Abstract
Severely stressed, with their resources depleted, and their cellular machinery working beyond capacity, the host cells that are used for heterologous protein production have no option but to activate their stress response pathways in order to mitigate the accumulating effects of expressing a foreign, possibly toxic, protein at vast quantities. The result is lower protein yield and quality, with many products being misfolded or part of inclusion bodies that need further processing. Recently, new techniques aim to shift the control of protein production from humans to cells and empower the latter to regulate the production process, thus leading to increased protein quality. Herein we provide a perspective on how integrative synthetic biology can be applied to traditional biotechnological applications with potentially transformative results.
Keywords: synthetic biology, bioengineering, self-regulation, stress induced response, computational modeling, heterologous protein, recombinant protein production, adaptive control
From early on, the production of recombinant proteins in microbial cells has revolutionized our ability to produce industrial and medical products in vast quantities. Human insulin, human growth hormone (HGH), α/β/γ interferons are some examples of the mass-produced products in the multi-billion dollar recombinant protein production industry. Several organisms, such as bacteria, yeasts, plants, insect and mammalian cells, currently serve as production systems for heterologous proteins. Escherichia coli is arguably one of the most widely used hosts for recombinant protein production since it can achieve high yields, it is fast and inexpensive to grow and it can be easily modified. Remarkably, E. coli can accumulate up to 80% of its dry weight in recombinant proteins,1 and thus has the potential for very high protein yields that is desired in an industrial scale process.
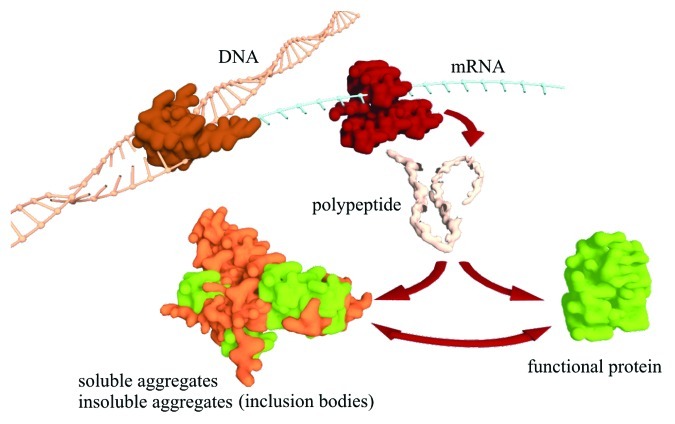
Unfortunately, recombinant protein expression can be detrimental for cells, as it constitutes a metabolic burden through the depletion of precursor metabolites.2 Additionally, high levels of expression lead to cellular stress that involves the induction of chaperones, foldases and proteases.3 With the cellular machinery over its capacity, recombinant proteins form inclusion bodies, which represent insoluble protein aggregates (Fig. 1) and necessitate downstream processing with expensive and time-consuming de/re-naturation methods.4 Finding the optimal balance between protein yield and quality is always a challenge, as these two variables tend to have opposing dynamics during protein production.
Figure 1. Overview of the possible outcomes during recombinant protein production. Figure credits: Athanasios Tsoukalas.
To this end, several of the available expression platforms depend on the constitutive expression or manual fine-tuning of target and auxiliary protein expression in order to achieve the desired balance. For example, since high protein expression leads to lower protein quality and to highly stressed cells, molecular chaperones and foldases are usually co-expressed from accessory plasmids with no further control.5 Too much or too little expression of these proteins results in drastically decreased recombinant protein quality and yield. This leads to time-consuming fine-tuning, that is not robust to process modifications (e.g., a change in temperature, medium, or target protein) that may shift the optimal operating point. In addition, control of the protein production systems can be achieved through inducible promoters that drive the recombinant protein expression, although this necessitates the use of expensive chemicals that can interfere with the process.6
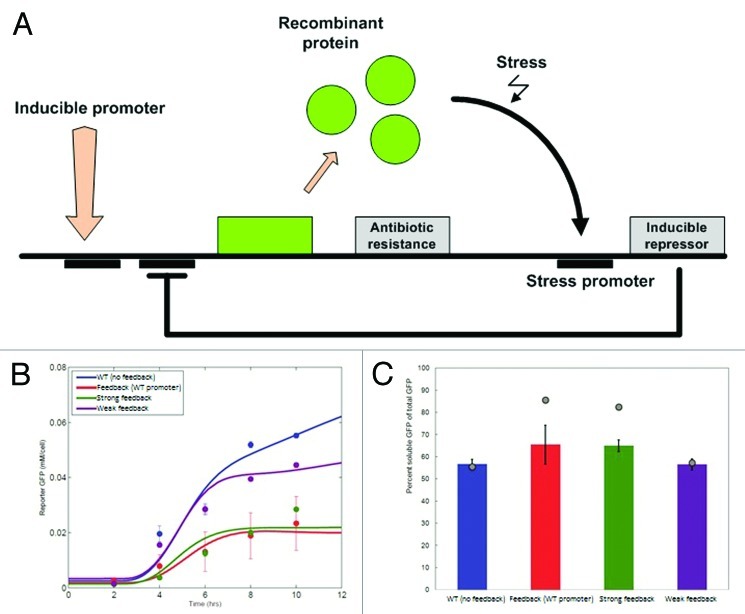
In our recent effort to empower the host with control of the protein production process, we introduced a synthetic feedback loop that decreases the production of the recombinant protein, once cellular stress is detected.7 As shown in Figure 2, the expression of the recombinant protein, which was GFP in our case, induced a stress response that activated expression of the downstream gene from a stress-inducible promoter. We selected the promoter of the IbpAB operon, as it encodes inclusion binding proteins that are upregulated during the expression of recombinant products. The downstream gene is a repressor (TetR here) that binds to the recombinant protein promoter and significantly decreases its expression.
Figure 2. (A) Overview of a self-regulatory system for recombinant protein production. An inducible promoter is used to turn-on recombinant protein production. When the cell is stressed, a strong repressor that is under the control of a stress reporter will be expressed, which will decrease or shut down the protein production. In our proof-of-principle experiments, we placed the recombinant protein (GFP) under a T7 promoter that would be repressed by LacI. The stress-induced promoter for heat shock proteins IbpA and IbpB was used to drive the TetR expression, which in turn adaptively regulated the production of GPF by binding to its promoter. (B) Simulation (solid lines) and experimental (dots) results for the non-feedback system (blue line) and three feedback variations based on different stress promoter variants (engineered through site-directed mutagenesis). Interestingly, the wild-type stress promoter exhibited large variation in its expression, while the engineered promoters are fairly invariant in their effect. (C) Soluble vs. insoluble fraction of the recombinant protein (GPF). Both experimental results (bars) and computational predictions (gray points) are shown for the three representative variations depicted in (B). The difference between the experimentally measured and computationally derived values likely stem from the fact that in the model we don’t account for the effect of TetR proteins in the depletion of the cellular resources and folding machinery.
To engineer a system with the characteristics mentioned above, we used a synthetic biology approach: First, we created a model of the proposed circuit topology to gain an insight on its dynamics and obtain the operation point for the different combinations of the key circuit components. Then we used the Registry of Standard Biological Parts to acquire the basic building blocks for each of the circuit elements and to build biobrick-compatible constructs. To fine-tune the circuit behavior we altered the ribosomal binding site affinity and stress promoter strength through random mutagenesis and subsequent screening. Cells were able to modulate their protein production and achieved much higher soluble protein fractions relative to those lacking the self-regulatory control (80% vs. 55% soluble fraction) and the system operated without modifications in different media and temperature ranges. In our proof-of-concept, “flight-to-quality,” system, this increase in the solubility was at the expense of protein yield (in some cases more than 50% reduction) and further experimentation is needed to bring the proposed method to a production scale. Despite this shortcoming, these promising results argue that adaptive and robust control of protein production processes at a cellular level is within reach. Our work also demonstrates the importance of integrating computational modeling and experimentation, as our models predicted the desired dynamic range of operation for the different components that we had to select, and our experimental results validated the fairly impressive predictive capacity of the computational methods used (Fig. 2B and C).
Self-regulatory and adaptive control does not have to be exhausted in the direct repression of the heterologous protein expression, as there are several other applications that have significant potential for process optimization. The currently available systems may have either low inducibility or leaky expression, which is non-optimal in the case that the recombinant product is toxic to the host cells. Toxicity of the inducer (e.g., IPTG) also limits these systems from being used for therapeutic products,8 while the need for a constant induction at high cell densities considerably adds to costs in large scale production processes. For these reasons a pH inducible system9 and more recently a temperature-inducible system have been proposed,10 but both these systems rely on variable environmental parameters and as such are limited to a narrow range of cultivation conditions. One solution would be to use a pulse inducible promoter system: the promoter that drives the T7 polymerase gene (and is critical as the recombinant protein is under a T7 promoter) can be engineered to include a T7 binding site next to the inducible binding site and thus introduce positive feedback. In this system, after the initial T7 induction from an inducer pulse, the circuit would lock in a T7-on state and would not need any further exogenous control. Another interesting avenue is to exploit the bacterial quorum sensing system so that recombinant protein expression starts only when the cell density reaches a threshold, and there has been some work in that direction.11 Control of the accessory genes (foldases, chaperones, etc.) from auxiliary constructs so that they are produced only under stressful conditions will also increase protein yield, as this will lower the metabolic burden of their otherwise constant expression.
Finally, integration of all these concepts in a single protein production system is a particularly challenging problem as the system complexity increases exponentially with each additional component that we introduce. Here is where the structured synthetic biology approach of model abstractness, standardization and automation is of paramount importance. Well-characterized and standardized parts will allow us to create computational models of dynamic circuit behavior with high predictive capacity, although efficiently capturing the dynamics in the complex bioreactor environment will always be a challenge. Despite the recent advances in computational tools for computer-aided biological circuit design and optimization, the capacity of these tools to come up with optimal designs for rational engineering of biological systems in real conditions is still very limited. If synthetic biology and rational computer-aided circuit construction aspires to advance to synthetic systems that go beyond a handful of interacting parts that can be adopted in industrial applications, a methodological approach to create the necessary experimental and computational infrastructure is needed. This includes the creation of quantitatively characterized libraries for various inducible promoter systems in widely used host organisms, the development of standardized, modular vectors, and the development of open-source, freely-available software tools for the automated design and optimization of synthetic circuits that adhere to user-defined constraints. This is a very exciting time for synthetic biologists, computational scientists and biotechnology professionals since synergistic interactions among researchers in these disciplines are likely to result in transformative advances in the field.
Footnotes
Previously published online: www.landesbioscience.com/journals/bioe/article/21935
References
- 1.Demain AL, Vaishnav P. Production of recombinant proteins by microbes and higher organisms. Biotechnol Adv. 2009;27:297–306. doi: 10.1016/j.biotechadv.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Gill RT, Valdes JJ, Bentley WE. A comparative study of global stress gene regulation in response to overexpression of recombinant proteins in Escherichia coli. Metab Eng. 2000;2:178–89. doi: 10.1006/mben.2000.0148. [DOI] [PubMed] [Google Scholar]
- 3.Allen SP, Polazzi JO, Gierse JK, Easton AM. Two novel heat shock genes encoding proteins produced in response to heterologous protein expression in Escherichia coli. J Bacteriol. 1992;174:6938–47. doi: 10.1128/jb.174.21.6938-6947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrer-Miralles N, Domingo-Espín J, Corchero JL, Vázquez E, Villaverde A. Microbial factories for recombinant pharmaceuticals. Microb Cell Fact. 2009;8:17. doi: 10.1186/1475-2859-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gasser B, Saloheimo M, Rinas U, Dragosits M, Rodríguez-Carmona E, Baumann K, et al. Protein folding and conformational stress in microbial cells producing recombinant proteins: a host comparative overview. Microb Cell Fact. 2008;7:11. doi: 10.1186/1475-2859-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jana S, Deb JK. Strategies for efficient production of heterologous proteins in Escherichia coli. Appl Microbiol Biotechnol. 2005;67:289–98. doi: 10.1007/s00253-004-1814-0. [DOI] [PubMed] [Google Scholar]
- 7.Dragosits M, Nicklas D, Tagkopoulos I. A synthetic biology approach to self-regulatory recombinant protein production in Escherichia coli. J Biol Eng. 2012;6:2. doi: 10.1186/1754-1611-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesley SA, Graziano J, Cho CY, Knuth MW, Klock HE. Gene expression response to misfolded protein as a screen for soluble recombinant protein. Protein Eng. 2002;15:153–60. doi: 10.1093/protein/15.2.153. [DOI] [PubMed] [Google Scholar]
- 9.Chou CH, Aristidou AA, Meng SY, Bennett GN, San KY. Characterization of a pH-inducible promoter system for high-level expression of recombinant proteins in Escherichia coli. Biotechnol Bioeng. 1995;47:186–92. doi: 10.1002/bit.260470210. [DOI] [PubMed] [Google Scholar]
- 10.Valdez-Cruz NA, Caspeta L, Pérez NO, Ramírez OT, Trujillo-Roldán MA. Production of recombinant proteins in E. coli by the heat inducible expression system based on the phage lambda pL and/or pR promoters. Microb Cell Fact. 2010;9:18. doi: 10.1186/1475-2859-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsao CY, Hooshangi S, Wu HC, Valdes JJ, Bentley WE. Autonomous induction of recombinant proteins by minimally rewiring native quorum sensing regulon of E. coli. Metab Eng. 2010;12:291–7. doi: 10.1016/j.ymben.2010.01.002. [DOI] [PubMed] [Google Scholar]