Abstract
The classical view of DNA posits that DNA must be stiff below the persistence length (<150 bp) but recent studies addressing this have yielded contradictory results. We developed a fluorescence-based, protein-free assay for studying the cyclization of single DNA molecules in real time. The assay samples the equilibrium population of a sharply bent, transient species which is entirely suppressed in single molecule mechanical measurements and is biologically more relevant than the ligated species sampled in the traditional ligase-based assay. The looping rate has a remarkably weak length dependence between 67 and 106 bp that can not be described by the worm-like chain model. Many biologically significant protein-DNA interactions that involve looping and bending of DNA below 100 bp likely use this intrinsic bendability of DNA.
Bending and looping of lengths of DNA below 100 base pair (bp) is ubiquitous in cellular processes such as regulated gene expression in bacteria and eukaryotes (1, 2), packaging of DNA in viral capsids and DNA storage complexes in eukaryotes (3). Quantifying the intrinsic bendability of DNA at these biologically important length scales is essential for understanding DNA-protein interactions. According to a widely used approximation, DNA duplex is modeled as an elastic rod and its mechanical properties are described by the worm-like chain (WLC) model. Persistence length (lp) is a measure of the bending rigidity of DNA and for a DNA molecule that is several kilobasepair (kbp) or longer, lp can be readily measured using single molecule manipulation tools and is about 50 nm or 150 basepairs (4). In this framework, formation of DNA loops or sharp bends over length scales shorter than lp incurs a large energetic cost which makes the probability of their spontaneous formation vanishingly small.
Many approaches have been developed to quantify and model the inherent flexibility and bendability of DNA at short length scales. In the cyclization assay, the ligase protein traps DNA molecules in the looped conformation and then the looped and unlooped populations are separated based on their different gel mobility (5). Recent experiments using this assay and other techniques have challenged the classical picture of DNA as an elastic rod (6). Cyclization of DNA fragments of ~100 bp using the ligase assay yielded up to four orders of magnitude higher cyclizability (j-factor) compared to the prediction of the WLC model (7). However this extraordinary result was questioned and attributed to too high a ligase concentration used in the experiments (8). Small angle x-ray scattering was used to measure end-to-end distance variations of short DNA fragments labeled with gold nanoparticles. These experiments suggested a cooperative stretching behavior over two helical turns (9), however some aspects of the data are still paradoxical (10-12). Analysis of DNA images acquired using atomic force microscopy also deduced lower bending energies than what WLC predicts (13). However, this method is indirect, is based on surface absorption of DNA molecules and does not provide any dynamic information.
Contradictory and inconclusive results from these measurements call for a more direct approach to quantify flexibility of DNA on short length scales. In addition, most existing bulk approaches suffer from inherent limitations such as limited range of physical conditions and formation of byproducts other than monomer DNA circles, which limit their applicability to other systems. For example, due to non-specific interactions of DNA and the ligase protein, the ligase-based assay may not be suitable for studying cyclization of very short DNA molecules (14). Moreover, looping events cannot be detected in real time using bulk techniques. Due to geometrical and technical limitations, single molecule DNA stretching approaches cannot be used to study the mechanics of very short DNA molecules. Even for a moderate length of DNA with several hundred basepairs, many corrections are required to account for the finite chain length and the boundary conditions (15). In addition, because of the relatively long persistence length of dsDNA, even a hundred femto Newton of force makes configurations with sharp local bending inaccessible. Therefore, DNA responses measured via mechanical stretching would not include any contribution from such sharply bent conformations even if they existed in a relaxed DNA.
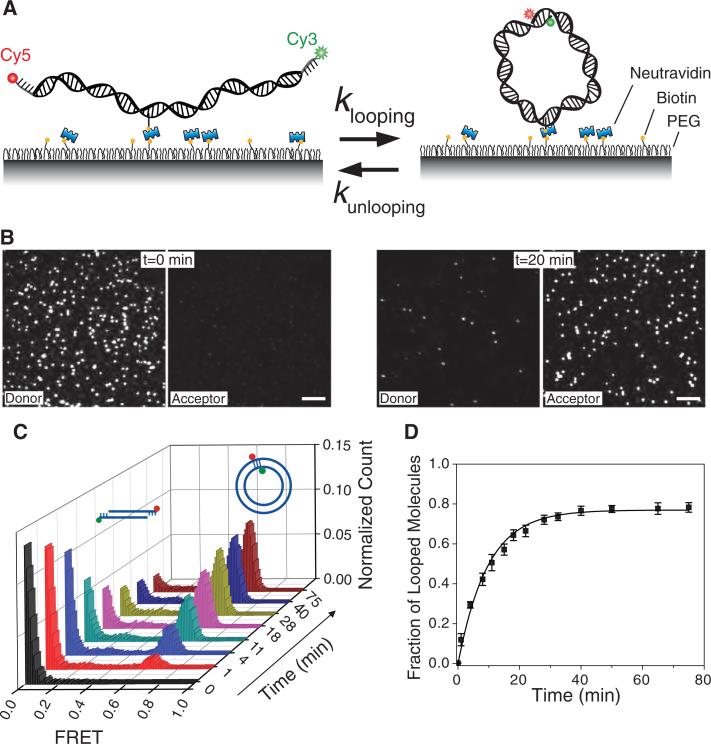
We developed a cyclization assay, based on single molecule fluorescence resonance energy transfer (smFRET) (16, 17), for directly monitoring the cyclization of single DNA molecules (Fig. 1A) (18). To avoid dimer formation during long-term observation, DNA molecules are immobilized on a polymer-coated surface through a biotin linker attached to a base at an internal DNA location. We avoided motifs such as A-tracts which are known to induce considerable intrinsic curvature (19). The DNA probe is a duplex with single stranded extensions on both 5’ ends. Each DNA molecule is labeled with Cy3 (donor) and Cy5 (acceptor) fluorophores at the 5’ end of the strands. Single stranded overhangs are complementary so that hybridization will trap the DNA molecules in the looped state. In the unlooped state the donor and acceptor are distant from each other and the molecules show zero FRET. Looping brings the dyes close to each other and the DNA molecules exhibit a high FRET signal. Therefore, the looped state can be clearly distinguished from the unlooped state based on the FRET value and the relative intensities of donor and acceptor (Fig. 1B).
Figure 1.
(A) Donor (Cy3) and acceptor (Cy5) labeled DNA molecules were immobilized on the surface via biotin-neutravidin interaction. (B) Fluorescence images of single 91 bp DNA molecules in corresponding donor and acceptor channels are shown before (left panels) and 20 minutes after adding high salt (1 M NaCl) buffer (right panels). Scale bar is 5 μm. (C) Histograms of FRET efficiency as a function of time (t=0 is when high salt was introduced) show the evolution of looped (high FRET) and unlooped (low FRET) populations. (D) Fraction of looped DNA (high FRET population) as a function of time, measured from the histograms in C. An exponential fit to this curve gives the looping rate R. Data are means ± SEM (N = 5).
The experiment starts in a buffer without added ions in order to strongly favor the unlooped state. Introducing a buffer containing high concentration of Na+ or Mg2+ can stabilize the looped state. Depending on the lengths of the DNA duplex and the single stranded overhangs, different behaviors were observed during the probing time window, typically ranging from less than 1 minute to up to 4 hours. DNA molecules formed stable loops, showed dynamics between looped and unlooped conformations, or exhibited no looping events. For example, for a 91 bp initial dsDNA with 10 nt overhangs, looping was nearly irreversible and the looped high FRET state accumulated to saturation within about 20 min (Fig. 1C, D). In this case, the looping rate R could be determined by fitting the time evolution of the looped population with a single exponential function (Fig. 1D).
We measured the looping rate R for a series of DNA molecules with a 10 nucleotide overhang on each end and with the circular size ranging from 67 to 106 bps (the circular size is the circumference of the DNA circle formed after looping, and is therefore the sum of the initial dsDNA length and the overhang length). The measured looping time, 1/R, varied from less than 10 minutes to more than 200 minutes (Fig. 2A, black squares). This 20 fold change in the looping times is surprisingly small, because we expected that DNA significantly shorter than the persistent length would take dramatically longer to form a loop. However, the result is qualitatively consistent with the observations that short DNA loops induced by the lac repressor and AraC protein can form efficiently in vivo and in vitro (2, 20-22), suggesting that the protein-induced looping may use the intrinsic flexibility of the DNA. Also, our result is consistent with a recent study which predicted that bending energy for short DNA loops would be independent of the loop length (13). In addition to the length dependence, a variation in looping rate depending on the angular phase between the two cohesive ends would be expected. Indeed, our data displays oscillation in R with a period of about one helical turn (see data points between 93 bp and 106 bp).
Figure 2.
(A) Looping time as a function of DNA circular length for surface tethered DNA (black squares) and vesicle encapsulated DNA molecules (red squares). (B) Looping time for 7 DNA sequences with 63 basepair duplex length and 10 nt overhang. R73 is the standard sequence used in A. Poly-A constructs were constructed by inserting n=10, 17, 26 and 38 consecutive A bases in the middle of a random sequence (E8). Data are means ± SEM (N≥3).
We performed a variety of controls to confirm that the surprisingly weak length dependence of the looping rates was not an artifact of our experimental scheme. First, to rule out possible contributions from surface tethering and internal biotin labeling, we repeated the experiments for DNA molecules without any internal modification and confined in 200 nm diameter phospholipid vesicles that are permeable to ions (23). Because infusion of 1 M NaCl ruptured the vesicles, we instead used 10 mM Mg2+ to stabilize the looped state. This is the same ionic condition used in the standard ligase assay. For five DNA constructs ranging from 94 to 105 bp, the looping times were similar between the vesicle encapsulated measurements and the surface measurements (Fig. 2A, red squares). Because DNA bending and torsional rigidity are sequence dependent (24), it remained possible that the high flexibility we observed was due to an extraordinarily flexible sequence. Therefore, we measured the looping times for a series of DNA constructs with identical loop length and overhang sequence but different internal base composition (Fig. 2B). We found that our standard sequence (R73) is not an outlier in terms of the looping rate. The DNA derived from a nucleosome positioning sequence (TA) (25) showed much faster (35 fold) looping than our standard sequence R73 (7). We also examined the looping behavior of sequences with potentially curvature inducing A tracks, (A)n, where n=0, 10, 17, 26, 38 embedded in otherwise randomly chosen sequence and found that the looping rate varied from more than 30 fold higher (n=0) to 2 fold lower (n=26) than that of R73. The two orders of magnitude difference in the looping time for these sequences despite the fact that the final 12 bp of these duplexes on both ends are similar, rules out duplex end opening as a possible mechanism for the rapid looping observed in our assay.
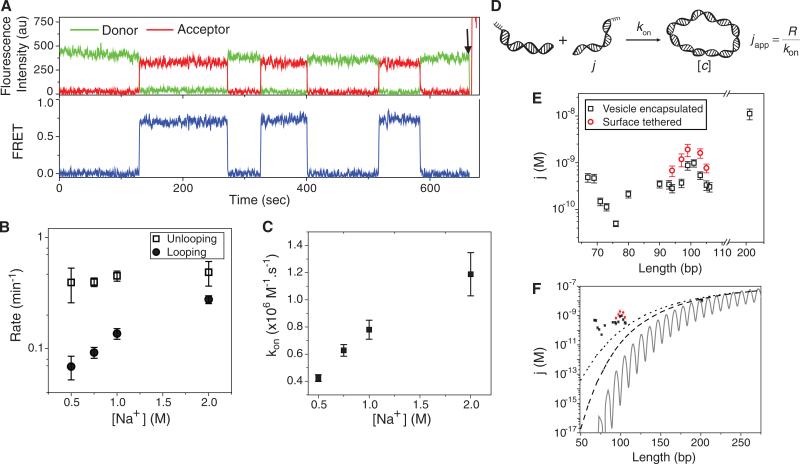
Changing the length of overhang to 8 nt allowed us to observe real time looping-unlooping dynamics (Fig. 3A) and evaluate the rates directly as a functional of salt. While the unlooping rate did not change between 0.5 M to 2 M Na+, the looping rate increased 3 fold in this range (Fig. 3B). However, since we observed the same increase in the bimolecular annealing rate (Fig. 3C) we can attribute the acceleration in looping at higher salt solely to the annealing enhancement. Therefore, monovalent ion concentrations above 0.5 M do not have a detectable effect on dsDNA flexibility (3).
Figure 3.
(A) Representative fluorescence intensities (top, green for donor and red for acceptor) and corresponding FRET efficiency (bottom, blue) time traces measured from a single DNA molecule in 750 mM NaCl. The DNA has 91 bp initial dsDNA with 8 nt single stranded overhangs. The arrow indicates a direct acceptor excitation to verify the acceptor has not photobleached. (B) Looping and unlooping rates as a function of [NaCl]. The DNA has 91 bp initial dsDNA with 8 nt ss-overhangs. (C) Bimolecular association rate (kon) measured as shown in Fig. s2 shows the same 3 fold increase as the looping rate with increasing [NaCl]. Data are means ± SEM (N≥300 molecules). (D) The model to relate looping rate (R), bimolecular association rate (kon) and apparent j-factor. (E) j-factor for surfaced tethered DNA (black squares) and vesicle encapsulated DNA (red circles). (F) Measured j-factor for surface tethered DNA (black squares) and vesicle encapsulated DNA (red squares). Solid black curve is the Shimada-Yamakawa prediction for DNA cyclization. Dashed line and dotted line are the WLC predictions for the j-factor of DNA circles with free boundary condition and for DNA molecules with 5 nm capture radius, respectively. Data are means ± SEM (N≥3).
We also investigated the effect of the single stranded overhang length on the stability of DNA loops. Decreasing the overhang length from 10 nt to 9 nt or 8 nt while maintaining the initial duplex, sharply increased the unlooping rate by about two orders of magnitude without an appreciable change in the looping rate (fig. S1A). Likewise, the equilibrium fraction of the looped state became progressively smaller by shortening the overhang (fig. S1B). Therefore, the main effect of longer overhangs in our assay compared to the 4 nt overhangs typical in the ligase assay is to increase the lifetime of the looped state. When a DNA loop forms, internal elastic energy stored in the loop is expected to provide a shear force that promotes unlooping. Indeed we found that 8 bp of duplex melts 20 times faster in a DNA circle than in a DNA dimer, likely due to internal tension in a circle which is absent in a dimer (supplementary text and fig. S1C).
The looping rate, R, can be calculated as the product of the bimolecular association rate kon between 10 nt long complementary strands and the effective concentration of one end of DNA in the vicinity of the other end, which we call the apparent j factor, japp (5) (Fig. 3D). Using a similar surface-based assay but for intermolecular annealing (supplementary text and fig. S2), kon was measured to be 0.78±0.07×106M–1s–1 in 1 M NaCl and 0.26±0.04×106M–1s–1 in 10 mM Mg2+, both consistent with an earlier estimate for short oligonucleotide annealing (26). The corresponding apparent j-factors, calculated using japp = R/kon are shown in Fig. 3E. Our calculated apparent j-factors along with the prediction of the WLC model (27, 28) are plotted in Fig. 3F. The solid line and the dashed line are the j-factor for a semiflexible polymer with parallel and free boundary conditions and the dotted line is the j-factor for a polymer with free boundary condition and 5 nm capture radius (the two ends anneal when they are closer than 5 nm) (29, 30). The measured japp values matched the theoretical prediction for 201 bp DNA but deviated from the theoretical values by orders of magnitude for the shortest lengths examined even under the most liberal boundary condition.
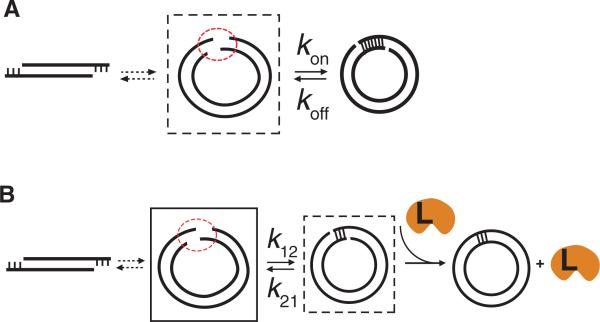
Our observation that the looping rate does not drop precipitously with decreasing DNA length is in stark contrast to the steep drop in the j-factor predicted by the WLC model. In many biologically relevant protein-DNA interactions such as in some genetic switches, the DNA bendability plays an important role in determining the state of the switch by controlling the concentration of one protein binding site in the vicinity of the other binding site (1, 31). Our assay samples such an equilibrium in which two DNA ends are in close proximity, but not annealed (dashed box in Fig. 4A). In contrast, the ligase assay samples the equilibrium of the annealed state (dashed box in Fig. 4B) (5). The equilibrium looped population, which is the substrate for ligase protein, is very sensitive to the unlooping rate (figs. S1A and S1B). The looping rate is biologically more relevant because it reports on how quickly two regions of DNA are brought into close proximity whereas the unlooping rate is additionally influenced by the melting rate of the short duplex formed. Our assay could independently measure the looping rate without being affected by the loop instability caused by internal tension in the short DNA circles. Many DNA binding proteins may have evolved to use the extraordinary flexibility of the DNA to capture and further stabilize transiently bent or looped DNA conformations.
Figure 4.
(A) Our assay reports on the equilibrium population of the intermediate state (dashed box) with the two DNA ends in close proximity. (B) Schematic representation of DNA cyclization reaction steps in the ligase assay. The intermediate state with the two DNA ends in close proximity (solid box) does not get sampled in this assay. Instead, the ligase samples the equilibrium population of the annealed state (dashed box). Ligase protein is labeled L.
Extended WLC models have previously been developed to explain the remarkable flexibility of short DNA by allowing for the formation of temporary bubbles (32) or kinks (33), and molecular dynamics simulations observed emergence of kinks in small DNA minicircles (34, 35). Others accommodate high bendability of short DNA by introducing non-harmonic elastic behavior (13). To gain insight into the mechanism of facile looping, we performed experiments on DNA constructs with a single backbone nick, double nicks or a single basepair mismatch in the middle and observed one to two orders of magnitude higher looping rate compared to our original DNA constructs (figs. S3A and S3B). This significant enhancement in the looping rate confirms that stable defects such as a single basepair mismatch or a nick can enhance global cyclizability of DNA, suggesting that similar but transient defects, if they are frequent enough, may explain the extreme bendability observed here. However, whether the high bendability of DNA at short length scales comes from transient kinks or bubbles or stems from anharmonic elasticity of DNA require improved computational methods and further studies.
Supplementary Material
One sentence summary.
DNA molecules below the persistence length exhibit significant intrinsic bendability.
Acknowledgments
We thank R. Phillips, H. Garcia, K. Rangunathan, R. Zhou, S. Doganay, A. Jain, K. Lee, G. Lee and S. Arslan for helpful discussions. We would like to dedicate this paper to the memory of late Jonathan Widom whose generous comments on an earlier version greatly inspired us; and whose untimely death is a tremendous loss for the field. This work was supported by US National Science Foundation grants 0646550 and 0822613 and US National Institutes of Health grant GM065367 to T.H. T.H. is an employee of the Howard Hughes Medical Institute.
References
- 1.Blackwood EM, Kadonaga JT. Science. 1998;281:60. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 2.Wong OK, Guthold M, Erie DA, Gelles J. PLoS Biol. 2008;6:e232. doi: 10.1371/journal.pbio.0060232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richmond TJ, Davey CA. Nature. 2003;423:145. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 4.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6185. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shore D, Langowski J, Baldwin RL. Proc. Natl. Acad. Sci. U.S.A. 1981;78:4833. doi: 10.1073/pnas.78.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters JP, Maher LJ. Q. Rev. Biophys. 2010;43:23. doi: 10.1017/S0033583510000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cloutier TE, Widom J. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3645. doi: 10.1073/pnas.0409059102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du Q, Smith C, Shiffeldrim N, Vologodskaia M, Vologodskii A. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5397. doi: 10.1073/pnas.0500983102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathew-Fenn RS, Das R, Harbury PAB. Science. 2008;322:446. doi: 10.1126/science.1158881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker NB, Everaers R. Science. 2009;325:538. doi: 10.1126/science.1168786. [DOI] [PubMed] [Google Scholar]
- 11.Mathew-Fenn RS, Das R, Fenn TD, Schneiders M, Harbury PAB. Science. 2009;325:538. [Google Scholar]
- 12.Mastroianni AJ, Sivak DA, Geissler PL, Alivisatos AP. Biophys. J. 2009;97:1408. doi: 10.1016/j.bpj.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiggins PA, et al. Nat. Nanotech. 2006;1:137. [Google Scholar]
- 14.Yuan C, Lou XW, Rhoades E, Chen H, Archer LA. Nucleic Acids Res. 2007;35:5294. doi: 10.1093/nar/gkm582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seol Y, Li J, Nelson P, Perkins T, Betterton MD. Biophys. J. 2007;93:4360. doi: 10.1529/biophysj.107.112995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy R, Hohng S, Ha T. Nat. Meth. 2008;5:507. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ha T, et al. Proc. Natl. Acad. Sci. U.S.A. 1996;93:6264. [Google Scholar]
- 18.Materials and methods and additional information are available as supporting material on Science Online.
- 19.Burkhoff AM, Tullius TD. Nature. 1988;331:455. doi: 10.1038/331455a0. [DOI] [PubMed] [Google Scholar]
- 20.Han L, et al. PLoS ONE. 2009;4:e5621. doi: 10.1371/journal.pone.0005621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker NA, Kahn JD, James Maher L., III J. Mol. Biol. 2005;349:716. doi: 10.1016/j.jmb.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 22.Lee DH, Schleif RF. Proc. Natl. Acad. Sci. U.S.A. 1989;86:476. doi: 10.1073/pnas.86.2.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cisse I, Okumus B, Joo C, Ha T. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12646. doi: 10.1073/pnas.0610673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagerman PJ. Annu. Rev. Biophys. Biophys. Chem. 1988;17:265. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- 25.Lowary PT, Widom J. Proc. Natl. Acad. Sci. U.S.A. 1997;94:1183. doi: 10.1073/pnas.94.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pörschke D, Eigen M. J. Mol. Biol. 1971;62:361. doi: 10.1016/0022-2836(71)90433-5. [DOI] [PubMed] [Google Scholar]
- 27.Shimada J, Yamakawa H. Macromolecules. 1984;17:689. [Google Scholar]
- 28.Towles KB, Beausang JF, Garcia HG, Phillips R, Nelson PC. Physical Biology. 2009;6:22. doi: 10.1088/1478-3975/6/2/025001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan J, Kawamura R, Marko JF. Physical Review E. 2005;71:061905. doi: 10.1103/PhysRevE.71.061905. [DOI] [PubMed] [Google Scholar]
- 30.Douarche N, Cocco S. Physical Review E. 2005;72:061902. doi: 10.1103/PhysRevE.72.061902. [DOI] [PubMed] [Google Scholar]
- 31.Schleif R. Annl. Rev. Biochem. 1992;61:199. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- 32.Yan J, Marko JF. Physical Review Letters. 2004;93:108108. doi: 10.1103/PhysRevLett.93.108108. [DOI] [PubMed] [Google Scholar]
- 33.Wiggins PA, Phillips R, Nelson PC. Physical Review E. 2005;71:021909. doi: 10.1103/PhysRevE.71.021909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lankas F, Lavery R, Maddocks JH. Structure. 2006;14:1527. doi: 10.1016/j.str.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 35.Curuksu J, Zacharias M, Lavery R, Zakrzewska K. Nucleic Acids Res. 2009;37:3766. doi: 10.1093/nar/gkp234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.