Abstract
Shewanella oneidensis cytochrome c nitrite reductase (soNrfA), a dimeric enzyme that houses five c-type hemes per protomer, carries out the six-electron reduction of nitrite and the two-electron reduction of hydroxylamine. Protein film voltammetry (PFV) has been used to study the cytochrome c nitrite reductase from Escherichia coli (ecNrfA) previously, revealing catalytic reduction of both nitrite and hydroxylamine substrates by ecNrfA adsorbed to a graphite electrode that is characterized by ‘boosts’ and attenuations in activity depending on the applied potential. Here, we use PFV to investigate the catalytic properties of soNrfA during both nitrite and hydroxylamine turnover and compare those properties to ecNrfA. Distinct differences in both the electrochemical and kinetic characteristics of soNrfA are observed, e.g., all detected electron transfer steps are one-electron in nature, contrary to what has been observed in ecNrfA (Angove, H. C., Cole, J. A., Richardson, D. J., and Butt, J. N. (2002) Protein film voltammetry reveals distinctive fingerprints of nitrite and hydroxylamine reduction by a cytochrome C nitrite reductase, J Biol Chem 277, 23374-23381). Additionally, we find evidence of substrate inhibition during nitrite turnover and negative cooperativity during hydroxylamine turnover, neither of which have previously been observed in any cytochrome c nitrite reductase. Collectively these data provide evidence that during catalysis, potential pathways of communication exist between the individual soNrfA monomers comprising the native homodimer.
Cytochrome c nitrite reductase (NrfA) carries out a key step in the bacterial nitrate respiration pathway under anaerobic conditions.(1) The periplasmic enzyme catalyzes the six-electron reduction of nitrite to ammonia, the second step in the two-step conversion of nitrate to ammonia.(2) During this process, NrfA draws electrons from the quinol pool, which are provided by the oxidation of a non-fermentable substrate such as formate or H2, and thereby facilitates the generation of an electrochemical proton motive force.(1, 3, 4) In addition to catalyzing the reduction of nitrite, NrfA is also capable of carrying out the two-electron reduction of hydroxylamine and the five-electron reduction of nitric oxide. The physiological function of the latter two reductions is not well understood, but it has been speculated that the hydroxylamine reduction activity may play a role in cellular detoxification processes.(5, 6) Additionally, because both nitric oxide and hydroxylamine are readily reduced by NrfA, it has been hypothesized that they are intermediates along the nitrite reduction pathway.(7, 8)
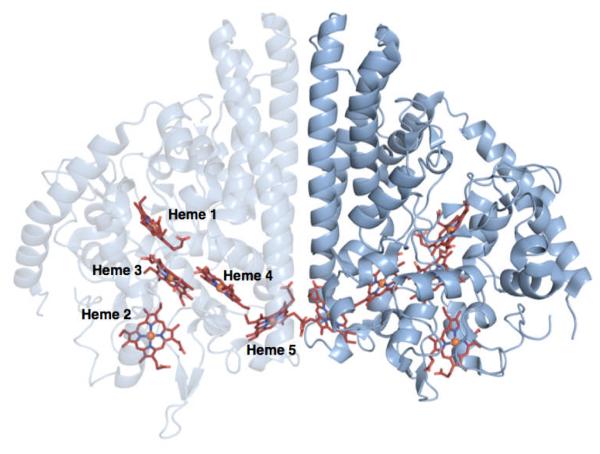
NrfA purifies as a homodimer with an extensive dimer interface and contains five c-type hemes per monomer (Figure 1). The crystal structures for NrfA from several different organisms are available, e.g., Escherichia coli(7), Wolinella succinogenes(9), Sulfurospirillum deleyianum(10), and most recently Shewanella oneidensis(11). The overall arrangement of active site residues and hemes are conserved across all structures. Each monomer contains four bis-His ligated hemes which have been proposed to act as electron relays, as well as a unique, lysine-ligated active site heme.
Figure 1.
Crystal structure of soNrfA. One monomer of the complete homodimer is transparent to highlight the positions of c-type hemes. Image rendered in PyMol from PDB file 3UBR.pdb.
The catalytic mechanism of NrfA is largely unknown; however at least one mechanism has been proposed based on crystal structures of NrfA with bound intermediates.(8) While this study has been useful as a starting point for suggesting potential intermediates along the catalytic cycle, it has yet to be experimentally shown how electrons and protons are delivered to the active site of the enzyme.(12) In particular, it is still not known if electrons are delivered to the active site one at a time, or if there are coordinated two-electron transfers, which could be achieved by electronically coupled hemes that could effectively deliver two electrons simultaneously: spectroscopic studies have provided some evidence for coordinated transfer.(7, 13)
One strategy to probe the mechanistic chemistry of NrfA has been through the use of protein film voltammetry (PFV), an electrochemical technique that has proven quite useful in studying the mechanisms of redox enzymes (see refs 14,15 for reviews). This method provides the ability to modulate the applied potential while monitoring catalytic activity in the form of current passed by the enzyme. Studying the Escherichia coli nitrite reductase (ecNrfA) Butt and co-workers have found that ecNrfA will electrocatalytically reduce nitrite and hydroxylamine at pyrolytic graphite edge (PGE) electrodes.(12, 13, 16, 17) These studies have produced an electrochemical ‘fingerprint’ for NrfA, where catalytic waves produced in PFV experiments are composed of multiple features. These features include a ‘boost’ in activity visible at high nitrite concentrations, as well as a decrease in activity at low potentials and low nitrite concentrations. Additionally, catalytic waves observed during hydroxylamine turnover are also complex, containing multiple features at both low and high concentrations. It has been suggested that this electrochemical fingerprint should be common to all cytochrome c nitrite reductases(12), a hypothesis tested in this report.
Shewanella oneidensis MR-1 is a facultative anaerobe from the γ-proteobacteria capable of reducing a broad array of metals and organic compounds, which may prove useful in bioremediation and bioenergy applications.(18-20) Recently, the cytochrome c nitrite reductase from S. oneidensis (soNrfA) has been purified, crystallized, and its redox centers electrochemically characterized. (4, 11) The structural arrangement of the hemes and active site residues are largely consistent with the other NrfA structures reported in the literature. Additionally, cyclic voltammograms recorded using PFV with films of soNrfA in the absence of substrate yielded a broad envelope of signal, corresponding to the reduction and subsequent reoxidation of the five heme cofactors within soNrfA, which can be assigned potentials of (−295, −230, −166, −105, and −36 mV at pH 6).(11) These results are consistent with results of spectropotentiometric titrations of soNrfA, and other NrfA enzymes that have shown that redox transformations occur over a broad range of potential.(7, 21-23) There was no evidence of electronic coupling between hemes, as the envelope of signal can be fit to a model corresponding to the reduction of five one-electron centers. This response is similar to what has been reported for ecNrfA adsorbed to an electrode.(13)
Here, we report the characterization of the catalytic properties of soNrfA using PFV during both nitrite and hydroxylamine turnover, and compare those catalytic properties to ecNrfA. Despite the similarity in structure to ecNrfA(11), we have found distinct differences in both the electrochemical and kinetic characteristics of soNrfA that address the stoichiometry of electron transfer steps in the catalytic cycle, highlight the relative rates of activation/inactivation at different potentials, and provide evidence that the individual monomers that comprise a NrfA homodimer are not isolated from one another during catalysis.
MATERIALS AND METHODS
Protein Purification
Shewanella oneidensis cytochrome c nitrite reductase was purified from a high yield expression system as described previously by Youngblut and colleagues.(11) Protein stocks were stored in aliquots at −80 °C and working stocks were stored at −20 °C.
Protein Film Voltammetry Experiments
SoNrfA was immobilized on a pyrolytic graphite edge (PGE) electrode by pipetting 3 μL 175 μM enzyme solution onto a freshly polished electrode, waiting approximately 20 seconds, then pipetting off excess enzyme solution. Prior to film generation electrodes were polished with an aqueous slurry of 1.0 μm alumina, then the alumina was removed by sonicating for several minutes. After generation of an enzyme film, electrodes were immersed in a mixed-buffer with 5 mM each of sodium acetate, MES, MOPS, HEPES, TAPS, CHES, and CAPS, 100 mM NaCl and 2 mM CaCl2 which allowed buffering over a range of pH values. For hydroxylamine turnover experiments, 10 mM of each of the buffering salts were used with 100 mM NaCl and 2 mM CaCl2. The pH of stock solutions of hydroxylamine were adjusted to pH 8.3. Additionally, the final pH of the cell solution was confirmed to be pH 8.3 after each complete hydroxylamine titration. A three electrode electrochemical cell was used, which used a platinum wire as a counter electrode and a saturated calomel reference electrode. All potentials are corrected by +242 mV and reported relative to the standard hydrogen electrode. The reaction cell was water-jacketed, which allowed regulation of temperature.
Experiments were carried out anaerobically in an MBruan Labmaster glove box under a nitrogen atmosphere at 20°C, unless otherwise noted. PFV experiments were performed using a PGSTAT 12 or PGSTAT 30 AutoLab (Ecochemie) potentiostat, equipped with ECD and FRA modules. The working electrode was rotated during catalytic experiments using a EG&G electrode rotator. All PFV data were collected using the GPES software package (Ecochemie). All cyclic voltammograms were analyzed using the SOAS package.(24) For both catalytic and non-turnover signals, background electrode capacitance was subtracted from the raw data. All reported limiting currents or midpoint potentials were also measured using SOAS. In the case of catalytic PFV experiments, only the reductive (cathodic) scan was used for measurements of midpoint potentials and limiting currents.
RESULTS
Overview of electrochemical nitrite reduction by soNrfA
Upon addition of nitrite to the electrochemical cell solution, cyclic voltammograms produced by soNrfA at PGE electrodes are converted into large, reducing sigmoidal current waves, indicating the reduction of nitrite by a catalytically active enzyme (Figure 2A). As the electrode potential is lowered, the catalytic current eventually reaches a limiting value (ilim); the magnitude of the limiting current value becomes larger with increasing substrate concentration in a manner analogous to Michaelis-Menten kinetics. In a similar fashion to ecNrfA, the limiting currents and shape of the catalytic wave are somewhat dependent on the rotation rate of the working electrode, due to the very fast turnover numbers of the enzyme, which allow it to quickly deplete substrate in the direct proximity of the electrode surface. Nevertheless, current is minimally dependent upon rotation rates at values greater than 2000 rpm (Figure S1), and at rotations of 3000 rpm, diffusional contributions to the limiting current are small (less than 2-3%).
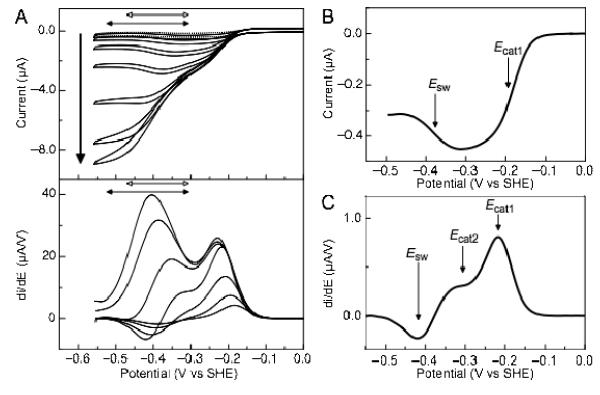
Figure 2.
Current-potential profile of soNrfA nitrite reduction. (A) Cyclic voltammograms of soNrfA in the presence of increasing concentrations of nitrite. Reactions were carried out at 20°C, pH ~8.3, scan rate 10 mV/s, electrode rotation rate 3000 rpm. Bold arrow indicates direction of increasing nitrite concentration (1.4, 5.5, 17, 54, 160, 490, and 1460 μM). Full scans are shown in top panel. Filled horizontal double-headed arrows indicate the potential range where the boost is observed, −300 to −525 mV. Open horizontal double-headed arrows indicate the potential range where the switch is observed, −300 to −470 mV. First derivative of reductive scans of catalytic waves are shown in bottom panel. (B) Baseline-subtracted reductive catalytic wave for soNrfA at 1.37 μM nitrite showing the decrease in activity at potentials below ~ −300 mV. (C) First derivative of reductive catalytic wave for soNrfA at 54.1 μM nitrite showing that three distinct features can be simultaneously observed.
Previous catalytic PFV experiments with ecNrfA reported by Butt and colleagues(12), as well as experiments with other other related cytochrome c nitrite reductases, have established a ‘fingerprint’ for nitrite turnover, characterized by a number of distinct features that can be observed in the catalytic voltammograms.(12, 13) Cyclic voltammograms of soNrfA display comparable features. Distinct features in catalytic waves produced in a PFV experiment are best illustrated by plotting the first derivate of the catalytic waves (Figure 2A, bottom panel), where peaks are centered on a midpoint potential, termed Ecat. A positive peak indicates an increase in activity while a negative peak indicates a decrease in activity. Notably, like ecNrfA, soNrfA exhibits differential activity within ranges of potentials: after the onset of catalysis, in the presence of low concentrations of nitrite current magnitude steadily increases until a potential value of approximately −300 mV is reached (Figure 2B). When the electrode potential is swept to lower potentials the activity of the enzyme is attenuated, which is observed as a decrease in current magnitude with decreasing potential. The decrease in activity is centered on a potential termed the ‘switch’ (Esw)(12, 13), and the steepness of this feature is consistent with a one-electron process.
As the concentration of nitrite is increased a new feature begins to develop in the catalytic wave which is observed in the −300 to −525 mV potential window in the catalytic current-potential profile (Figure 2A, filled horizontal arrows). This boost in activity at higher nitrite concentrations is also observed in ecNrfA.(12)
It is important to note that under certain conditions such as pH > 8 and ~50 μM nitrite, the ‘switch’ and the ‘boost’ can be observed simultaneously in soNrfA PFV experiments. At pH values > 8, the magnitude of each component varies as a function of substrate in a non-linear manner suggesting that these two features represent two independent processes (Figure 2C). Hence, the onset of the boost is not necessarily related to the diminished prominence of the switch at higher nitrite concentrations. Thus, a total of three separate features are observed during nitrite turnover by soNrfA: Ecat1, the primary catalytic feature which is observed at the onset of catalysis; Ecat2, the boost in activity observed at higher nitrite concentrations; and Esw, the attenuation of activity observed at lower potentials.
As outlined above, the overall current-potential profile of soNrfA is similar to ecNrfA. However, there are key differences between the enzymes upon closer inspection of soNrfA voltammetry. Unlike ecNrfA, the boost in activity (Ecat2) is barely visible in soNrfA at pH 7, even at saturating nitrite concentrations. Increasing the pH of the cell solution to >8 results a catalytic ‘fingerprint’ much more similar to that of ecNrfA (See Figure 3 in ref 12), with the boost developing as nitrite concentration increases until it eventually dominates the waveform (Figure 2). Because the current-potential profile is more similar to ecNrfA at higher pH, we have further characterized soNrfA at pH ~8.3. There are other notable differences between the behaviors of soNrfA and ecNrfA: in some experiments with soNrfA, the decrease in activity associated with Esw is observable even at nitrite concentrations well above Km (~500 μM) at near neutral pH values, while the same feature is only observed at very low concentrations of nitrite in ecNrfA (< 25 μM) in the same pH range.
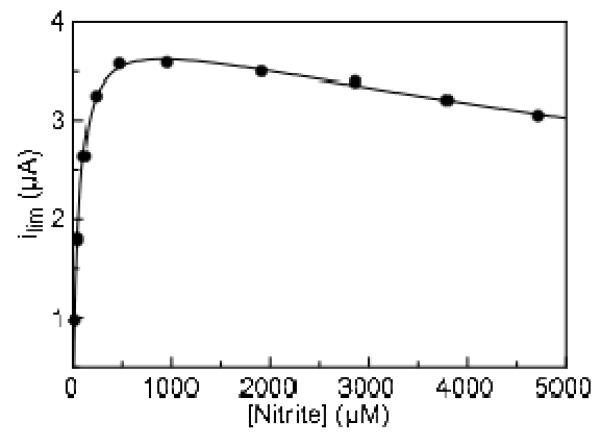
Figure 3.
Variation in limiting current as a function of the concentration of nitrite. Experiment carried out at pH 8.3, 20°C, 20 mV/s. Data is fit to a substrate inhibition model (Equation 1). Limiting current was measured at −550 mV.
An additional difference is the steepness of the catalytic waves centered at Ecat1 and Ecat2 in soNrfA, which is related to the number of electrons associated with that catalytic wave. Catalytic waves produced during nitrite turnover fit best to models accounting for one-electron processes (Figure S2). This is true at both low and high nitrite concentrations. Taking the first derivative of catalytic waves produces peak-like signals that can be analyzed in a similar manner to that of non-turnover signals. Thus, a catalytic wave associated with a simple one-electron process, displays a derivative with a predicted peak-width of ~89 mV at half height, while a two-electron feature will similarly yield a derivative with a much sharper peak-width (~44.5 mV). Analysis of the first derivatives of catalytic waves produced during turnover in the presence of excess nitrite showed that Ecat1 and Ecat2 have half-height peak widths of 82 ± 4 mV and 137 ± 2 mV respectively. At low concentration, when the switch is apparent, the catalytic wave is 84 ± 8 mV, while the switch is 88 ± 6 mV at half height. These peak widths are consistent with one-electron processes, however the width of Ecat2, which is observed during turnover of excess nitrite, is broader than expected. Nevertheless, both are consistent with one-electron processes while at the onset of nitrite reduction ecNrfA yielded catalytic waves that were found to be two-electron in nature.(12)
At pH values greater than 8.5 and less than 6.5 and at scan rates up to 20 mV/s, the reductive and oxidative scan of catalytic waves during nitrite reduction are essentially superimposable, behavior that is expected for an electroactive catalyst subjected to a complete, cyclic set of potential sweep increases (Figure 2). However, between pH 7 and 8, at high nitrite concentrations (~500 μM) the reductive and oxidative scan are not superimposable and hysteresis is observed, such that the return oxidative sweep becomes separated from the reductive sweep and the two scans do not superimpose. This indicates that the activity of the enzyme is attenuated after exposure to very low potentials. This behavior appears to be associated with the appearance of the ‘switch’, which occurs in the pH 7 range, as mentioned above. Similar behavior is observed with ecNrfA but the specifics of this behavior appear to be quite different: first, it has been reported that adding 2 mM CaCl2 to the electrochemical buffer improves this non-superimposable nature for ecNrfA voltammograms.(12, 16) While adding CaCl2 to the cell solution results in a perturbation of the overall shape of the cyclic voltammogram for soNrfA nitrite turnover, the inequality between the oxidative and reductive scans was still significant even at concentrations up to 10 mM CaCl2. (Nevertheless, 2 mM CaCl2 was routinely included in the cell solution for all experiments for all experiments reported here.) Second, the reversibility of ecNrfA catalytic voltammograms are improved by increasing the scan rate to values above or equal to 20 mV/s, which corresponds to the enzyme being exposed to potentials below −400 mV for approximately 20 seconds or less. This has been attributed to a time dependence for exposure of the enzyme film to lower potentials; longer exposure (slower scan rate) leads to more inactivation and more irreversibility in catalytic voltammograms. Here the opposite effect is observed with soNrfA: lowering the scan rate to values below 2 mV/s significantly improves the superimposability of the oxidative and reductive scans, and the scans are most reversible at 0.5 mV/s (Figure S3). The sweep from −600 to −400 mV at 2 mV/s takes ~100 seconds (compared to just 10 seconds at 20 mV/s), suggesting that the process responsible for the reactivation of the enzyme that occurs as the electrode potential is swept back to oxidative potentials is quite slow. After the enzyme activity is attenuated as the potential is lowered, the scan rate must be sufficiently slow to allow the attenuation process to be reversed otherwise the activity of the enzyme would remain attenuated.
Determination of kinetic parameters for soNrfA nitrite reduction
Determining Michaelis-Menten parameters for soNrfA using limiting currents (at −550 mV) revealed what appears to be atypical kinetics during nitrite reduction by this enzyme (Figure 3). After reaching concentrations of nitrite in excess of 1 mM, the magnitude of the limiting current (enzyme activity) begins to decrease with increasing nitrite concentration. This behavior has not been reported for either ecNrfA or soNrfA previously; however it is likely that prior nitrite titrations were not carried out to high enough concentrations to observe this phenomenon. We are now investigating the soNrfA-catalyzed reduction of nitrite by methyl viologen mono-cation radical, and preliminary studies suggest that above pH 8 but not below, enzyme activity decreases at very high nitrite concentrations (data not shown). A common cause for decreased activity in PFV experiments is “film-loss”, a decreasing population of active enzymes adsorbed to the electrode.(25) However, the amount of film loss observed from scan to scan at high nitrite concentrations was negligible, and could not account for the decrease in current observed under these conditions (Figure S4). Additionally, increasing the concentration of NaCl in the buffer to 150 mM did not affect the kinetic parameters (Km, Vmax) or the overall shape of the nitrite titration data, excluding the possibility that the observed inhibition is due to a generic ionic strength effect (Figure S5). Thus, it seems that that soNrfA experiences substrate inhibition at higher nitrite concentrations.
In order to obtain accurate estimates of kinetic values for the Fig. 3 data, these data were fit to a model in which a second substrate molecule binds so as to inhibit the enzyme (Eq. 1). Ki is then the dissociation constant
| (1) |
of an inhibitory ternary complex.(26) Product inhibition seems unlikely since only picomolar concentrations of enzyme are present, and the amount of product that accumulates in solution during an experiment is therefore very small. Using the substrate inhibition model we have determined the Km for nitrite reduction to be 54 ± 12 μM, with a Ki of 18 ± 4 mM. Additionally, because of our ability to obtain robust non-turnover signals with soNrfA we can calculate enzyme concentration on the electrode and therefore determine a value for kcat. We obtain an average enzyme coverage of 2.3 pmol/cm2, which was calculated by measuring the peak current resulting from an enzyme film. Using PFV we determined the apparent k with respect to nitrite to be 7 ± 2 s−1 cat, which translates to 42 ± 10 electrons s−1. These values are much lower than values reported for soNrfA using solution assays (4944 electrons s−1), yet represent current magnitudes and surface coverages that are similar to the ecNrfA system.(12, 13) The difference between the apparent kcat determined by PFV may be the result of a number of factors. First we characterized soNrfA at pH ~8.3, to maximize our ability to observe all three components of the catalytic signatures simultaneously, whereas solution assays were performed at pH 7(11, 12) (the pH optimum is ~7.5(6)). Alternatively, the difference may indicate that only a small sub-population of the enzymes detected in non-turnover experiments actually participate in catalysis, which would lead to an overestimation of surface coverage, as has been proposed for the ecNrfA, where as few as 4% of enzymes in a monolayer of ecNrfA were catalytically competent.(15, 27) Whatever the reason for this discrepancy, our value for K is very consistent with the value of 23 ± 4 μM determined for soNrfA by solution assays,(11) M indicating that adsorption to the electrode surface does not negatively impact catalysis for the sub-population that is active.
Overview of Hydroxylamine Reduction by soNrfA
As reported for ecNrfA, the concentration of hydroxylamine needed to observe turnover by soNrfA is higher than concentrations required for nitrite reduction. Hydroxylamine turnover for soNrfA was been observed at concentrations as low as 30 μM, an order of magnitude lower than the minimum hydroxylamine concentrations required to see turnover by ecNrfA.
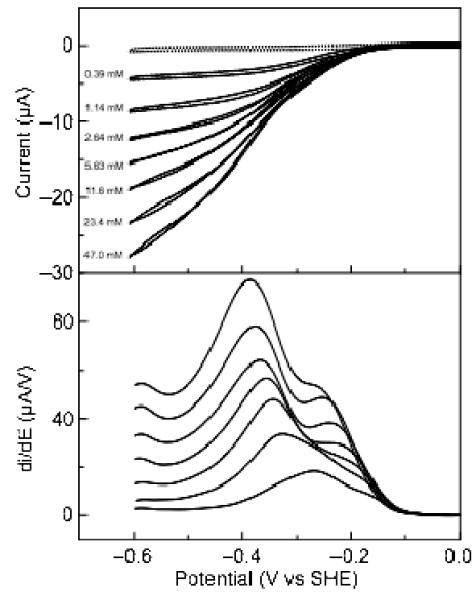
As seen in the case of nitrite reduction, the behavior of soNrfA during hydroxylamine turnover was most similar to that of ecNrfA (collected at pH 7) when the soNrfA data were collected at pH 8.3. Rotation rate did not appear to affect the catalytic response of soNrfA, so we performed all hydroxylamine-turnover experiments at 3000 rpm (Figure S6). The catalytic fingerprint of hydroxylamine catalysis is similar to that reported for ecNrfA, with three features within the catalytic waveforms (Figure 4). The waveforms are dominated by a feature positioned at intermediate potential (Ecat2), which overlaps with a second, higher potential feature (Ecat1). The third feature (Ecat3) occurs at low potentials, and is very small compared to the other two features. The extent of overlap between Ecat1 and Ecat2 prevented the direct measurement of the half-height widths of the first-derivates of the catalytic waves. Despite the relatively broad nature of the catalytic waves observed during hydroxylamine turnover, the waves observed at low hydroxylamine concentrations (1.2 mM) are clearly better fit by a model that incorporates two one-electron waves than by a model incorporating a pair of two-electron feature (Figure S2). Additionally, catalytic waves produced at 47 mM hydroxylamine appeared similar to those produced at high nitrite concentrations indicating that these features are also not likely the result of two-electron processes. This contrasts with hydroxylamine turnover by ecNrfA, where catalytic waves produced at low hydroxylamine concentrations (~1 mM) were well fitted by a model incorporating a two-electron feature.(12) Taken together, these data indicate that only one-electron steps are being detected during hydroxylamine turnover, under the conditions tested.
Figure 4.
Catalytic current-potential profile for hydroxylamine turnover by soNrfA. Cyclic voltammograms of soNrfA in the presence of increasing concentrations of hydroxylamine. Reactions were carried out at 20°C, pH ~8.3, scan rate 20 mV/s, electrode rotation rate 3000 rpm. Nitrite concentrations are listed to the left of each voltammogram. Full scans are shown in the top panel and first derivatives of reductive scans of the catalytic waves are shown in bottom panel. The dotted line shows soNrfA on an electrode in the absence of hydroxylamine.
As is clearly seen in Figure 4 (top) hysteresis is observed in the catalytic-current potential profile for hydroxylamine turnover by soNrfA at high concentrations of substrate, which was not reported for ecNrfA hydroxylamine reduction. Interestingly, the hysteresis observed during hydroxylamine reduction is markedly different from the hysteresis observed during nitrite reduction: during the return scan in the oxidative direction, the enzyme appears to be more active than during the reductive scan. This is observed in Figure 4 as a crossover that occurs between the reductive and oxidative scans that occurs immediately after the electrode potential begins to be swept back in the oxidative direction. During this time the two scans do not appear to be superimposable. The scans cross again at higher potentials, after which the oxidative and reductive scans become superimposable. This phenomenon is present at scan rates ranging between 0.5 and 30 mV/s. Because lowering the scan rate did not appear to improve the reversibility of the oxidative and reductive catalytic scans, a scan rate of 20 mV/s was routinely used for analyses reported here. We also found that adding CaCl2 to the cell solution did not improve the reversibility of the oxidative and reductive scans, in parallel to the reported insensitivity to Ca2+ concentration for the current-potential profile of hydroxylamine reduction by ecNrfA.(12)
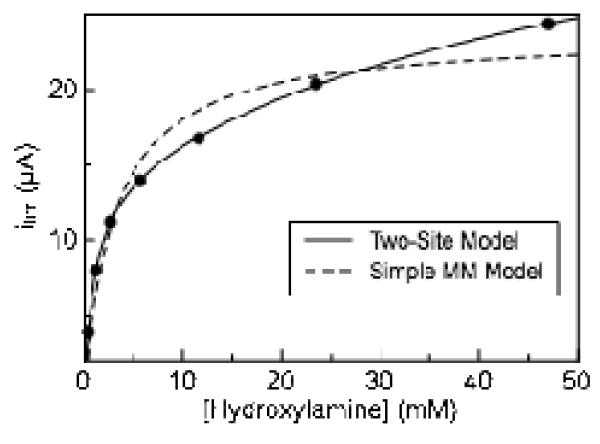
As in the case of nitrite reduction, plots of limiting current (taken at −550 mV) versus hydroxylamine concentration appear to deviate from a simple Michaelis-Menten model of enzyme kinetics, though without apparent substrate inhibition (Figure 5). Enzyme activity does not reach a plateau with increasing hydroxylamine concentration as rapidly as predicted by simple Michaelis-Menten kinetics. Instead, the magnitude of the limiting current continues to steadily increase as the hydroxylamine concentration is raised. Ignoring non-hyperbolic data and fitting to a simple Michaelis-Menten model thus leads to inaccurate kinetic parameter estimates.(28) The contribution to the limiting current at −550 mV due to background reaction of hydroxylamine with graphite electrodes was found to be nearly zero at concentrations less than 20 mM hydroxylamine and approximately 0.2 μA at 60 mM hydroxylamine, eliminating simple background reaction of substrate with the electrode as an explanation the biphasic nature of the kinetic plots (Figure S7). Therefore, in order to obtain more accurate kinetics parameters, a modified Michaelis-Menten model (Eq. 2) was used to account for the non-hyperbolic nature of the hydroxylamine kinetic data, which includes a second set of kinetic parameters:
| (2) |
Figure 5.
Variation in limiting current with hydroxylamine concentration Experiment carried out at pH 8.3, 20°C, 20 mV/s. Data is fitted with a two site model (Eq 2), and a simple model for Michaelis-Menten kinetics Limiting current measured at −550 mV.
Fitting to equation 2 yields a KM1 and KM2 values for soNrfA hydroxylamine reduction to be 0.43 ± 0.1 mM and 10.0 ± 1 mM respectively at pH 8.3. Corresponding values kcat1 and kcat2 of 64 ± 14 and 266 ± 57 electrons s−1 respectively were also determined at this pH. These value are also lower than the flux of 4760 electrons s−1 reported by spectrophotometric assays for soNrfA and is much lower than the value of 31000 electrons s−1 determined for ecNrfA by PFV. (7, 11) The discrepancies are likely due to the same issues addressed above for nitrite turnover. Importantly, the range of our values of KM (~0.43 and 10 mM, determined by fitting to a two-site model) are in excellent agreement with the the value of KM (8.3 ± 2.4 mM) determined by solution assays for soNrfA(11), indicating that the behavior of soNrfA adsorbed to an electrode is similar to its behavior in solution assays.
DISCUSSION
Here we have presented the first characterization of electrocatalysis by the cytochrome c nitrite reductase from Shewanella oneidensis, enabling a comparison with the well examined ecNrfA. Overall, the catalytic-current potential profile of soNrfA appears grossly similar to that of ecNrfA. However, upon closer examination, differences between the two enzymes are observed that provide new insights about the processes being observed in catalytic waves produced in PFV studies of NrfA. In particular, we provide evidence that catalysis is likely mediated by a series of one-electron steps, that the process responsible for reactivation after the ‘switch’ off at excessive overpotentials is relatively slow, and that the individual monomers that make up a soNrfA homodimer are most likely not acting independent of one another during catalysis.
Overview of the soNrfA catalytic ‘fingerprint’
The mechanism responsible for the ‘switch’ in ccNiRs (Esw, Figure 2) is currently not well understood; however several hypotheses have been proposed based on PFV studies of ecNrfA.(12, 16, 17) The general working hypothesis is that the attenuation of activity associated with Esw observed at lower potentials is the result of a conformational rearrangement driven by the reduction of the lower potential hemes 4 or 5.(17) Reduction of one (or both) of these heme centers results in uncompensated negative charge, which triggers the inactivation process.(17) This hypothesis was supported by the pH dependence of the switch, which occurs to a much larger extent at more alkaline pH values for ecNrfA, when there are fewer protonated side chains to compensate for the increased negative charge. In addition to the loss in activity associated with Esw, a further depletion in activity has been observed for the ecNrfA in the form of hysteresis (an additional loss of current in the return potential sweeps) under some conditions. (12, 16) As noted in Figure S3, this is also found for soNrfA, though under different conditions. Here we attempt to combine these observations into a single model for the possible basis of deactivation of NrfA at low potentials, that accounts for both the Esw behavior and the observation of hysteresis.
For soNrfA hysteresis is observed at faster (>10 mV/s) scan rates, pH 7-8, and nitrite concentrations above KM, such that a transformation reducing activity of the enzyme must occur at the electrode. The reverse, reactivation process that would return the enzyme to the more active form upon reoxidation is slow, which causes the return oxidative scan to deviate substantially from the reductive scan (i.e., hysteresis). When the scan rate is sufficiently slow (< 10 mV/s), the enzyme appears to have sufficient time to return to the more active form upon reoxidation, and the oxidative and reductive scans become superimposable (Figure S3).
In the case of nitrite reduction by ecNrfA, the scan rate dependence of hysteresis in ecNrfA appears to behave in a manner opposite to that of soNrfA; for ecNrfA hysteresis was only observed at scan rates slower than 20 mV/s during nitrite reduction, and above these scan rates, the oxidative and reductive scans were fully reversible, yet hysteresis is observed for the soNrfA enzyme (Figure S3). Why a difference exists in the timescales associated with hysteresis for the two different enzymes is puzzling; however, it may be that the process responsible for inactivation in ecNrfA is slower than in soNrfA. In such a model, at higher scan rates the inactivating process is out-run and therefore not observed, and would only manifest in voltammograms where the scan rate is sufficiently slow (< 1 mV/s). However the effect of scanning at very slow scan rates (<1 mV/s) has not been reported for ecNrfA. Similarly, here we cannot “out-run” the hysteresis with the soNrfA (as was the case for the ecNrfA) as timescales required (> 100 mV/s) interfere with steady-state catalysis. While the hysteresis is most visible at nitrite concentrations above KM, the deactivation process appears to be synonymous to the process identified by Esw: i.e., hysteresis only seems possible when Esw is observed. It is therefore reasonable to assume that the same process governs both the attenuation in activity that results in hysteresis, and the attenuation in activity observed at sub-KM nitrite concentrations (Esw). Thus, our data supports the hypothesis that Esw is associated with an inactivating conformational rearrangement that is experienced by NrfA at low potentials, where the reactivation process is slower than inactivation. Compared to the ecNrfA, the deactivation rate for soNrfA must be faster. This type of behavior has been observed in Ni-Fe hydrogenases, where large degrees of hysteresis in catalytic waves are the result of an inequality between the rates of inactivation and reactivation.(29-33)
The current-potential profiles for both nitrite and hydroxylamine reduction by soNrfA both exhibit two main catalytic features: a high potential feature (Ecat1) and a low potential boost (Ecat2), which is most prominent at higher substrate concentrations (Figures 2A, 4). The electron transfer steps represented by these features are not currently well understood. It has been suggested that the boost must be a change in the rate limiting step as the concentration of substrate is raised.(17) An additional possibility is that the boost may be the result of a shift in the path of electron flow through the enzyme, such as in the case of fumarate reductase (FRD), a multi-centered enzyme responsible for catalyzing the reduction of fumarate in the bacterial respiratory chain.(34, 35) From analysis of the potential at which the boost was centered in FRD, and the shape of the waves under different conditions, it was concluded that the two catalytic waves are the result of two distinct electronic relays to the active site. Only at high substrate concentrations, when turnover is fast and the demand for electrons is high, does the second relay contribute to the rate of catalysis.(36) The increased prominence of the boost in soNrfA as substrate concentration is raised, combined with the correlation between the position of the boost and the reduction potentials of the lower potential hemes (Figure 2 and Figure 5 in ref 11) suggests that the boost may be the result of a similar mechanism. In this regard an attractive hypothesis is that one relay path brings electrons into the active site via hemes on a single monomer, whereas the second path allows input of electrons into the active site from hemes on the second monomer of the NrfA homodimer, which is known to be highly stable.(11)
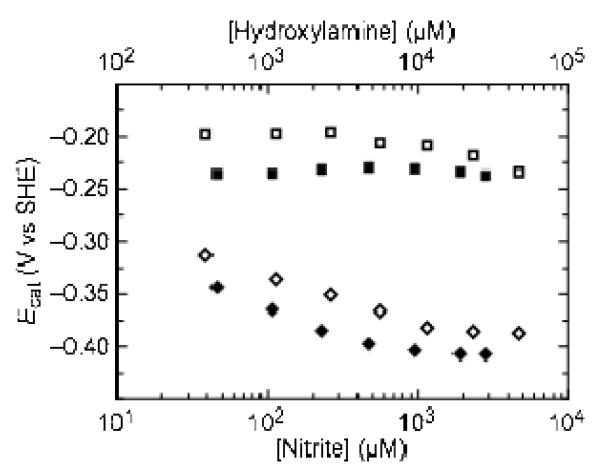
In such a model, heme 5 could provide an alternate relay to the active site that only engages as the substrate concentration is raised and the demand for electrons is high. This hypothesis is supported by the dependence of the positions of Ecat values on the concentration of substrate (Figure 6). Butt and coworkers recently suggested(13) that when the value of the rate constant for intra-molecular electron transfer between a relay and an active site center is approximately equal to the rate of conversion of substrate to product, the value of Ecat will tend toward the reduction potential of the electron relay as substrate concentration increases and the rate of turnover becomes more dependent on the rate of electron transfer from the relay to the active site. During turnover of both substrates, the position of Ecat2 tends toward more negative potentials and plateaus at approximately −375 mV. The shift toward lower potential suggests that as substrate concentration increases, the rate of turnover becomes more dependent on the lowest potential heme (heme 5), consistent with the hypothesis that it is acting as an alternate path for electrons to the active site. Additionally, our kinetic data supports the hypothesis that the monomers of NrfA are not isolated from one another during catalysis (see the discussion of the soNrfA kinetic results).
Figure 6.
Variation of the position of Ecat1 (squares) and Ecat2 (diamonds) with the concentration of nitrite (filled symbols) and hydroxylamine (open symbols). Data were collected at pH 8.3, 20°C, 3000 rpm, 20 mV/s, 2 mM CaCl2. Nitrite concentration is plotted on the lower x-axis and hydroxylamine concentration is plotted on the upper x-axis.
Whatever the cause of the boost, our data clearly indicate that this process is distinct from the process responsible for the switch during nitrite turnover since the two processes can be observed simultaneously (Figure 2C). Thus, the switch is not converted into a boost as nitrite concentration is raised; these separate phenomena simply can be observed under different conditions.
The data presented here also addresses the hypothesis that hydroxylamine is an intermediate along the nitrite reduction pathway.(8) The fact that Ecat1 and Ecat2 are observed within the same window of potential for both nitrite and hydroxylamine reduction (Figures 2A, 4) suggests that the detected electron transfer steps represented by Ecat1 and Ecat2 represent similar processes. This could indicate that hydroxylamine is indeed an intermediate in the nitrite reduction pathway. Further characterization of these features is currently underway.
Stoichiometry of detected electron transfer steps
With ecNrfA, turnover of sub-KM concentrations of nitrite and hydroxylamine on an electrode are characterized by sharp n = 2 main waves, which were interpreted as being the result of electronic coupling between the active site heme and heme 3.(12, 15) This was also supported by the results of EPR potentiometric titrations of ecNrfA, where upon reduction, signals at g = 3.5 and 10.8 disappeared without the formation of signals which would be expected to arise if the hemes were uncoupled.(7, 12) The titration of the g = 3.5 and 10.8 signals, however, could not be fitted to n = 2 Nernstian curves.(7) Additionally, results of spectroelectrochemical and MCD titrations provided no evidence for an n = 2 process in the thermodynamic properties of the enzyme.(37) It is therefore not known why PFV studies of ecNrfA revealed n = 2 steps but it has been suggested that this is the result of a slow electrochemical step occurring immediately after two one-electron steps whose thermodynamics are such that they appear as a cooperative process.(15)
A longstanding question about the catalytic mechanism of ccNiRs, has been the stoichiometry by which electrons are delivered to substrate during catalysis. Einsle and coworkers have proposed a mechanism on the basis of crystallography and DFT calculations where electrons are delivered to substrate in a combination of one- and two-electron steps.(8) Here we have shown that all catalytic waves observed during both nitrite and hydroxylamine turnover by soNrfA (at low and high substrate concentrations) are consistent with one-electron processes under all of the conditions tested. Our data is also consistent with non-turnover PFV studies as well as results from spectroelectrochemistry of soNrfA which found no evidence of heme centers being reduced in cooperative two-electron steps (11) — the cooperative two-electron reduction of a pair of heme centers would presumably be the mechanism by which electrons could be delivered to substrate in coordinated two-electron transfers. Our inability to detect catalytic features consistent with two-electron processes, combined with all other available data, suggests that during catalysis electrons are not delivered to substrate in coordinated transfers of two-electrons, and are instead delivered in a series of one-electron steps.
Modest Effect of Ca2+ on shape on the shape of catalytic waves
We have found that adding CaCl2 to the electrochemical cell solution during catalytic PFV experiments has only a modest effect on the overall shape of catalytic waves observed during both nitrite and hydroxylamine turnover by PFV. In catalytic PFV experiments involving ecNrfA, it was found that increasing the CaCl2 in the reaction mixture from 0 to 0.5 mM significantly improved the reversibility catalytic waves, such that the reductive and oxidative scans became more superimposable.(12, 16) The origin of the sensitivity to CaCl2 has been hypothesized as being the result of an exchangeable Ca2+ ion, which binds to one of two structurally conserved Ca2+ binding sites within each enzyme monomer, one of which is close to the active site heme.(12, 16) While these Ca2+ binding sites are present in the recently reported crystal structure of soNrfA, the presence of either Ca2+ or chelators had no effect on the activity of soNrfA in solution assays.(11) The reason for the difference in Ca2+ sensitivity between the ec and so enzymes is not currently known, but it is clear that including Ca2+ does not significantly affect catalysis in the case of soNrfA.
Atypical kinetics during nitrite and hydroxylamine reduction
We have found that titration of soNrfA with either nitrite or hydroxylamine, when monitored using PFV, yields atypical kinetic profiles (Figure 3 and 5). In the case of nitrite turnover, increasing substrate concentration beyond 1 mM leads to a decrease in the activity of soNrfA with increasing nitrite concentration. This apparent substrate inhibition has not previously been reported for any cytochrome c nitrite reductase. Additionally, titrations of soNrfA with hydroxylamine lead to data that does not reach a constant velocity as rapidly as would be expected for simple Michaelis-Menten kinetics.
Based on our data, we believe that a simple Michaelis-Menten model of enzyme kinetics is insufficient to describe catalysis in soNrfA, and possibly other ccNiRs. Failure to account for the atypical portion of kinetic plots, by truncating the data for example, can lead to incorrect estimates of kinetic values. In the case of nitrite turnover for example, previously reported values of KM and kcat may actually be a sum of the increasing (normal activity) and decreasing (substrate inhibition) components of these values, or differential reactivity of the two monomers in a NrfA homodimer.(38, 39) Thus, by accounting for these atypical kinetic profiles, more complex models are required to provide accurate estimates of soNrfA kinetic values.
There are a number of possible reasons why substrate inhibition is observed during nitrite reduction. The data we have obtained fits reasonably well to a model describing an inactive ternary complex that forms upon binding of a second substrate molecule to a monomer (Eq 1). This is an interesting result, because it suggests that soNrfA contains a second nitrite binding site in each monomer to which it binds very weakly (mM range). An alternative explanation is that there is an allosteric interaction responsible for the substrate inhibition.
In contrast, the hydroxylamine data fits well to a model which incorporates two values each for KM and kcat (Eq 2). This suggests that there could be two binding sites within each soNrfA monomer: one with high activity and high affinity, and one with low activity and low affinity. While there is no obvious place for the binding of a second molecule of hydroxylamine to the substrate, this hypothesis would need to be tested by crystallizing soNrfA in the presence of high concentrations of substrate. This data could also be explained by an interaction between monomers in an soNrfA dimer, which leads to a form of negative cooperativity. It seems that in the case of soNrfA, if there were an interaction between monomers, it affects both substrate binding as well as enzyme activity. Alternatively, the biphasic Michaelis-Menten behavior can be explained by the presence of two distinct enzyme populations upon the electrode, capable of catalyzing the same reaction. We feel that this is unlikely, given the purity of the enzyme samples used and that if there were multiple conformations of soNrfA existing on the electrode surface, it seems unlikely that there would be exactly two populations, instead of a distribution of differing states.
Considering the steady-state kinetics observed in soNrfA for the two substrates, a number of hypotheses can be developed. Because hydroxylamine is presumably an intermediate along the nitrite reduction pathway, the atypical kinetics of both nitrite and hydroxylamine reduction are likely interrelated. One explanation is that there is a second substrate binding site in each monomer of soNrfA which binds both nitrite and hydroxylamine. This substrate binding site has an inhibitory effect when nitrite is bound, but increases activity and lowers substrate affinity when hydroxylamine is bound. Insight into this possibility could be gained by experiments using both nitrite and hydroxylamine as substrates simultaneously, where under certain conditions one substrate may be predominately bound to the allosteric site and the reaction of the second substrate would be affected. However, this type of experiment is challenging, as it would require a way to selectively monitor turnover of just one substrate — a task which is difficult with PFV due to the similar reduction potentials of ccNiR turnover of nitrite and hydroxylamine. Alternatively, if the kinetic profile during hydroxylamine reduction were the result of an interaction between monomers in soNrfA via cooperativity, it is tempting to propose that this interaction between ccNiR monomers is also responsible for the substrate inhibition kinetics observed during nitrite turnover. If this were the case, it is not a second binding site in each monomer that is the result of the inhibition, it is instead the two substrate-bound form of the dimer, where one substrate is bound to each monomer, that is the inactive form of soNrfA.
Such a structurally-based model is attractive as the two monomers interact via an extensive dimer interface made up primarily of long alpha-helices that span the length of one side of each monomer.(7) In addition, heme 5 in each monomer lies very close to this interface, such that the two heme 5 Fe atoms are located within 12 Å of each other, suggesting efficient intra-dimer electron transfer is possible. This has led to the proposal that the two monomers interact with one another electronically, such that electrons can be shuttled between the two monomers during catalysis via these closely interacting hemes.(9, 17, 41) Here, we add to this model, by suggesting the possibility that active sites in each monomer in an soNrfA dimer are not isolated from one another. The precise nature of this communication between active sites is not known, but it appears to be a form of negative cooperativity, where increasing substrate decreases the affinity of the enzyme for that substrate while simultaneously causing a slight increase in activity. Why binding of hydroxylamine and nitrite produce different effects - biphasic kinetics in the former and substrate inhibition in the latter - is not known. However, this difference may be due to different conformations that result from the binding of each substrate.
CONCLUSIONS
Here we have characterized the cytochrome c nitrite reductase from Shewanella oneidensis using protein film voltammetry. We have found distinct characteristics in its electrochemical fingerprint that set it apart from the previously characterized E. coli enzyme. In particular, we detect only one-electron electron transfer steps during soNrfA catalysis of nitrite and hydroxylamine which, combined with other data, suggest during soNrfA turnover electrons are delivered to substrate in a series of one-electron transfers. Additionally, we found that the reactivation step that occurs after the switch is quite slow. Despite the differences that we have observed between the soNrfA and ecNrfA enzymes, we have found that the current-potentials profile for both nitrite and hydroxylamine turnover by these enzymes are globally similar. This suggests that catalysis in both the soNrfA and ecNrfA enzymes, and possibly other ccNiRs, are likely governed by similar rate-defining events, as suggested previously by Butt and colleagues.(12) Importantly, we have also found evidence of substrate inhibition during nitrite turnover, as well as biphasic kinetics during hydroxylamine turnover. The mechanism for these atypical kinetics is not known; however these results suggest the possibility of an interaction between monomers that make up a functional soNrfA.
Supplementary Material
Acknowledgments
Funding Source Statement This research was supported by the National Institutes of Health (S.J.E. GM072663, E.T.J. grant F31-GM099416) and the National Science Foundation (A.A.P. NSF-0843459 and NSF-1121770)
Abbreviations
- NrfA
cytochrome c nitrite reductase
- soNrfA
Shewanella oneidensis NrfA
- ecNrfA
Escherichia coli NrfA
- PFV
protein film voltammetry
- PGE
pyrolytic graphite edge
- FRD
fumarate reductase
Footnotes
SUPPORTING INFORMATION AVAILABLE Rotation rate dependence of nitrite and hydroxylamine turnover, fits of catalytic waves to one and two electron models and a description of the fitting methods, dependence of hysteresis on scan rate during nitrite turnover, PFV data showing negligible film loss during nitrite turnover, variable ionic strength experiments, and PFV controls for hydroxylamine reactivity are included. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Simon J. Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol Rev. 2002;26:285–309. doi: 10.1111/j.1574-6976.2002.tb00616.x. [DOI] [PubMed] [Google Scholar]
- 2.Potter LC, Millington P, Griffiths L, Thomas GH, Cole JA. Competition between Escherichia coli strains expressing either a periplasmic or a membrane-bound nitrate reductase: does Nap confer a selective advantage during nitrate-limited growth? Biochem J. 1999;344(Pt 1):77–84. [PMC free article] [PubMed] [Google Scholar]
- 3.Pope NR, Cole JA. Generation of a membrane potential by one of two independent pathways for nitrite reduction by Escherichia coli. J Gen Microbiol. 1982;128:219–222. doi: 10.1099/00221287-128-1-219. [DOI] [PubMed] [Google Scholar]
- 4.Gao H, Yang ZK, Barua S, Reed SB, Romine MF, Nealson KH, Fredrickson JK, Tiedje JM, Zhou J. Reduction of nitrate in Shewanella oneidensis depends on atypical NAP and NRF systems with NapB as a preferred electron transport protein from CymA to NapA. ISME J. 2009;3:966–976. doi: 10.1038/ismej.2009.40. [DOI] [PubMed] [Google Scholar]
- 5.Stach P, Einsle O, Schumacher W, Kurun E, Kroneck PM. Bacterial cytochrome c nitrite reductase: new structural and functional aspects. J Inorg Biochem. 2000;79:381–385. doi: 10.1016/s0162-0134(99)00248-2. [DOI] [PubMed] [Google Scholar]
- 6.Kajie S, Anraku Y. Purification of a hexaheme cytochrome c552 from Escherichia coli K 12 and its properties as a nitrite reductase. Eur J Biochem. 1986;154:457–463. doi: 10.1111/j.1432-1033.1986.tb09419.x. [DOI] [PubMed] [Google Scholar]
- 7.Bamford VA, Angove HC, Seward HE, Thomson AJ, Cole JA, Butt JN, Hemmings AM, Richardson DJ. Structure and spectroscopy of the periplasmic cytochrome c nitrite reductase from Escherichia coli. Biochemistry. 2002;41:2921–2931. doi: 10.1021/bi015765d. [DOI] [PubMed] [Google Scholar]
- 8.Einsle O, Messerschmidt A, Huber R, Kroneck PM, Neese F. Mechanism of the six-electron reduction of nitrite to ammonia by cytochrome c nitrite reductase. J Am Chem Soc. 2002;124:11737–11745. doi: 10.1021/ja0206487. [DOI] [PubMed] [Google Scholar]
- 9.Einsle O, Stach P, Messerschmidt A, Simon J, Kroger A, Huber R, Kroneck PM. Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs. J Biol Chem. 2000;275:39608–39616. doi: 10.1074/jbc.M006188200. [DOI] [PubMed] [Google Scholar]
- 10.Einsle O, Messerschmidt A, Stach P, Bourenkov GP, Bartunik HD, Huber R, Kroneck PM. Structure of cytochrome c nitrite reductase. Nature. 1999;400:476–480. doi: 10.1038/22802. [DOI] [PubMed] [Google Scholar]
- 11.Youngblut M, Judd ET, Srajer V, Sayyed B, Goelzer T, Elliott SJ, Schmidt M, Pacheco AA. Laue crystal structure of Shewanella oneidensis cytochrome c nitrite reductase from a high-yield expression system. J Biol Inorg Chem. 2012;17:647–662. doi: 10.1007/s00775-012-0885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angove HC, Cole JA, Richardson DJ, Butt JN. Protein film voltammetry reveals distinctive fingerprints of nitrite and hydroxylamine reduction by a cytochrome C nitrite reductase. J Biol Chem. 2002;277:23374–23381. doi: 10.1074/jbc.M200495200. [DOI] [PubMed] [Google Scholar]
- 13.Gates AJ, Kemp GL, To CY, Mann J, Marritt SJ, Mayes AG, Richardson DJ, Butt JN. The relationship between redox enzyme activity and electrochemical potential-cellular and mechanistic implications from protein film electrochemistry. Phys Chem Chem Phys. 2011;13:7720–7731. doi: 10.1039/c0cp02887h. [DOI] [PubMed] [Google Scholar]
- 14.Leger C, Elliott SJ, Hoke KR, Jeuken LJ, Jones AK, Armstrong FA. Enzyme electrokinetics: using protein film voltammetry to investigate redox enzymes and their mechanisms. Biochemistry. 2003;42:8653–8662. doi: 10.1021/bi034789c. [DOI] [PubMed] [Google Scholar]
- 15.Leger C, Bertrand P. Direct electrochemistry of redox enzymes as a tool for mechanistic studies. Chem Rev. 2008;108:2379–2438. doi: 10.1021/cr0680742. [DOI] [PubMed] [Google Scholar]
- 16.Burlat B, Gwyer JD, Poock S, Clarke T, Cole JA, Hemmings AM, Cheesman MR, Butt JN, Richardson DJ. Cytochrome c nitrite reductase: from structural to physicochemical analysis. Biochem Soc Trans. 2005;33:137–140. doi: 10.1042/BST0330137. [DOI] [PubMed] [Google Scholar]
- 17.Gwyer JD, Richardson DJ, Butt JN. Diode or tunnel-diode characteristics? Resolving the catalytic consequences of proton coupled electron transfer in a multi-centered oxidoreductase. J Am Chem Soc. 2005;127:14964–14965. doi: 10.1021/ja054160s. [DOI] [PubMed] [Google Scholar]
- 18.Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, Nealson KH, Osterman AL, Pinchuk G, Reed JL, Rodionov DA, Rodrigues JL, Saffarini DA, Serres MH, Spormann AM, Zhulin IB, Tiedje JM. Towards environmental systems biology of Shewanella. Nat Rev Microbiol. 2008;6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 19.Hau HH, Gralnick JA. Ecology and biotechnology of the genus Shewanella. Annu Rev Microbiol. 2007;61:237–258. doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- 20.Beg QK, Zampieri M, Klitgord N, Collins SB, Altafini C, Serres MH, Segre D. Detection of transcriptional triggers in the dynamics of microbial growth: application to the respiratorily versatile bacterium Shewanella oneidensis. Nucleic Acids Res. 2012;40:7132–7149. doi: 10.1093/nar/gks467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eaves DJ, Grove J, Staudenmann W, James P, Poole RK, White SA, Griffiths I, Cole JA. Involvement of products of the nrfEFG genes in the covalent attachment of haem c to a novel cysteine-lysine motif in the cytochrome c552 nitrite reductase from Escherichia coli. Mol Microbiol. 1998;28:205–216. doi: 10.1046/j.1365-2958.1998.00792.x. [DOI] [PubMed] [Google Scholar]
- 22.Pereira IA, LeGall J, Xavier AV, Teixeira M. Characterization of a heme c nitrite reductase from a non-ammonifying microorganism, Desulfovibrio vulgaris Hildenborough. Biochim Biophys Acta. 2000;1481:119–130. doi: 10.1016/s0167-4838(00)00111-4. [DOI] [PubMed] [Google Scholar]
- 23.Costa C, Moura JJ, Moura I, Wang Y, Huynh BH. Redox properties of cytochrome c nitrite reductase from Desulfovibrio desulfuricans ATCC 27774. J Biol Chem. 1996;271:23191–23196. [PubMed] [Google Scholar]
- 24.Fourmond V, Hoke K, Heering HA, Baffert C, Leroux F, Bertrand P, Leger C. SOAS: a free program to analyze electrochemical data and other one-dimensional signals. Bioelectrochemistry. 2009;76:141–147. doi: 10.1016/j.bioelechem.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 25.Fourmond V, Lautier T, Baffert C, Leroux F, Liebgott P-P, Dementin S, Rousset M, Arnoux P, Pignol D, Meynial-Salles I, Soucaille P, Bertrand P, LeÃÅger C. Correcting for Electrocatalyst Desorption and Inactivation in Chronoamperometry Experiments. Analytical Chemistry. 2009;81:2962–2968. doi: 10.1021/ac8025702. [DOI] [PubMed] [Google Scholar]
- 26.Copeland RA. Enzymes : a practical introduction to structure, mechanism, and data analysis. 2nd ed Wiley-VCH; New York: 2000. [Google Scholar]
- 27.Gwyer JD, Zhang J, Butt JN, Ulstrup J. Voltammetry and in situ scanning tunneling microscopy of cytochrome C nitrite reductase on Au(111) electrodes. Biophys J. 2006;91:3897–3906. doi: 10.1529/biophysj.106.089755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutzler JM, Tracy TS. Atypical kinetic profiles in drug metabolism reactions. Drug Metab Dispos. 2002;30:355–362. doi: 10.1124/dmd.30.4.355. [DOI] [PubMed] [Google Scholar]
- 29.Jones AK, Lamle SE, Pershad HR, Vincent KA, Albracht SP, Armstrong FA. Enzyme electrokinetics: electrochemical studies of the anaerobic interconversions between active and inactive states of Allochromatium vinosum [NiFe]-hydrogenase. J Am Chem Soc. 2003;125:8505–8514. doi: 10.1021/ja035296y. [DOI] [PubMed] [Google Scholar]
- 30.Pandelia ME, Fourmond V, Tron-Infossi P, Lojou E, Bertrand P, Leger C, Giudici-Orticoni MT, Lubitz W. Membrane-bound hydrogenase I from the hyperthermophilic bacterium Aquifex aeolicus: enzyme activation, redox intermediates and oxygen tolerance. Journal of the American Chemical Society. 2010;132:6991–7004. doi: 10.1021/ja910838d. [DOI] [PubMed] [Google Scholar]
- 31.Parkin A, Cavazza C, Fontecilla-Camps JC, Armstrong FA. Electrochemical investigations of the interconversions between catalytic and inhibited states of the [FeFe]-hydrogenase from Desulfovibrio desulfuricans. Journal of the American Chemical Society. 2006;128:16808–16815. doi: 10.1021/ja064425i. [DOI] [PubMed] [Google Scholar]
- 32.Lamle SE, Albracht SP, Armstrong FA. Electrochemical potential-step investigations of the aerobic interconversions of [NiFe]-hydrogenase from Allochromatium vinosum: insights into the puzzling difference between unready and ready oxidized inactive states. Journal of the American Chemical Society. 2004;126:14899–14909. doi: 10.1021/ja047939v. [DOI] [PubMed] [Google Scholar]
- 33.Leger C, Dementin S, Bertrand P, Rousset M, Guigliarelli B. Inhibition and aerobic inactivation kinetics of Desulfovibrio fructosovorans NiFe hydrogenase studied by protein film voltammetry. Journal of the American Chemical Society. 2004;126:12162–12172. doi: 10.1021/ja046548d. [DOI] [PubMed] [Google Scholar]
- 34.Armstrong FA, Heering HA, Hirst J. Reactions of complex metalloproteins studied by protein-film voltammetry. Chem Soc Rev. 1997;26:169–179. [Google Scholar]
- 35.Sucheta A, Ackrell BA, Cochran B, Armstrong FA. Diode-like behaviour of a mitochondrial electron-transport enzyme. Nature. 1992;356:361–362. doi: 10.1038/356361a0. [DOI] [PubMed] [Google Scholar]
- 36.Heering HA, Weiner JH, Armstrong FA. Direct Detection and Measurement of Electron Relays in a Multicentered Enzyme:‚Äâ Voltammetry of Electrode-Surface Films of E. coli Fumarate Reductase, an Iron,àíSulfur Flavoprotein. Journal of the American Chemical Society. 1997;119:11628–11638. [Google Scholar]
- 37.Marritt SJ, Kemp GL, Xiaoe L, Durrant JR, Cheesman MR, Butt JN. Spectroelectrochemical characterization of a pentaheme cytochrome in solution and as electrocatalytically active films on nanocrystalline metal-oxide electrodes. J Am Chem Soc. 2008;130:8588–8589. doi: 10.1021/ja802641a. [DOI] [PubMed] [Google Scholar]
- 38.LiCata VJ, Allewell NM. Is substrate inhibition a consequence of allostery in aspartate transcarbamylase? Biophys Chem. 1997;64:225–234. doi: 10.1016/s0301-4622(96)02204-1. [DOI] [PubMed] [Google Scholar]
- 39.Lin Y, Lu P, Tang C, Mei Q, Sandig G, Rodrigues AD, Rushmore TH, Shou M. Substrate inhibition kinetics for cytochrome P450-catalyzed reactions. Drug Metab Dispos. 2001;29:368–374. [PubMed] [Google Scholar]
- 40.Cleland WW. [20] Substrate inhibition. In: Daniel LP, editor. Methods in Enzymology. Academic Press; 1979. pp. 500–513. [PubMed] [Google Scholar]
- 41.Clarke TA, Mills PC, Poock SR, Butt JN, Cheesman MR, Cole JA, Hinton JC, Hemmings AM, Kemp G, Soderberg CA, Spiro S, Van Wonderen J, Richardson DJ. Escherichia coli cytochrome c nitrite reductase NrfA. Methods Enzymol. 2008;437:63–77. doi: 10.1016/S0076-6879(07)37004-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.