome the boundary between the two basins of attraction, of (i) the stable steady state and (ii) the stable limit cycle, respectively. As shown in figure 2c, this can be achieved by increasing the external AHL concentration, [Ã], above a certain threshold. We find a bifurcation structure similar to figure 3a when varying other parameters in the model, namely (kpli, kpaA, δ1, δ2, f, KMaÃ, τA). This finding supports further the oscillatory robustness of the model and reinforces the conclusions that oscillatory behaviour is intrinsic to the system.
), this stable family of (nearly zero) fixed point solutions coexists with the stable family of periodic orbits that are born and die in the pair of supercritical Hopf bifurcation points, HB1 and HB2, respectively. In biological terms, this means that in order for oscillations to occur, the cell must overcome the boundary between the two basins of attraction, of (i) the stable steady state and (ii) the stable limit cycle, respectively. As shown in figure 2c, this can be achieved by increasing the external AHL concentration, [Ã], above a certain threshold. We find a bifurcation structure similar to figure 3a when varying other parameters in the model, namely (kpli, kpaA, δ1, δ2, f, KMaÃ, τA). This finding supports further the oscillatory robustness of the model and reinforces the conclusions that oscillatory behaviour is intrinsic to the system.
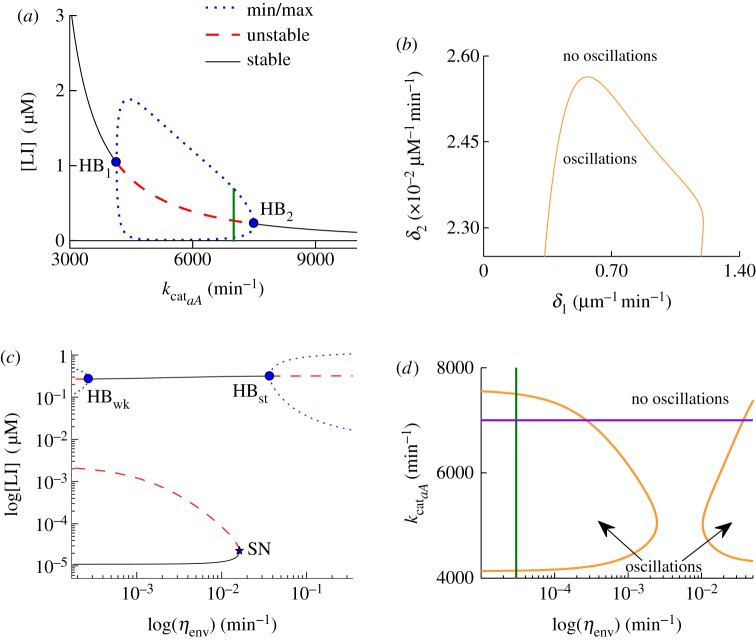
Figure 3.
Bifurcation diagrams for a single cell. For bifurcation diagrams (a) and (c) maxima and minima of the stable limit cycle solutions are shown by a dotted line. Unstable steady-state solutions are shown by a dashed line. Stable steady-state solutions by a solid line. (a) Bifurcation diagram depicting the dynamics of variable [LI] as a function of the parameter kcataA. Oscillations are due to a Hopf bifurcation. Hopf bifurcation points are labelled as HB1 and HB2 respectively and indicated by a dot. The value of kcataA used in the model simulations is shown by a vertical line. (b) Two-parameter bifurcation diagram depicting the Hopf bifurcation HB1/2 in the (δ1, δ2) plane. Oscillations occur when the parameters take values inside the area shown by the Hopf loci. (c) Bifurcation diagram depicting the dynamics of the variable [LI] as a function of the parameter ηenv. Hopf bifurcation points are labelled as HBwk/st for weak/strong coupling and indicated by a dot. A saddle–node (SN) bifurcation point is labelled as SN and indicated with a star. (d) Two-parameter bifurcation diagram showing the regions where oscillatory behaviour is possible as a function of ηenv and kcataA. The vertical line indicates the value of ηenv used in the model simulations. The horizontal line shows the value of kcataA used in the model simulations. Note that their intersection lies within the oscillatory boundary of the low ηenv value. (Online version in colour.)
It is important to note that stable oscillatory behaviour is observed when we introduce an order of magnitude difference in the effective degradation rates of aA and LI, δ1 and δ2, in the model, as can be seen in the supplementary material, table S1. The dependence of oscillatory behaviour on the degradation rates, which represent the normalized maximum catalytic rate of the ClpX/P complex for LI and aA, respectively, is summarized in the two-parameter bifurcation diagram shown in figure 3b. The orange curve depicted in figure 3b represents the loci of the Hopf bifurcation points found in the model and discussed earlier, and therefore outlines the regions in two-parameter space, i.e. the (δ1, δ2) plane, where the system will oscillate, provided that there is sufficient external AHL present. The order of magnitude difference that we find sufficient for the existence of oscillations effectively allows for time-scale separation between the slow component of the system aA and the rest of the network components. Similar time-scale separation conditions have already been identified as a requirement for oscillations in other studies [20,21].
4. Cell population behaviour
4.1. Mean field approximation
The single cell analysis demonstrates that oscillatory behaviour is dependent on the concentration of external AHL, the system's autoinducer. To better characterize this dependence at a population level, we begin by considering a mean field approximation of the system, i.e. by representing the interaction of all the cells present in the system as an average effective coupling. Although this approach reduces the many-cell system into a single cell system, it allows us to study macroscopic properties such as the cell density represented by the parameter ηenv [16], which is inversely proportional to the volume of the external medium and affects AHL concentration.
In figure 3c, we depict a projection of the bifurcation diagram for ηenv plotted in the ([LI], ηenv) plane. We find that for low concentrations of LI, there is a stable family (branch) of fixed points in the system that loses stability in a SN bifurcation as ηenv increases. At this point, the branch of now unstable fixed points turns around (to the left), the parameter ηenv begins to decrease and eventually undergoes another SN bifurcation (not shown) turning to the right, so that ηenv starts to increase again. Importantly, the concentration of LI has also increased substantially. Further increase of ηenv allows the branch of fixed points to regain stability in a supercritical Hopf bifurcation (labelled HBwk in figure 3c).
The occurrence of a Hopf bifurcation at HBwk gives rise to a family of stable periodic solutions at non-zero concentrations of LI that coexists with the family of stable fixed points for near-zero concentrations. As the parameter ηenv increases further, the branch of stable fixed points at non-zero LI concentration loses stability in another Hopf bifurcation (labelled HBst in figure 3c). HBst is again supercritical and gives rise to another stable family of periodic solutions. However, in this parameter range, the system is no longer bistable, and the family of periodic solutions (oscillations) is the only stable attractor of the system.
This is an important observation because increase in the parameter ηenv reflects an increase in cell density; the number of cells per unit volume. Our analysis demonstrates that if the size of the population is sufficiently large within a contained volume environment, as is the case of the experimental set up in Danino et al. [2], the only stable state of the system would be an oscillatory one. However, when ηenv is small, it can be difficult for the system to maintain a sufficient concentration of external AHL to support stable oscillatory behaviour. This in turn can lead to a ‘switched off’ cellular state represented by the near-zero concentration of LI. This is seen as a stable branch of fixed points in both figure 3a,c).
The oscillatory behaviour of the system that is related to cell density is best summarized in the two-parameter bifurcation diagram of kcataA and ηenv, as shown in figure 3d. Here, we plot the loci of the two Hopf bifurcation points, HBwk (left curve) and HBst (right curve) in the (kcataA, ηenv) plane. Stable oscillations in the system exist to the left of the curve that represents HBwk points and to the right of the curve that depicts the loci of HBst. In addition, the stable oscillations that belong to the left half-plane (associated with low cell density) coexist with stable steady-state solutions in the model. Interestingly, the model predicts that there is an intermediate range of cell density where the system could be either in a low or high (yet non-oscillatory) stable steady state. This is dependent on the concentration of external AHL and is represented by the region between the two HB curves in figure 3d. Because the coupling in the model is mediated via external AHL concentration, the increase in cell density also results in an increased coupling strength between cells of the same population, whereas low density corresponds to weakly coupled cells.
4.2. The effect of increasing the number of coupled cells
Next, we investigate the effect of a growing population in an agent-based model setting. Because we would like to be able to use continuation tools to perform bifurcation analysis, we study various small populations of explicitly coupled cells. This setting also allows us to study the modes of coordination between individual-coupled cells. We find that the bifurcation structure of the coupled cells system is qualitatively the same as the one shown in figure 3a. Continuation of the higher stable fixed point solutions in the model with respect to important parameters (kcataA, kpli, kpaA, δ1, δ2, f, KMaA, τA, ηenv) shows that there is a slight reduction in the range of stable oscillatory behaviour, as depicted in figure 4. This is due to the fact that the Hopf bifurcation pairs (HB1 and HB2) come closer together as the number of cells increases.
Figure 4.
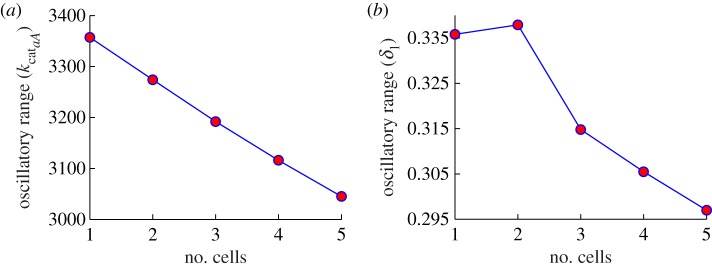
Reduction in oscillatory range with increased number of coupled cells. Dependence of the oscillatory range on the number of coupled cells for ηenv = 3E−5. The two panels show the difference between the two bifurcation points, HB1 and HB2, for parameter (a) kcataA (y-axis, see figure 3a for single-cell bifurcation diagram) and (b) δ1 (y-axis), as a function of increasing coupled cells (x-axis). (Online version in colour.)
Direct numerical simulations with various small populations (2–10 cells) of all-to-all coupled cells (figure 5a) indicate that the oscillatory threshold of external AHL concentration is lower in the coupled cells system than in the single cell system. This observation can be explained by a shift in the boundary between attractors in the phase space in favour of the limit cycle attractor. The increase in population size affects not only the boundary between attracting states, but also the waveform of oscillations in the model. We observe a decrease in the period of oscillations, from approximately 410–420 min in a single cell to approximately 380–390 min in a population of 10 coupled cells. In addition, the oscillations are characterized by a smaller amplitude, as shown in figure 5b.
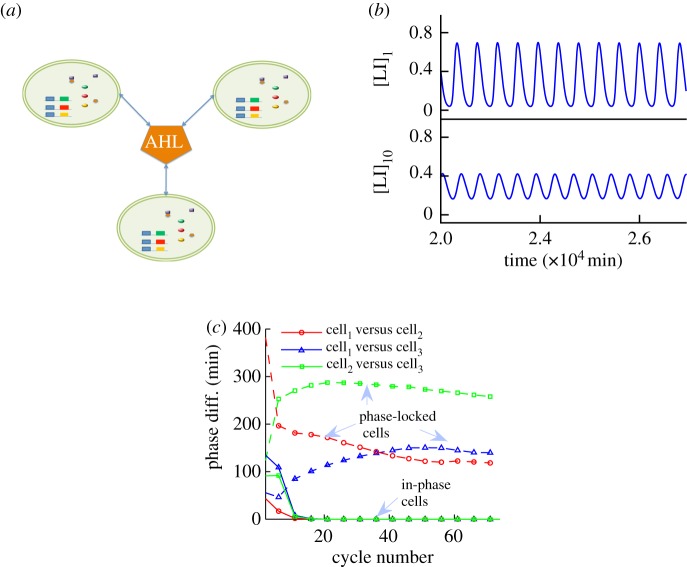
Figure 5.
Small population and its oscillatory behaviour. (a) AHL couples all cells in the environment, in an all-to-all network. Each cell carries the same GRN as the single cell model. (b) Comparison of the periodicity and amplitude difference of a single cell ([LI]1) and a cell part of a 10-cell population ([LI]10). An increase in number of cells decreases the period by 30 min and also decreases the overall amplitude of oscillations. (c) Phase difference plot of a coupled population of three cells for ηenv = 3E−5, dashed lines, where the cells slowly drift to a phase-locked state. This is not the case when the population is strongly coupled, as a result of lower external volume (ηenv = 3E−2, solid lines). In this case, the cells quickly converge to perfectly synchronized (in-phase) oscillatory state. (Online version in colour.)
Finally, we simulate various population sizes with heterogeneous initial conditions in order to investigate if and when in-phase (synchronized) oscillations across population members can be obtained. An example of the phase difference between cells observed in a three-cell population is shown in figure 5c. In the case where cell density (represented by ηenv) corresponds to weaker coupling, as described in the mean field approximation of the system (i.e. oscillations born from the HBwk loci), the oscillations among agents are coordinated as shown by the constant phase difference obtained in the simulations. As might be expected, increasing the coupling strength between cells by setting ηenv at a larger value (i.e. oscillations born from the HBst loci) results in perfectly synchronized (in-phase) oscillations, as indicated by phase difference convergence to zero.
5. Spatio-temporal dynamics
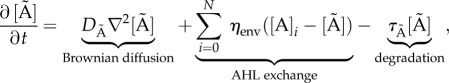
In order to study a more realistically sized population of cells in an explicit spatial context, we model a static (non-dividing) population of varying size (1–4800 cells) in microfluidic chambers (the population upper limit is restricted by the available computational power). The chamber dimensions are consistent with [2]. The model is implemented in BSim [13,14], a three-dimensional framework developed to study bacterial populations. BSim is an agent-based simulator, where each bacterial cell is defined as an agent that has explicit spatial positions in the three-dimensional environment and contains a GRN allowing simulation of intracellular dynamics. In particular, each agent in the simulation contains the GRN given in equations (2.1)–(2.4) and the external AHL chemical field is modelled with the reaction–diffusion PDE
 |
5.1 |
describing AHL Brownian diffusion with a diffusion constant DÃ, the exchange of AHL between agents and the environment with rate ηenv, and AHL degradation rate τÃ. The agent metabolic states, the agents' spatial position and environmental factors are coupled as illustrated in figure 1.
By using the same parameter set as with the ODE model (reported in the supplementary material, table S1), we find damped oscillations with the concentrations of GRN components going to a non-zero stable steady state, indicating the existence of a focus. Brownian diffusion of the external chemical field allows AHL to diffuse away from the source, i.e. the cell. This means that the exchange between intracellular and extracellular AHL is delayed. This is different from the non-spatially explicit model where extracellular AHL build-up is immediate and uniform in the entire volume, which in turn leads to immediate exchange between cells and environment. To obtain oscillations in the spatial version of the model, it was necessary to alter the parameters from those used in the single cell model (all parameter changes are given the supplementary material, table S1). This modified parameter set was used to investigate population behaviour. Note that parameter values are still within the bounds given in the literature and are of biological relevance.
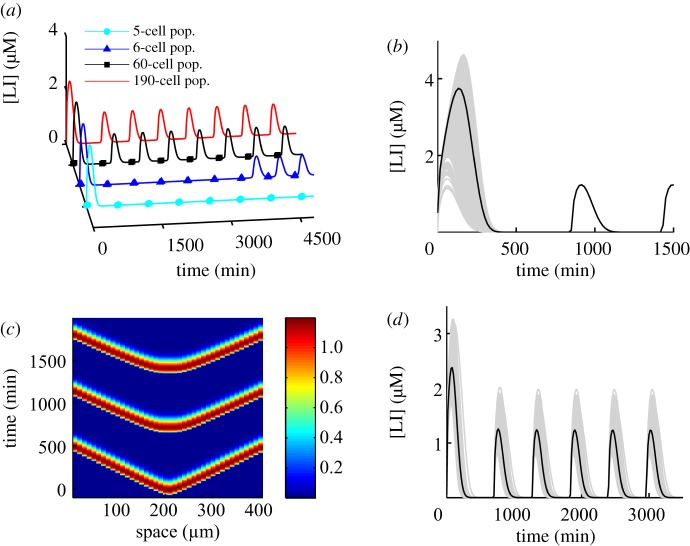
As shown in figure 6a, for chamber dimensions consistent with [2], sustained oscillations occur for a population with a minimum size of six cells and are maintained throughout the various population sizes we model. The model predicts that given sufficient time for the external AHL to build up even a small population of cells will oscillate. This could not be seen experimentally as in the physiological system, the population size increases exponentially owing to continuous cell divisions [2]. This is illustrated in figure 6a, where increase in the population allows for faster onset of oscillatory behaviour and marginally increases the amplitude of oscillations. Generally, the amplitude of oscillations is higher with respect to the non-spatially explicit version of the model. Above a certain threshold with respect to population size, a further increase does not affect the period of oscillations. Above this threshold, the observed period remains the same at approximately 416 min.
Figure 6.
How the population affects oscillatory behaviour in large-scale BSim simulations. (a) A plot of perfectly synchronized cells part of a population. Population sizes are indicated in the legend. An increasing population brings about onset of oscillations earlier owing to earlier accumulation of sufficient external AHL concentration. Cells are in a 200 × 50 × 1 µm3 chamber. (b) A population of 1139 cells with different initial conditions in a 200 × 50 × 1 µm3 chamber synchronized in the first few minutes of the simulation and then oscillated in-phase. Time series of individual cells are plotted for the entire population in grey. The black line indicates the population average. (c) A space–time plot illustrating a row of identical cells (part of a 1200 strong population) in the middle of a 400 × 300 × 1 µm3 chamber with DÃ = 0.009. The plot illustrates wave propagation through the chamber as cells become fully active sequentially in a neighbour-to-neighbour fashion. The gradient key illustrates the value of the variable [LI] in micromolar. (d) Time series illustrating synchrony of a heterogeneous population of 1139 cells in a 200 × 50 × 1 µm3 chamber. Time series of individual cells are plotted for the entire population in grey. The black line indicates the population average. Heterogeneity was introduced by modifying parameter values (production and degradation rates), using a Gaussian distribution with 5% standard deviation around the mean value of each parameter.
On the basis of analysis of the mean-field model described earlier, we expect that the strength of the coupling between cells can affect the length of the transient to synchronization. We conducted a series of numerical experiments varying parameters that could affect synchronization and oscillatory behaviour. We varied population size, chamber dimensions, cell motility, variation of the Brownian diffusion coefficient DÃ, GRN and chemical field initial conditions. Variation of DÃ affected the onset of oscillations. In simulations where a few non-motile cells were present in a 200 × 50 × 1 µm3 chamber, higher DÃ values delayed the onset of oscillations. Beyond DÃ = 10 µm3 s−1, no further delay was seen in oscillatory onset. Example of effects of local concentration gradients is shown in the supplementary material. The value of the diffusion coefficient also affected the synchronization properties of the population. Higher values resulted in faster synchronization of the population with cells having identical phase.
Motility affected only small populations (e.g. 5–15 cells) and had the same effect as increasing the diffusion coefficient, DÃ (results not shown). This is not surprising as motion of AHL-producing cells allows for more rapid AHL equilibriation throughout the chamber. Populations of greater than 15 cells were always perfectly synchronized for DÃ > 10 µm3 s−1 even when cells had different initial conditions as shown in figure 6b, where a 1139 non-motile cell population synchronizes within the first cycle of oscillation.
Our model has qualitatively similar behaviour to the experiments presented in figure 3 of Danino et al. [2] showing waves of bacterial population's activity. Figure 6c depicts a space–time plot illustrating waves of bacterial population's activity in our model that is similar to the experiments presented in figure 3c,d of [2] (see also accompanying video of simulation in the supplementary material). Simulations of the experiment shown in figure 3a,b of Danino et al. [2] is presented in the supplementary information, figure S2.
To further investigate population effects, we also simulate a heterogeneous population of cells in BSim. In a biological setting, cells will have slight variation in the rates of reaction of various processes as a result of cell-to-cell variability during cell division [22]. This may affect their ability to oscillate. We model cell-to-cell variability by varying the production and degradation rates of each cell across the population using a narrow Gaussian distribution around the mean value of a parameter (where mean value is the value illustrated the electronic supplementary material, table S1). Narrow distributions are chosen because variability should be minimal in a clonal population growing in a tightly regulated experimental environment. The results presented in figure 6d assume a Gaussian distribution with standard deviation of 5 per cent and shows that heterogeneous populations maintain synchrony in the spatially resolved version of the model.
6. Conclusions and discussion
In this study, we investigate whether macroscopic behaviour reflects individual cell properties, and how the population size affects this behaviour, by focusing on the example of oscillations across a population of quorum-coupled cells [2]. We show the effect on oscillatory behaviour with respect to increasing population size, using numerical simulation and bifurcation analysis. We also demonstrate how spatio-temporal dynamics affect the oscillatory behaviour of the population.
GRN design in the system under investigation [2] includes positive and negative feedback loops. Such loops, such as activation–inhibition pairs, are an important ingredient in GRN models, that allow for the occurrence of oscillations [20,21,23], including oscillations that arise from a Hopf bifurcation. GRN models introduce such feedback at the level of the genetic code by affecting gene expression at the level of the promoter (well-known examples are Gardner et al. [8] and Elowitz & Leibler [9]). In the model, we present in this study, positive feedback occurs at the level of the promoter, via L : A, but negative feedback is indirect and involves only the post-translational component aA.
We show that the GRN model in this study allows oscillations to be born in a Hopf bifurcation, provided that there is sufficient time scale separation among the model variables. Time-scale separation is a sufficient condition for oscillations in negative feedback systems, as explained in Novák & Tyson [21] and first illustrated by Goodwin's oscillator [24]. This separation allows the system to have a memory of previous states that can affect the current state leading to oscillations. In Danino et al. [2], such delays are implemented explicitly using DDEs that separate transcription and translation processes temporally and prevent the system from settling to a steady state. However, the lack of compartmentalization in bacteria prevents significant temporal separation of transcription and translation processes [17–19]. Our analysis indicates that oscillations are also possible without temporally separating transcription and translation.
In our model, we introduce time-scale separation between the dynamics of LI and aA, the post-translational modules responsible for activation and inhibition in the system by varying their degradation rates δ1 and δ2, respectively. Although necessary from a mathematical perspective, the biological basis of this is not immediately apparent. Both proteins, aA and LI, are substrates for the same degradation enzyme, ClpX/P, and would be expected to have similar degradation rates. However, a possible biological explanation can be given involving the mechanics of the enzyme ClpX/P—a molecular complex with two major functions [15,25]. Substrate is unfolded by ClpX and processively passed to the ClpP protease for degradation. In our model, we take into account both molecular functions via the rate parameter δx. Because the substrates of ClpX/P, aA and LI, are of similar size [26,27], it is likely that the difference in the rates of degradation can be attributed to an increased stability of aA during the unfolding process. To the best of our knowledge, this suggestion has not been confirmed experimentally.
The GRN has an interesting positive feedback design via LI and AHL, whose concentration values are affected by the production rate constants kpLI and kp2, respectively. Bifurcation analysis, in these parameters, indicates defined ranges where oscillations exist (see electronic supplementary material, figure S1). Increasing values within each range increases the amplitude of oscillations offering additional control over the oscillatory waveform [20]. The AHL production rate constant is an order of magnitude bigger than LI. Even if aA and LI, whose actions are opposing in the network, are produced at similar rates, positive feedback is sufficiently amplified via AHL for oscillations to exist.
Stable oscillatory behaviour in the model is dependent on the amount of AHL present in the system. This is consistent with quorum behaviour. GRN components become activated, once a certain threshold concentration of an activator is exceeded in the cellular medium [3,28], as previously shown with other mathematical models [29,30]. Of greater biological interest is the existence of two separate Hopf bifurcation loci. At high cell densities, as described by an increase in parameter ηenv and shown in figure 3d, there exists only a single stable attractor, the oscillatory one. By contrast, the cells can oscillate at low cell-density only if there is sufficiently high concentration of external AHL present. However, in realistic settings, it is more likely that this concentration is low initially but following a number of cell divisions, the required cell density for oscillations is eventually achieved. High cell density also increases cell-to-cell coupling as it modulates the communication mechanism—the external AHL concentration. Increased coupling allows for synchronized (in-phase) oscillations in the system.
Our results are consistent with the experiments of Danino et al. [2]. However, our theoretical analysis offers an alternative explanation from the theoretical conclusions in Danino et al. [2] that individual cells would oscillate independently of cell density. Our analysis suggests that the experimentally observed behaviour of bacterial populations is an intrinsic population-level property, and not simply the result of individually oscillating cells becoming synchronized. As such, an increase in cell density not only affects the waveform and synchrony of oscillations, but is also responsible for driving the system from a bistable to a monostable oscillatory regime, as shown in the bifurcation diagram of figure 3c. Experimentally, this can be tested by dilution assays and external AHL concentration modulation possibly by the introduction of the aA enzyme extracellularly.
Introduction of an explicit spatial component facilitates the analysis of the system's behaviour by studying additional features that can affect the population-wide oscillations. In this study, we showed that in a microfluidic environment, synchrony and oscillatory behaviour are also affected by local and global concentrations of AHL as a result of Brownian diffusion. Strongly coupled cells will oscillate synchronously with the same phase throughout the population, whereas weakly coupled cells will oscillate locally (in synchrony only with their neighbours). Coupling is affected by cell density and the diffusion of AHL within the environment, both of which are features essential for quorum-sensing.
To answer our original question regarding the nature of macroscopic oscillations in Danino et al. [2], we conclude that the system is able to provide robust and stable oscillations at the population level as a result of high cell density. It would be of great interest to investigate whether such behaviour exists in higher organisms; whether groups of cells use communication mechanisms intimately associated with cell density to (re)enforce a specific population behaviour. In our particular example [2], it is possible to interrupt the communal response by reducing cellular density and limiting external AHL concentration. Such strategies have already been observed among competing bacterial species where quorum-quenching mechanisms are used to disrupt communication and suppress quorum effects in the opposing community [3,4,31,32]. Our results also highlight the importance of using microfluidic technology to approximate the effects of large population sizes in a manageable, observable and accessible environment. The predictive power of models may be reduced if population and spatio-temporal effects are not taken into account, as indicated by the results obtained at different levels of abstraction.
Acknowledgements
The authors declare no competing interests. P.M. was supported by EPSRC grant no. EP/E501214/1 and KT-A by EPSRC grant no. EP/I018638/1. P.M. would like to thank Thomas Gorochowski and Antoni Matyjaszkiewicz for discussions regarding BSim models, David Barton for discussions on particle swarm optimization algorithms and Jakub Nowacki for help regarding the xpppy python suite for drawing bifurcation diagrams using python.
References
- 1.Milias-Argeitis A, Summers S, Stewart-Ornstein J, Zuleta I, Pincus D, El-Samad H, Khammash M, Lygeros J. 2011. In silico feedback for in vivo regulation of a gene expression circuit. Nat. Biotechnol. 29, 1114–1116 10.1038/nbt.2018 (doi:10.1038/nbt.2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danino T, Mondragón-Palomino O, Tsimring L, Hasty J. 2010. A synchronized quorum of genetic clocks. Nature 463, 326–330 10.1038/nature08753 (doi:10.1038/nature08753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346 10.1146/annurev.cellbio.21.012704.131001 (doi:10.1146/annurev.cellbio.21.012704.131001) [DOI] [PubMed] [Google Scholar]
- 4.Bassler BL, Losick R. 2006. Bacterially speaking. Cell 125, 237–246 10.1016/j.cell.2006.04.001 (doi:10.1016/j.cell.2006.04.001) [DOI] [PubMed] [Google Scholar]
- 5.Nealson K, Platt T, Hastings J. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104, 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitehead N, Barnard AM, Slater H, Simpson NJ, Salmond GP. 2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25, 365–404 10.1111/j.1574-6976.2001.tb00583.x (doi:10.1111/j.1574-6976.2001.tb00583.x) [DOI] [PubMed] [Google Scholar]
- 7.Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64, 2240–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner TS, Cantor CR, Collins JJ. 2000. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 10.1038/35002131 (doi:10.1038/35002131) [DOI] [PubMed] [Google Scholar]
- 9.Elowitz MB, Leibler S. 2000. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 10.1038/35002125 (doi:10.1038/35002125) [DOI] [PubMed] [Google Scholar]
- 10.Dubnau D, Losick R. 2006. Bistability in bacteria. Mol. Microbiol. 61, 564–572 10.1111/j.1365-2958.2006.05249.x (doi:10.1111/j.1365-2958.2006.05249.x) [DOI] [PubMed] [Google Scholar]
- 11.Ullner E, Zaikin A, Volkov E, García-Ojalvo J. 2007. Multistability and clustering in a population of synthetic genetic oscillators via phase-repulsive cell-to-cell communication. Phys. Rev. Lett. 99, 1–4 10.1103/PhysRevLett.99.148103 (doi:10.1103/PhysRevLett.99.148103). [DOI] [PubMed] [Google Scholar]
- 12.Kuznetsov IUA. 1998. Elements of applied bifurcation theory, vol. 112 Berlin, Germany: Springer [Google Scholar]
- 13.BSim source code. See http://bsim-bccs.sourceforge.net/ . (updated 2012; cited October 11) [Google Scholar]
- 14.Gorochowski TE, et al. 2012. BSim: an agent-based tool for modeling bacterial populations in systems and synthetic biology. PLoS ONE 7, e42790. 10.1371/journal.pone.0042790 (doi:10.1371/journal.pone.0042790) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGinness KE, Baker TA, Sauer RT. 2006. Engineering controllable protein degradation. Mol. Cell 22, 701–707 10.1016/j.molcel.2006.04.027 (doi:10.1016/j.molcel.2006.04.027) [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Ojalvo J, Elowitz MB, Strogatz SH. 2004. Modeling a synthetic multicellular clock: repressilators coupled by quorum sensing. Proc. Natl Acad. Sci. USA 101, 10 955–10 960. 10.1073/pnas.0307095101 (doi:10.1073/pnas.0307095101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young R, Bremer H. 1975. Analysis of enzyme induction in bacteria. Biochem. J. 152, 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J, Xiao J, Ren X, Lao K, Xie XS. 2006. Probing gene expression in live cells, one protein molecule at a time. Science 311, 1600–1603 10.1126/science.1119623 (doi:10.1126/science.1119623) [DOI] [PubMed] [Google Scholar]
- 19.Proshkin S, Rahmouni AR, Mironov A, Nudler E. 2010. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328, 504–508 10.1126/science.1184939 (doi:10.1126/science.1184939) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai TYC, Choi YS, Ma W, Pomerening JR, Tang C, Ferrell JE. 2008. Robust, tunable biological oscillations from interlinked positive and negative feedback loops. Science (New York, NY) 321, 126–129 10.1126/science.1156951 (doi:10.1126/science.1156951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novák B, Tyson JJ. 2008. Design principles of biochemical oscillators. Nat. Rev. Mol. Cell Biol. 9, 981–991 10.1038/nrm2530 (doi:10.1038/nrm2530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang S. 2009. Non-genetic heterogeneity of cells in development: more than just noise. Development (Cambridge, England) 136, 3853–3862 10.1242/dev.035139 (doi:10.1242/dev.035139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell O, Savery NJ, Grierson CS, di Bernardo M. 2010. A comparative analysis of synthetic genetic oscillators. J. R. Soc. Interface 7, 1503–1524 10.1098/rsif.2010.0183 (doi:10.1098/rsif.2010.0183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodwin BC. 1963. Temporal organization in cells. A dynamic theory of cellular control processes. New York, NY: Academic Press [Google Scholar]
- 25.Farrell CM, Grossman AD, Sauer RT. 2005. Cytoplasmic degradation of ssrA-tagged proteins. Mol. Microbiol. 57, 1750–1761 10.1111/j.1365-2958.2005.04798.x (doi:10.1111/j.1365-2958.2005.04798.x) [DOI] [PubMed] [Google Scholar]
- 26.UniProt Consortium Protein sequence and functional information for AiiA. See http://www.uniprot.org/uniprot/Q8KWS6. (updated January 2012; cited October 11, 2012) [Google Scholar]
- 27.UniProt Consortium Protein sequence and functional information for LuxI. See http://www.uniprot.org/uniprot/P12747. (updated January 2012; cited October 11, 2012) [Google Scholar]
- 28.Fuqua C, Parsek MR, Greenberg EP. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35, 439–468 10.1146/annurev.genet.35.102401.090913 (doi:10.1146/annurev.genet.35.102401.090913) [DOI] [PubMed] [Google Scholar]
- 29.Goryachev AB, Toh DJ, Lee T. 2006. Systems analysis of a quorum sensing network: design constraints imposed by the functional requirements, network topology and kinetic constants. Bio Syst. 83, 178–187 10.1016/j.biosystems.2005.04.006 (doi:10.1016/j.biosystems.2005.04.006) [DOI] [PubMed] [Google Scholar]
- 30.Klapper I, Dockery J. 2010. Mathematical description of microbial biofilms. IAM Rev. 52, 221–265 [Google Scholar]
- 31.Shapiro J. 1998. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52, 81–104 10.1146/annurev.micro.52.1.81 (doi:10.1146/annurev.micro.52.1.81) [DOI] [PubMed] [Google Scholar]
- 32.Kaufmann GF, et al. 2005. Revisiting quorum sensing: discovery of additional chemical and biological functions for 3-oxo-N-acylhomoserine lactones. Proc. Natl Acad. Sci. USA 102, 309–314 10.1073/pnas.0408639102 (doi:10.1073/pnas.0408639102) [DOI] [PMC free article] [PubMed] [Google Scholar]