The successful advancement of biology and medicine necessitates a comprehensive and integrative understanding of the proteome in a healthy individual, and through inference, the identification of those proteins considered to be abnormal and having the potential to herald disease. Protein markers, complementary to gene markers, constitute the molecular basis for personalized medicine. Given the enormity of the cardiovascular disease burden in modern societies, application of a large-scale protein analysis to cardiac physiology and pathophysiology is important for current and future therapeutic approaches. After nearly a decade of investigation within the realm of cardiovascular proteomics, the daunting question remains: “Is proteomics ready to help elucidate normal physiology in health and pathophysiology in cardiovascular disease?”
To this end, the National Heart, Lung, and Blood Institute of the National Institutes of Health had implemented two Proteomics Programs. The first of these programs, the NHLBI Proteomics Initiative, was instrumental in the establishment of ten multi-disciplinary centers to enhance and develop innovative protein based technologies and apply them to relevant biological questions in order to advance our knowledge of heart, lung, blood and sleep disorders. A parallel second program, the NHLBI Clinical Proteomics Program, was initiated with the goal promoting a systematic, comprehensive and large-scale validation of existing and new candidate protein markers that are appropriate for use in the diagnosis and management of heart, lung, blood and sleep disorders. Additionally, a multimarker approach was encouraged to enhance the sensitivity, specificity and predictive value of existing and new markers. These programs have been at the forefront of research and technology in cardiovascular medicine, championing the application of proteomic technologies such as mass spectrometry and protein arrays for cardiovascular research, thus paving the way for the translation of proteomic technologies into the clinical arena.
Salient technological advancements resulting from the NHLBI efforts include: the construction of a cryogenic Fourier Transform MS platform for high sensitivity mass spectra 1, 2; microfluidic chips for high throughput proteomics 3–5, annotated gel markup language open source software for 2-D electrophoresis based proteomics 6, a Trans-Proteomic Pipeline (TPP) for all steps of MS based proteomics 7; a PeptideAtlas for data basing and statistical validation of mass spectrometry data 8; high content, single-cell, phospho-specific flow cytometry 9; and peptoid based microarrays for protein high throughput capture 10. Furthermore, NHLBI efforts have facilitated the transition of recent biomarker discoveries into the validation pipeline. Highlights include the demonstration that ST2-, a marker for biomechanical stress, complements NT-proBNP in predicting cardiovascular death and congestive heart failure in STEMI patients 8–9; the demonstration that thrombus precursor protein, a marker of activated coagulation, in conjunction with established cardiovascular risk factors provides complementary information for risk assessment in acute coronary syndromes 10; the demonstration in stable CAD patients that elevated levels of Lp-PLA2 were highly predictive of nonfatal adverse outcomes independent of traditional CV risk factors 11.
While significant progress has been made in the past decade, proteomic research in the cardiovascular field is now presented with new milestones and exciting goals. Cardiovascular proteomics must overcome a number of challenges to fulfill its’ promises. There is an apparent disconnection between discoveries on the technological front and their effective translation to advance cardiovascular biology and medicine. Major challenges include an inadequate source of high quality reagents (e.g., high quality antibodies), insufficient development of standardized protocols/procedures (e.g., SOPs for biomarker quantification and validation), and limited infrastructure for data organization and data analyses. Current technologies are also pale to discern transient changes; they are not conducive to inferring real time protein network information, nor do they facilitate the capture of temporal changes at high resolution in proteomes. The backbone of the current proteomic instrumentation, the mass spectrometer, has traditionally been more adept at qualitative measurements than quantitative measurements and significant improvements are essential before proteomics can be adopted into the clinical laboratory. In parallel, there is a demand to optimize reagent design, to provide reproducible quantitative measurements, and to optimize sample consumption/throughput to refine multiplex immunoassay platforms. In a more global sense, additional challenges include our limited understanding of which proteins and their combinations are useful to measure, and our inherent need to go beyond correlation to understand biology. A significant barrier for advancing putative biomarkers into clinics is the lack of infrastructure for validation of candidate proteins in well-characterized human samples that are also associated with high quality clinical data, and an insufficient number of investigators with the appropriate expertise.
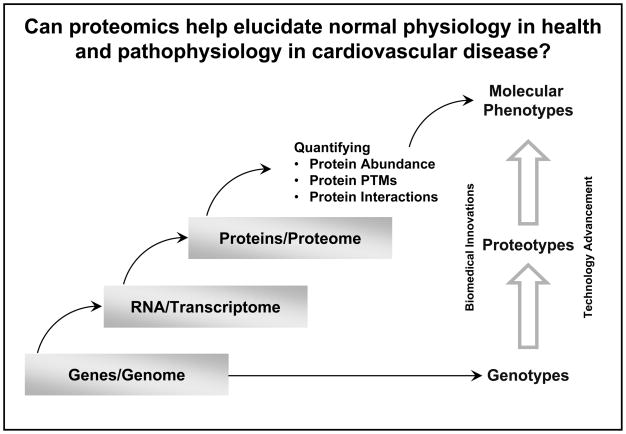
The current objectives of cardiovascular proteomic research include understanding the proteome at multiple domains and at multiple levels of the cardiovascular system. These efforts are as diverse as the physiological effects and signaling networks that operate in response to environmental factors, the reflection of environmental influences in protein compartmentalization, and the ability to detect pathologies prior to decompensation and eruption into symptoms and illness. The ever-growing technological capability to characterize the proteotype of cells and organs, e.g., defining protein post translational modifications, quantifying protein abundance, as well as determining protein interactions, affords new opportunities to advance our understanding in molecular phenotypes (Figure 1). The NHLBI supported technological developments have also vastly increased the capacity for discovery of novel candidate biomarkers or a panel of biomarkers to formulate a “disease fingerprint”. Given the complexity of the discovery, validation and implementation of these discriminatory markers in the clinical arena, requires a sustained effort of the cardiovascular community. This long term commitment is also essential in counteracting the declining rate of introduction of biomarker assays into the clinic.
Figure 1.
Quantitative information on protein abundance, protein post-translational modifications (PTMs), and protein interactions are essential to advance our understanding of molecular phenotypes in cardiovascular biology and medicine.
The accomplishment and experience gained through the NHLBI Proteomics Programs have helped us to identify critical issues in cardiovascular proteomics that we must address. First, proteomic technologies should be applied to enhance our current understanding of cardiovascular biology, function, and molecular phenotypes to help understand causes of heart disease that cannot be explained by known risk factors. Secondly, proteomic technologies are developed to address the dynamics and temporal features with respect to disease pathogenesis–which stage of cardiovascular disease should we be targeting for biomarker discovery? Early detection, diagnosis, risk stratification, and therapy monitoring are a few windows of time that would benefit from reliable markers; but the implementation of these markers needs to be balanced with practical issues. Third, advancement in cardiovascular medicine would be greatly facilitated by a more rigorous examination of the existing biomarkers, in addition to the discovery of those that are novel.
We are confident, however, that through the continued development of proteomic technologies and validation/implementation of easily accessible platforms, a proteomic knowledgebase supporting biological and medical advances will emerge. This knowledge base will drive the successful integration of cardiovascular proteome biology into the clinical setting and it will enable the community to reliably and reproducibly link proteins with specific stages of molecular phenotypes in cardiovascular disease. Our ability to achieve the next phase of goals are supported by the continuation of fruitful partnerships that the NHLBI has nurtured, including investigators, professional scientific societies, voluntary health organizations, patient advocacy groups, community organizations, foundations, corporations, federal-state-local agencies, and international organizations. This collaborative effort drives productivity and propels innovation as we move forward, ensuring success in an ever evolving environment.
Acknowledgments
Funding Sources: NIH/NHLBI
Footnotes
Conflict of Interest Disclosures:
None
References
- 1.Lin C, Mathur R, Aizikov K, O’Connor PB. First signal on the cryogenic Fourier-transform ion cyclotron resonance mass spectrometer. J Am Soc Mass Spectrom. 2007;18:2090–2093. doi: 10.1016/j.jasms.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ro KW, Liu J, Knapp DR. Plastic microchip liquid chromatography-matrix-assisted laser desorption/ionization mass spectrometry using monolithic columns. J Chromatogr A. 2006;1111:40–47. doi: 10.1016/j.chroma.2006.01.105. [DOI] [PubMed] [Google Scholar]
- 3.Stanislaus R, Arthur JM, Rajagopalan B, Moerschell R, McGlothlen B, Almeida JS. An open-source representation for 2-DE-centric proteomics and support infrastructure for data storage and analysis. BMC Bioinformatics. 2008;9:4. doi: 10.1186/1471-2105-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005;1:2005.0017. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deutsch EW, Lam H, Aebersold R. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO. 2008;9:429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krutzik PO, Crane JM, Clutter MR, Nolan GP. High-content single-cell drug screening with phosphospecific flow cytometry. Nat Chem Biol. 2008;4:132–142. doi: 10.1038/nchembio.2007.59. [DOI] [PubMed] [Google Scholar]
- 7.Udugamasooriya DG, Dineen SP, Brekken RA, Kodadek T. A peptoid “antibody surrogate” that antagonizes VEGF receptor 2 activity. J Am Chem Soc. 2008;130:5744–5752. doi: 10.1021/ja711193x. [DOI] [PubMed] [Google Scholar]
- 8.Shimpo M, Morrow DA, Weinberg EO, Sabatine MS, Murphy SA, Antman EM, Lee RT. Serum levels of the interleukin-1 receptor family member ST2 predict mortality and clinical outcome in acute myocardial infarction. Circulation. 2004;109:2186–2190. doi: 10.1161/01.CIR.0000127958.21003.5A. [DOI] [PubMed] [Google Scholar]
- 9.Sabatine MS, Morrow DA, Higgins LJ, MacGillivray C, Guo W, Bode C, Rifai N, Cannon CP, Gerszten RE, Lee RT. Complementary roles for biomarkers of biomechanical strain ST2 and N-terminal prohormone B-type natriuretic peptide in patients with ST-elevation myocardial infarction. Circulation. 2008;117:1936–1944. doi: 10.1161/CIRCULATIONAHA.107.728022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mega JL, Morrow DA, de Lemos JA, Mohanavelu S, Cannon CP, Sabatine MS. Thrombus Precursor Protein and clinical outcomes in patients with acute coronary syndromes. J Amer Coll Cardiol. 2008;51:2422–2429. doi: 10.1016/j.jacc.2008.01.069. [DOI] [PubMed] [Google Scholar]
- 11.Sabatine MS, Morrow DA, O’Donoghue M, Jablonksi KA, Rice MM, Solomon S, Rosenberg Y, Domanski MJ, Hsia J PEACE Investigators. Prognostic utility of lipoprotein-associated phospholipase A2for cardiovascular outcomes in patients with stable coronary artery disease. Arterioscler Thromb Vasc Biol. 2007;27:2463–2469. doi: 10.1161/ATVBAHA.107.151670. [DOI] [PubMed] [Google Scholar]