Abstract
We have identified novel nuclear bodies, which we call pearls, in the giant oocyte nuclei of Xenopus laevis and X. tropicalis. Pearls are attached to the lampbrush chromosomes at specific loci that are transcribed by RNA polymerase III, and they disappear after inhibition of polymerase III activity. Pearls are enriched for small Cajal body-specific RNAs (scaRNAs), which are guide RNAs that modify specific nucleotides on splicing snRNAs. Surprisingly, snRNAs themselves are not present in pearls, suggesting that pearls are not functionally equivalent to Cajal bodies in other systems, which contain both snRNAs and scaRNAs. We suggest that pearls may function in the processing of RNA pol III transcripts, such as tRNA, 5S rRNA, and other short non-coding RNAs.
Keywords: Cajal bodies, histone locus bodies, pearls, RNA polymerase III, scaRNAs
Introduction
Because of its large size and wealth of structural detail, the amphibian oocyte nucleus or germinal vesicle (GV) has long been a favorite object for both cytological and molecular studies. In addition to the lampbrush chromosomes (LBCs), the GV contains hundreds to thousands of extrachromosomal bodies that can be classified into a relatively few types based on morphology, molecular composition, and association with specific loci on the LBCs. Three of these – nucleoli, histone locus bodies (HLBs), and speckles – constitute the vast majority of extrachromosomal bodies in Xenopus and each represents a different relationship between the body and the locus or loci with which they are associated. We will not discuss the speckles here, because they have been studied the least (Gall et al. 2004).
Best known and most prominent during much of oocyte development are the hundreds of extrachromosomal nucleoli. They are unique in that they contain amplified copies of rDNA (Brown and Dawid 1968; Gall 1968) and they transcribe their own RNA from this DNA. In both X. laevis and X. tropicalis, there is a single nucleolus organizer locus on one LBC, although for unknown reasons a nucleolus is rarely found there. Nevertheless, it is clear that both the chromosomal and extrachromosomal nucleoli represent hypertrophied gene loci, in that they contain reiterated DNA templates and the transcripts derived from them (Fig. 1A). They are functionally equivalent to LBC loops, and in some salamanders, such as Ambystoma and Plethodon, even take the form of loops or rings (Callan 1966; Kezer and Macgregor 1973).
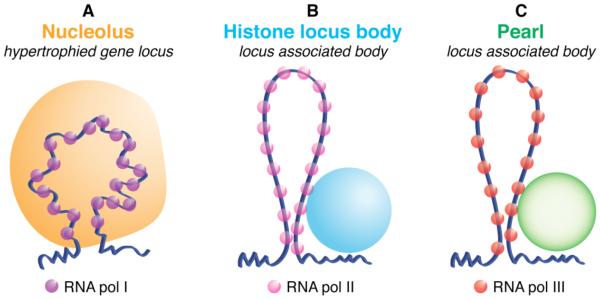
Fig. 1.
Nuclear bodies and their relationship to gene loci. A) The nucleolus contains the entire RNA polymerase I (pol I) machinery for transcribing rRNA and assembling the two subunits of the ribosome. It is a hypertrophied gene locus structurally and functionally equivalent to a LBC loop. B) The histone locus body (HLB) is tightly associated with the histone gene locus, but does not contain histone genes or their transcripts, which are located on LBC loops at this locus. The HLB itself contains factors involved in processing histone transcripts. C) Pearls are located at sites of pol III transcription. However, pol III transcription does not occur in the pearls, but on loops associated with them. In this respect HLBs and pearls are similar. We speculate that factors found in pearls are used for processing pol III transcripts.
HLBs are superficially similar to nucleoli. Like the nucleoli, they occur free in the nucleoplasm and attached to specific loci on the chromosomes, in this case the histone gene loci (Gall et al. 1981; Callan et al. 1991). However, HLBs do not contain the genes coding for histones nor the transcripts derived from them, and in this respect they differ fundamentally from nucleoli (Stephenson et al. 1981; Gall et al. 1983) (Fig. 1B). There has been much confusion about the terminology of HLBs, which over the years have been called spheres (Callan and Lloyd 1960), coiled bodies (Gall et al. 1995), Cajal bodies (Gall et al. 1999), and finally HLBs (Nizami et al. 2010). Our lab must take major blame for this confusion, the excuse being that increased knowledge has forced us to distinguish what were thought at first to be the same or similar bodies. The culprit is the protein coilin. Coilin is a protein of unknown function, originally described from the nuclear coiled bodies of mammalian cells (Andrade et al. 1991). Coiled bodies were later renamed Cajal bodies (CBs), and coilin became accepted as a specific marker for CBs. A problem arose when we identified Drosophila coilin and showed that it occurred in two distinctly different bodies (Liu et al. 2009). One of these bodies, the Drosophila CB, shares molecular markers found in CBs of other organisms, particularly snRNAs and the guide RNAs that modify them. These guide RNAs were themselves named small Cajal body specific (sca) RNAs because of their colocalization with coilin in CBs of mammalian cells (Darzacq et al. 2002; Richard et al. 2003). The second coilin-positive body in Drosophila cells is the HLB, which we named from its invariant association with the histone gene locus. In addition to coilin, the Drosophila HLB contains the U7 snRNP and other factors involved in histone pre-mRNA processing (Liu et al. 2006; Godfrey et al. 2009; White et al. 2011).
From our studies on Drosophila it became clear that the coilin-positive bodies we had called CBs in the amphibian GV were, in fact, HLBs (Nizami et al. 2010). Indeed, their association with the histone loci had been recognized decades earlier (Gall et al. 1981; Callan et al. 1991). With the major coilin-positive bodies now recognized as HLBs, the question arises whether there are CBs at all in the amphibian GV. Here we describe novel coilin-positive bodies from the GV of X. laevis and X. tropicalis, which we call pearls. In addition to coilin, pearls contain scaRNAs and WDR79, a protein known to associate with scaRNAs in other cell types (Tycowski et al. 2009; Venteicher et al. 2009), at first suggesting that pearls are the “real” CBs in the GV. However, unlike CBs in other cell types, pearls do not contain splicing snRNAs. Moreover, they are specifically associated with RNA polymerase (pol) III loci on the chromosomes (Fig. 1C), a feature not shared with CBs in other organisms. For these reasons we refer to pearls as “CB-like” organelles. Here we describe the molecular composition of pearls, their association with the LBCs, their dependence on pol III transcription, and their dynamic behavior.
Materials and Methods
Oocytes, oocyte injections, and GV spreads
Pieces of ovary were surgically removed from X. tropicalis and X. laevis adult females and kept in OR2 medium (Wallace et al. 1973) at room temperature (~22° C). Oocytes remain in usable condition for up to one week. For injection of RNA and DNA constructs, precise volumes were injected into oocytes using the Nanoject semi-micro injector (Drummond). GV spread preparations were made from mature oocytes (~0.8 mm diameter in X. tropicalis and ~1.2 mm in X. laevis) as described previously (Gall and Wu 2010). Spreads were fixed in 2-4 % paraformaldehyde for 20 min-1 h before being immunostained for 1 h each in primary and secondary antibodies.
Whole mount immunostaining and fluorescent in situ hybridization (FISH)
Immunostaining and FISH for Xenopus whole-mount tissues were performed as described (Liu et al. 2009). Briefly, a whole ovary was removed from 3-5 cm X. tropicalis or X. laevis froglets and fixed in 4% paraformaldehyde in OR2 for 10 min. Tissue was stored long-term in phosphate-buffered saline (PBS) at 4° C. Antibody staining was performed overnight at room temperature in 10% horse serum. FISH was carried out in in situ mix at 42° C for 4-18 hr. In situ mix consists of 50% formamide, 5X SSC, 50 μg/mL heparin, 500 μg/mL yeast tRNA, 9 mM citric acid pH6, 0.1% Tween-20.
Antibodies
Primary antibodies were as follows: rabbit polyclonal serum C236 against X. laevis coilin (from Zheng’an Wu) used at 1:500-1:1000 on GV spreads; mouse mAb H1 against X. laevis coilin (Tuma et al. 1993), used at 1:200 on small pieces of whole ovary; mouse mAb against human symplekin (BD Transduction Laboratories) used at 1:1000; affinity purified rabbit polyclonal serum against Drosophila melanogaster FLASH, used at 1:1000 (Yang et al. 2009), a gift from Z. Dominski; mouse mAb 72B9 against fibrillarin (Reimer et al. 1987), used at 1:5-1:10; rat mAb 3F10 against the hemagglutinin (HA) tag (Roche) used at 1 μg/mL; rabbit polyclonal serum against RPB6 used at 1:5,000-1:10,000, a gift from Robert Roeder; mouse mAb Y12 against symmetric dimethylarginine (sDMA), used at 1:25; mouse mAb K121 against the trimethylguanosine (TMG) cap on snRNAs (Oncogene Science), used 1 μg/mL. Secondary antibodies were goat or donkey anti-rabbit, anti-mouse, or anti-rat labeled with Alexa 488 or Alexa 594 (Invitrogen).
Clones and in vitro transcribed RNA
Clones used for making antisense in situ hybridization probes were X. laevis U3 snoRNA in pBluescript (from Rocco Savino) and X. laevis U85 scaRNA in pCRII (cloned by Christine Murphy from genomic DNA). Clones for making sense transcripts for injection were X. laevis U92 scaRNA (pugU2-34/44) in pGEM3Z (Zhao et al. 2002); human mgU2-25/61 scaRNA (Tycowski et al. 2004); GFP X. laevis coilin in pCS2 (Handwerger et al. 2002); X. laevis U7 in pBS (Wu et al. 1996); D. melanogaster mgU2-28 scaRNA wild type and ΔCAB mutant in pGEM-T (Svetlana Deryusheva). DNA encoding X. laevis telomerase RNA was cloned from genomic DNA into the pGEM-T Easy vector following a protocol modified from (Chen et al. 2000), using oligos ZN27 (AAT CAG CGT TTA AAG CTC AAT GTG G, contains T3 RNA polymerase promoter) and ZN28 (ACA TGT CGG GGA CTG GCT GA). The cloned sequence was validated against the NCBI entry for X. laevis telomerase RNA, NR 003556. X. tropicalis WDR79-HA was cloned from Xenopus ovary RNA into the pGEM-T Easy vector. The oligo pair used for cloning included a Kozak sequence in the forward oligo (ZN51: GCC GCC ACC ATG TTG GGC AGA GAG GAG GAA GA) and an HA-tag sequence in the reverse oligo (ZN52: TTT ATT TTT CTA AGC GTA ATC TGG CAC ATC ATA TGG GTA CTC ACG TGC CGC CCC CCC). In vitro transcribed RNA was generated using standard protocols (Gall et al. 1999). In brief, ~ 1 μg plasmid DNA or 100 ng PCR product was used as templates in a 20 μL reaction (1X Stratagene transcription buffer, 1-2 mM NTPs, 40 units of RNase inhibitor, 50 units of Strategene T3/T7/SP6 RNA polymerase). For mRNA, 2.5 mM 7-methyl guanosine cap was included in the reaction (NEB). Non-coding RNAs (scaRNAs, snRNAs and snoRNAs) were directly labeled by including 250 μM fluorescein-UTP (Boehringer Mannheim), Cy3-CTP, Cy5-UTP or Cy5-CTP (GE Healthcare). Template DNA was subsequently digested using DNase I (Roche), which was then heat inactivated and chelated with 25 mM EDTA. RNA was purified using Illustra G-50 microspin purification columns (GE Healthcare). RNAs used for injection/targeting experiments were transcribed from templates that included a T3 or T7 promoter immediately upstream of the 5′ end of the molecule, which minimized the presence of non-genomic sequence elements.
Molecular beacons
Molecular beacons (MBs) for X. laevis U85 scaRNA were generously provided by Salvatore Marras and Sanjay Tyagi (Mhlanga and Tyagi 2006). The probe sequences were as follows: MB Xenopus large 5′ TMR-CCGU CCCUGCCUGGUUUUCCAAU ACGG-BHQ-2 3′; MB Xenopus small #1 5′ TMR-CAGC CCUGCCUGGUUUUC GCUG-BHQ-2 3′; MB Xenopus small #2: 5′ TMR-CGAA CCCUGCCUGGUU UUCG-BHQ-2 3′; MB scrambled control M8.2 5′ TMR-CTTC GTCCACAAACACAACTCCT GAAG – BHQ-2 3′. The underlined sequences are the complementary arm sequences of the MB probes; the non-underlined regions hybridize to target sequences leading to unquenching of the fluorophore (TMR = tetramethylrhodamine, BHQ-2 = Black Hole Quencher 2). MBs were injected into mature X. laevis oocytes either into the cytoplasm (23 nL at 50 ng/μL) or into the GV (4.6 nL at 5 ng/μL). Cytoplasmic injection worked as efficiently as GV injection and was used for most experiments.
Western blots
Extracts of GV proteins were prepared by first isolating intact GVs in cold isolation medium (83 mM KCl, 17 mM NaCl, 6.5 mM Na2HPO4, 3.5 mM KH2PO4, 1mM MgCl2, 1mM dithiothreitol). GVs were then dissolved in standard protein sample buffer (0.35M Tris, pH 6.8, 30% glycerol, 10% sodium dodecyl sulfate, 0.6M dithiothreitol, 0.012% bromophenol blue). Samples were boiled before loading onto a 12% polyacrylamide resolving gel. After electrophoresis, the proteins were transferred to a polyvinylidene difluoride membrane and HA-tagged proteins were detected with rat mAb 3F10 against the HA tag (Roche) used at 50 ng/mL, followed by horseradish peroxidase-labeled antibody against rat IgG (Jackson) used at 1:40,000 for 1 h. Chemiluminescence was generated by an ECL reagent (Pierce SuperSignal ECL) and the blot was exposed to film for 20 sec-5 min. “Rainbow” marker (GE Healthcare RPN 800E) was used for molecular weight calibration.
Fluorescence recovery after photobleaching (FRAP)
Mature X. tropicalis oocytes were injected with 9.2 nL of GFP-labeled X.l. coilin mRNA (~50 pg). Two days later, individual GVs were isolated in oil just prior to each FRAP experiment. FRAP was performed on a single pearl, followed by FRAP on an HLB from the same GV. FRAP was conducted on a Leica SP5 laser-scanning confocal microscope using the FRAP Wizard program. The protocol was as follows. Scanning was conducted at 400 Hz with a line average of 2 with a 63X oil objective at optical zoom 4. The argon laser power was set to 30% and the 488 nm laser intensity to 20%. The scanning order was: pre-bleach; bleach (the region of interest (ROI) was a 10 μm circle around the body, scanned 5 times at higher laser intensity); post bleach 1 (2 scans at the fastest possible setting, 2.62 sec/frame); post bleach 2 (4 scans at 15 sec/frame); post bleach 3 (30 scans at 60 sec/frame). Images were exported as TIFF files and processed with Image J software (Rasband, W.S., http://imagej.nih.gov/ij/). The mean fluorescence intensity of each frame was normalized by comparing two ROIs of the same size, one with a body and one with background only. Graphs were generated in Microsoft Excel.
Drug treatments
For inhibition of RNA pol III, oocytes were injected with either Tagetin (Epicentre) or α-amanitin (Sigma). Tagetin was injected into 0.8 mm oocytes to a final concentration of 5-50 μM (23 nL of 60-600μM Tagetin). This corresponds to the concentration range (~ 3-10 μM) that specifically inhibits RNA pol III but not RNA pol II activity on reporter constructs in X. laevis oocytes (Steinberg et al. 1990). We could monitor the appropriate concentration by visual inspection of LBC loop morphology – the vast majority of loops are transcribed by RNA pol II and they contract or disappear entirely when RNA pol II is inhibited. To inhibit both RNA pol II and RNA pol III with α-amanitin, we injected 4.6 ng of drug into each oocyte (23 nL of 200 μg/mL α-amanitin). This is 1000X more than is needed to inhibit RNA pol II (Steinberg et al. 1990). GVs were isolated from drug-treated oocytes 1-2 days after injection.
Results
Pearls are novel coilin-positive structures in the Xenopus germinal vesicle
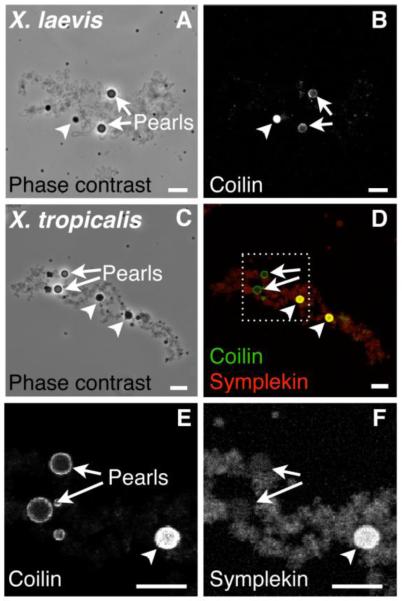
The morphology, composition and chromosomal position of nuclear bodies can be determined from GV spread preparations (Gall and Wu 2010). Using this technique for X. tropicalis and X. laevis GVs, we have identified a novel class of nuclear body that we term pearls. Pearls are physically attached to LBCs at specific loci and are named after their distinct appearance in the phase-contrast microscope. They are perfectly spherical, highly refractile, and have a shell-like structure reminiscent of cultured pearls (Fig. 2). Compositionally, there is no unique marker for pearls, but they are enriched for coilin and can be distinguished from other nuclear bodies by using multiple markers. HLBs in GVs from mature oocytes are enriched for both coilin and histone pre-mRNA processing components, whereas pearls contain coilin but not the processing components (Fig. 2). Coilin does not label any other bodies in the GV. Furthermore, because of their shell-like spherical structure, pearls can usually be distinguished from HLBs by immunostaining for coilin alone; pearls have a ring-like appearance in optical section, whereas HLBs appear as uniformly labeled circles (Fig. 2E).
Fig. 2.
Pearls are novel coilin-positive bodies attached to specific loci on lampbrush chromosomes. (A-D) Single LBCs in GV spreads from mature oocytes of X. laevis (A-B) and X. tropicalis (C-D). (A and C) Phase contrast images show that pearls are phase-refractile bodies. (B) Antibody stain of coilin, which has a shell-like localization in pearls and a uniform label in HLBs. (D) Antibody stain of coilin (green) and an HLB component, symplekin (red). (E-F) Enlargement of boxed region in panel D, separated into single color channels for coilin (E) and symplekin (F). Arrows point to pearls, arrowheads to HLBs. Bar = 10 μm.
Pearls are consistently present in early diplotene stage Xenopus oocytes
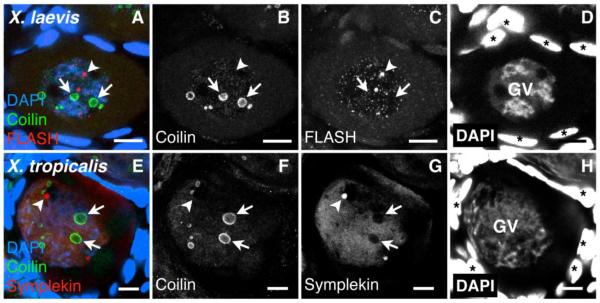
We detected pearls in the early stages of oocyte development by immunostaining fragments of whole ovaries from juvenile frogs. These ovaries contain immature yolk-less oocytes that allow for direct visualization of the intact GV. In immature oocytes of both X. tropicalis and X. laevis, an antibody against coilin labels ring-like spherical bodies that resemble pearls and are distinct from HLBs (Fig. 3). In contrast to later stages, coilin is not present in HLBs of young oocytes (Nizami et al. 2010), and is thus a unique marker for pearls (Fig. 3B, F). Pearls are consistently present in immature oocytes in the Xenopus ovary (14/14 X. laevis juveniles from several different broods). In addition, they have a stereotypic distribution within the nucleus during oocyte development (Supplemental Fig. 1). They first appear in early diplotene of meiosis, when the chromosomes enter the lampbrush stage (~50 μm diameter X. laevis oocyte, Supplemental Fig. 1C). As both the GV and the oocyte begin to grow, the nucleoplasmic coilin level increases, with a gradual loss of coilin from the pearls (100 μm – 150 μm X. laevis oocytes, Supplemental Fig. 1D, E). We conclude that pearls are a consistent feature of GVs in early-stage Xenopus oocytes.
Fig. 3.
Pearls in pre-vitellogenic Xenopus oocytes. Whole mounts of immature oocytes from X. laevis (A-D) and X. tropicalis (E-H). (A and E) Immunostaining shows that coilin (green) and HLB component FLASH (red) are not colocalized. DAPI stains DNA (blue). (B-D and F-H) Single channel images. Arrows point to pearls, arrowheads to HLBs. Asterisks indicate follicle cell nuclei (DAPI overexposed). Bar = 10 μm.
Pearls contain U85 scaRNA and U3 snoRNA
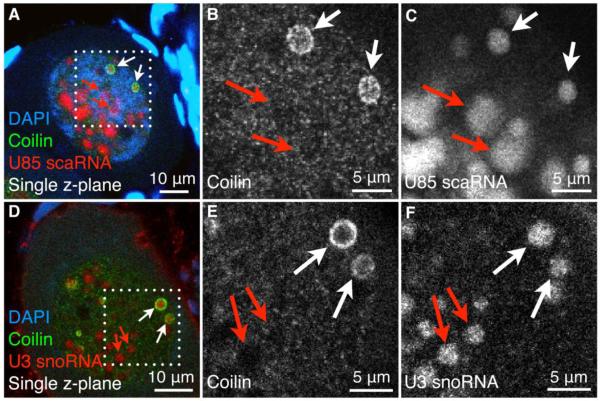
Because pearls contain coilin, a canonical CB component, we asked whether they contain other typical CB components. We first examined U85 scaRNA, a guide RNA for the U5 splicing snRNA. U85 and other scaRNAs were named because of their high concentration in CBs of mammalian tissue culture cells (Darzacq et al. 2002). Using fluorescent in situ hybridization (FISH) on whole immature X. laevis oocytes we found that pearls contain U85 (Fig. 4A-C). Unexpectedly, we also observed U85 localization in nucleoli (Fig. 4A, C), an issue to which we will return later.
Fig. 4.
Endogenous U85 scaRNA and U3 snoRNA are localized in pearls and nucleoli. Whole mounts of immature oocytes from X. laevis. (A-C) Immunostaining for coilin (green) followed by FISH for U85 scaRNA (red, antisense probe) and DAPI stain for DNA (blue). (D-F) Immunostaining for coilin (green) followed by FISH for U3 snoRNA (red, antisense probe) and DAPI stain for DNA (blue). White boxes outline the regions that are magnified in the gray-scale panels. White arrows point to pearls, red arrows to nucleoli. The GV has numerous spherical and irregularly shaped nucleoli resulting from extrachromosomal amplification of the rDNA.
The key molecular feature that distinguishes scaRNAs from snoRNAs is the presence of a short motif (CAB box) on scaRNAs (Richard et al. 2003), which is recognized by the CB-targeting protein, WDR79 (Tycowski et al. 2009; Venteicher et al. 2009). It is generally believed that snoRNAs are absent from CBs because they lack the CAB box. Instead, snoRNAs concentrate in nucleoli, with the well-established exception that U3 snoRNA is present in both nucleoli and CBs in mammalian cells (Verheggen et al. 2002). We therefore used a FISH probe for U3 on whole immature oocytes of X. laevis. We observed that U3 is enriched in both nucleoli and pearls (Fig. 4D-F). Both U3 snoRNA and U85 scaRNA occur in the core region of pearls and are excluded from the shell-like surface (Fig. 4).
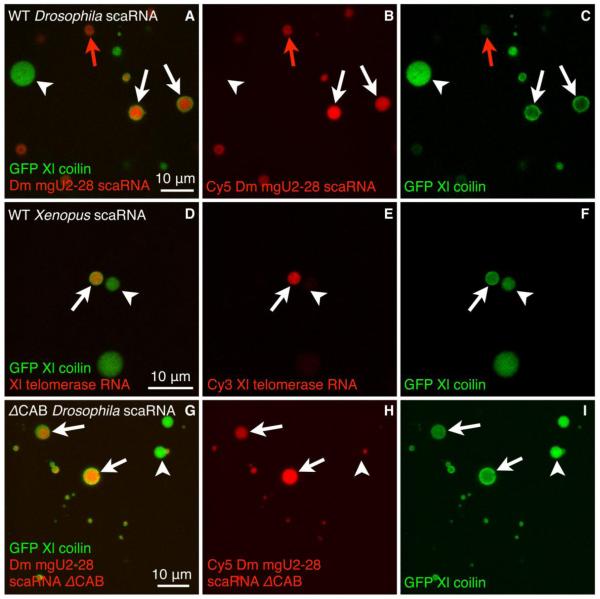
Because endogenous U85 scaRNA occurs in both pearls and nucleoli of early-stage oocytes, we tested whether scaRNAs target to these compartments when exogenously introduced into mature oocytes. We co-injected an in vitro transcribed, fluorescently labeled Drosophila scaRNA (Dm mgU2-28) and GFP-X. laevis coilin mRNA into the cytoplasm of mature X. tropicalis oocytes (stage V). We then isolated intact GVs in oil to analyze the intensity of the fluorescence signals. Injected Drosophila mgU2-28 scaRNA targeted to pearls and nucleoli, but was excluded from HLBs (Fig. 5A-C). We could distinguish pearls from HLBs, because coilin targets to HLBs uniformly, whereas it targets to the outer shell of pearls (Fig. 5C). Furthermore, the scaRNA preferentially targets to the core of the pearls (Fig. 5A), mimicking the endogenous pattern of U85 scaRNA and U3 snoRNA in immature oocytes (Fig. 4D). We found a similar targeting pattern (to pearls and nucleoli but not HLBs) for another scaRNA, X. laevis telomerase RNA (Fig. 5D-F). Furthermore, we observed targeting to nucleoli for multiple additional scaRNAs (Supplemental Fig. 2).
Fig. 5.
The CAB motif is dispensable for scaRNA localization to pearls and nucleoli. Confocal images of X. tropicalis GVs isolated in oil ~ 5 hours after oocytes were co-injected with X. laevis GFP-coilin mRNA (green) and the following fluorescently-labeled scaRNAs (red): (A-C) Drosophila mgU2-28 scaRNA; (D-F) X. laevis telomerase RNA; (G-I) Drosophila mgU2-28 scaRNA lacking the CAB motif (ΔCAB). White arrows point to pearls (ring-like coilin), white arrowheads to HLBs (uniform coilin), and red arrows to nucleoli (dense fibrillar component, verified by examination in DIC channel). Both wild-type and ΔCAB mutant scaRNAs target to pearls and nucleoli, but not to HLBs. Bar = 10 μm.
To exclude the possibility that the nucleolar localization of the injected scaRNAs is an artifact of overloading, we examined the endogenous localization of X.l. U85 scaRNA by injecting a “molecular beacon” probe into mature X. laevis oocytes. Molecular beacons are labeled hairpin constructs that fluoresce only when they hybridize to their target nucleic acid (Mhlanga and Tyagi 2006). Nucleoli of mature oocytes were labeled by the U85 molecular beacon, confirming the presence of this scaRNA (Supplemental Fig. 2H), as we had observed for immature oocytes by in situ hybridization (Fig. 4A-C).
The CAB motif is dispensable for scaRNA targeting to pearls and nucleoli
To determine a possible role of the CAB box in targeting scaRNAs to pearls and nucleoli in the amphibian oocyte, we prepared a Drosophila mgU2-28 construct that lacks the CAB box. We then injected labeled RNA transcribed from this construct into X. tropicalis oocytes. We found that there was no change in the targeting pattern for the CAB deletion mutant, which concentrated in pearls (Fig. 5G-I) and nucleoli (not shown) like the original construct. We conclude that scaRNAs in the Xenopus GV occur in both pearls and nucleoli, and that their localization is not dependent on the CAB motif.
The CAB-binding protein WDR79 targets to pearls and HLBs
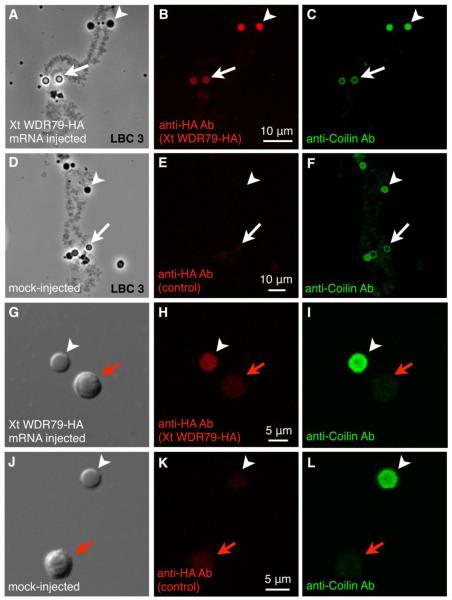
To further understand scaRNA localization in the Xenopus GV and particularly the role of the CAB motif, we examined the localization of the CAB-binding protein, WDR79. We could not assess the distribution of endogenous WDR79 protein because commercially available anti-human WDR79 antibodies did not label any structures in the Xenopus GV, nor did they bind X.t. WDR79 protein on a western blot. As an alternative, we injected HA-tagged X.t. WDR79 mRNA into the cytoplasm of X. tropicalis oocytes, where it was translated into a protein of appropriate size and localized in the GV (Supplemental Fig. 3B). X.t. WDR79 specifically targeted to pearls and HLBs, but not to nucleoli or other nuclear compartments (Fig. 6). Thus, WDR79 targets to a body that does not contain scaRNAs (HLBs) and to only one of the two bodies that do contain scaRNAs (pearls, not nucleoli). These results further suggest that localization of scaRNAs in Xenopus GVs is not governed entirely by WDR79 binding to the CAB motif.
Fig. 6.
X. tropicalis scaRNA-binding protein WDR79 targets to pearls and HLBs but not to nucleoli. GV spreads from X. tropicalis oocytes injected with either X. tropicalis WDR79-HA mRNA (A-C, G-I) or water (D-F, J-L). Preparations were immunostained with antibodies against the HA-tag (red) and X.l. coilin (green). (A-C) LBC 3 with attached HLBs (arrowheads) and pearls (arrows). X.t. WDR79-HA protein is localized specifically in HLBs and pearls. (D-F) LBC 3 from mock-injected oocyte shows no red label (HA-tag) in HLBs or pearls. (G-I) The HLB contains injected X.t. WDR79-HA protein (red) and endogenous coilin (green). The nucleolus is not labeled. (J-L) In the mock-injected oocyte there is no HA-tag (red). The HLB can be identified by its endogenous coilin (green). A, D, G, and J are phase contrast images. White arrows point to pearls, white arrowheads to HLBs, red arrows to nucleoli.
Pearls lack splicing snRNPs
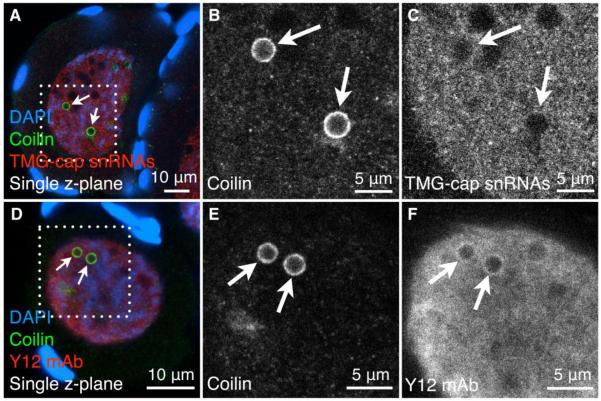
To continue the comparison of pearls to CBs, we immunostained whole immature X. laevis oocytes with antibodies that recognize two different components of splicing snRNPs: mAb K121, which recognizes the trimethylguanosine cap on the snRNAs themselves (Krainer 1988); and mAb Y12, which recognizes an epitope found on several snRNP proteins, specifically symmetric dimethylarginine (Lerner et al. 1981; Brahms et al. 2000). We found that splicing snRNPs are enriched in the nucleoplasm, but are excluded from pearls and nucleoli (Fig. 7). To verify the absence of splicing snRNAs from pearls, we used FISH probes for U2 and U4 snRNA on immature oocytes from X. laevis. We saw localization of the probes primarily in the nucleoplasm (Supplemental Fig. 4). In some oocytes, we observed enrichment of snRNAs in amorphous masses that could represent the matrix of particularly prominent LBC loops (Wu et al. 1991). We conclude that splicing snRNPs are abundant in early stage GVs but are absent from pearls.
Fig. 7.
Splicing snRNPs are excluded from pearls. Whole mounts of immature oocytes from X. laevis. (A-C) Immunostaining for coilin (green) and snRNAs (red, mAb K121, which recognizes the TMG-cap). DAPI stains DNA (blue). (D-F) Immunostaining for coilin (green) and snRNP proteins (red, mAb Y12, which recognizes the symmetric dimethylarginine epitope found on at least some snRNP proteins). DAPI stains DNA (blue). White boxes outline the regions that are magnified in the gray-scale panels. White arrows point to pearls, which do not contain splicing snRNPs in their core region. There is a slight enrichment of mAb Y12 in the outer shell of pearls (F). This probably does not represent assembled snRNPs, because a similar pattern is not seen for the TMG-cap (C)
Live dynamics of coilin in pearls
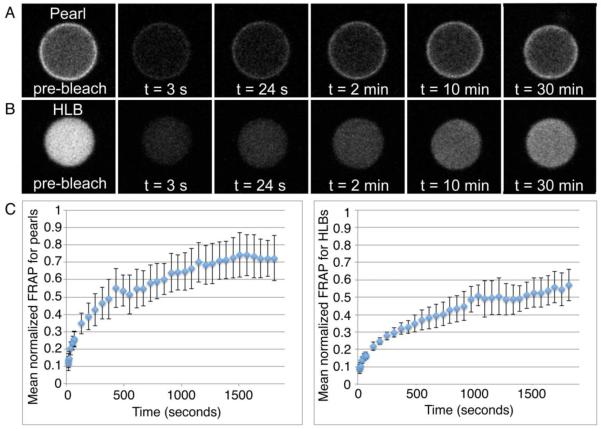
To determine whether pearls are static storage organelles or dynamic steady-state structures, we injected GFP-tagged X. laevis coilin mRNA into X. tropicalis oocytes, removed the GV under oil, and measured the fluorescence recovery after photobleaching (FRAP) (Fig. 8). In each GV we bleached one pearl, which we distinguished from HLBs based on the shell-like localization of coilin. We found that the coilin in pearls recovers after photobleaching, reaching a plateau of approximately 70% recovery (Fig. 8A, C). For each pearl that we bleached, we also bleached an HLB from the same GV to control for the integrity of the GV and the proper activity of the GFP-X.l. coilin protein. We found that coilin in HLBs reached a recovery plateau on a time scale similar to that of coilin in pearls, albeit at a slightly lower normalized level (approximately 60% instead of 70%, Fig. 8B, C). Furthermore, the recovery time for coilin in pearls and HLBs in X. tropicalis corresponded well to values from previous FRAP experiments for multiple HLB markers in X. laevis (Handwerger et al. 2003).
Fig. 8.
FRAP experiments demonstrate that pearls are dynamic nuclear structures. X. tropicalis oocytes were injected with GFP-labeled X.l. coilin mRNA. After at least 4h and up to 48h, GVs were isolated in oil. Pearls and HLBs both accumulated labeled coilin. (A) Representative time course of FRAP on a single pearl. (B) Representative time course of FRAP on an HLB from the same GV. (C) Mean normalized FRAP curves for bleached pearls (left) and HLBs (right). FRAP was performed on a pearl first, and then an HLB from the same GV. Error bars are standard deviations. For pearls, each point on the FRAP curve is the mean fluorescence intensity for 7 complete time courses from independent GVs, and 7 partial time courses that were stopped because the body went out of focus. For the HLBs, each point on the FRAP curve is the mean fluorescence intensity for 6 complete time courses from independent GVs, and 2 additional partial time courses. Four different frogs were sampled during these experiments on 4 different days.
We also observed some fusion events of pearls-to-pearls and HLBs-to-HLBs (Supplemental Fig. 5). Fusion events have been reported previously for several organelles that lack investing membranes, such as CBs, nucleoli, and P granules (Boudonck et al. 1999; Platani et al. 2000; Brangwynne et al. 2009; Brangwynne et al. 2011). Interestingly, fusing pearls could be found in physical contact with an HLB. This indicates that unlike bodies can be physically associated without fusing, but when like bodies touch, a fusion event can follow. We conclude that pearls are dynamic steady-state structures that exchange components with the nucleoplasm and can fuse with other pearls.
Pearls map to specific gene loci on X. tropicalis lampbrush chromosomes
To identify a potential function for pearls based on association with specific gene loci, we mapped the attachment sites of pearls on LBCs of X. tropicalis. We found that pearls are attached to specific loci on LBC 3, LBC 4 and LBC 10 (Supplemental Fig. 6). We did not observe extra-chromosomal pearls. Although pearls occur at specific and reproducible positions on LBCs, they are not invariably present at these loci in mature GVs (Supplemental Fig. 7A). Furthermore, the frequency of attachment differs among the different LBCs. Pearls are attached almost invariably at a specific locus on LBC 10 (95% of GVs), but occur less frequently and with greater variability at two prominent loci on LBCs 3 and 4 (~ 60% of GVs, with a large standard deviation). There are additional unmapped loci in frogs that have pearls on LBCs 3 and 4 (e.g., frogs 5 and 6, Supplemental Fig. 7B).
Pearls are dependent on RNA polymerase III transcription
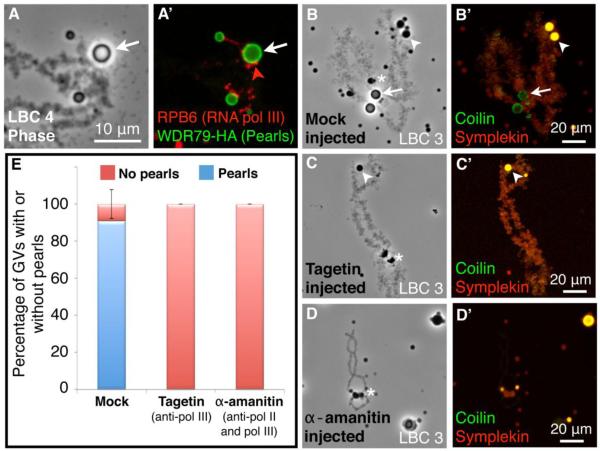
The fact that pearls occupy specific loci on several LBCs suggests that these loci either contain repeated sequences or share some activity in common. Because LBC maps do not provide sequence-level resolution of pearl attachment sites, we used a cytological approach to narrow down the functional class of gene(s) with which pearls are associated. Antibodies against RNA polymerase III (pol III) subunits were previously used to map approximately 90 RNA pol III loci on LBCs in X. laevis (Murphy et al. 2002). We now find that pearls occur only at pol III loci in both X. tropicalis and X. laevis mature oocytes (Fig. 9A, A’ and Supplemental Fig. 8A-D). Pol III is not in the pearls themselves but on short loops immediately adjacent to them (Fig. 9A, A’). Whole mounts of immature oocytes also show association of pearls with pol III (Supplemental Fig. 8B, D).
Fig. 9.
Pearls disappear from RNA pol III loci when transcription is inhibited. (A and A’) A LBC from a mature X. tropicalis oocyte injected with X.t. WDR79-HA mRNA to label pearls (anti-HA antibody, green) and immunostained for a pol III subunit, RPB6 (red). Pearls occur only at loci that are labeled with RNA pol III. (B-B’,C-C’,D-D’) Images of X. tropicalis LBC 3. Left panels are phase contrast images, right panels are confocal images of antibody stain for coilin (green, in pearls and HLBs) and symplekin (red, in HLBs and LBC loops). White arrows point to pearls on the pearl locus, white arrowheads to HLBs at the histone gene locus, and white stars to “lumpy loops” that were used to identify LBC 3. (B-B’) Mock injected oocyte. (C-C’) Oocyte injected with RNA pol III-specific drug inhibitor, Tagetin. LBCs lose pearls but retain RNA pol II loops that label with symplekin. (D-D’) Oocyte injected with a high concentration of alpha-amanitin (200 μg/mL), which inhibits both RNA pol II and RNA pol III transcription. LBCs are stripped of all loops and also lose pearls. (E) Mean loss of pearls from GVs after inhibition of RNA pol III transcription. A high percentage of GVs from mock-injected oocytes had at least one pearl (~85%, n = 43 GVs), whereas none of the GVs from Tagetin-injected oocytes (n = 25 GVs) or α-amanitin-injected oocytes (n = 26 GVs) had pearls. Error bars are the standard deviations between frogs (n = 4) for each experimental condition.
To test whether pearl maintenance is dependent on ongoing pol III transcription, we injected mature X. tropicalis oocytes with two commonly used inhibitors of pol III activity: tagetitoxin (commercially available as Tagetin) and α-amanitin. Tagetin specifically inhibits pol III transcription in various eukaroyotes, including X. laevis oocytes (Steinberg et al., 1990). Unlike Tagetin, α-amanitin does not specifically inhibit pol III. Instead, it inhibits pol II at low concentration (~1 μg/mL) and both pol II and pol III at high (~100 – 200 μg/mL). We injected X. tropicalis oocytes with water (mock injection), Tagetin, or α-amanitin. One day later we isolated individual GVs and scored for the presence or absence of pearls, counting only those GV spreads in which all 10 chromosomes could be identified. For these experiments we used frogs that displayed a high frequency of pearls before injection. 85% of mock-injected oocytes had pearls at one or more of the pearl loci, whereas none of the oocytes injected with Tagetin or α-amanitin had pearls (Fig. 9E). The appearance of the LBCs after drug treatment confirmed the different specificities of the two drugs. Tagetin-injected oocytes had normal sized pol II loops that stained with the pre-mRNA processing component symplekin (Fig. 9C, C’). In contrast, oocytes injected with α-amanitin had contracted LBCs that were stripped of loops, a hallmark of pol II inhibition (Fig. 9D, D’). We conclude that pearls are dependent on ongoing RNA polymerase III transcription.
Discussion
Pearls are novel nuclear bodies in Xenopus GVs at RNA polymerase III loci
We have identified pearls as a novel class of nuclear bodies in the Xenopus GV. They are attached specifically to loci that are transcribed by pol III, and they disappear after inhibition of pol III activity. We suggest that pearls may function in processing pol III transcripts, which include 5S ribosomal RNA, tRNAs, 7SK RNA, 7SL RNA and U6 snRNA (Paule and White 2000). Because the X. tropicalis genome has been sequenced and genetically mapped to 10 chromosomes (Kochan et al. 2003; Khokha et al. 2009; Hellsten et al. 2010; Wells et al. 2011), it should eventually be possible to use in situ hybridization to identify the genetic loci at which pearls occur.
Pol III activity has been previously linked to the perinucleolar compartment (PNC) in mammalian cell lines (Matera et al. 1995; Wang et al. 2003; Pollock and Huang 2010). It will be instructive to compare pearls and the PNC, since much is known about the composition and dynamics of the PNC. There is also an intriguing correlation between PNC prevalence and malignant tumors (Norton et al. 2008). There are, however, differences between the PNC and pearls: unlike the PNC, pearls are not known to contain pol III transcripts. Moreover, they contain scaRNAs and coilin, which are absent from the PNC.
Pearls are CB-like bodies
CBs were originally defined by the presence of the protein coilin (Andrade et al. 1991; Raska et al. 1991). Moreover, coilin is essential for the integrity of CBs in organisms as evolutionarily divergent as Arabidopsis, Drosophila, Danio and mice (Tucker et al. 2001; Collier et al. 2006; Liu et al. 2009; Strzelecka et al. 2010). Because pearls contain coilin, we considered the possibility that pearls are the oocyte equivalent of CBs in other tissues (summarized in Table 1). Similarities between pearls and CBs include composition (coilin, WDR79, U3 snoRNA and scaRNAs), spherical morphology and steady-state dynamics. However, there are two key dissimilarities: the lack of splicing snRNPs in pearls, and the lack of specificity of scaRNAs for pearls. The composition of pearls is thus incompatible with the proposed functional role for CBs in spliceosome assembly (Darzacq et al. 2002; Schaffert et al. 2004; Stanek and Neugebauer 2004). We conclude that pearls are not functionally equivalent to CBs from other systems, and have adopted the term “CB-like” to describe them. Non-canonical CB-like structures also occur in the GVs of pigeon oocytes. These bodies contain coilin and have a shell-like structure similar to that of pearls. However, they differ from pearls in containing splicing snRNPs and lacking fibrillarin (the enzymatic component of box C/D sca/snoRNPs) (Khodyuchenko et al. 2012). In total, there are at least four prominent coilin-positive bodies in different systems: CBs, HLBs, pearls, and the pigeon GV bodies. Thus coilin can associate with a variety of RNP complexes to form macromolecular assemblies with similar physical properties and dynamics but with different molecular composition and function.
Table 1.
Comparison of pearls with other nuclear organelles. Pearls differ from CBs by their lack of splicing snRNPs and their invariant association with pol III loci. Oocyte nucleoli contain scaRNAs, unlike somatic nucleoli. Literature references for each characteristic are included within the body of this paper, except for the association of CBs with snRNA loci (Frey and Matera 1995; Smith et al. 1995).
| Component or property |
Xenopus GV pearls |
Xenopus GV nucleoli |
CBs | Nucleoli | HLBs |
|---|---|---|---|---|---|
| Coilin | + | - | + | - | + |
| WDR79 | + | - | + | - | + |
| scaRNAs | + | + | + | - | - |
| U3 snoRNA | + | + | + | + | - |
| Splicing snRNPs | - | - | + | - | - |
| Dynamic body | + | + | + | + | + |
| Locus association | RNA pol III loci |
rDNA locus |
snRNA gene loci* |
rDNA locus |
Histone gene locus |
scaRNA localization in pearls and nucleoli of the Xenopus GVs
The localization of scaRNAs in the Xenopus GV is unusual. Not only are they found in pearls but they also occur in nucleoli and can be targeted to nucleoli after exogenous injection. These results are inconsistent with the generally accepted model of scaRNA localization, according to which the CAB box of scaRNAs targets them specifically to CBs (Richard et al. 2003; Tycowski et al. 2009). Earlier observations on scaRNA localization in the GV are also puzzling. Telomerase RNA, a non-guide scaRNA, targeted to nucleoli in Xenopus GVs (Narayanan et al. 1999). However, no specific localization was found in Xenopus GVs for a guide scaRNA that modifies U2 snRNA (Zhao et al. 2002). Furthermore, there are data suggesting that splicing snRNAs can be post-transcriptionally modified in the nucleoli of Xenopus GVs (Ganot et al. 1999; Yu et al. 2001).
If the Xenopus GV does not, in fact, contain a functional CB, then the unusual distribution of scaRNAs can be partially explained by the lack of their usual target. Under these circumstances scaRNAs might localize by default to nucleoli due to the inherent nucleolar-localization motifs present in box C/D and H/ACA RNAs. However, the lack of a canonical CB cannot explain all of the localization data. We suggest that the CAB box and its binding protein WDR79 are not the only factors that determine how scaRNAs reach their ultimate targets in the nucleus. In particular, we find that elimination of the CAB motif from scaRNAs has no effect on their localization in the GV. Furthermore, there is a discrepancy between the localization of scaRNAs in Xenopus GVs (pearls and nucleoli, but not HLBs) and the localization of WDR79, the protein that binds the CAB box (pearls and HLBs, but not nucleoli). Relevant to this issue, a recent study shows that human WDR79 binds to a CAB box on small Alu RNA transcripts. These Alu transcripts are structurally very similar to H/ACA scaRNAs, but they localize to the nucleoplasm, not to CBs (Jady et al. 2012). These results suggest that RNA complexes that contain WDR79 have novel functions unrelated to post-transcriptional modification of splicing snRNAs. In this light, we speculate that the colocalization of scaRNAs and WDR79 in pearls could reflect a role for scaRNAs in regulating pol III transcripts.
Concluding remarks
Although the molecular composition of pearls suggests a relationship to CBs in other cell types, and hence to snRNP biogenesis, we believe that the similarities between pearls and CBs may be somewhat misleading. Instead, we suggest that the association of pearls with pol III loci is the key to understanding their molecular function(s). In this respect it is more useful to compare pearls with HLBs, not because of similarities in composition, which are minimal, but because of the relationship of the body to the locus at which it is attached (Fig. 1). HLBs were named from their association with the histone gene locus (Liu et al. 2006). After their initial identification in Drosophila, it became clear that similar bodies occur at the histone gene loci in other organisms. In particular, we realized that the bodies we had called CBs in the amphibian GV were, in fact HLBs (Nizami et al. 2010). Terminology aside, the two most important features of HLBs in Drosophila and in the amphibian oocyte are the following. First, although attached to the chromosomes at the histone gene loci, they do not contain histone genes or histone transcripts (Stephenson et al. 1981; Gall et al. 1983). Second, they contain specific processing factors that convert the nascent transcripts into mature histone mRNA (Wu and Gall 1993; Bellini and Gall 1998; Abbott et al. 1999). We suggest that pearls may bear the same relationship to pol III loci as HLBs do to the histone gene loci. We have shown that antibodies against pol III stain loops at approximately 90 loci in X. laevis (Murphy et al. 2002) and about half that number in X. tropicalis (unpublished data). Although the pol III loops at a given locus may be intimately associated with a pearl (when present), the body of the pearl does not stain with the pol III antibody. Like HLBs, pearls are associated with specific loci, but probably do not contain transcripts from these loci. If we carry the analogy further, we may suppose that pearls contain the factors required for processing pol III transcripts.
These considerations point the way to further studies on pearls. A high priority will be to identify the specific loci at which they occur. This can be done in principle by bioinformatic analysis, once there is better correlation between the X. tropicalis genetic and mitotic chromosome maps (Kochan et al. 2003; Khokha et al. 2009; Wells et al. 2011), the LBC cytological maps (Penrad-Mobayed et al. 2009) and the annotated genome (Hellsten et al. 2010). Even without information for all pearl sites, some could be identified by in situ hybridization with specific probes for pol III transcripts (Callan et al. 1988). An equally high priority will be to identify additional components of pearls, particularly known pol III processing factors. If pearls do, indeed, contain such factors, then the analogy with HLBs would be complete: they would contain processing factors for transcripts from the gene loci to which they are attached.
Supplementary Material
Acknowledgements
We thank Svetlana Deryusheva (Carnegie Institution) for fluorescently labeled wild type and mutant Drosophila mgU2-28 scaRNAs. The Xenopus U85 scaRNA molecular beacon was generously made for us by Salvatore Marras and Sanjay Tyagi (Public Health Research Institute, New Jersey). The Xenopus U92 scaRNA construct was a gift from Yi-Tao Yu (University of Rochester Medical Center). The human mgU2-25/61 scaRNA construct was a gift from Kazimierz Tycowski (Yale University). Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01 GM33397. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. JGG is American Cancer Society Professor of Developmental Genetics.
Abbreviations
- CAB
Cajal body box
- CB
Cajal body
- EDTA
ethylene diamine tetraacetic acid
- FISH
fluorescent in situ hybridization
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- GV
germinal vesicle
- HA
hemagglutinin
- HLB
histone locus body
- LBC
lampbrush chromosome
- mAb
monoclonal antibody
- MB
molecular beacon
- NTP
nucleotide triphosphate
- OR2
Oak Ridge 2
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- rDNA
ribosomal DNA
- rRNA
ribosomal RNA
- scaRNA
small CB-specific RNA
- sDMA
symmetric dimethylarginine
- snRNA
small nuclear RNA
- SSC
saline-sodium citrate
- tRNA
transfer RNA
- TMG
trimethylguanosine
- UTP
uridine triphosphate
References
- Abbott J, Marzluff WF, Gall JG. The stem-loop binding protein (SLBP1) is present in coiled bodies of the Xenopus germinal vesicle. Mol Biol Cell. 1999;10:487–499. doi: 10.1091/mbc.10.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade LE, Chan EK, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini M, Gall JG. Coilin can form a complex with the U7 small nuclear ribonucleoprotein. Mol Biol Cell. 1998;9:2987–3001. doi: 10.1091/mbc.9.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudonck K, Dolan L, Shaw PJ. The movement of coiled bodies visualized in living plant cells by the green fluorescent protein. Mol Biol Cell. 1999;10:2297–2307. doi: 10.1091/mbc.10.7.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Lührmann R. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J Biol Chem. 2000;275:17122–17129. doi: 10.1074/jbc.M000300200. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci USA. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Dawid IB. Specific gene amplification in oocytes. Oocyte nuclei contain extrachromosomal replicas of the genes for ribosomal RNA. Science. 1968;160:272–280. doi: 10.1126/science.160.3825.272. [DOI] [PubMed] [Google Scholar]
- Callan HG. Chromosomes and nucleoli of the axolotl, Ambystoma mexicanum. J Cell Sci. 1966;1:85–108. doi: 10.1242/jcs.1.1.85. [DOI] [PubMed] [Google Scholar]
- Callan HG, Gall JG, Murphy C. The distribution of oocyte 5S, somatic 5S and 18S + 28S rDNA sequences in the lampbrush chromosomes of Xenopus laevis. Chromosoma. 1988;97:43–54. [Google Scholar]
- Callan HG, Gall JG, Murphy C. Histone genes are located at the sphere loci of Xenopus lampbrush chromosomes. Chromosoma. 1991;101:245–251. doi: 10.1007/BF00365156. [DOI] [PubMed] [Google Scholar]
- Callan HG, Lloyd L. Lampbrush chromosomes of crested newts Triturus cristatus (Laurenti) Philos Trans R Soc Lond B Biol Sci. 1960;243:135–219. [Google Scholar]
- Chen J-L, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- Collier S, Pendle A, Boudonck K, van Rij T, Dolan L, Shaw P. A distant coilin homologue Is required for the formation of Cajal bodies in Arabidopsis. Mol Biol Cell. 2006;17:2942–2951. doi: 10.1091/mbc.E05-12-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. Differential synthesis of the genes for ribosomal RNA during amphibian oogenesis. Proc Natl Acad Sci USA. 1968;60:553–560. doi: 10.1073/pnas.60.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Diaz MO, Stephenson EC, Mahon KA. The transcription unit of lampbrush chromosomes. In: Subtelny S, Kafatos F, editors. Gene Structure and Regulation in Development. Alan R. Liss; New York: 1983. pp. 137–146. [Google Scholar]
- Gall JG, Stephenson EC, Erba HP, Diaz MO, Barsacchi-Pilone G. Histone genes are located at the sphere loci of newt lampbrush chromosomes. Chromosoma. 1981;84:159–171. doi: 10.1007/BF00399128. [DOI] [PubMed] [Google Scholar]
- Gall JG, Tsvetkov A, Wu Z, Murphy C. Is the sphere organelle/coiled body a universal nuclear component? Dev Genet. 1995;16:25–35. doi: 10.1002/dvg.1020160107. [DOI] [PubMed] [Google Scholar]
- Gall JG, Wu Z. Examining the contents of isolated Xenopus germinal vesicles. Methods. 2010;51:45–51. doi: 10.1016/j.ymeth.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Wu Z, Murphy C, Gao H. Structure in the amphibian germinal vesicle. Exp Cell Res. 2004;296:28–34. doi: 10.1016/j.yexcr.2004.03.017. [DOI] [PubMed] [Google Scholar]
- Ganot P, Jady BE, Bortolin ML, Darzacq X, Kiss T. Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol Cell Biol. 1999;19:6906–6917. doi: 10.1128/mcb.19.10.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey AC, White AE, Tatomer DC, Marzluff WF, Duronio RJ. The Drosophila U7 snRNP proteins Lsm10 and Lsm11 are required for histone pre-mRNA processing and play an essential role in development. RNA. 2009;15:1661–1672. doi: 10.1261/rna.1518009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerger KE, Murphy C, Gall JG. Steady-state dynamics of Cajal body components in the Xenopus germinal vesicle. J Cell Biol. 2003;160:495–504. doi: 10.1083/jcb.200212024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerger KE, Wu Z, Murphy C, Gall JG. Heat shock induces mini-Cajal bodies in the Xenopus germinal vesicle. J Cell Sci. 2002;115:2011–2020. doi: 10.1242/jcs.115.10.2011. [DOI] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Ketele A, Kiss T. Human intron-encoded Alu RNAs are processed and packaged into Wdr79-associated nucleoplasmic box H/ACA RNPs. Genes Dev. 2012;26:1897–1910. doi: 10.1101/gad.197467.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kezer J, Macgregor HC. The nucleolar organizer of Plethodon cinereus cinereus (Green). II. The lampbrush nucleolar organizer. Chromosoma. 1973;42:427–444. doi: 10.1007/BF00399410. [DOI] [PubMed] [Google Scholar]
- Khodyuchenko T, Gaginskaya E, Krasikova A. Non-canonical Cajal bodies form in the nucleus of late stage avian oocytes lacking functional nucleolus. Histochem Cell Biol. 2012;138:57–73. doi: 10.1007/s00418-012-0938-z. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Krylov V, Reilly MJ, Gall JG, Bhattacharya D, Cheung CYJ, Kaufman S, Lam DK, Macha J, Ngo C, et al. Rapid gynogenetic mapping of Xenopus tropicalis mutations to chromosomes. Dev Dyn. 2009;238:1398–1346. doi: 10.1002/dvdy.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochan KJ, Wright DA, Schroeder LJ, Shen J, Morizot DC. Genetic linkage maps of the West African clawed frog Xenopus tropicalis. Dev Dyn. 2003;226:99–102. doi: 10.1002/dvdy.10193. [DOI] [PubMed] [Google Scholar]
- Krainer AR. Pre-mRNA splicing by complementation with purified human U1, U2, U4/U6 and U5 snRNPs. Nucleic Acids Res. 1988;16:9415–9429. doi: 10.1093/nar/16.20.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Jr., Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-L, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG. The Drosophila melanogaster Cajal body. J Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-L, Wu Z, Nizami Z, Deryusheva S, Rajendra TK, Beumer KJ, Gao H, Matera AG, Carroll D, Gall JG. Coilin Is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell. 2009;20:1661–1670. doi: 10.1091/mbc.E08-05-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Frey MR, Margelot K, Wolin SL. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhlanga MM, Tyagi S. Using tRNA-linked molecular beacons to image cytoplasmic mRNAs in live cells. Nat Protoc. 2006;1:1392–1398. doi: 10.1038/nprot.2006.242. [DOI] [PubMed] [Google Scholar]
- Murphy C, Wang Z, Roeder RG, Gall JG. RNA polymerase III in Cajal bodies and lampbrush chromosomes of the Xenopus oocyte nucleus. Mol Biol Cell. 2002;13:3466–3476. doi: 10.1091/mbc.E02-05-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A, Lukowiak A, Jady BE, Dragon F, Kiss T, Terns RM, Terns MP. Nucleolar localization signals of Box H/ACA small nucleolar RNAs. EMBO J. 1999;18:5120–5130. doi: 10.1093/emboj/18.18.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizami ZF, Deryusheva S, Gall JG. Cajal bodies and histone locus bodies in Drosophila and Xenopus. Cold Spring Harbor Symp Quant Biol. 2010;75:313–320. doi: 10.1101/sqb.2010.75.005. [DOI] [PubMed] [Google Scholar]
- Norton JT, Pollock CB, Wang C, Schink JC, Kim JJ, Huang S. Perinucleolar compartment prevalence is a phenotypic pancancer marker of malignancy. Cancer. 2008;113:861–869. doi: 10.1002/cncr.23632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paule MR, White RJ. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penrad-Mobayed M, El Jamil A, Kanhoush R, Perrin C. Working map of the lampbrush chromosomes of Xenopus tropicalis: A new tool for cytogenetic analyses. Dev Dyn. 2009;238:1492–1501. doi: 10.1002/dvdy.21930. [DOI] [PubMed] [Google Scholar]
- Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol. 2000;151:1561–1574. doi: 10.1083/jcb.151.7.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock C, Huang S. The perinucleolar compartment. Cold Spring Harbor Perspectives in Biology: The Nucleus. 2010;2:267–276. doi: 10.1101/cshperspect.a000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I, Andrade LE, Ochs RL, Chan EK, Chang CM, Roos G, Tan EM. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res. 1991;195:27–37. doi: 10.1016/0014-4827(91)90496-h. [DOI] [PubMed] [Google Scholar]
- Reimer G, Pollard KM, Penning CA, Ochs RL, Lischwe MA, Busch H, Tan EM. Monoclonal autoantibody from a (New Zealand black x New Zealand white) F1 mouse and some human scleroderma sera target an Mr 34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheum. 1987;30:793–800. doi: 10.1002/art.1780300709. [DOI] [PubMed] [Google Scholar]
- Richard P, Darzacq X, Bertrand E, Jady BE, Verheggen C, Kiss T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 2003;22:4283–4293. doi: 10.1093/emboj/cdg394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffert N, Hossbach M, Heintzmann R, Achsel T, Lührmann R. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 2004;23:3000–3009. doi: 10.1038/sj.emboj.7600296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KP, Carter KC, Johnson CV, Lawrence JB. U2 and U1 snRNA gene loci associate with coiled bodies. J Cell Biochem. 1995;59:473–485. doi: 10.1002/jcb.240590408. [DOI] [PubMed] [Google Scholar]
- Stanek D, Neugebauer KM. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J Cell Biol. 2004;166:1015–1025. doi: 10.1083/jcb.200405160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg TH, Mathews DE, Durbin RD, Burgess RR. Tagetitoxin: a new inhibitor of eukaryotic transcription by RNA polymerase III. J Biol Chem. 1990;265:499–505. [PubMed] [Google Scholar]
- Stephenson EC, Erba HP, Gall JG. Histone gene clusters of the newt Notophthalmus are separated by long tracts of satellite DNA. Cell. 1981;24:639–647. doi: 10.1016/0092-8674(81)90090-8. [DOI] [PubMed] [Google Scholar]
- Strzelecka M, Trowitzsch S, Weber G, Lührmann R, Oates AC, Neugebauer KM. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol. 2010;17:403–409. doi: 10.1038/nsmb.1783. [DOI] [PubMed] [Google Scholar]
- Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EKL, Lafarga M, Conlon RA, Matera AG. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol. 2001;154:293–308. doi: 10.1083/jcb.200104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma RS, Stolk JA, Roth MB. Identification and characterization of a sphere organelle protein. J Cell Biol. 1993;122:767–773. doi: 10.1083/jcb.122.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Aab A, Steitz JA. Guide RNAs with 5′ caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr Biol. 2004;14:1985–1995. doi: 10.1016/j.cub.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Shu M-D, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell. 2009;34:47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C, Lafontaine DLJ, Samarsky D, Mouaikel J, Blanchard J-M, Bordonné R, Bertrand E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 2002;21:2736–2745. doi: 10.1093/emboj/21.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RA, Jared DW, Dumont JN, Sega MW. Protein incorporation by isolated amphibian oocytes III. Optimum incubation conditions. J Exp Zool. 1973;184:321–333. doi: 10.1002/jez.1401840305. [DOI] [PubMed] [Google Scholar]
- Wang C, Politz JC, Pederson T, Huang S. RNA polymerase III transcripts and the PTB protein are essential for the integrity of the perinucleolar compartment. Mol Biol Cell. 2003;14:2425–2435. doi: 10.1091/mbc.E02-12-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DE, Gutierrez L, Xu Z, Krylov V, Macha J, Blankenburg KP, Hitchens M, Bellot LJ, Spivey M, Stemple DL, et al. A genetic map of Xenopus tropicalis. Dev Biol. 2011;354:1–8. doi: 10.1016/j.ydbio.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AE, Burch BD, Yang X-C, Gasdaska PY, Dominski Z, Marzluff WF, Duronio RJ. Drosophila histone locus bodies form by hierarchical recruitment of components. J Cell Biol. 2011;193:677–694. doi: 10.1083/jcb.201012077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Gall JG. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc Natl Acad Sci USA. 1993;90:6257–6259. doi: 10.1073/pnas.90.13.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Murphy C, Gall JG. The Sm binding site targets U7 snRNA to coiled bodies (spheres) of amphibian oocytes. RNA. 1996;2:811–823. [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Murphy C, Callan HG, Gall JG. Small nuclear ribonucleoproteins and heterogeneous nuclear ribonucleoproteins in the amphibian germinal vesicle: loops, spheres, and snurposomes. J Cell Biol. 1991;113:465–483. doi: 10.1083/jcb.113.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X-C, Burch BD, Yan Y, Marzluff WF, Dominski Z. FLASH, a proapoptotic protein involved in activation of caspase-8, Is essential for 3′ end processing of histone pre-mRNAs. Mol Cell. 2009;36:267–278. doi: 10.1016/j.molcel.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y-T, Shu M-D, Narayanan A, Terns RM, Terns MP, Steitz JA. Internal modification of U2 small nuclear (snRNA) occurs in nucleoli of Xenopus oocytes. J Cell Biol. 2001;152:1279–1288. doi: 10.1083/jcb.152.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li Z-H, Terns RM, Terns MP, Yu Y-T. An H/ACA guide RNA directs U2 pseudouridylation at two different sites in the branchpoint recognition region in Xenopus oocytes. RNA. 2002;8:1515–1525. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.