Abstract
Mitochondrial functioning and morphology are known to be connected through cycles of organelle fusion and fission that depend upon mitochondrial membrane potential (ΔΨM); however, we lack an understanding of the features and dynamics of natural mitochondrial populations. Using data from our recent study of univariate mitochondrial phenotypic variation in Caenorhabditis briggsae nematodes, we analyzed patterns of phenotypic correlation for 24 mitochondrial traits. Our findings support a role for ΔΨM in shaping mitochondrial dynamics, but no role for mitochondrial ROS. Further, our study suggests a novel model of mitochondrial population dynamics dependent upon cellular environmental context and with implications for mitochondrial genome integrity.
Keywords: complex I, electron transport chain, membrane potential, mitochondrial dynamics, nad5Δ, reactive oxygen species
1. Introduction
Mitochondria are dynamic organelles that participate in continuous cycles of fusion, fission and autophagy within the cells of nearly all eukaryotic organisms. These cycles serve to link mitochondrial shape to organelle function (Chen et al., 2005; Duvezin-Caubet et al., 2006) as well as each mitochondrion to the larger mitochondrial population (Hyde et al., 2010). Mitochondria perform several functions vital to eukaryotic life, including bioenergy (ATP) production and regulation of calcium homeostasis and apoptosis, nearly all of which depend upon the process of oxidative phosphorylation at the mitochondrial electron transport chain (ETC). Electron transfer through functional protein complexes of the ETC is coupled to the pumping of protons across the mitochondrial inner membrane, which establishes a mitochondrial membrane potential (ΔΨM). This ΔΨM provides the potential energy to generate ATP and serves to control fusion-fission cycles (Twig, Elorza, et al., 2008), both of which are necessary for mitosis, fuel sensing, autophagy and other processes (Graef & Nunnari, 2011; Mitra et al., 2009; Molina et al., 2009). A natural consequence of ETC function is the occasional leakage of electrons onto molecular oxygen, which can generate reactive oxygen species (ROS) (Raha & Robinson, 2000). Under normal circumstances, excess ROS are scavenged by various antioxidants before they can damage important macromolecules (Imlay, 2008; Sedensky & Morgan, 2006); however, impairment of the ETC often results in elevated ROS production (Dingley et al., 2009; Grad & Lemire, 2004; Verkaart et al., 2007) and oxidative damage of proteins and nucleic acids (Wanagat et al., 2001; Yang et al., 2007), along with depressed ΔΨM (Gaskova et al., 2007; Lemire et al., 2009; Ventura et al., 2006) and altered mitochondrial dynamics (Ichishita et al., 2008).
Findings like those above highlight the integration between mitochondrial function, morphology and the fusion-fission cycle, and many recent studies have aimed to reveal the mechanistic bases of these relationships (Chen et al., 2005; Chen & Chan, 2005; Palermo et al., 2007; Pham et al., 2004; Westermann, 2012; Wikstrom et al., 2009; Yasuda et al., 2011). Mitochondrial ROS level does not appear to be related consistently to mitochondrial morphology or dynamics. For example, elevated mitochondrial ROS levels have been associated with both increased (Koopman et al., 2005) and decreased mitochondrial branching (Grünewald et al., 2010; Pletjushkina et al., 2006). Conversely, studies examining mitochondrial form and function in isolated cells and/or mutant organisms reveal a direct link between ΔΨM and mitochondrial morphology, such that higher ΔΨM induces organellar elongation (Ishihara et al., 2003; Legros et al., 2002) and loss of ΔΨM causes severe fragmentation of the mitochondrial network (Duvezin-Caubet et al., 2006; Song et al., 2007). Many of these mitochondrial shape changes are mediated by an altered balance between mitochondrial fusion and fission (Chen et al., 2005; Okamoto & Shaw, 2005), which is increasingly appreciated to have a role in human disease. For instance, abnormal fusion-fission cycles are characteristic of neurodegenerative disorders including Parkinson’s and Alzheimer’s disease (Irrcher et al., 2010; Su et al., 2010; Trimmer et al., 2000; Wang et al., 2008; Winklhofer & Haass, 2010). Many of the genes and cellular intermediates involved in mitochondrial dynamics (and its imbalance) have now been identified and characterized (Dimmer et al., 2002; Griffin et al., 2006; Ishihara et al., 2003; Lee et al., 2004; Meeusen et al., 2004; Scorrano, 2005; Yaffe, 1999). Based on such work, Twig and colleagues proposed that the fusion-fission-apoptosis cycle creates a “quality control axis” that acts to maintain mitochondrial integrity (Twig, Hyde, et al., 2008). In their model, persistently depolarized mitochondria (those with low ΔΨM) are segregated from the functional group by their inability to fuse. In this way low-functioning mitochondria – and perhaps, damaged mitochondrial genomes – are weeded out and an overall healthier organelle population is thus maintained (Bess, et al., 2012; Hyde et al., 2010; Kowald & Kirkwood, 2011; Meyer & Bess, 2012; Twig, Hyde, et al., 2008).
Despite the abundance of research focused on the dynamics of individual mitochondria (i.e., fusion and fission cycles), less attention has been devoted to the population-level behaviors of these organelles. A recent review highlights various “global” (cellular) and “local” (individual mitochondrion) controls thought to influence mitochondrial fusion and fission, suggesting that the collective mitochondrial population can indeed respond to cellular cues (Hyde et al., 2010). For example, mitochondria have been observed to undergo concerted hyper-fusion during G1-S phase of the cell cycle, and subsequent hyper-fragmentation as the cell progresses into S phase (Hyde et al., 2010). Still, we have a limited understanding of the biological roles of mitochondrial fission-fusion cycling and its organism-level consequences, and little information regarding the features and dynamics of mitochondrial populations and how these might influence individual mitochondrial form and function. Further, although we have some information about the patterns of relationship between certain mitochondrial phenotypes (e.g., ΔΨM and organelle elongation), no comprehensive survey of such phenotypes has been conducted within live organisms. Finally, the extent to which research on cell lines and genetic fusion-fission mutants will apply to natural populations of organisms remains unknown.
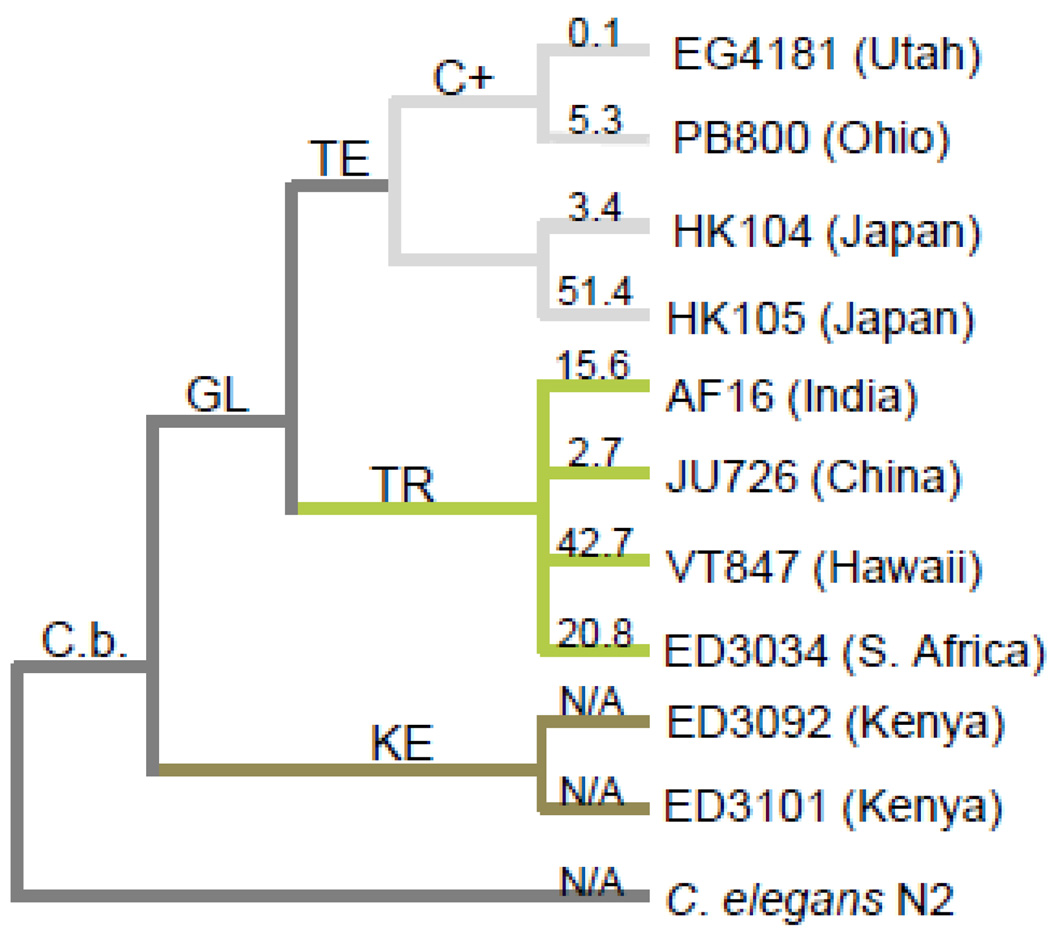
Caenorhabditis nematodes have emerged as important models for studying the underlying causes of mitochondrial ETC dysfunction and its associated biological consequences. Mitochondrial metabolism and ETC function are known to be extremely similar in worms and mammals (reviewed in Dimmer et al., 2002; Westermann, 2010). Also, nematodes have highly differentiated tissues and a transparent cuticle that make them amenable to live imaging studies. Caenorhabditis briggsae in particular offers many advantages for mitochondrial biology research including its substantial mitochondrial genetic (Howe & Denver, 2008) and phenotypic (Clark et al., 2012; Cutter et al., 2010; Estes et al., 2011; Hicks et al., 2012; Raboin et al., 2010; Ross et al., 2011) diversity. C. briggsae exhibit a cosmopolitan distribution and mitochondrial genetic analyses group its known natural isolates into three major phylogeographic clades corresponding to latitude of origin (Figure 1 in Howe & Denver, 2008) (Fig. 1). Recent work indicates that isolates within these clades are likely adapted to local thermal regimes (Jovelin & Cutter, 2011; Prasad et al., 2011). We found that phylogenetic membership also accounts for among-isolate variation in several mitochondrial form and function traits; this was particularly true for ΔΨM, which was an extremely reliable predictor of clade membership (Hicks et al., 2012). Further, C. briggsae appear especially prone to acquiring mitochondrial deletion mutations (Howe et al., 2010), a process that has likely contributed to its high levels of standing mitochondrial genetic diversity. Indeed, many natural populations of C. briggsae harbor a large deletion (nad5Δ) within their mitochondrial genomes that removes half of the NADH-dehydrogenase 5 (nad5) gene (Figure 1 in Howe & Denver, 2008), which encodes an integral subunit of ETC complex I. nad5Δ-bearing genomes were recently shown to behave as selfish genetic elements (Clark et al., 2012) and levels of nad5Δ heteroplasmy (the average number of deletion-bearing genomes per individual) are known to vary from zero to over 50% among geographically-segregated isolates of C. briggsae (Estes et al., 2011; Howe & Denver, 2008). Recent work showed that nad5Δ level was unrelated to isolate-specific variation in ΔΨM, ROS, and aspects of mitochondrial morphology (Hicks et al., 2012), but that it is likely to be detrimental to nematode health and fitness at high (< ~40%) heteroplasmy levels (Estes et al., 2011; Howe & Denver, 2008). In summary, its extensive genetic and subcellular phenotypic variation makes C. briggsae a promising natural system in which to investigate individual- and population-level mitochondrial dynamics.
Figure 1. C. briggsae natural isolates.
Phylogenetic relationship, geographic origin and average nad5Δ heteroplasmy level (numbers above branches) of C. briggsae isolates included in this study (modified from Howe & Denver, 2008). nad5Δ heteroplasmy levels were determined using qPCR and describe the average percentage of nad5Δ-deletion bearing genomes within individual worms from each isolate (Howe & Denver, 2008). GL = global superclade; KE = Kenya clade; TE and TR = temperate and tropical subclades of GL; C(+) = isolates bearing the compensatory Ψnad5Δ-2 allele. Note that we assayed the natural HK104 isolate here rather than the inbred line reported in Estes et al. (2011), which evolved high nad5Δ levels in the lab (see Section 2.1).
We present a reanalysis of data from our recent study of variation in C. briggsae mitochondrial form and function (Hicks et al., 2012), which quantified 24 mitochondrial phenotypes including ROS level, ΔΨM and aspects of organelle morphology on replicate live worms from 10 natural isolates of C. briggsae (Fig. 1). This work reported univariate analyses of these traits to describe standing levels of phenotypic variation among clades and isolates and focused its interpretation on phylogeographic patterns of phenotypic variation. By contrast, the current study reports a systematic evaluation of the bivariate relationships between all mitochondrial phenotypes from the combined dataset. Our aim was to examine the connections between mitochondrial physiology and dynamics within a natural system.. Our findings support a major role for ΔΨM in shaping mitochondrial dynamics. Based on previous studies and current models of mitochondrial dynamics, we expected to observe more punctate morphologies among low-ΔΨM mitochondria due to their reduced rates of fusion. Conversely, we expected that mitochondria with high ΔΨM would maintain the canonical elongated shape. Our findings were in agreement with both of these expectations and provide general support for Twig’s model (Twig, Hyde, et al., 2008) of mitochondrial dynamics. Furthermore, our results suggest an addition to this model in which individual organelles respond to their functional environment; i.e., the average ΔΨM of the surrounding mitochondrial population.
2. Materials and Methods
2.1. Nematode strains
We use data from our recent study (Hicks et al., 2012) in which an array of mitochondrial phenotypes were measured for ten natural isolates of Caenorhabditis briggsae nematodes (Fig. 1). Nuclear (Cutter et al., 2006; Jovelin & Cutter, 2011) and mitochondrial (Raboin et al., 2010) phylogenetic analyses place most C. briggsae natural isolates into three major phylogeographic clades that are latitudinally distinct, referred to as the Kenyan, Temperate, and Tropical clades (Fig. 1). Tropical clade isolates are found in tropical latitudes and contain substantial genetic diversity (Cutter et al., 2006), while temperate clade isolates inhabit northern latitudes and exhibit little genetic diversity (Cutter et al., 2006). Several nuclear polymorphisms distinguish Kenyan clade isolates from both temperate and tropical clade strains (Dolgin et al., 2008).The isolates used in this study were chosen to represent these three major phylogeographic clades, and to encompass the full range of known nad5Δ heteroplasmy level – from zero to ~50% deletion-bearing genomes. Briefly, the appearance of nad5Δ depends upon the presence of a mitochondrial pseudogene - Ψnad5-2 (see Figure 1 in Howe & Denver, 2008). The two Temperate clade isolates (PB800 and EG4181) harbor a compensatory Ψnad5-2 allele that limits the recurrent formation of nad5Δ; the two Kenyan clade isolates (ED3101 and ED3092) completely lack Ψnad5-2, which precludes formation of nad5Δ (Howe & Denver, 2008). C. briggsae strains and the evolutionary genetics of nad5Δ have been described in further detail elsewhere (Estes et al., 2011; Hicks et al., 2012; Howe & Denver, 2008).
2.2. Sample preparation and image analysis
For detailed methods regarding nematode sample preparation, image acquisition and analysis, please refer to (Dingley et al., 2009; Estes et al., 2011; Hicks et al., 2012). Briefly, data for all mitochondrial traits were obtained by analyzing confocal images of the pharyngeal bulb region of young adult nematodes. Worms were incubated with 10 μm concentrations of the mitochondria-targeted fluorescent dye(s) appropriate for each experiment (below). After 24 hours, worms were washed free of dye, paralyzed using levamisole, and imaged using a high-resolution wide-field confocal microscope (Advanced Light Microscopy Core, Oregon Health and Science University). All images were deconvolved prior to analysis and all image analysis was performed using ImageJ software (NIH).
The relative intensity of MitoSox Red (Molecular Probes, Eugene, OR) fluorescence from the terminal pharyngeal bulb of each worm was used to quantify relative ROS levels. Zielonka and Kalyanaraman (2010) determined that MitoSOX Red quantifies total levels of mitochondrial oxidants when used in conjunction with microscopic analysis (Zielonka & Kalyanaraman, 2010). Final ROS levels for each isolate were calculated as the difference between pharyngeal bulb intensity in labeled and unlabeled control worms from each isolate. Dye-based ROS measurements reflect both the rates of ROS generation and ROS scavenging by antioxidant enzymes or small molecules, and thus give a comprehensive view of the level of oxidative stress experienced by an organism. Supporting this claim, a comparison of our ROS measurements with a survey of oxidative DNA damage (frequency of 8-oxo-dG lesions) conducted on a set of C. elegans mutation-accumulation lines (Denver et al., 2009; Denver et al., 2012) was highly positively correlated (Spearman’s ρ1,6=0.943, P<=0.05) (J. Joyner Matos, K. Hicks, D. Denver, S. Estes, C. Baer, unpubl.). Finally, we find no relationship between pharyngeal pumping rates and ROS or ΔΨM (Estes et al., 2011; Hicks et al., 2012) indicating that our measures are not biased by variation in the rates of dye uptake by feeding.
Relative ΔΨM levels were quantified using MitoTracker Red CMXRos (Molecular Probes), a dye that localizes exclusively to polarized organelles (Pendergrass et al., 2004). The ΔΨM assays were performed concurrently with those of mitochondrial morphology by co-labeling worms with the ΔΨM-dependent probe MitoTracker Red CMXRos, and with MitoTracker Green FM (Molecular Probes), which accumulates within all mitochondria regardless of their respiration state (Pendergrass et al., 2004). This experimental setup allowed us to detect state-specific mitochondrial traits, such as shape changes occurring only in depolarized mitochondria, and to directly correlate mitochondrial ΔΨM and morphology traits. Unfortunately, the spectral similarities between the ROS and ΔΨM probes make it necessary to use separate images for ROS analysis. Thus, associations between ROS and all other mitochondrial traits should be interpreted with caution.
Finally, as previously discussed (Hicks et al., 2012), we failed to co-label nematodes treated as above with either DAPI or Hoechst 33342 (Sigma), which would have allowed us to visualize cell nuclei and thereby assess the intracellular distributions of mitochondria. (Appropriate GFP fusions are not yet available for C. briggsae.) Both dyes noticeably interfered with the fluorescence of the above MitoTracker dyes in C. briggsae (Hicks, pers. obs.). Our study therefore focuses on properties of individual mitochondria and mitochondrial populations within the pharyngeal bulb organ.
2.3. Trait descriptions and statistical analysis
A total of 24 mitochondrial form and function traits were analyzed (Table S1). Briefly, relative mitochondrial membrane potential (ΔΨM max) served as an indicator of mitochondrial functionality. Functional mitochondria were distinguished by their quantifiable uptake of MitoTracker Red CMXRos, while nonfunctional mitochondria took up untraceable amounts of MitoTracker Red CMXRos. Relative reactive oxygen species level (ROS max) further characterized mitochondrial activity. Maximum rather than mean ΔΨM and ROS levels were used because we previously found a significant effect of levamisole (the cholinergic agonist used to paralyze nematodes for image acquisition) on mean but not maximum ROS levels (Hicks et al., 2012). Additionally, we scored ten traits that describe aspects of either the functional (subscript F), nonfunctional (subscript N), or the total (subscript T) pharyngeal mitochondrial population: the combined area of the mitochondrial population (AFP, ANP, ATP), the ratio of the area of functional to nonfunctional mitochondria (AFP/NP), and the percentage of the total mitochondrial area that is functional (AFP/TP), the number of organelles (NF, NN, NT), the ratio of functional to nonfunctional organelles (NF/N), and the percentage of functional mitochondria (NF/T). Differences in mitochondrial morphology were measured using the area (AF, AN), aspect ratio (ARF, ARN), and circularity (CF, CN) of individual mitochondria. Aspect ratio quantifies elongation and has a minimal value of 1, which corresponds to a perfect circle (Russ, 2002). Circularity will also equal 1 when the measured object is a perfect circle, but decreases to 0 as the object becomes more branched (Russ, 2002). Because circularity cannot accurately be measured for extremely small objects (ImageJ website), we omitted from all analyses mitochondria smaller than 2 pixels. Finally, the variance (subscript V) in circularity (CFV, CNV) and aspect ratio (ARFV, ARNV) measured the degree of heterogeneity within the mitochondrial population of each nematode. See Hicks, et al. (2012) for further description of image processing.
Because our data often violated assumptions of the Pearson product-moment correlation (e.g., normally distributed data, monotonic bivariate relationships), we characterized correlations among mitochondrial form and function characters by calculating Spearman rank-order correlation coefficients between each pair of traits as in Huang et al. (2004). Because ROS levels were measured on different sets of nematodes than all other traits, we measured isolate-mean correlations for these pairs of traits. All statistical analysis was performed in JMP 9 (SAS Institute, Cary, NC).
3. Results
3.1. Mitochondrial trait associations
We analyzed the relationships between pairs of mitochondrial traits (Table S2), individual measurements of which were originally obtained by Hicks et al. (2012), for all natural isolates following (Estes et al., 2011). First, no significant correlations between ROS and any other mitochondrial trait were revealed (data not shown). Maximum ΔΨM was, however, statistically related to a number of other characters. ΔΨM was positively related to aspect ratio of functional mitochondria (ARF) (ρ = 0.208, P<=0.01), meaning that isolates with higher maximum ΔΨM tended to have more elongated mitochondria. Similarly, maximum ΔΨM was also weakly negatively correlated to the circularity of functional mitochondria (CF) (ρ = −0.248, P<=0.01), suggesting that worms with higher maximum ΔΨM fluorescence also tended to have less circular – or more branched – organelles. It is important to note that maximum ΔΨM was necessarily positively correlated to traits related to functional mitochondrial area (AF, AFP, and ATP) since individuals with higher scores for these traits had necessarily taken up more membrane-potential dependent dye and thus had higher values for ΔΨM.
A number of the other correlations were expected due to the nature of the measurements (e.g., between traits describing the number of mitochondria and those describing the combined area of mitochondria populations); however, a systematic survey of the remaining (statistically significant) correlations revealed consistent patterns of relationship between the major classes of mitochondrial traits (shape, area, and number), which can be summarized as follows:
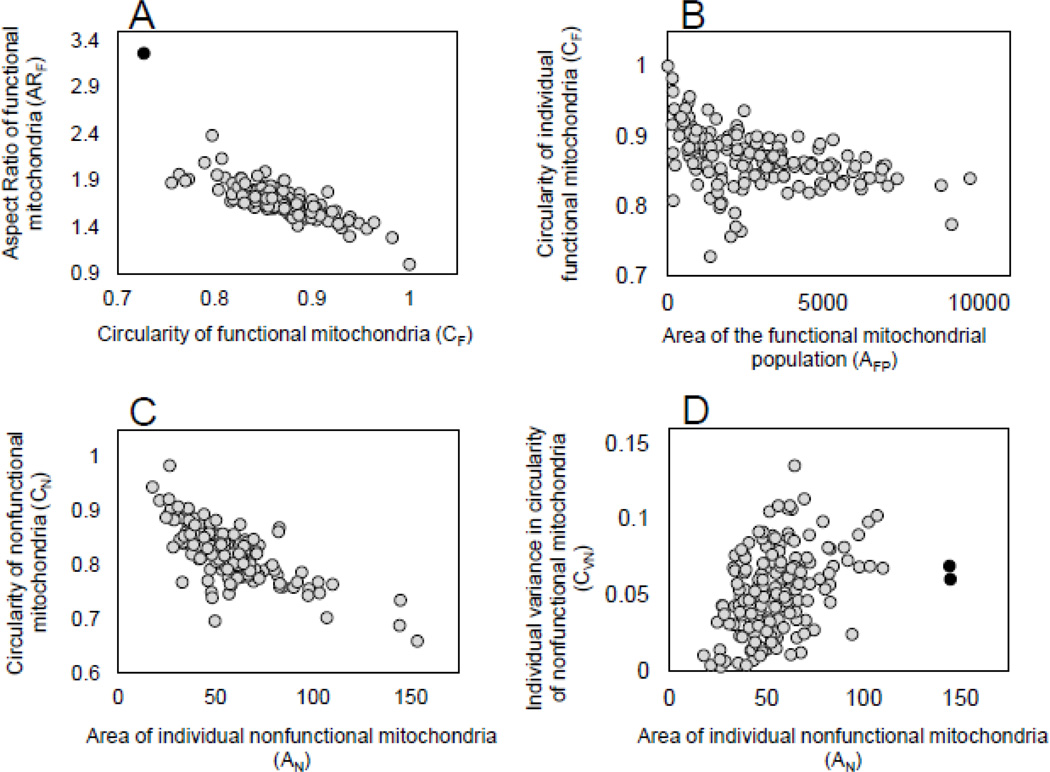
(1) As expected, circularity (CN and CF) demonstrated a strong negative correlation with aspect ratio (ARN and ARF) for both nonfunctional and functional mitochondria (ρ = −0.811, P<=0.001 and ρ = −0.801, P<=0.001, respectively). Figure 2A shows the relationship between these two traits for functional mitochondria. Circularity responds to changes in surface irregularities (or the amount of branching) of each mitochondrion whereas aspect ratio responds to the elongation of organelles (Koopman et al., 2005; Russ, 2002). This negative relationship must certainly owe itself largely to the fact that more circular mitochondria are less elongate; however, it also implies that mitochondrial branching was rare and that deviations from perfect circularity were most often achieved by elongation rather than by branching for all organelles regardless of their functional status.
Figure 2. Examples of bivariate relationships of mitochondrial phenotypes.
Patterns of relationship between traits describing mitochondrial size, morphology, and within-individual variance are shown. All measurements were made on the same set of confocal images (see Section 2.2) and each point represents the bivariate phenotype for an individual nematode (N=167–170). A) Aspect ratio is negatively related to circularity within the functional mitochondria of individual worms (ρ=-0.806, P<=0.0001). Removing one outlier (black symbol) decreases the correlation slightly (ρ=-0.797, P<=0.0001). B) As the two-dimensional area of the total functional mitochondrial population increases, functional mitochondria become less circular (ρ=-0.509, P<=0.0001). C) As the two-dimensional area of individual nonfunctional mitochondria increases, these mitochondria become less circular (ρ=-0.698, P<=0.0001). D) Individual nonfunctional mitochondria become more variable with respect to circularity as their two-dimensional area increases (ρ=-0.409, P<=0.0001). When two outliers (black symbols) are removed, the correlation remains unchanged. Outliers were determined by calculating Mahalanobis distances for the correlation between traits of each sample and visually identifying extreme outliers.
(2) In isolates containing a higher ratio of polarized mitochondria (higher scores for NF/N or NF/T), all mitochondria were larger (NF/N and AF: ρ = 0.317, P<=0.001; NF/N and AN: ρ = 0.168, P<=0.05), less circular (NF/N and CF: ρ = −0.276, P<=0.001; NF/N and CN: ρ = −0.196, P<=0.05) (e.g., Fig. 2B shows this pattern for functional mitochondria), and more elongate (NF/N and ARF: ρ = 0.183, P<=0.05; NF/N and ARN: ρ = 0.188, P<=0.05) regardless of their functional state. (Correlations between NF/T and other listed traits follow similar trends; Table S2.) However, only the depolarized mitochondria in these isolates were significantly more variable with regard to circularity (NF/N and CNV: ρ = 0.190, P<=0.05). Similarly, as the area of individual functional mitochondria (AF) or the combined area of the functional mitochondrial population (AFP) increased, all mitochondria became less circular (AFP and CF: ρ = −0.509, P<=0.001; AFP and CN: ρ = −0.216, P<=0.001) and more elongate (AFP and ARF: ρ = 0.256, P<=0.001; AFP and CN: ρ = 0.160, P<=0.05. (Correlations between AF and other listed traits follow similar trends; Table S2).
(3) In isolates with more nonfunctional/depolarized mitochondria (higher scores for NN), the nonfunctional mitochondria in these isolates become less elongate and more uniform with respect to this trait (NN and ARN: ρ = −0.200, P<=0.01; NN and ARNV: ρ = −0.241, P<=0.01). Conversely, the polarized mitochondria in these isolates were more variable with regard to elongation (NN and ARFV: ρ = 0.154, P<=0.05). As the area of individual nonfunctional mitochondria (AN) or the area of the nonfunctional mitochondrial population (ANP) increased, nonfunctional mitochondria responded by deviating from circularity, (ANP and CN: ρ= −0.316, P<=0.001, AN and CN: ρ = −0.698, P<=0.001; Fig. 2C) in ways indicative of elongation or branching (c.f., Koopman et al., 2005), and by becoming more variable with respect to circularity (AN and CNV: ρ= 0.409, P<=0.001, Fig. 2D). Because we omitted mitochondria smaller than 2 pixels from all analyses (Hicks et al., 2012 and Materials and Methods), the strength of these correlations and the fact that they apply only to depolarized mitochondria suggest that they should not be influenced by any size-related bias.
Finally, we note that regressions of average mitochondrial trait correlations for each isolate on isolate-specific nad5Δ heteroplasmy revealed no evidence that any of the mitochondrial phenotypic associations were related to nad5Δ level; however, because our estimates of average nad5Δ for each C. briggsae isolate were obtained from a different set of worms than those phenotyped (Hicks et al., 2012), the biological meaning of these tests is questionable.
4. Discussion
4.1. Implications for mitochondrial dynamics
We performed a systematic evaluation of phenotypic correlations among mitochondrial traits originally reported in (Hicks et al., 2012) with the aim of uncovering patterns that describe the relationships between mitochondrial physiology and morphology. While some studies have indicated that ROS production can be associated with dramatic ultra-structural transformations in mitochondria (Koopman et al., 2005; Liot et al., 2009; Wang et al., 2008), our analyses failed to clearly relate net ROS levels with any alterations in mitochondrial shape or population structure. Again, a caveat prohibiting further interpretation of this result is that ROS was necessarily measured on different individual nematodes than all other mitochondrial traits (see section 2.2). Thus, estimates of ROS were obtained from different sets of mitochondria than those describing ΔΨM and morphology traits.
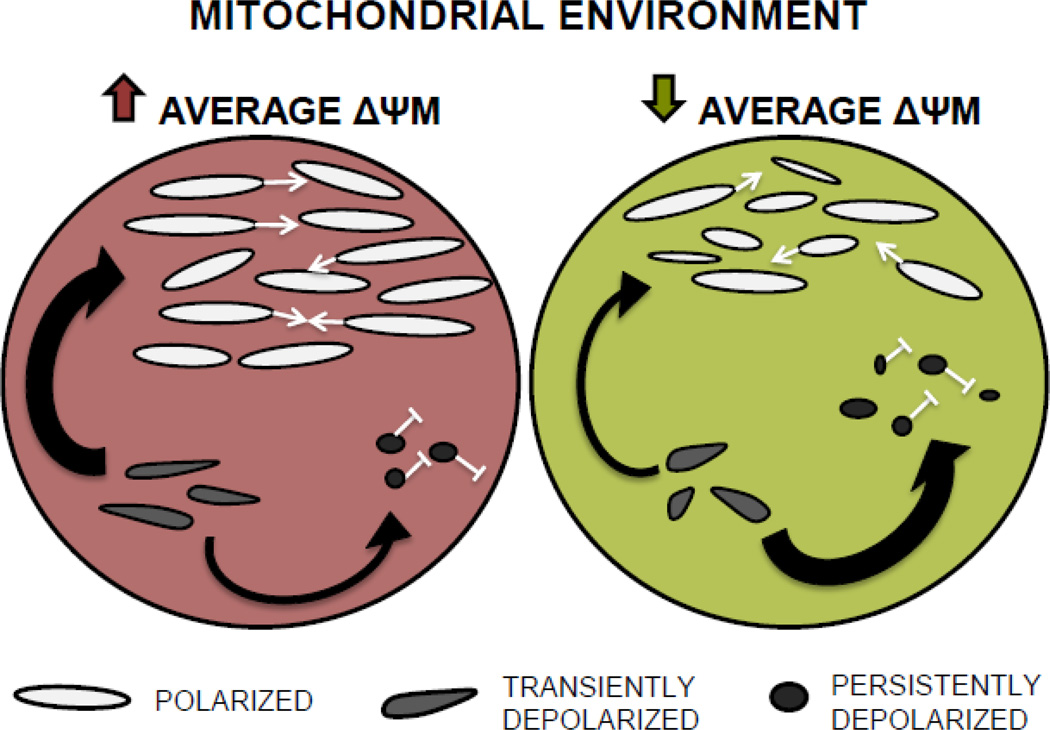
In agreement with previous studies (Ishihara et al., 2003; Mattenberger et al., 2003; Twig, Elorza, et al., 2008), our findings suggest a central role for ΔΨM in shaping mitochondrial morphology, manifested in its relationship with several aspects of mitochondrial shape and population structure (Table S2). Further, our analysis of mitochondrial trait associations revealed that, regardless of their functional state, mitochondria appear to respond differently depending on their functional neighborhood; i.e., whether they co-occur with other organelles within the nematode pharynx that are mainly polarized or depolarized. The morphology and physiological state of individual mitochondria is dependent on ΔΨM (Ishihara et al., 2003; Miceli et al., 2011; Twig & Shirihai, 2011). This makes sense as several critical organellar functions are contingent upon ΔΨM, including mitochondrial fusion and ATP and ROS production rates (Gaskova et al., 2007; Ishihara et al., 2003; Murphy, 2009). Recent work has shown that mitochondrial fusion is brief and accompanied by fission (Twig, Elorza, et al., 2008; Wikstrom et al., 2009) and that normal cycles of fusion and fission are necessary to maintain the canonical ovoid mitochondrial shape (Chen & Chan, 2005; Kageyama et al., 2011). The model of mitochondrial life cycles proposed by Twig, et al. (2008) connects mitochondrial morphology and function by suggesting that, following a fusion-fission cycle, one daughter mitochondrion remains polarized while the other is transiently depolarized. The transiently depolarized daughter will either regain ΔΨM (if it contains a sufficient number of functional ETC components) and resume its participation in the fusion-fission cycle, or it will remain depolarized and undergo fission and eventual mitophagic degradation (Fig. 1 in Twig et al., 2008). Our data suggest that such fusion-cycling occurs within the context of a larger mitochondrial population that is itself either more or less polarized (Fig. 3). Specifically, we find that both polarized and depolarized mitochondria are more elongate (less fragmented) in C. briggsae isolates containing more mitochondria with high ΔΨM (higher values for NF/N and AFP), although depolarized organelles are slightly more variable in shape than polarized organelles. Conversely, when they inhabit less functional isolates (those with more mitochondria with low ΔΨM), both polarized and depolarized mitochondria become increasingly heterogeneous in shape, but depolarized mitochondria become overall more fragmented (Table S2). Placing these data within the context of Twig et al.’s (2008) model, we propose that a majority of the mitochondria in isolates with higher average ΔΨM will exhibit the “typical” ovoid shape by maintaining normal rates of fusion-fission cycles. Transiently depolarized mitochondria in this environment will be more likely to recover membrane polarization after fission, helping to maintain a large polarized mitochondrial population. Conversely, isolates with lower ΔΨM will suffer a reduced frequency of fusion-fission cycling and display increased shape heterogeneity in the entire mitochondrial population. Transiently depolarized mitochondria in this environment will be less likely to harbor functional ETC products and will more often join the persistently depolarized population, which is unable to undergo fusion. Polarized mitochondria in these isolates will then experience reduced numbers of fusion “mates” – in essence, an intracellular Allee effect (Allee, 1931), which will contribute to further reduced rates of fusion-fission cycling and lead to increased shape heterogeneity of all mitochondrial morphs (Fig. 3).
Figure 3. A context-dependent model of mitochondrial dynamics in which mitochondria respond to the functional state of their intracellular environment.
We propose that an organism has three types of mitochondrial populations, polarized, transiently depolarized, and persistently depolarized. The polarized population is capable of undergoing fusion (white arrows) while the persistently depolarized population is not (white blunted arrow). The transiently depolarized mitochondria are produced after a fusion-fission cycle and will either regain sufficient ΔψM and join the polarized/fusing population (gray arrow) or, if they are unable to regain ΔψM, join the persistently depolarized population (black arrow) (Twig et al., 2008). We propose that mitochondria in a more functional environment (higher ΔψM, at left) are more likely to regain ΔψM and join the polarized/fusing population. Normal fusion-fission cycles will maintain a majority of mitochondria in the canonical ovoid morph. Here, many depolarized mitochondria are destined to recover ΔψM and rejoin the fusing population. Conversely, mitochondria in a less functional environment (lower ΔψM, at right) are less likely to regain ΔψM and will therefore join the persistently depolarized/non-fusing population resulting in fewer polarized mitochondria. Here, lower than normal rates of fusion and fission will increase the shape heterogeneity of all mitochondria.
4.2. Implications for mitochondrial genome integrity and evolution
It has been suggested that damaged mitochondrial genomes may be preferentially shunted to the depolarized daughter organelle – the one more often destined for degradation (Kowald & Kirkwood, 2011; Twig, Hyde, et al., 2008). If this is the case, mitochondrial fusion-fission cycling may have a critical role to play in maintaining mtDNA genome stability and could conceivably contribute to intra- and interspecific differences in mtDNA mutation rates and heteroplasmy levels. Recent work shows that the removal of damaged mtDNA in C. elegans requires mitochondrial fusion (Bess et al., 2012; Meyer & Bess, 2012). Because the fusion-fission cycle relies on ΔΨM, alterations to ΔΨM that are unrelated to mtDNA quality (i.e., that weaken the link between mitochondrial genotype and phenotype) could reduce the efficacy of the selective process (Twig, Hyde, et al., 2008). Hicks, et al. (2012) determined that much of the measured variation in mitochondrial phenotypes – and especially that of ΔΨM - related to the phylogeographic clade membership of particular C. briggsae isolates, rather than to nad5Δ level (Tables S1 and S2 in Hicks et al., 2012). Specifically, Tropical isolates tended to have the lowest values for ΔΨM followed by Kenyan isolates, and Temperate isolates exhibited the highest ΔΨM (Figure 3G in Hicks et al., 2012). This led to the hypothesis that Tropical C. briggsae isolates may have adaptively reduced their ΔΨM in order to counter increased ROS levels brought on by high temperatures (Brand, 2000; Hicks et al., 2012). Because C. briggsae are ectotherms, the external temperature can directly influence their physiology; higher temperatures can increase nematode metabolism and ROS generation. Additionally, Tropical clade worms contain fewer total mitochondria (NT and NF) within the focal area (pharyngeal bulb) compared to both the Temperate and Kenyan clades (Tukey HSD, α = 0.05; (Figure 3H in Hicks et al., 2012)). Since cold-adapted ectotherms often exhibit increased mitochondrial density (Morley et al., 2009), the reduction in mitochondrial number within Tropical clade isolates is also consistent with an adaptive response to heat. If it is indeed the case that Tropical C. briggsae isolates have adaptively reduced their ΔΨM, they may experience a reduced efficiency of selection allowing the amplification of damaged mtDNA molecules – such as those bearing nad5Δ. Such a dynamic could help to explain the counter-intuitive finding that Tropical C. briggsae isolates display higher average nad5Δ heteroplasmy levels despite having larger effective population sizes (and presumably more efficient natural selection) than Temperate clade isolates that have fixed compensatory mutations preventing nad5Δ accumulation (Howe & Denver, 2008). In other words, the Tropical isolate’s adaptive reduction of ΔΨM in response to high temperature may interfere with the selective removal of nad5Δ by the mitochondrial fusion-fission cycle. Alternatively, because nad5Δ affects a component of mitochondrial ETC complex I that is putatively involved in H+ pumping (Janssen et al., 2006; Lenaz et al., 2006), the deletion may itself reduce ΔΨM and provide a ROS avoidance mechanism that does not directly produce heat (unlike mitochondrial uncoupling) - thus conferring a direct benefit to Tropical C. briggsae isolates (Brand, 2000; Iser et al., 2005). Either scenario implies that different evolutionary pressures may be shaping the subcellular phenotypes of different C. briggsae populations and suggests fruitful avenues for experimental work.
4.3. Conclusions
Our analysis suggests that ΔΨM, but not mitochondrial ROS level, has a major role in shaping mitochondrial dynamics within natural populations of C. briggsae nematodes. We also identified a set of correlations that may describe a global control mechanism for mitochondrial dynamics. In particular, our findings suggest a model of mitochondrial population dynamics in which cellular environmental context dependency – and in particular, whether the mitochondrial population is mainly polarized or depolarized – is a key feature. To our knowledge, ours is the first study to connect natural variation in subcellular phenotypes to a model of mitochondrial dynamics. The model is also congruent with recent work highlighting the importance of both organellar and cellular influence on mitochondrial fusion-fission processes (Hyde et al., 2010; Kowald & Kirkwood, 2011), but robust tests will require experimental confirmation of several assumptions, including whether ΔΨM correlates linearly to fission-fusion ability. The natural phenotypic and genetic variation within the C. briggsae system will be advantageous for further study in this area (e.g., partitioning of the genetic and cellular environmental components of observed mitochondrial phenotypic variation). With particular regard to C. briggsae evolution, our findings highlight the need for future work to understand what if any role mitochondrial fission-fusion dynamics play in mediating transmission of nad5Δ-bearing genomes and adaptation to local thermal and other conditions.
Supplementary Material
Table S1. Designated labels and descriptions of all mitochondrial traits measured in C. briggsae natural isolates. Subscripts N, F, and T denote whether the trait refers to Nonfunctional, Functional, or Total (all) mitochondria. Subscripts P and V indicate that the measure refers to the entire mitochondrial population or the average within-individual variance in that trait, respectively.
Table S2. Spearman’s correlation coefficients between mitochondrial traits measured in C. briggsae natural isolates. (See section 3.1 and Table S1 for trait descriptions.) *,**, and *** indicate statistical significance at the 0.05, 0.01, and 0.001 levels, respectively. Because ROS levels were measured on different sets of nematodes than all other traits and required that we calculate isolate-mean correlations for these pairs of traits, correlations involving ROS (none of which were significantly different from zero) are not included above (N =164–170).
Highlights.
Mitochondrial morphology depends on ΔψM but not on reactive oxygen species level
Individual mitochondria are larger and more elongated when surrounded by high-ΔψM mitochondrial populations
Mitochondrial populations with low ΔψM exhibit greater morphological heterogeneity than those with high ΔψM
Individual organelle morphology depends upon the functional state of the larger mitochondrial population
Acknowledgements
We thank A. Snyder (Advanced Light Microscopy Core, Oregon Health and Science University) for generous technical advice on confocal imaging and analysis. Thanks also to A. Coleman-Hulbert and S. Smith for lab assistance. This work was funded by National Institutes of Health grant 5 R01 GM087628-02 to DRD and SE, PSU Faculty Enhancement grant to SE, and American Heart Association Predoctoral Fellowship 11PRE4880069 to KAH.
Abbreviations
- ATP
adenosine triphosphate
- ETC
electron transport chain
- ΔΨM
mitochondrial membrane potential
- nad5
NADH-dehydrogenase 5
- ROS
reactive oxygen species
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kiley A. Hicks, Email: khicks@pdx.edu.
Dee R. Denver, Email: denvedee@cgrb.oregonstate.edu.
References
- Allee WC. Co-operation among animals. Am. J. Sociol. 1931;37:386–398. [Google Scholar]
- Brand MD. Uncoupling to survive? The role of mitochondrial inefficiency in ageing. Exp. Gerontol. 2000;35:811–820. doi: 10.1016/s0531-5565(00)00135-2. [DOI] [PubMed] [Google Scholar]
- Bess AS, Crocker TL, Ryde IT, Meyer JN. Mitochondrial dynamics and autophagy aid in removal of persistent mitochondrial DNA damage in Caenorhabditis elegans. Nucleic Acids Res. 2012;40:7916–7931. doi: 10.1093/nar/gks532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC. Emerging functions of mammalian mitochondrial fusion and fission. Hum. Mol. Genet. 2005;14(Spec No):R283–R289. doi: 10.1093/hmg/ddi270. [DOI] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- Clark KA, Howe DK, Gafner K, Kusuma D, Ping S, Estes S, Denver DR. Selfish little circles, transmission bias and evolution of large deletion-bearing mitochondrial DNA in Caenorhabditis briggsae nematodes. PLoS One. 2012;7:e41433. doi: 10.1371/journal.pone.0041433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Félix MA, Barriere A, Charlesworth D. Patterns of nucleotide polymorphism distinguish temperate and tropical wild isolates of Caenorhabditis briggsae. Genetics. 2006;173:2021–2031. doi: 10.1534/genetics.106.058651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Yan W, Tsvetkov N, Sunil S, Felix MA. Molecular population genetics and phenotypic sensitivity to ethanol for a globally diverse sample of the nematode Caenorhabditis briggsae. Mol. Ecol. 2010;19:798–809. doi: 10.1111/j.1365-294X.2009.04491.x. [DOI] [PubMed] [Google Scholar]
- Denver DR, Dolan PC, Wilhelm LJ, Sung W, Lucas-Lledo JI, Howe DK, Lewis SC, Okamoto K, Thomas WK, Lynch M, Baer CF. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc. Natl. Acad. Sci. U S A. 2009;106:16310–16314. doi: 10.1073/pnas.0904895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Wilhelm LJ, Howe DK, Gafner K, Dolan PC, Baer CF. Variation in base-substitution mutation in experimental and natural lineages of Caenorhabditis nematodes. Genome Biology and Evolution. 2012;4:513–522. doi: 10.1093/gbe/evs028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer KS, Fritz S, Fuchs F, Messerschmitt M, Weinbach N, Neupert W, Westermann B. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:847–853. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingley S, Polyak E, Lightfoot R, Ostrovsky J, Rao M, Greco T, Ischiropoulos H, Falk MJ. Mitochondrial respiratory chain dysfunction variably increases oxidant stress in Caenorhabditis elegans. Mitochondrion. 2009;10:125–136. doi: 10.1016/j.mito.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin ES, Félix MA, Cutter AD. Hakuna Nematoda: genetic and phenotypic diversity in African isolates of Caenorhabditis elegans and C. briggsae. Heredity. 2008;100:304–315. doi: 10.1038/sj.hdy.6801079. [DOI] [PubMed] [Google Scholar]
- Duvezin-Caubet S, Jagasia R, Wagener J, Hofmann S, Trifunovic A, Hansson A, Chomyn A, Bauer MF, Attardi G, Larsson NG, Neupert W, Reichert AS. Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J. Biol. Chem. 2006;281:37972–37979. doi: 10.1074/jbc.M606059200. [DOI] [PubMed] [Google Scholar]
- Estes S, Coleman-Hulbert AL, Hicks KA, de Haan G, Martha SR, Knapp JB, Smith SW, Stein KC, Denver DR. Natural variation in life history and aging phenotypes is associated with mitochondrial DNA deletion frequency in Caenorhabditis briggsae. BMC Evol. Biol. 2011;11:11. doi: 10.1186/1471-2148-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskova D, DeCorby A, Lemire BD. DiS-C3(3) monitoring of in vivo mitochondrial membrane potential in C. elegans. Biochem. Biophys. Res. Commun. 2007;354:814–819. doi: 10.1016/j.bbrc.2007.01.073. [DOI] [PubMed] [Google Scholar]
- Grad LI, Lemire BD. Mitochondrial complex I mutations in Caenorhabditis elegans produce cytochrome c oxidase deficiency, oxidative stress and vitamin-responsive lactic acidosis. Hum. Mol. Genet. 2004;13:303–314. doi: 10.1093/hmg/ddh027. [DOI] [PubMed] [Google Scholar]
- Graef M, Nunnari J. Mitochondria regulate autophagy by conserved signalling pathways. EMBO J. 2011;30:2101–2114. doi: 10.1038/emboj.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin EE, Detmer SA, Chan DC. Molecular mechanism of mitochondrial membrane fusion. Biochim. Biophys. Acta. 2006;1763:482–489. doi: 10.1016/j.bbamcr.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Grünewald A, Voges L, Rakovic A, Kasten M, Vandebona H, Hemmelmann C, Lohmann K, Orolicki S, Ramirez A, Schapira AHV, Pramstaller PP, Sue CM, Klein C. Mutant Parkin impairs mitochondrial function and morphology in human fibroblasts. PloS ONE. 2010;5:e12962. doi: 10.1371/journal.pone.0012962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Howe DK, Leung A, Denver DR, Estes S. In Vivo Quantification Reveals Extensive Natural Variation in Mitochondrial Form and Function in Caenorhabditis briggsae. PLoS ONE. 2012;7:e43837. doi: 10.1371/journal.pone.0043837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe DK, Baer CF, Denver DR. High rate of large deletions in Caenorhabditis briggsae mitochondrial genome mutation processes. Genome Biology and Evolution. 2010;2010:29–38. doi: 10.1093/gbe/evp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe DK, Denver DR. Muller’s Ratchet and compensatory mutation in Caenorhabditis briggsae mitochondrial genome evolution. BMC Evol. Biol. 2008;8:62. doi: 10.1186/1471-2148-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Xiong C, Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U S A. 2004;101:8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde BB, Twig G, Shirihai OS. Organellar vs. cellular control of mitochondrial dynamics. Sem. Cell Dev. Biol. 2010;21:575–581. doi: 10.1016/j.semcdb.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichishita R, Tanaka K, Sugiura Y, Sayano T, Mihara K, Oka T. An RNAi screen for mitochondrial proteins required to maintain the morphology of the organelle in Caenorhabditis elegans. J. Biochem. 2008;143:449–454. doi: 10.1093/jb/mvm245. [DOI] [PubMed] [Google Scholar]
- Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev.of Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irrcher I, Aleyasin H, Seifert EL, Hewitt SJ, Chhabra S, Phillips M, Lutz AK, Rousseaux MW, Bevilacqua L, Jahani-Asl A, Callaghan S, MacLaurin JG, Winklhofer KF, Rizzu P, Rippstein P, et al. Loss of the Parkinson’s disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum. Mol. Genet. 2010;19:3734–3746. doi: 10.1093/hmg/ddq288. [DOI] [PubMed] [Google Scholar]
- Iser WB, Kim D, Bachman E, Wolkow C. Examination of the requirement for ucp- 4, a putative homolog of mammalian uncoupling proteins, for stress tolerance and longevity in C. elegans. Mech. of Ageing Dev. 2005;126:1090–1096. doi: 10.1016/j.mad.2005.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Jofuku A, Eura Y, Mihara Katsuyoshi. Regulation of mitochondrial morphology by membrane potential, and DRP1-dependent division and FZO1-dependent fusion reaction in mammalian cells. Biochem. Biophys. Res. Commun. 2003;301:891–898. doi: 10.1016/s0006-291x(03)00050-0. [DOI] [PubMed] [Google Scholar]
- Janssen RJ, Nijtmans LG, van den Heuvel LP, Smeitink JA. Mitochondrial complex I, structure, function and pathology. J. Inherit. Metab. Dis. 2006;29:499–515. doi: 10.1007/s10545-006-0362-4. [DOI] [PubMed] [Google Scholar]
- Jovelin R, Cutter AD. MicroRNA sequence variation potentially contributes to within-species functional divergence in the nematode Caenorhabditis briggsae. Genetics. 2011;189:967–976. doi: 10.1534/genetics.111.132795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Zhang Z, Sesaki H. Mitochondrial division, molecular machinery and physiological functions. Curr. Opin. Cell Biol. 2011;23:427–434. doi: 10.1016/j.ceb.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott AB, Bossy-Wetzel E. Impairing the mitochondrial fission and fusion balance, a new mechanism of neurodegeneration. Ann. NY Acad. Sci. 2008;1147:283–292. doi: 10.1196/annals.1427.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman WJ, Verkaart S, Visch HJ, van der Westhuizen FH, Murphy MP, van den Heuvel LW, Smeitink JA, Willems PH. Inhibition of complex I of the electron transport chain causes O2-. -mediated mitochondrial outgrowth. Am. J. Physiol. Cell Physiol. 2005;288:1440–1450. doi: 10.1152/ajpcell.00607.2004. [DOI] [PubMed] [Google Scholar]
- Kowald A, Kirkwood TB. Evolution of the mitochondrial fusion-fission cycle and its role in aging. Proc. Natl. Acad. Sci. U S A. 2011;108:10237–10242. doi: 10.1073/pnas.1101604108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Jeong S-Y, Karbowski M, Smith CL, Youle RJ. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol. Biol. Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros F, Lombès A, Frachon P, Rojo M. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol. Biol. Cell. 2002;13:4343–4354. doi: 10.1091/mbc.E02-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire BD, Behrendt M, DeCorby A, Gaskova D. C. elegans longevity pathways converge to decrease mitochondrial membrane potential. Mech. Ageing Dev. 2009;130:461–465. doi: 10.1016/j.mad.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Lenaz G, Fato R, Genova ML, Bergamini C, Bianchi C, Biondi A. Mitochondrial Complex I, structural and functional aspects. Biochim. Biophys. Acta. 2006;1757:1406–1420. doi: 10.1016/j.bbabio.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Liot G, Bossy B, Lubitz S, Kushnareva Y, Sejbuk N, Bossy-Wetzel E. Complex II inhibition by 3-NP causes mitochondrial fragmentation and neuronal cell death via an NMDA- and ROS-dependent pathway. Cell Death and Differ. 2009;16:899–909. doi: 10.1038/cdd.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattenberger Y, James DI, Martinou JC. Fusion of mitochondria in mammalian cells is dependent on the mitochondrial inner membrane potential and independent of microtubules or actin. FEBS Lett. 2003;538:53–59. doi: 10.1016/s0014-5793(03)00124-8. [DOI] [PubMed] [Google Scholar]
- Meeusen S, McCaffery JM, Nunnari J. Mitochondrial fusion intermediates revealed in vitro. Science. 2004;305:1747–1752. doi: 10.1126/science.1100612. [DOI] [PubMed] [Google Scholar]
- Meyer JN, Bess AS. Involvement of autophagy and mitochondrial dynamics in determining the fate and effects of irreparable mitochondrial DNA damage. Autophagy. 2012;8:12. doi: 10.4161/auto.21741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miceli MV, Jiang JC, Tiwari A, Rodriguez-Quiñones JF, Jazwinski SM. Loss of mitochondrial membrane potential triggers the retrograde response extending yeast replicative lifespan. Frontiers in Genetics. 2011;2:102. doi: 10.3389/fgene.2011.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc. Natl. Acad. Sci. U S A. 2009;106:11960–11965. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina AJ, Wikstrom JD, Stiles Linsey, Las Guy, Mohamed Hibo, Elorza A, Walzer Gil, Twig G, Katz Steve, Corkey Barbara E, Shirihai OS. Mitochondrial networking protects beta-cells from nutrient-induced apoptosis. Diabetes. 2009;58:2303–2315. doi: 10.2337/db07-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley SA, Lurman GJ, Skepper JN, Portner HO, Peck LS. Thermal plasticity of mitochondria, a latitudinal comparison between Southern Ocean molluscs. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2009;152:423–430. doi: 10.1016/j.cbpa.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Murphy MP. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Palermo V, Falcone C, Mazzoni C. Apoptosis and aging in mitochondrial morphology mutants of S. cerevisiae. Folia. Microbiol. (Praha) 2007;52:479–483. doi: 10.1007/BF02932107. [DOI] [PubMed] [Google Scholar]
- Pendergrass W, Wolf N, Poot M. Efficacy of MitoTracker Green and CMX rosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry A. 2004;61:162–169. doi: 10.1002/cyto.a.20033. [DOI] [PubMed] [Google Scholar]
- Pham NA, Richardson T, Cameron J, Chue B, Robinson BH. Altered mitochondrial structure and motion dynamics in living cells with energy metabolism defects revealed by real time microscope imaging. Microsc. Microanal. 2004;10:247–260. doi: 10.1017/S143192760404005X. [DOI] [PubMed] [Google Scholar]
- Pletjushkina OY, Lyamzaev KG, Popova EN, Nepryakhina OK, Ivanova OY, Domnina LV, Chernyak BV, Skulachev VP. Effect of oxidative stress on dynamics of mitochondrial reticulum. Biochim. Biophys. Acta. 2006;1757:518–524. doi: 10.1016/j.bbabio.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Prasad A, Croydon-Sugarman MJF, Murray RL, Cutter AD. Temperaturedependent fecundity associates with latitude in Caenorhabditis briggsae. Evolution. 2011;65:52–63. doi: 10.1111/j.1558-5646.2010.01110.x. [DOI] [PubMed] [Google Scholar]
- Raboin MJ, Timko AF, Howe DK, Félix MA, Denver DR. Evolution of Caenorhabditis mitochondrial genome pseudogenes and Caenorhabditis briggsae natural isolates. Mol. Biol. Evol. 2010;27:1087–1096. doi: 10.1093/molbev/msp318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raha S, Robinson BH. Mitochondria, oxygen free radicals, disease and ageing. Trends Biochem. Sci. 2000;25:502–508. doi: 10.1016/s0968-0004(00)01674-1. [DOI] [PubMed] [Google Scholar]
- Ross JA, Koboldt DC, Staisch JE, Chamberlin HM, Gupta BP, Miller RD, Baird SE, Haag ES. Caenorhabditis briggsae recombinant inbred line genotypes reveal inter-strain incompatibility and the evolution of recombination. PLoS Genetics. 2011;7:e1002174. doi: 10.1371/journal.pgen.1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ JC. The Image Processing Handbook. Boca Raton: CRC Press, LLC; 2002. [Google Scholar]
- Scorrano L. Proteins that fuse and fragment mitochondria in apoptosis, con-fissing a deadly con-fusion? J. Bioenerg. Biomembr. 2005;37:165–170. doi: 10.1007/s10863-005-6572-x. [DOI] [PubMed] [Google Scholar]
- Sedensky MM, Morgan PG. Mitochondrial respiration and reactive oxygen species in C. elegans. Exp. Gerontol. 2006;41:957–967. doi: 10.1016/j.exger.2006.06.056. [DOI] [PubMed] [Google Scholar]
- Song Z, Chen H, Fiket M, Alexander C, Chan DC. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B, Wang Xinglong, Zheng L, Perry G, Smith MA, Zhu X. Abnormal mitochondrial dynamics and neurodegenerative diseases. Biochim. Biophys. Acta. 2010;1802:135–142. doi: 10.1016/j.bbadis.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer PA, Swerdlow RH, Parks JK, Keeney P, Bennett JP, Jr, Miller SW, Davis RE, Parker WD., Jr Abnormal mitochondrial morphology in sporadic Parkinson’s and Alzheimer's disease cybrid cell lines. Exp. Neurol. 2000;162:37–50. doi: 10.1006/exnr.2000.7333. [DOI] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Hyde BB, Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis, the bioenergetic view. Biochim. Biophys. Acta. 2008;1777:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Shirihai OS. The interplay between mitochondrial dynamics and mitophagy. Antiox. Redox Signaling. 2011;14:1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura N, Rea SL, Testi R. Long-lived C. elegans Mitochondrial mutants as a model for human mitochondrial-associated diseases. Exp. Gerontol. 2006;41:974–991. doi: 10.1016/j.exger.2006.06.060. [DOI] [PubMed] [Google Scholar]
- Verkaart S, Koopman WJ, van Emst-de Vries SE, Nijtmans LGJ, van den Heuvel LWPJ, Smeitink JAM, Willems PHGM. Superoxide production is inversely related to complex I activity in inherited complex I deficiency. Biochim. Biophys. Acta-Molecular Basis of Disease. 2007;1772:373–381. doi: 10.1016/j.bbadis.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Wanagat J, Cao Z, Pathare P, Aiken JM. Mitochondrial DNA deletion mutations colocalize with segmental electron transport system abnormalities, muscle fiber atrophy, fiber splitting, and oxidative damage in sarcopenia. FASEB J. 2001;15:322–332. doi: 10.1096/fj.00-0320com. [DOI] [PubMed] [Google Scholar]
- Wang W, Fang H, Groom L, Cheng A, Zhang W, Liu J, Wang X, Li K, Han P, Zheng M, Yin J, Mattson MP, Kao JP, Lakatta EG, Sheu SS, et al. Superoxide flashes in single mitochondria. Cell. 2008;134:279–290. doi: 10.1016/j.cell.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Xinglong, Su B, Fujioka H, Zhu X. Dynamin-like protein 1 reduction underlies mitochondrial morphology and distribution abnormalities in fibroblasts from sporadic Alzheimer’s disease patients. The Am. J. Pathol. 2008;173:470–482. doi: 10.2353/ajpath.2008.071208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. Bioenergetic role of mitochondrial fusion and fission. Biochim. Biophys. Acta. 2012;10:1833–1838. doi: 10.1016/j.bbabio.2012.02.033. [DOI] [PubMed] [Google Scholar]
- Westermann B. Mitochondrial dynamics in model organisms, What yeasts, worms and flies have taughtus about fusion and fission of mitochondria. Sem. Cell Dev. Biol. 2010;21:542–549. doi: 10.1016/j.semcdb.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Wikstrom JD, Twig G, Shirihai OS. What can mitochondrial heterogeneity tell us about mitochondrial dynamics and autophagy? The Int. J. Biochem. Cell Biol. 2009;41:1914–1927. doi: 10.1016/j.biocel.2009.06.006. [DOI] [PubMed] [Google Scholar]
- Winklhofer KF, Haass C. Mitochondrial dysfunction in Parkinson’s disease. Biochim. Biophys. Acta. 2010;1802:29–44. doi: 10.1016/j.bbadis.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Yaffe MP. The Machinery of Mitochondrial Inheritance and Behavior. Science. 1999;283:1493–1497. doi: 10.1126/science.283.5407.1493. [DOI] [PubMed] [Google Scholar]
- Yang W, Li J, Hekimi S. A Measurable increase in oxidative damage due to reduction in superoxide detoxification fails to shorten the life span of long-lived mitochondrial mutants of Caenorhabditis elegans. Genetics. 2007;177:2063–2074. doi: 10.1534/genetics.107.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Hartman PS, Ishii T, Suda H, Akatsuka A, Shoyama T, Miyazawa M, Ishii N. Interrelationships between mitochondrial fusion, energy metabolism and oxidative stress during development in Caenorhabditis elegans Biochem. Biophys. Res. Commun. 2011;404:751–755. doi: 10.1016/j.bbrc.2010.12.017. [DOI] [PubMed] [Google Scholar]
- Zielonka J, Kalyanaraman B. Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation, another inconvenient truth. Free. Radic. Biol. Med. 2010;48:983–1001. doi: 10.1016/j.freeradbiomed.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Designated labels and descriptions of all mitochondrial traits measured in C. briggsae natural isolates. Subscripts N, F, and T denote whether the trait refers to Nonfunctional, Functional, or Total (all) mitochondria. Subscripts P and V indicate that the measure refers to the entire mitochondrial population or the average within-individual variance in that trait, respectively.
Table S2. Spearman’s correlation coefficients between mitochondrial traits measured in C. briggsae natural isolates. (See section 3.1 and Table S1 for trait descriptions.) *,**, and *** indicate statistical significance at the 0.05, 0.01, and 0.001 levels, respectively. Because ROS levels were measured on different sets of nematodes than all other traits and required that we calculate isolate-mean correlations for these pairs of traits, correlations involving ROS (none of which were significantly different from zero) are not included above (N =164–170).