Abstract
Detection of the phytopathogen Pseudomonas cannabina pv alisalensis, the causal agent of bacterial blight of crucifers is essential for managing this disease. A phage-based diagnostic assay was developed that detects and identifies P. cannabina pv alisalensis from cultures and diseased plant specimens. A recombinant “light-tagged” reporter phage was generated by integrating the luxAB genes into the P. cannabina pv alisalensis phage PBSPCA1 genome. PBSPCA1::luxAB is viable, stable and detects P. cannabina pv alisalensis within minutes and with high sensitivity by conferring a bioluminescent signal. Detection is dependent on cell viability since cells treated with a bactericidal disinfectant are unable to elicit a signal. Importantly, the reporter phage detects P. cannabina pv alisalensis from diseased plant specimens indicating the potential of the diagnostic for disease identification. The reporter phage displays promise for the rapid and specific diagnostic detection of cultivated isolates, and infected plant specimens.
Keywords: disease, phytopathogen, crucifers, reporter phage, diagnostic, bioluminescence
Bacteriophages (phages) specifically infect and lyse their bacterial host and cell lysis has been used for many years as a method for specifically identifying target bacteria.1-3 Phage typing schemes exist for the majority of bacterial species and are sometimes used to identify specific strains within a species.4,5 The phage typing process is relatively simple; phage dilutions are spotted onto a bacterial lawn, and if the bacteria are sensitive to the phage, lysis results in an area of clearing. Although robust, the methodology requires the maintenance of a large number of phage stocks and bacterial propagating strains. Because pure bacterial cultures must be employed with phage typing, this methodology is not amenable to complex clinical or environmental samples. To address this limitation, phages are being exploited in a variety of technologies including phage-labeling, phage amplification, and reporter phage.6,7 The latter approach relies on integrating reporter genes into the phage genome to create a genetically engineered phage capable of emitting a detectable signal (usually colorimetric, fluorescent or bioluminescent). In the absence of a target cell, the reporter phage cannot produce a signal. If the target cell is present, the reporter phage binds to specific receptors on the bacterial cell wall, and infects the cell. Phage and reporter gene expression ensues and the subsequent signal provides a positive identification of the bacterium. The genes encoding the bacterial luciferase (luxAB) are commonly used as the reporter of choice as following the addition of an aldehyde substrate, luciferase catalyzes a reaction of which one of the products is “light” (Fig. 1A and B). The reporter phage system requires minimal processing of samples and although naturally occurring autobioluminescent environmental or clinical samples can occur, they are extremely rare. Consequently, “light-tagged phage” have been shown useful for detection of a variety of human pathogenic bacteria in vitro, and from complex food and clinical specimens.8-13 We describe the development of a “luxAB-tagged” reporter phage for the detection of an important phytopathogen, and demonstrate that the reporter phage holds promise as a disease diagnostic for bacterial blight.14
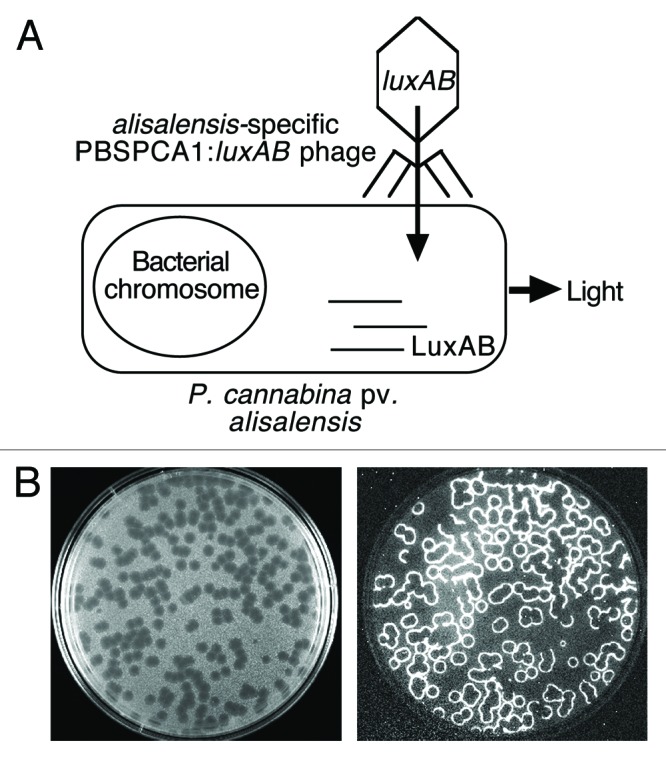
Figure 1. (A) Detection overview. The bacterial luxAB reporter genes (encoding the luciferase protein) were integrated into the genome of P. cannabina pv alisalensis phage PBSPCA1 to create a recombinant “luxAB-tagged” reporter phage. In the presence of the target cell, the reporter phage binds to a specific receptor and infects the cell. The luxAB genes, which are controlled by phage promoters, are expressed using the host’s transcriptional and translational machinery to produce the luciferase enzyme; following the addition of the substrate n-decanal and in the presence of oxygen and a flavin mononucleotide, the luciferase catalyzes a complex reaction resulting in light emission. (B) “Bioluminescent plaques.” A “luxAB-tagged” reporter phage was titered on its host strain using soft agar overlays and the plaques were examined under light (left panel) and dark field (right panel) illumination. Note the bioluminescence at the plaque periphery (phage/cell interface) which is indicative of phage-infected cells.
The phytopathogen Pseudomonas cannabina pv alisalensis (formerly known as Pseudomonas syringae pv alisalensis15) is the causal agent of bacterial blight, which is a disease afflicting cruciferous vegetables. Cruciferous vegetables are a dominant food crop worldwide and thus a valuable commodity. The disease, which can render the crop unmarketable, is transmissible and there is anecdotal evidence that P. cannabina pv alisalensis can be a seed contaminant. Phage lysis assays using phage PBSPCA1 (formerly known as PBS1) is used as a standard for the identification of P. cannabina pv alisalensis.16-20 PBSPCA1 genome was partially sequenced in order to facilitate molecular engineering of luxAB integration. The phage genome is predicted to encode for ~60 proteins, one of which was determined to be a non-essential putative phoH-like (phosphate starvation-inducible) protein. This gene was therefore replaced with a luxAB gene cassette using homologous recombination. Although most phage can tolerate small size increases in their genome, a replacement strategy was chosen because the resulting recombinant genome would only be ~1.3 kb larger than its parent, and was deemed unlikely to result in the generation of defective reporter phage.
PBSPCA1::luxAB phage were isolated following a PCR screening process using successively higher dilutions of phage until single plaques could be picked and propagated. The recombinant genome was confirmed using PCR, and the ability of DNase-1-treated lysates to transduce a bioluminescent signal response to target cells (DNase-1 cannot degrade phage DNA protected by the capsid). The ‘fitness’ of the reporter phage is comparable to the parental wild-type phage as both exhibited similar lysis curves and lysate titers. PBSPCA1::luxAB detects cultured P. cannabina pv alisalensis within 20 min upon reporter phage addition (Fig. 2), indicating that it infects, and produces luciferase rapidly. Incubations for 60 min results in a 103-fold increase in signal, but more extended incubations (> 120 min) lead to a gradual signal decline due to cell-mediated lysis. The sensitivity limits of detection are approximately 103 CFU/mL, sufficient to detect P. cannabina pv alisalensis in diseased plant specimens (see Fig. 4), but possibly inadequate to reliably detect P. cannabina pv alisalensis from contaminated seeds. Improvements in sensitivity may be achieved by engineering 2nd generation reporters that maximize the signal. This can be achieved in several ways, including codon optimizing of luxAB reporter genes for expression in Pseudomonas and incorporating Pseudomonas transcriptional and translational signals for luxAB expression. For example, in a 2nd generation Yersinia pestis reporter phage, we integrated luxAB downstream of the major capsid gene, resulting in a 10-fold increase in assay sensitivity.21 Assay sensitivity may also be improved by utilizing phage that are defective in host cell lysis (resulting in greater signal accumulation). Using these or comparable strategies, a detection sensitivity of ~102 CFU/mL may be achieved. At very low concentrations (i.e., < 102 CFU/mL), the limiting step in detection is unlikely to be the signal strength, but instead the reduced likelihood of phage infection. Low level detection strategies may include collection and concentration of cells prior to phage infection.
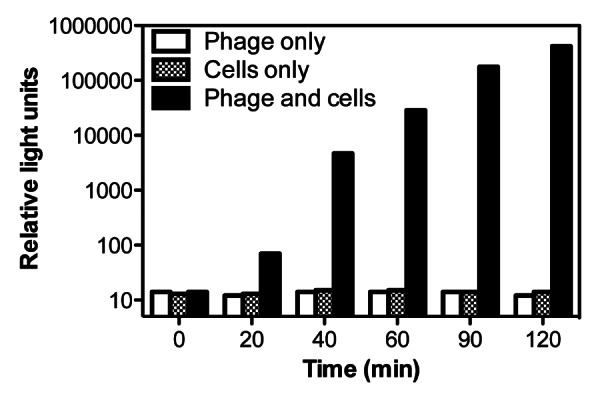
Figure 2. Phage-mediated transduction of a bioluminescent response to P. cannabina pv alisalensis. Cultures were grown at 28°C in NBY broth until an A600 of 0.15–2.0, mixed with the PBSPCA1::luxAB reporter phage, and bioluminescence (RLU) was measured over time. Numbers are the mean (n = 3) ± SD.
In contrast to other detection methodologies, e.g., PCR, hybridization, and immunoanalysis, reporter phage technology will only detect viable cells, while the others will detect live and dead cells. This ability to specifically detect only viable cells is critical if being used in a seed-based detection assay. For example, if a seed lot was suspected of bacterial infection, then treated by a prescribed seed treatment, the ability to verify if the treatment was effective (i.e., no viable cells remaining) is of considerable importance to the seed company. Reporter phages require the host’s transcriptional and translational machinery to express luciferase, and elicit bioluminescence; thus, signal generation is strictly dependent on host metabolic activity (Fig. 3A). To demonstrate this attribute, the reporter phage-mediated signal response of metabolically active cells was compared with the response elicited by compromised cells. A P. cannabina pv alisalensis culture was either left untreated (control) or treated with 70% ethanol for 30 min which resulted in a 105-106 reduction in viable cell counts (to < 102 CFU/mL). As expected, ethanol-treated cells were unable to elicit a bioluminescent signal upon incubation with the reporter phage (Fig. 3B). In contrast, for example, PCR detects DNA released from cells into the environment or inside dead cells, potentially resulting in false-positives that lead to unnecessary treatments or loss of seed lots.
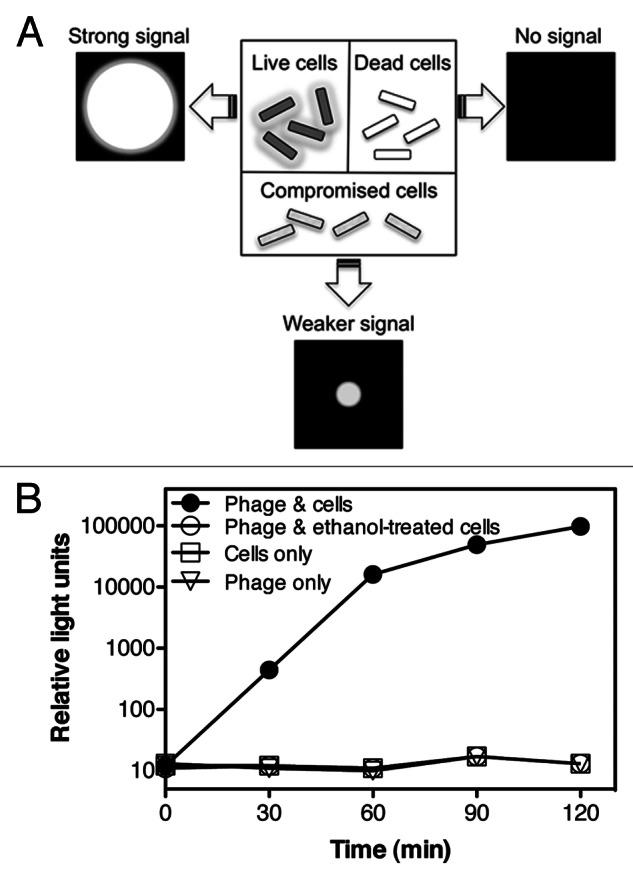
Figure 3. Viable cells are required for a bioluminescent signal response. (A) Viable, metabolically active cells are required for phage infection and reporter gene expression. Thus, metabolically active cells should elicit strong signal responses while dead cells will not elicit a signal response. (B) A P. cannabina pv alisalensis culture was divided equally and either left untreated, or incubated with 70% ethanol for 30 min. Following removal of the ethanol, control and treated cells were incubated with the reporter phage and bioluminescence (RLU) was measured over time. Numbers are the mean (n = 3) ± SD.
We reported previously that the reporter phage detects P. cannabina pv alisalensis in plants,14 indicating the ability of this system to be used in a phytopathogen diagnostic setting without the need for extensive and tedious pre-isolation of the bacterium. Brassica rapa (turnip greens), when inoculated and grown in a controlled greenhouse environment, developed bacterial blight as indicated by large chlorotic and necrotic areas on leaves. Diseased tissue, when incubated with the reporter phage, elicited a strong bioluminescent signal within 4 h of harvesting. Bacterial blight afflicts many different species of brassicas. B. rapa, along with Brassica juncea (mustard greens) and Brassica oleracea (collards) account for the majority of vegetable brassicas produced in the US. Each of these species is unique and may pose individual challenges in reporter phage diagnostics. For example, B. juncea and B. rapa have similar leaves, yet B. juncea produces large amounts of the thioethers glucosinolate and isothiocyanate. Although small amounts of these compounds can be found in most leafy greens, high amounts may inhibit phage gene expression. Further, the leaf of B. oleracea is thick and quite fibrous, features that may interfere with phage infection.
We tested whether PBSPCA1::luxAB can detect P. cannabina pv alisalensis in infected B. juncea and B. oleracea. P. cannabina pv alisalensis-infected B. juncea samples were obtained through experimental inoculation, while B. oleracea samples exhibiting characteristics of bacterial blight were provided by a South Carolina commercial grower.
B. juncea was grown in the field at the US Vegetable Laboratory in Charleston, South Carolina and then inoculated with P. cannabina pv alisalensis. Plants were then maintained in the field under normal weather conditions for 2 weeks before leaves, which exhibited characteristic signs of infection, were harvested and then analyzed with the reporter phage. As a control, a B. juncea breeding line, which exhibits resistance to P. cannabina pv alisalensis, was grown and inoculated under analogous conditions. These plants did not display signs of infection, and as expected, these control tissues did not elicit a bioluminescent signal response with the reporter phage (Fig. 4). In contrast, a bioluminescent signal response was evident from the infected leaves following a 2 or 4 h elution and outgrowth period.
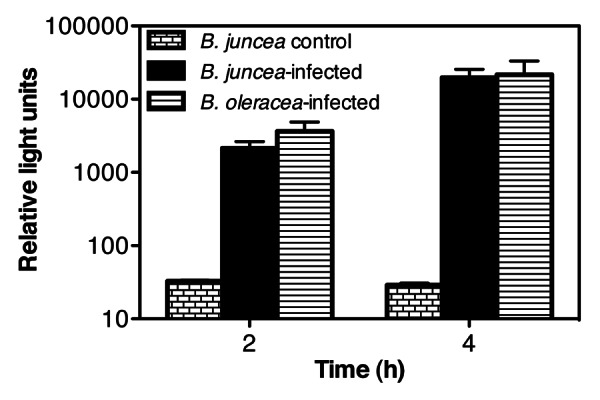
Figure 4. Reporter phage detection of P. cannabina pv alisalensis from infected Brassica. B. juncea were grown in the field at the US Vegetable laboratory (Charleston, SC), inoculated with P. cannabina pv alisalensis, and incubated for 14 d before the infected leaves were harvested. Commercially grown B. oleracea, showing symptoms of blight, were sent from a South Carolina grower to the US vegetable laboratory in Charleston for disease identification. An infection resistant cell line of B. juncea, grown under the same conditions, was used as a control. One cm discs of control and infected leaf parts were mixed with media, and incubated at 28°C to allow for bacterial elution and outgrowth. At various time points after outgrowth (2 or 4 h), 900 µL of the leaf eluate was placed in a fresh tube, and the reporter phage was added. Bioluminescence was measured 120 min after the addition of the reporter phage. Numbers represent the mean ± SE of 3 independent leaf samples.
B. oleracea leaves showing symptoms of bacterial blight, were sent from the commercial grower to the US Vegetable Laboratory in Charleston, SC for disease identification. In this particular case, over 60% of the grower’s 200 acre crop exhibited disease symptoms. Bacteria were isolated from the infected leaf samples using standard culturing methods. The bacteria displayed a blue fluorescent phenotype under UV exposure when grown in King’s B medium, a characteristic of P. cannabina. Analysis of DNA fragment banding patterns using published BOX-PCR protocols identified the isolate as P. cannabina pv alisalensis.22 These same infected leaves were then tested directly with the reporter phage for a bioluminescent signal response. These samples elicited a bioluminescent response as expected (Fig. 4). Collectively, this data indicates the ability of the reporter phage to detect P. cannabina pv alisalensis from different brassicas whether they were artificially infected in the greenhouse, or under field conditions. In addition, the reporter phage was able to detect P. cannabina pv alisalensis from “authentic” diseased specimens provided by a commercial grower.
Our development of PBSPCA1::luxAB represents the first bioluminescent reporter phage assay for an important plant pathogen. The assay functions well with both cultured isolates and naturally diseased specimens. Although there are a number of phage-based diagnostic assays that have been developed and marketed for the detection of human pathogens, it is surprising that phage-based diagnostic assays have not yet been developed within the agricultural industry. In contrast, there are a number of phage diagnostic assays that have been developed and marketed for the detection of human pathogens. For example, phage ϕA1122 is used for confirming identification of Y. pestis,23,24 phage γ is FDA-approved as a standard for the identification of Bacillus anthracis,25 FASTPlaqueTBTM is used for the detection of Mycobacterium tuberculosis, and a cocktail of phages (KeyPathTM) are FDA-cleared for the detection of methicillin-sensitive, or -resistant Staphylococcus aureus. Although none of these assays utilize genetically engineered reporter phage, the use and implementation of phage diagnostic assays are more developed for the clinical setting. A limiting factor in developing a diagnostic test is the time required for the isolation and selection of a phage that displays the desired host-range, i.e., a phage that displays species specificity and broad-strain infectivity. However, phages are the most abundant entities on earth with estimates ranging from 1030 to 1032 (ref. 26). Although it is likely that species-specific phages exist that may be used to target any bacterial pathogen of choice, identifying a candidate diagnostic phage can be a time consuming, laborious, and sometimes futile endeavor. Nevertheless, the potential of using phages as diagnostic tools, as in vitro biocontrol agents, or as therapeutic agents in vivo, will undoubtedly result in the development of more and more applications that utilize phage.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This research was supported by the National Science Foundation Small Business Innovative Research grant 1012059 and in part by the US Department of Education and the US Department of Agriculture. Any opinions, findings, conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the funding agencies.
Footnotes
Previously published online: www.landesbioscience.com/journals/bioe/article/22159
References
- 1.Frost JA, Kramer JM, Gillanders SA. Phage typing of Campylobacter jejuni and Campylobacter coli and its use as an adjunct to serotyping. Epidemiol Infect. 1999;123:47–55. doi: 10.1017/S095026889900254X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hickman-Brenner FW, Stubbs AD, Farmer JJ., 3rd Phage typing of Salmonella enteritidis in the United States. J Clin Microbiol. 1991;29:2817–23. doi: 10.1128/jcm.29.12.2817-2823.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loessner MJ, Busse M. Bacteriophage typing of Listeria species. Appl Environ Microbiol. 1990;56:1912–8. doi: 10.1128/aem.56.6.1912-1918.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castro D, Morińigo MA, Martinez-Manzanares E, Cornax R, Balebona MC, Luque A, et al. Development and application of a new scheme of phages for typing and differentiating Salmonella strains from different sources. J Clin Microbiol. 1992;30:1418–23. doi: 10.1128/jcm.30.6.1418-1423.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demczuk W, Soule G, Clark C, Ackermann HW, Easy R, Khakhria R, et al. Phage-based typing scheme for Salmonella enterica serovar Heidelberg, a causative agent of food poisonings in Canada. J Clin Microbiol. 2003;41:4279–84. doi: 10.1128/JCM.41.9.4279-4284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smartt AE, Ripp S. Bacteriophage reporter technology for sensing and detecting microbial targets. Anal Bioanal Chem. 2011;400:991–1007. doi: 10.1007/s00216-010-4561-3. [DOI] [PubMed] [Google Scholar]
- 7.Smartt AE, Xu T, Jegier P, Carswell JJ, Blount SA, Sayler GS, et al. Pathogen detection using engineered bacteriophages. Anal Bioanal Chem. 2011;12:732–52. doi: 10.1007/s00216-011-5555-5. [DOI] [PubMed] [Google Scholar]
- 8.Schofield DA, Westwater C. Phage-mediated bioluminescent detection of Bacillus anthracis. J Appl Microbiol. 2009;107:1468–78. doi: 10.1111/j.1365-2672.2009.04332.x. [DOI] [PubMed] [Google Scholar]
- 9.Ulitzur S, Kuhn J. Introduction of lux genes into bacteria, a new approach for the specific determination of bacteria and their antibiotic susceptibility. In: Schlomerich J, Andreesen R, Kapp A, Ernst M, Woods WG, eds. Bioluminescence and chemiluminescence new perspectives. New York: Wiley, 1987. [Google Scholar]
- 10.Schofield DA, Molineux IJ, Westwater C. Diagnostic bioluminescent phage for detection of Yersinia pestis. J Clin Microbiol. 2009;47:3887–94. doi: 10.1128/JCM.01533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loessner MJ, Rees CE, Stewart GS, Scherer S. Construction of luciferase reporter bacteriophage A511:luxAB for rapid and sensitive detection of viable Listeria cells. Appl Environ Microbiol. 1996;62:1133–40. doi: 10.1128/aem.62.4.1133-1140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loessner MJ, Rudolf M, Scherer S. Evaluation of luciferase reporter bacteriophage A511:luxAB for detection of Listeria monocytogenes in contaminated foods. Appl Environ Microbiol. 1997;63:2961–5. doi: 10.1128/aem.63.8.2961-2965.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ripp S, Jegier P, Johnson CM, Brigati JR, Sayler GS. Bacteriophage-amplified bioluminescent sensing of Escherichia coli O157:H7. Anal Bioanal Chem. 2008;391:507–14. doi: 10.1007/s00216-007-1812-z. [DOI] [PubMed] [Google Scholar]
- 14.Schofield DA, Bull CT, Rubio I, Wechter WP, Westwater C, Molineux IJ. Development of an engineered bioluminescent reporter phage for detection of bacterial blight of crucifers. Appl Environ Microbiol. 2012;78:3592–8. doi: 10.1128/AEM.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bull CT, Manceau C, Lydon J, Kong H, Vinatzer BA, Fischer-Le Saux M. Pseudomonas cannabina pv. cannabina pv. nov., and Pseudomonas cannabina pv. alisalensis (Cintas Koike and Bull, 2000) comb. nov., are members of the emended species Pseudomonas cannabina (ex Sutic & Dowson 1959) Gardan, Shafik, Belouin, Brosch, Grimont & Grimont 1999. Syst Appl Microbiol. 2010;33:105–15. doi: 10.1016/j.syapm.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Koike ST, Kammeijer K, Bull CT, O'Brien D. First report of bacterial blight of romanesco cauliflower (Brassica oleracea var. botrytis) caused by Pseudomonas syringae pv. alisalensis in California. Plant Dis. 2006;90:1551. doi: 10.1094/PD-90-1551B. [DOI] [PubMed] [Google Scholar]
- 17.Koike ST, Kammeijer K, Bull CT, O'Brien D. First report of bacterial blight of Rutabaga (Brassica napus var. napobrassica) caused by Pseudomonas syringae pv. alisalensis in California. Plant Dis. 2007;91:112. doi: 10.1094/PD-91-0112C. [DOI] [PubMed] [Google Scholar]
- 18.Bull CT, Goldman PH, Koike ST. Bacterial blight on arugula, a new disease caused by Pseudomonas syringae pv. alisalensis in California. Plant Dis. 2004;88:1384. doi: 10.1094/PDIS.2004.88.12.1384A. [DOI] [PubMed] [Google Scholar]
- 19.Bull CT, Goldman PH, Smith RF, Koike ST. Pseudomonas syringae pv. alisalensis and Pseudomonas syringae pv. maculicola cause disease on crucifers used in cover crop mixtures. Phytopathology. 2004;94:S150. [Google Scholar]
- 20.Cintas NA, Koike ST, Bull CT. A new pathovar, Pseudomonas syringae pv. alisalensis pv. nov., proposed for the casual agent of bacterial blight of broccoli and broccoli raab. Plant Dis. 2002;86:992–8. doi: 10.1094/PDIS.2002.86.9.992. [DOI] [PubMed] [Google Scholar]
- 21.Schofield DA, Molineux IJ, Westwater C. Rapid identification and antibiotic susceptibility testing of Yersinia pestis using bioluminescent reporter phage. J Microbiol Methods. 2012;90:80–2. doi: 10.1016/j.mimet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wechter WP, Keinath AP, Farnham MW, Smith JP. First report of bacterial leaf blight on broccoli and cabbage caused by Pseudomonas syringae pv. alisalensis in South Carolina. Plant Dis. 2010;94:132. doi: 10.1094/PDIS-94-1-0132C. [DOI] [PubMed] [Google Scholar]
- 23.Dennis DT, Gage KL, Gratz N, Poland JD, Tikhomirov E. Plague Manual: Epidemiology, Distribution, Surveillance, and Control. World Health Organization, Geneva., 1999. [Google Scholar]
- 24.Chu MC. Laboratory manual of plague diagnostic tests. CDC, Atlanta: Centers for Disease Control and Prevention., 2000. [Google Scholar]
- 25.Abshire TG, Brown JE, Ezzell JW. Production and validation of the use of gamma phage for identification of Bacillus anthracis. J Clin Microbiol. 2005;43:4780–8. doi: 10.1128/JCM.43.9.4780-4788.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kutter E, Sulakvelidze A. Introduction. In: Kutter E, Sulakvelidze A, eds. Bacteriophages: Biology and Applications. Boca Raton: CRC Press, 2005:1-4. [Google Scholar]


