Abstract
Phosphotyrosine-based signaling plays a vital role in cellular communication in multicellular organisms. Unexpectedly, unicellular choanoflagellates (the closest phylogenetic group to metazoans) possess numbers of tyrosine kinases that are comparable to those in complex metazoans. Here, we have characterized tyrosine kinases from the filasterean Capsaspora owczarzaki, a unicellular protist representing the sister group to choanoflagellates and metazoans. Two Src-like tyrosine kinases have been identified in C. owczarzaki (CoSrc1 and CoSrc2), both of which have the arrangement of SH3, SH2, and catalytic domains seen in mammalian Src kinases. In Capsaspora cells, CoSrc1 and CoSrc2 localize to punctate structures in filopodia that may represent primordial focal adhesions. We have cloned, expressed, and purified both enzymes. CoSrc1 and CoSrc2 are active tyrosine kinases. Mammalian Src kinases are normally regulated in a reciprocal fashion by autophosphorylation in the activation loop (which increases activity) and by Csk-mediated phosphorylation of the C-terminal tail (which inhibits activity). Similar to mammalian Src kinases, the enzymatic activities of CoSrc1 and CoSrc2 are increased by autophosphorylation in the activation loop. We have identified a Csk-like kinase (CoCsk) in the genome of C. owczarzaki. We cloned, expressed, and purified CoCsk and found that it has no measurable tyrosine kinase activity. Furthermore, CoCsk does not phosphorylate or regulate CoSrc1 or CoSrc2 in cells or in vitro, and CoSrc1 and CoSrc2 are active in Capsaspora cell lysates. Thus, the function of Csk as a negative regulator of Src family kinases appears to have arisen with the emergence of metazoans.
Tyrosine kinases are abundant in animals, and they play critical roles in cell growth, differentiation, survival, and motility.1,2 A hallmark of tyrosine kinases is that their enzymatic activity is tightly regulated in normal cells. Aberrant activation of tyrosine kinases (by overexpression, mutation, or gene fusion events) is closely linked to human cancer.3–5 A well-known example is v-Src, the oncogenic tyrosine kinase encoded by Rous sarcoma virus. v-Src is closely related to the cellular Src protein (c-Src) but contains a divergent C-terminal sequence that disrupts c-Src regulation.2
c-Src is a member of a family of nine related nonreceptor tyrosine kinases.2,6,7 The Src family kinases (SFKs) have a conserved domain arrangement that is necessary for their regulation: they possess an N-terminal myristoylation sequence, followed by unique, SH3, SH2, and kinase catalytic domains. A C-terminal tail contains a tyrosine (Y527), which, when phosphorylated, binds to the SH2 domain to inhibit the enzyme. The SH3 domain is also involved in inhibitory interactions with a polyproline helix in the SH2 catalytic linker region.2,8–10 Activation of Src kinases occurs through autophosphorylation in the activation loop at Y416. This is promoted by disruption of the intramolecular inhibitory interactions, either through dephosphorylation of the C-terminal tyrosine or through binding of a ligand to the SH2 or SH3 domain.8,11,12 The conserved domains of Src are also involved in substrate recognition and targeting, so the enzyme is active only at the right localization, and when the correct substrates are present.13,14
The C-terminal Src kinase (Csk) specifically phosphorylates Src family members at Y527, to establish low basal activity.2,15 Csk has a domain structure similar to that of Src; it contains SH3, SH2, and kinase domains but lacks an N-terminal membrane-targeting region and does not contain a tyrosine corresponding to Y416 in the activation loop.16 The regulatory relationship between Src and Csk has been highly conserved throughout the evolution of animals.17,18 The biological necessity for Csk-mediated Src regulation is highlighted in mouse gene knockout studies, where deletion of Csk leads to early stage embryonic death and severe neural tube defects.19
Approximately 600 million years ago, the Cambrian explosion marked the sudden appearance of multicellular animals.20–22 A requirement for multicellularity is the ability of cells to interact and communicate. Signaling components such as tyrosine kinases were therefore necessary for the evolution from unicellular organisms to multicellular animals.23 Many protein domains involved in animal cell signaling, including canonical tyrosine kinases such as Src and Csk, are not present in bacteria, plants, or even fungi, the closest multicellular relatives to animals.24 Surprisingly, recent genome analyses have shown that components of phosphotyrosine-based signaling exist in unicellular choanoflagellates, the closest living relatives to metazoans, suggesting that these may have played a role in the transition to multicellularity.25–28 Choanoflagellates and Metazoa contain many of the same signaling molecules and domains, and their common ancestor might contain these genes. The closest known relative of these two groups is the filasterean Capsaspora owczarzaki, a unicellular protist identified in 1980.29–32 Capsaspora also has a large and diverse complement of tyrosine kinases.33 Here we report that C. owczarzaki has two Src-like kinases (CoSrc1 and CoSrc2), as well as a Csk homologue (CoCsk). Although CoSrc1 and CoSrc2 have a domain organization similar to that of their mammalian counterparts, are active, and share many functional properties, CoCsk displays no activity. The lack of Csk-mediated regulation of Src in Capsaspora suggests that this highly conserved system arose later at the onset of metazoan evolution. We hypothesize the tight regulation of Src was a necessary development in the evolution of proper cell-cell interaction in multicellular animals.
MATERIALS AND METHODS
C. owczarzaki
A culture of C. owczarzaki ATCC30864 was purchased from the American Type Culture Collection (ATCC) and maintained at 23 °C in ATCC medium 1034.
Antibodies and Reagents
Nickel-nitriloacetic acid resin was purchased from Qiagen. Anti-phosphotyrosine antibody (clone 4G10) was from Millipore. Anti-FLAG antibody, EZview Red Anti-FLAG M2 Affinity Gel, CNBr-linked Sepharose, leupeptin, aprotinin, PMSF, sodium vanadate, and ethanolamine were all from Sigma. Anti-V5 antibody and glutathione-agarose-linked beads were from Invitrogen.
cDNA Cloning and Alignments
C. owczarzaki CoSrc1, CoSrc2, and CoCsk were predicted in the draft genome sequence, which is available at http://www.broadinstitute.org/annotation/genome/multicellularity_project/, by the use of Augustus.34 Model parameters of the prediction were specifically optimized for C. owczarzaki. The cDNAs were amplified and cloned from a Capsaspora cDNA library by polymerase chain reaction with specific primers. We confirmed that the sequences have no errors that change amino acids. For expression in insect cells, CoSrc1 and CoSrc2 (residues 331–566 and 395–590, respectively) were cloned into the BamHI and XbaI sites of pFastbac-HtC (Invitrogen). N-terminally FLAG-tagged full-length CoSrc1 and CoSrc2 (566 and 590 residues long, respectively) were expressed in mammalian cells by being subcloned into the XbaI and BamHI restriction sites of p3x-FLAG-CMV (Sigma). The CoCsk cDNA (encoding a protein of 501 residues) was subcloned into pGex-4T-1 (GE Healthcare) by EcoRI and XhoI restriction sites. V5-tagged CoCsk was expressed in mammalian cells by being subcloned into the BamHI and XbaI sites of plasmid pEF1/V5-HisA (Invitrogen). Amino acid sequences were aligned using ClustalW and formatted by BOX-SHADE (version 3.3.1 by K. Hofmann and M. D. Baron) in UCSK workbench 3.2. Phylogenetic tree analysis was performed by the Maximum Likelihood (ML) method. RAxML35 was used with the WAG + Γ model of evolution.36 The amino acid substitution rate was shaped by four rate categories.37
Protein Expression and Purification
His-tagged CoSrc1 and CoSrc2 were expressed in Sf9 insect cells using the Bac-to-Bac system (Invitrogen). Sf9 cells (600 mL) were infected with recombinant CoSrc1 or CoSrc2 baculovirus for 72 h. Cells were lysed in a French pressure cell, and CoSrc proteins were purified on a 5 mL column of nickel-nitrilotriacetic acid resin. Purified proteins were stored in 40% glycerol at −20 °C. Full-length GST-tagged CoCsk was expressed in 1 L Escherichia coli cultures. Proteins were purified on glutathione-agarose-linked beads and eluted with 20 mM glutathione in 50 mM Tris (pH 8.0).
Mammalian Cell Transfection and Western Blotting
Src/Yes/Fyn-deficient (SYF) cells were grown in Dulbecco’s modified Eagle's medium with 10% fetal bovine serum at 30 °C in 5% CO2. Cells were transfected at 50% confluency with TransIT transfection reagent at a ratio of 1:3 (DNA:TransIT). Cells were harvested after 48 h and lysed in RIPA buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA (pH 8), 1 mM phenylmethanesulfonyl fluoride, 1% NP-40, 0.25% sodium deoxycholate, 1 mM Na3VO4, 1 mg/mL aprotinin, and 1 mg/mL leupeptin] for 1 h at 4 °C. Lysates were centrifuged at 14000g for 15 min at 4 °C, separated via 10% SDS-PAGE, and transferred onto a polyvinylidene difluoride membrane. Proteins were detected by Western blotting with anti-phosphotyrosine, anti-FLAG, and anti-V5 antibodies.
Tyrosine Kinase Assays
CoSrc1 and CoSrc2 activity assays were conducted with [γ-32P]ATP using a phosphocellu-lose paper binding assay.38 Reactions were performed in 20mM Tris-HCl (pH 7.4), 10 mM MgCl2, 0.25 mM ATP, varying concentrations of peptide substrate, and [γ-32P]ATP (100–500 cpm/pmol). Peptide substrates were purified by reverse-phase high-pressure liquid chromatography. CoSrc1 and CoSrc2 substrate specificity reactions were performed with the following peptides: Src peptide, AEEEIYGEFEAKKKKG; EGFR peptide, AEEEEYFELVAKKKG; Abl peptide, EAIYAA-PFAKKKG; and Kemptide, LRRASLG. Substrate targeting experiments were performed using the following peptides: SH2 substrate, RRLEDAIYAAGGGGGEPPQpYEEIG; SH2 control, RRLEDAIYAAGGGGGEPPQFEEIG; SH3 substrate, AEEEI-YGEFGGRGAAPPPPPVPRGRG; and SH3 control, AEEEIY-GEFGGRGAAAAAAAVPRGRG.39,40 To study the autophos-phorylation-dependent activity of CoSrc1 and CoSrc2, the enzymes were first treated with immobilized GST-tagged Yersinia tyrosine phosphatase (YOP) for 30 min at 30 °C, followed by incubation with 0.25 mM ATP in kinase assay buffer for 30 min at 30 °C. Samples were prepared in the same way for mass spectrometry analysis. CoSrc (50 pmol) was added to 0.1 M ammonium bicarbonate, 0.1 M DTT, and 0.2 M iodoacetamide. The sample was then digested overnight with trypsin. Results were analyzed on a Thermo Fisher Scientific LTQ Orbitrap CL ETD instrument.
Csk was assayed with [γ-32P]ATP and poly(Glu4-Tyr). The reactions were conducted for 30 min at 30 °C, and then the mixtures were spotted onto Whatman 3MM paper and washed with 5% trichloroacetic acid at 55 °C, followed by liquid scintillation counting. CoCsk activity toward CoSrc1 and CoSrc2 was tested by incubating the proteins in kinase buffer with [γ-32P]ATP. The reactions were stopped by addition of Laemmli buffer and the mixtures analyzed by 10% SDS-PAGE and autoradiography. To quantify phosphorylation, bands were excised, dissolved in hydrogen peroxide, and analyzed by scintillation counting.41
Production and Purification of CoSrc-Specific Antibodies
Specific antibodies recognizing CoSrc1 and CoSrc2 were developed. Peptides derived from their N-terminal unique regions were used to create rabbit antibodies (Genemed Synthesis Inc.). The peptides used to generate antibodies toward CoSrc1 were CQAQQPMLPGQIMAQQ and CSTL-PGQRPGGPGGRV; the peptides used for CoSrc2 were CSNSKPHDPHDSDFKVSPSG and CSTTFTAPTPSTNSLK. These antibodies were then isolated from sera through affinity purification. To prepare affinity matrices, synthetic peptides (corresponding to the sequences used to generate antibodies) were added at a concentration of 0.2 mg/mL to 0.8 g of CNBr-Sepharose that had been swelled for 10 min in 1 mM HCl. These reactions proceeded for 1 h at room temperature and then at 4 °C overnight. Beads were washed, incubated with 100 mM ethanolamine for 1 h at room temperature, washed again, and stored in 500 mM NaCl in PBS; 1–2 mL of affinity matrices was incubated with 20 mL of serum containing CoSrc1 or CoSrc2 antibodies and 30 mL of phosphate-buffered saline overnight at 4 °C. Beads were washed with 500 mM NaCl in phosphate-buffered saline, transferred to a column, and eluted with 0.2 M acetic acid (pH 2.7) and 500 mM NaCl in a volume of 10–20 mL. Antibodies were stored in 50% glycerol.
Localization Experiments
Cells were grown on a coverslip and fixed with 4% paraformaldehyde freshly prepared in an ATCC medium 1034 phosphate buffer solution (pH 6.5) for 10 min. Cells were incubated with rabbit anti-CoSrc antibodies (1:10 concentration), together with mouse anti-α-tubulin (Sigma T-9026; 1:500), overnight at 4 °C. Secondary antibodies [anti-mouse Alexa Fluor 488 and anti-rabbit Alexa Fluor 568 (Invitrogen) in a 1:500 ratio] were then applied after washing, and cells were incubated for 1 h at room temperature. After the secondary antibodies had been washed off, Hoechst 33342 (Sigma) was applied and washed two more times. Cells were then mounted with fluorescence mounting medium (Dako) and observed under a TCS SP5 confocal microscope (Leica). We conducted all these procedures on a coverslip, exchanging liquid gently by pipetting. Cells were kept attached on the glass during the staining and microscopy. Images were processed with ImageJ (http://imagej.nih.gov/ij/).
Immunoprecipitation Assays
FLAG-tagged CoSrc1 and CoSrc2 were expressed in mammalian SYF cells alone or together with CoCsk. Cells were lysed as described above, and CoSrc1 and CoSrc2 were isolated from lysates (1 mg) by incubation with 40 μL of anti-FLAG affinity gel for 2 h at 4 °C. Samples were centrifuged at 5000g for 10 min at 4 °C. After being washed with phosphate-buffered saline, the protein-bound beads were used to measure CoSrc1 and CoSrc2 activity toward a synthetic Src substrate peptide with the phospho-cellulose binding assay.
The CoSrc1- and CoSrc2-specific antibodies described above were used to immunoprecipitate the enzymes from SYF or C. owczarzaki cell lysates. CoSrc1 and CoSrc2 were overexpressed in SYF cells, and endogenous levels of the proteins were used from Capsaspora cells. Cells were lysed in RIPA buffer for 1 h at 4°C and then centrifuged for 10 min at the same temperature. Lysates (1 mg) were incubated with CoSrc1- and CoSrc2-specific antibodies for 1 h at 4 °C, followed by incubation with immobilized Protein A beads for an additional 2 h at 4 °C. The samples were centrifuged, washed with PBS, incubated with peptide substrate and [γ-32P]ATP, and assayed using the phosphocellulose binding assay.
RESULTS
Cloning CoSrc1 and CoSrc2 from C. owczarzaki
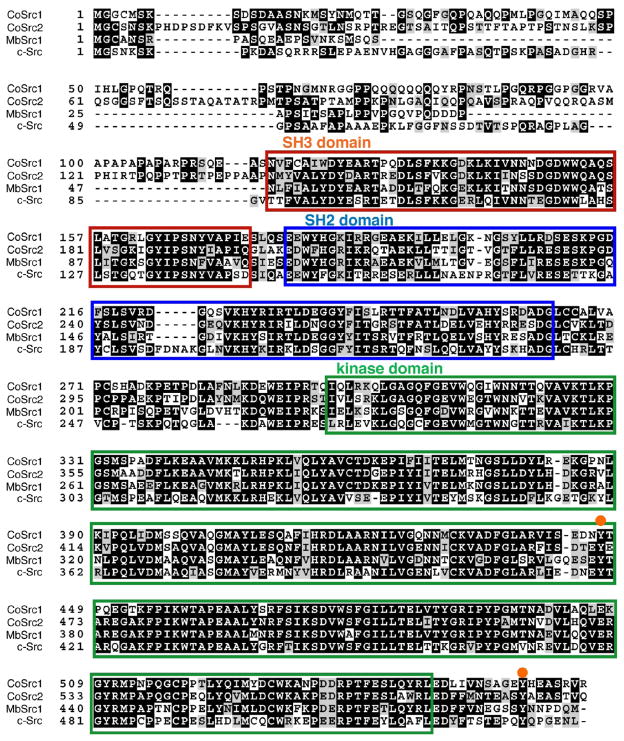
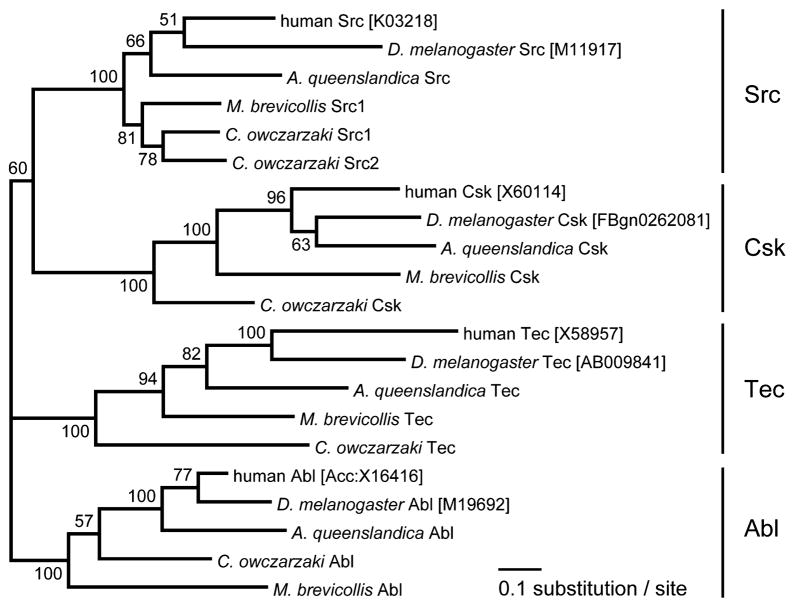
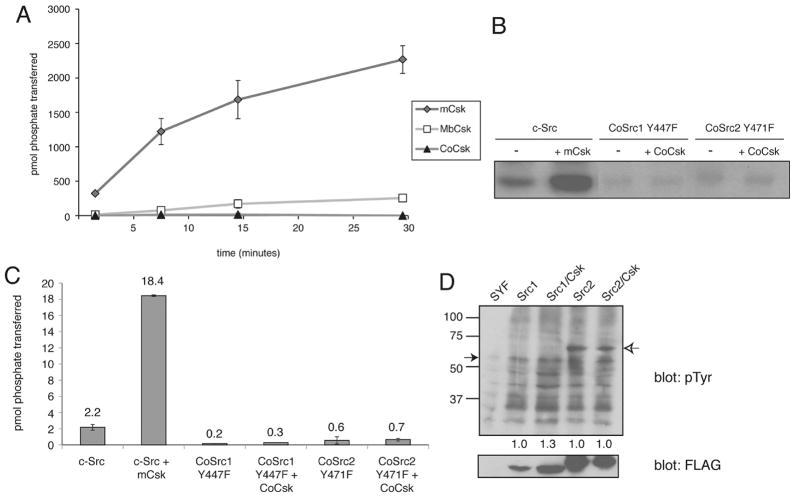
Two Src-like kinases were identified in the genome of C. owczarzaki, which we have designated CoSrc1 and CoSrc2. The Capsaspora Src-like kinases possess predicted SH3, SH2, and kinase catalytic domains in an arrangement similar to that of mammalian Src kinases and Monosiga brevicollis Src, MbSrc1 (Figure 1). A phylogenetic tree is shown in Figure 2, depicting the relationship among tyrosine kinases from C. owczarzaki, the choanoflagellate M. brevicollis, the sponge Amphimedon queens-landica, fly, and human. Sequence conservation in CoSrc1 and CoSrc2 is weakest in the N-terminal regions. The two critical regulatory tyrosines are conserved in Capsaspora (Figure 1): CoSrc1 and CoSrc2 both contain a tyrosine (Tyr-447 and Tyr-471, respectively) corresponding to the site of autophosphorylation in the c-Src activation loop (Y416). Phosphorylation of this residue in mammalian Src kinases increases activity. The conserved tyrosine in the mammalian C-terminal tail (Y527) is also present in CoSrc1 and CoSrc2 (Tyr-558 and Tyr-582, respectively). In mammalian Src kinases, this tyrosine is specifically phosphorylated by Csk, promoting an autoinhibited form of the kinase. In the choanoflagellates Monosiga ovata and M. brevicollis, Csk homologues phosphorylate the Src C-termini but fail to repress Src activity.42,43
Figure 1.
Amino acid sequence alignment of C. owczarzaki CoSrc1 and CoSrc2, M. brevicollis MbSrc1, and human c-Src. The SH3 domain is boxed in red, the SH2 domain boxed in blue, and the kinase domain boxed in green. The orange circles represent conserved regulatory tyrosines: Tyr-416 in the activation loop and Tyr-527 in the C-terminal tail.
Figure 2.
Phylogenetic tree of four cytoplasmic TK families. An alignment of 363 amino acid sites was used for the tree inference. The alignment covers SH3, SH2, and TK domains. GenBank accession numbers or Flybase IDs of sequences are given in brackets. Data for A. queenslandica, M. brevicollis, and C. owczarzaki were from the whole genome sequence. The numbers on branches indicate the statistical nodal support as obtained from 100 maximum likelihood bootstrap replicates using the WAG + Γ model of evolution.
CoSrc1 and CoSrc2 Are Active Tyrosine Kinases
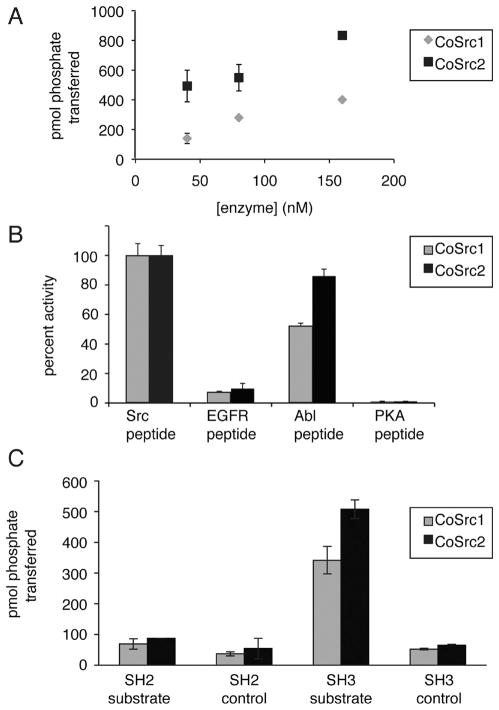
We expressed and purified CoSrc1 and CoSrc2 using the Sf9/ baculovirus system. CoSrc1 and CoSrc2 both showed concentration-dependent activity toward a synthetic Src substrate peptide. CoSrc2 was more active at all protein concentrations than CoSrc1 (Figure 3A). The specific activities of CoSrc1 and CoSrc2 (0.9 × 10−3 and 1.7 × 10−3 mol min−1 mg−1, respectively) were comparable to that of mammalian c-Src assayed in a similar manner (3.0 × 10−3 mol min−1 mg−1).39 Next, we assayed CoSrc1 and CoSrc2 using synthetic peptides containing substrate motifs for various protein kinases. The specificities of CoSrc1 and CoSrc2 were similar to each other, and to that of mammalian c-Src, in that they showed the strongest phosphorylation of the Src substrate peptide, followed by the Abl substrate. Neither showed any activity toward the protein kinase A substrate, consistent with the prediction that they are tyrosine-specific kinases (Figure 3B).
Figure 3.
CoSrc1 and CoSrc2 are active tyrosine kinases. (A) Activity of CoSrc1 and CoSrc2 at varying enzyme concentrations. Enzyme activities were measured with 0.5 mM Src peptide substrate and 0.25 mM ATP using the phosphocellulose paper binding assay. Reactions proceeded for 5 min at 30 °C. (B) Substrate specificities of CoSrc1 and CoSrc2 were investigated using synthetic peptides incorporating recognition motifs from four protein kinases.51 Enzymes were incubated with 0.25 mM [γ-32P]ATP and 200 μM peptide, and reactions proceeded for 20 min at 30 °C. Activities are presented relative to Src peptide (100%) for each enzyme. (C) Substrate targeting by CoSrc1 and CoSrc2. Enzymes (400 nM) were tested with synthetic peptide substrates containing SH3 or SH2 ligand sequences or matched controls (see Materials and Methods for peptide sequences). Reactions proceeded for 30 min at 30 °C, and mixtures were analyzed by the phosphocellulose paper assay. All assays were performed in duplicate, and error bars show standard deviations.
In addition to their involvement in autoinhibitory interactions, the SH2 and SH3 domains of Src family kinases play a role in substrate recognition. This can be recapitulated in vitro; mammalian Src family kinases show higher activity toward peptide substrates to which SH3 or SH2 ligands are attached.39,40 M. brevicollis MbSrc1 displays enhanced phosphorylation of peptides containing SH3 ligands, but not SH2 ligands.42 We tested Capsaspora CoSrc1 and CoSrc2 with substrates possessing SH2 or SH3 ligands (Figure 3C). CoSrc1 and CoSrc2 both phosphorylated the substrate sequence to a higher degree when the SH3 ligand was incorporated into the peptide. CoSrc1 showed a 6.5-fold enhancement (102.1 vs 15.5 μmol min−1 mg−1), whereas CoSrc2 showed a 7.8-fold enhancement (151.6 vs 19.4 μmol min−1 mg−1). Neither enzyme displayed enhanced recognition of a substrate with an SH2 ligand.
Mammalian Src autophosphorylates at the activation loop tyrosine (Tyr-416). This modification increases kinase activity by destabilizing the autoinhibited conformation. To test whether the Capsaspora Src kinases are regulated in a similar manner, we first dephosphorylated the purified Src proteins by treating them with YOP tyrosine phosphatase and then incubated them with ATP to promote autophosphorylation. While dephosphorylated CoSrc1 and CoSrc2 had no detectable activity, the ATP-treated enzymes were active (Figure 4A). We identified the major site of autophosphorylation in ATP-treated CoSrc2 by liquid chromatography and tandem mass spectrometry of tryptic peptides. As expected, the enzyme is autophosphorylated on Tyr-471, the equivalent of the activation loop Tyr-416 in mammalian Src family kinases (Figure S1 of the Supporting Information). To examine the activities of CoSrc1 and CoSrc2 in a cellular context, we expressed them in mammalian Src/Yes/Fyn-deficient fibroblasts. These cells lack all Src family kinases and present a low background level of tyrosine phosphorylation.44 We measured overall tyrosine kinase activity by anti-phosphotyrosine Western blotting. CoSrc2 (or mammalian c-Src) showed robust autophosphorylation and phosphorylation of additional SYF cell proteins, while CoSrc1 showed very little activity (Figure 4B).
Figure 4.
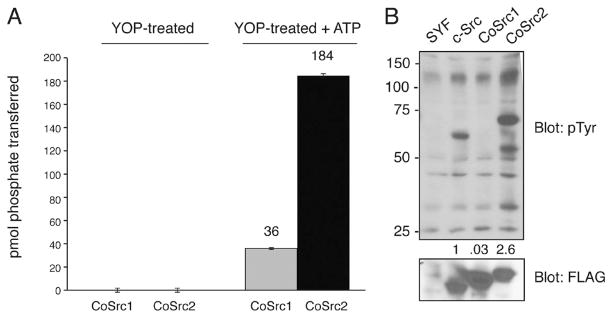
Autophosphorylation of CoSrc. (A) CoSrc1 and CoSrc2 were treated with immobilized GST-YOP for 30 min at 30 °C. The YOP was removed by centrifugation, and the mixture was assayed directly (left bars) or after incubation with ATP for 30 min at 30 °C (right bars). Activity measurements were performed in duplicate with the Src synthetic peptide and the phosphocellulose paper assay, as in Figure 2. (B) Whole cell lysates from untransfected SYF cells, or SYF cells expressing mammalian c-Src, CoSrc1, or CoSrc2, were analyzed by Western blotting with anti-phosphotyrosine antibody. The membrane was stripped and reprobed with anti-FLAG antibody to compare Src expression. The blots were quantitated by dividing the signal for autophosphorylated Src with the signal for total Src (ImageJ), and the value for c-Src was set to 1.0.
Cloning CoCsk from C. owczarzaki
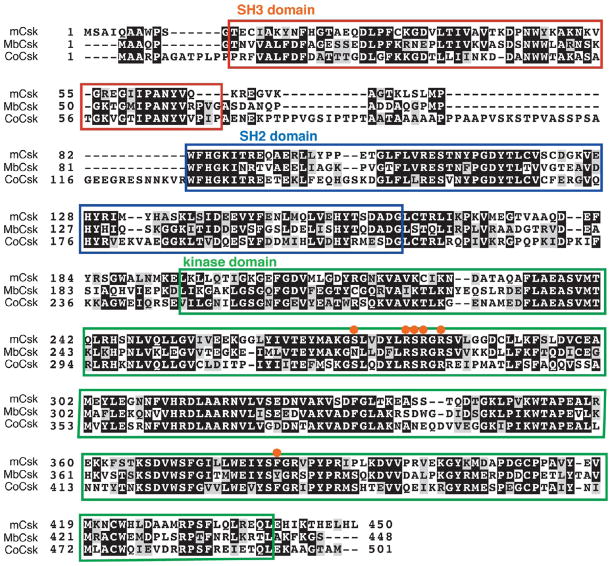
A C-terminal Src kinase (Csk) homologue was identified in the genome of C. owczarzaki. We aligned the sequence with the M. brevicollis Csk (MbCsk) and mammalian Csk (mCsk). CoCsk contains the conserved SH2, SH3, and kinase domain arrangement (Figure 5). A phylogenetic tree comparison is shown in Figure 2. CoCsk possesses all six of the residues previously identified as being important in Src recognition.45 MbCsk is lacking two of these Src recognition sites (Figure 5).
Figure 5.
Amino acid sequence alignment of C. owczarzaki CoCsk, M. brevicollis MbCsk, and human Csk. The SH3 domain is boxed in red, the SH2 domain boxed in blue, and the kinase domain boxed in green. The orange circles represent sites that are important for interaction with Src.45
CoCsk Has No Dectectable Activity
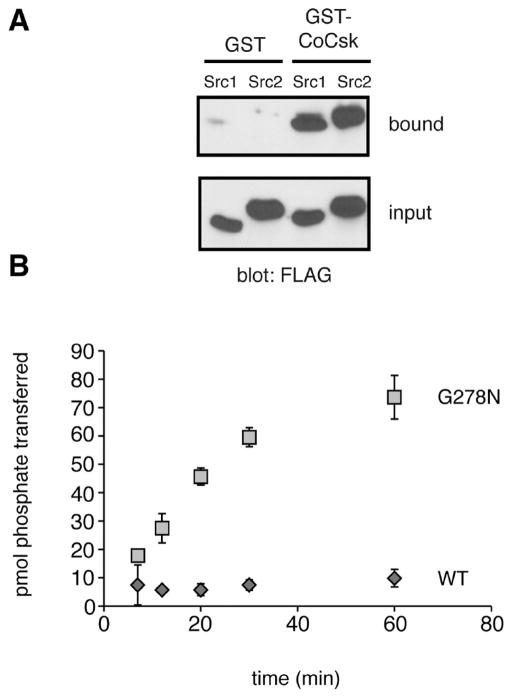
We amplified the cDNAs encoding the Capsaspora Csk homologue (CoCsk). Full-length CoCsk was cloned and expressed in bacterial cells as a fusion protein with glutathione S-transferase (GST). We also produced the corresponding mammalian and M. brevicollis GST-Csk constructs. We purified the GST-tagged proteins by glutathione-agarose chromatography and measured activity toward poly(Glu4Tyr), a general tyrosine kinase substrate (Figure 6A). In contrast to mammalian Csk or M. brevicollis Csk, CoCsk was completely inactive. CoCsk showed no activity toward the Src peptide substrate or any other substrates tested. We tested for phosphorylation of CoSrc1 and CoSrc2 by CoCsk using purified proteins and autoradiography. To reduce the level of background phosphorylation in these experiments, we produced mutant forms of CoSrc1 and CoSrc2 lacking the major sites of autophosphorylation (Y447F and Y471F, respectively). No CoCsk activity toward the Capsaspora Src family kinases, the presumed substrates of CoCsk, was detectable (Figure 6B,C). Mammalian Csk was also unable to phosphorylate CoSrc1 or CoSrc2 in vitro (Figure S2 of the Supporting Information). We also tested the activity of CoCsk toward CoSrc1 and CoSrc2 in mammalian cells. Autophosphorylation site mutant forms of CoSrc1 and CoSrc2 were expressed in mammalian SYF cells in the presence and absence of CoCsk. No additional phosphorylation of CoSrc1 or CoSrc2 was observed in cells co-expressing CoCsk (Figure 6D). To test for binding between CoCsk and CoSrc enzymes, we mixed immobilized GST-CoCsk with lysates from SYF cells expressing CoSrc1 or CoSrc2. Both forms of CoSrc bound to CoCsk, suggesting the possibility that CoCsk possesses a scaffolding function in Capsaspora (Figure 7A).
Figure 6.
Activity of CoCsk is undetectable. (A) Purified CoCsk, M. brevicollis Csk (MbCsk), and human Csk (200 nM) were assayed with poly(Glu4Tyr) (1 mg/mL). Reaction mixtures contained 500 μM [γ-32P]ATP. Activity was analyzed at various time points by scintillation counting. (B). CoCsk does not phosphorylate CoSrc in vitro. Purified CoCsk (400 nM) was incubated with purified CoSrc1 or CoSrc2 (activation loop mutants, 800 nM) in the presence of [γ-32P]ATP. Reactions were stopped by the addition of Laemmli buffer, and mixtures were analyzed by SDS-PAGE and autoradiography. (C) The bands shown in panel B were excised from the gel, dissolved, and analyzed by scintillation counting. (D) Whole cell lysates from SYF cells co-expressing the activation loop mutants (CoSrc1 Y447F or CoSrc2 Y471F) and CoCsk were analyzed by Western blotting with the anti-pTyr antibody. The filled arrow denotes the position of CoSrc1, and the empty arrow denotes the position of CoSrc2. The membrane was stripped and reprobed with anti-FLAG antibody to measure Src expression. The blots were quantitated by dividing the signal for autophosphorylated Src by the signal for total Src. Values in the lanes with CoCsk were compared to the values for CoSrc1 and CoSrc2 alone, which were each set to 1.0.
Figure 7.
Biochemical studies of CoCsk. (A) Binding of CoSrc1 and CoSrc2 to CoCsk. GST-CoCsk (or GST control) was immobilized on glutathione-agarose beads. Lysates (0.5 mg) from SYF cells expressing CoSrc1 or CoSrc2 were incubated with the resins for 1 h at 4 °C. After being washed, bound proteins were analyzed by anti-FLAG Western blotting. For comparison, 5% of the lysates were analyzed by anti-FLAG Western blotting (bottom). (B) The G278N mutation partially restores CoCsk activity. Purified wild-type GST-CoCsk and GST-CoCsk-G278N were assayed with poly(Glu4Tyr) (1 mg/mL). Reaction mixtures contained 500 μM [γ-32P]ATP. Activity was analyzed at various time points by scintillation counting.
Capsaspora CoCsk shows homology with its counterparts from mammals and choanoflagellates (Figure 5). Furthermore, the catalytic domain of CoCsk contains all of the conserved residues important for ATP binding and catalysis (Figure S3 of the Supporting Information). However, unlike the Src kinase domain, which is constitutively active without the presence of its regulatory domains, the kinase domain of Csk requires interactions with its SH2 and SH3 domains for activity.46 Mutagenesis experiments highlighted interactions between the β3-αC loop in the kinase domain and multiple amino acids in the SH2 domain that are necessary for Csk activity.47 The SH3-SH2 linker region is also crucial for Csk activation.48 In CoCsk, there are approximately 20 amino acids in these regions that differ from the residues conserved in mammalian, fly, sponge, and M. brevicollis Csks; these sequence variations could explain the lack of detectable CoCsk activity. We tested this possibility by site-directed mutagenesis of CoCsk Gly-278, which is in the kinase-SH2 interface. In mammalian, fly, sponge, and M. brevicollis Csks, an Asn residue is present at this position. An alanine substitution at this position in mammalian Csk led to a decrease in activity.48 We produced a G278N mutant form of CoCsk, expressed and purified the GST fusion protein, and tested its activity against poly(Glu4Tyr). The mutation led to a partial recovery of CoCsk activity (specific activity of 0.5 μmol min−1 mg−1) (Figure 7B). These results suggest that Gly-278 and other residues in the interface contribute to the low activity of CoCsk. Importantly, these results also argue that the low activity of CoCsk is an intrinsic feature of the protein, rather than (for example) an artifact of protein expression in E. coli.
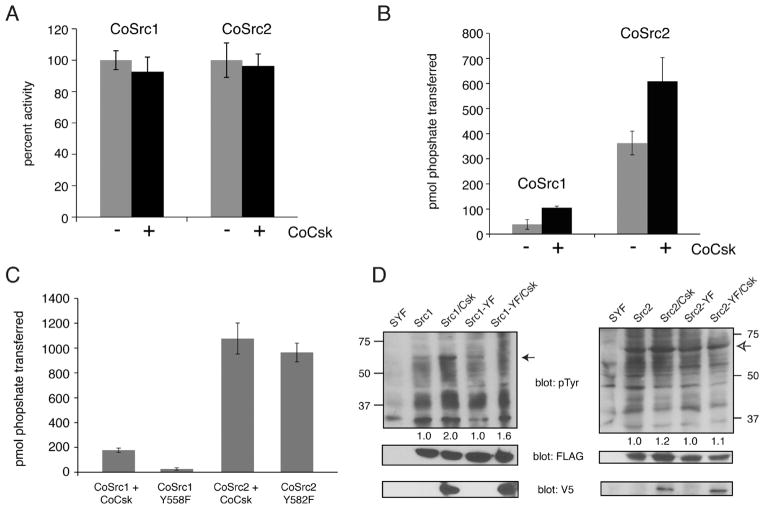
CoCsk Does Not Inhibit CoSrc
Phosphorylation of the C-termini of mammalian Src family kinases by Csk promotes the closed, inactive form. Although we were not able to detect any phosphorylation of CoSrc1 or CoSrc2 by CoCsk in vitro or in SYF cells, we tested for any inhibitory effect of CoCsk. No significant change in in vitro activity of purified CoSrc1 and CoSrc2 was seen upon incubation with CoCsk (Figure 8A). Next, we expressed CoSrc1 and CoSrc2 in SYF cells in the presence or absence of CoCsk. We immunoprecipitated CoSrc1 and CoSrc2 and measured their activities toward a synthetic Src substrate. In contrast to the inhibitory effect of Csk on mammalian Src kinases, co-expression of CoCsk led to modest increases in CoSrc activity (Figure 8B). We also tested CoSrc variants in which the C-terminal tyrosines had been mutated to phenylalanine, eliminating the possibility of negative regulation (CoSrc1 Y558F and CoSrc2 Y582F). The mutant forms of CoSrc1 and CoSrc2 were expressed in SYF cells, immunoprecipitated, and compared to wild-type CoSrc1 and CoSrc2 co-expressed with CoCsk. We observed no significant difference in CoSrc activity between the mutant (and presumably maximally active) forms and wild-type CoSrc isolated from CoCsk-expressing cells (Figure 8C). These results suggest that CoCsk is incapable of promoting CoSrc inhibition. Anti-phosphotyrosine Western blotting of SYF cell lysates showed comparable levels of overall cellular phosphorylation in the presence or absence of CoCsk, consistent with these findings (Figure 8D).
Figure 8.
CoCsk does not inhibit CoSrc Activity. (A) In vitro CoSrc kinase assays were performed in the presence or absence of purified CoCsk. Reaction mixtures contained 500 nM CoSrc1 or CoSrc2, CoCsk (600 nM), 500 μM [γ-32P]ATP, and RCM-lysozyme (0.4 mg/mL) as a Src substrate. After 20 min at 30 °C, reaction mixtures were analyzed by scintillation counting, as described in Materials and Methods. For each enzyme, activity without CoCsk was normalized to 100%. (B) CoSrc1 or CoSrc2 was expressed in SYF cells in the presence and absence of CoCsk. CoSrc proteins were immunoprecipitated with immobilized FLAG antibody and incubated with Src substrate peptide and [γ-32P]ATP. After 20 min at 30 °C, activity was measured using the phosphocellulose paper assay. (C) Similar experiments were conducted to compare wild-type CoSrc1 and CoSrc2 (isolated from SYF cells co-expressing CoCsk) with mutant forms lacking C-terminal tyrosines. In each case, equivalent immunoprecipitation of CoSrc kinases was verified by anti-FLAG Western blotting (data not shown). (D) Western blot analyses of CoSrc1 (left) or CoSrc2 (right) activity (wild-type or C-terminal tail YF mutants) in the presence or absence of CoCsk. Whole cell lysates of SYF cells were analyzed by anti-pTyr blotting. The filled arrow denotes the position of CoSrc1; the empty arrow denotes the position of CoSrc2. Equal expression of CoSrc1 and CoSrc2 was confirmed by anti-FLAG blotting and equal expression of CoCsk by anti-V5 blotting. Quantitation was performed as described in the legend of Figure 6.
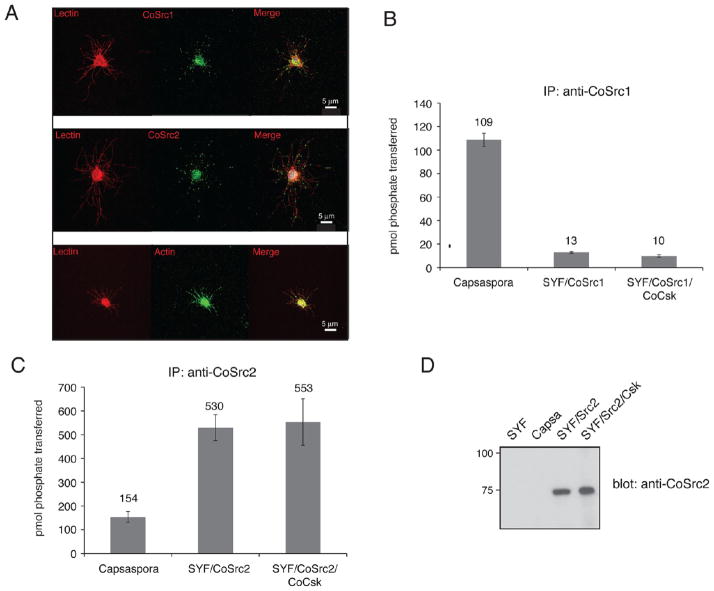
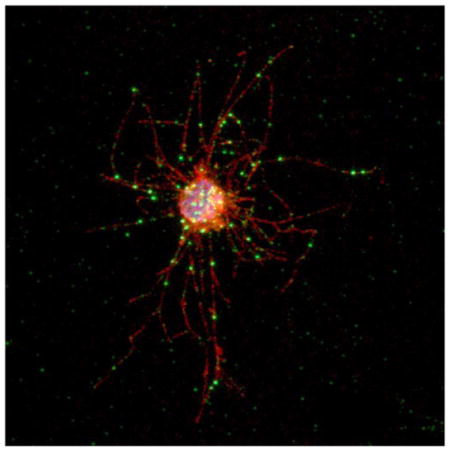
To study endogenous CoSrc1 and CoSrc2 in C. owczarzaki cells, we generated specific antipeptide antibodies. Immunofluorescence experiments showed that CoSrc1 and CoSrc2 had similar localization patterns, with both clustering at specific points along filopodia as well as in the cell body. These points of localization matched those seen along filopodia when cells were probed with an actin antibody (Figure 9A). We used the CoSrc1- and CoSrc2-specific antibodies to immunoprecipitate the enzymes from Capsaspora lysates. Endogenous CoSrc1 and CoSrc2 isolated from Capsaspora cells were active toward a synthetic Src substrate peptide (Figure 9B,C). The levels of activity (≈100–150 pmol of phosphate/1.5 mg of lysate) were on the same order of magnitude as that of CoSrc1 or CoSrc2 overexpressed in SYF cells (≈20 or ≈500 pmol of phosphate/ mg, respectively) (Figure 9B,C). We attempted to analyze the amounts of immunoprecipitated CoSrc by Western blotting. Overexpressed CoSrc1 and CoSrc2 in SYF cells could easily be visualized, while levels of endogenous CoSrc1 amd CoSrc2 protein in Capsaspora cells were undetectable (Figure 9D). These experiments suggest that the specific activities of CoSrc1 and CoSrc2 in Capsaspora cells are higher than those measured in mammalian cells, consistent with a lack of Csk-mediated regulation.
Figure 9.
CoSrc localization and activity in C. owczarzaki cells. (A) Cells were fixed and immunostained with CoSrc1-, CoSrc2-, or actin-specific antibodies prior to fluorescence microscopy. Lectin stains the entire cell as well as the filopodia. (B) Anti-CoSrc1 immunoprecipitation reactions were conducted with lysates from Capsaspora cells or SYF cells overexpressing CoSrc1 in the presence or absence of CoCsk. Activity measurements were performed by the phosphocellulose paper assay with the Src synthetic peptide and [γ-32P]ATP. (C) Anti-CoSrc2 immunoprecipitation reactions were conducted with lysates from Capsaspora cells or SYF cells overexpressing CoSrc2 in the presence or absence of CoCsk. (D) Representative Western blot to compare the levels of CoSrc2 in Capsaspora and SYF cells.
DISCUSSION
Functional studies of the ancestral counterparts to mammalian signaling molecules can provide a window into their evolution. Choanoflagellates, the closest known living relatives of metazoans, possess numerous tyrosine kinases. The Src and Csk family kinases from the choanoflagellates M. brevicollis and M. ovata are broadly similar to the mammalian families in their domain architectures and enzymatic function. In both species, Csk phosphorylates Src but does not tightly regulate activity.42,43 The M. brevicollis enzymes showed some additional differences from mammalian c-Src, including a lack of SH2-directed substrate recognition and a novel C2 lipid-binding domain in one isoform (MbSrc4).42,49 Because these signaling molecules most likely emerged before the split between metazoan and choanoffagellates, we have analyzed the genome of their sister lineage, C. owczarzaki. We have investigated the properties of its two Src-like kinases and one Csk, comparing them to their mammalian counterparts to discover their functional similarities and differences.
The specific activities of CoSrc1 and CoSrc2 toward a synthetic peptide containing an optimal Src recognition sequence are comparable to that of mammalian c-Src. The substrate specificities of CoSrc1 and CoSrc2 also resemble that of c-Src; both phosphorylated the Src substrate synthetic peptide with a greater efficiency than peptides with recognition sequences for other tyrosine kinase substrates (Figure 3B). CoSrc1 and CoSrc2 also displayed the same dependence on the phosphorylation of the activation loop tyrosine observed for mammalian c-Src (Figure 4A). In contrast, although CoSrc1 and CoSrc2 both showed evidence of SH3 substrate targeting as measured by synthetic peptides, the SH2 domain was not coupled to substrate recognition (Figure 3C). These results are similar to those seen for the M. brevicollis MbSrc1.42 The absence of SH2 targeting in both Monosiga and Capsaspora Src’s suggests that functional coupling of the SH2 and kinase domains and autoregulation did not develop until later in SFK evolution.
Genomic analysis has shown that C. owczarzaki contains almost all of the necessary components of mammalian focal adhesions, although these complexes were previously thought to exist in only metazoans. Indeed, Capsaspora is the only nonmetazoan group that possesses an integrin-mediated adhesion pathway with all the same components present in the integrin machinery of metazoans. Choanoflagellates have secondarily lost the integrin adhesion pathway and contain only some of the signaling and scaffolding proteins associated with the integrin adhesion apparatus such as Src and talin.50 This shows that the origins of the protein precursors of the mammalian integrin adhesion complexes predate the transition to multicellularity in the metazoan lineage. Localization experiments showed a general pattern of the presence of CoSrc1 and CoSrc2 in the filopodia of Capsaspora cells, colocalizing with actin (Figure 9A). Capsaspora cells use these filopodia to crawl along a substrate. Thus, a potential explanation is that they use focal adhesion-like complexes to generate force for moving. We hypothesize that ancestral metazoans co-opted this nearly intact machinery to create the modern focal adhesion system. Unfortunately, it is not at present possible to test the physiological roles of CoSrc and CoCsk in intact Capsaspora cells, because the technology does not yet exist to introduce genes or to block their function in this organism.
Functional studies of the Capsaspora Csk revealed an unexpected difference between this protein and all previously studied Csk kinases, including those from the choanoflagellates, M. ovata, and M. brevicollis. In mammalian cells, Csk plays a crucial role in regulating Src activity by phosphorylating a conserved tyrosine at the C-terminal tail. We found that CoCsk displayed no measurable activity toward a synthetic kinase substrate or toward CoSrc (Figure 6). CoCsk was expressed and purified using exactly the same methodology that was used for mammalian Csk46 and M. brevicollis Csk.42 Furthermore, a G278N mutation resulted in a partial recovery of CoCsk activity (Figure 7B), demonstrating that the experimental procedures used to express and assay CoCsk were compatible with the measurement of CoCsk activity. Using our protocolwith [γ-32P]ATP and poly(Glu4-Tyr) as substrates, we conservatively estimate the lower detection limit of our assay with wild-type CoCsk to be a specific activity of 2.5 nmol min−1 mg−1, which is approximately 50 times lower than the activity seen for mammalian Csk (121 nmol min−1 mg−1). CoCsk also had no inhibitory effect on CoSrc activity in vitro or in cells (Figure 8). In the immunoprecipitation-kinase assays from SYF cells (Figure 8B), the presence of CoCsk actually enhanced CoSrc activity, perhaps because of the formation of a complex between the two proteins (Figure 7A); such a complex could stabilize the open form of CoSrc. Immunoprecipitation-kinase assays from C. owczarzaki cells showed that CoSrc1 and CoSrc2 had activities comparable to those of the enzymes overexpressed in mammalian cells (Figure 9). The concentrations of CoSrc1 and CoSrc2 were undetectable in Capsaspora lysates, suggesting that the specific activities are high. This is consistent with a lack of endogenous CoCsk activity. A high basal level of Src activity was also observed in M. ovata.43 Csk may have evolved as an inactive kinase or alternatively as a kinase that did not phosphorylate Src; in the latter case, Csk could have lost its activity in the filasterean lineage. In metazoan cells, Csk is essential for preventing unbridled Src activity; it is likely that the development of the intimate Csk-Src relationship was crucial in the evolution of signaling systems in multicellular animals.
Supplementary Material
Acknowledgments
Funding
Work in W.T.M.’s laboratory was supported by Grant CA58530 from the National Institutes of Health. Work in I.R.-T.’s lab was supported by a European Research Council Starting Grant (ERC-2007-StG-206883) and a grant (BFU2008-02839/BMC) from Ministerio de Ciencia e Innovacion (MICINN).
We thank the Proteomic Facility at Stony Brook for acquisition of mass spectra. The genome sequence of C. owczarzaki is being determined by the Broad Institute of MIT/Harvard University under the auspicies of the National Human Genome Research Institute (NHGRI) and within the UNICORN initiative.
ABBREVIATIONS
- SH3
Src homology domain 3
- SH2
Src homology domain 2
- SFKs
Src family kinases
- SYF
Src, Yes, Fyn knockout cells
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- GST
glutathione S-transferase
- YOP
Yersinia tyrosine phosphatase
Footnotes
The authors declare no competing financial interest.
Additional data and sequence analysis and structural modeling of the CoCsk catalytic domain. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hubbard SR, Till JH. Protein tyrosine kinase structure and function. Annu Rev Biochem. 2000;69:373–398. doi: 10.1146/annurev.biochem.69.1.373. [DOI] [PubMed] [Google Scholar]
- 2.Brown MT, Cooper JA. Regulation, substrates and functions of src. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Kolibaba KS, Druker BJ. Protein tyrosine kinases and cancer. Biochim Biophys Acta. 1997;1333:F217–F248. doi: 10.1016/s0304-419x(97)00022-x. [DOI] [PubMed] [Google Scholar]
- 5.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 6.Bjorge JD, Jakymiw A, Fujita DJ. Selected glimpses into the activation and function of Src kinase. Oncogene. 2000;19:5620–5635. doi: 10.1038/sj.onc.1203923. [DOI] [PubMed] [Google Scholar]
- 7.Parsons SJ, Parsons JT. Src family kinases, key regulators of signal transduction. Oncogene. 2004;23:7906–7909. doi: 10.1038/sj.onc.1208160. [DOI] [PubMed] [Google Scholar]
- 8.Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee CH, Kuriyan J, Miller WT. Activation of the Sire-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 9.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal Structures of c-Src Reveal Features of Its Autoinhibitory Mechanism. Mol Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 10.Sicheri F, Kuriyan J. Structures of Src-family tyrosine kinases. Curr Opin Struct Biol. 1997;7:777–785. doi: 10.1016/s0959-440x(97)80146-7. [DOI] [PubMed] [Google Scholar]
- 11.Miller WT. Determinants of Substrate Recognition in Nonreceptor Tyrosine Kinases. Acc Chem Res. 2003;36:393–400. doi: 10.1021/ar020116v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briggs SD, Sharkey M, Stevenson M, Smithgall TE. SH3-mediated Hck Tyrosine Kinase Activation and Fibroblast Transformation by the Nef Protein of HIV-1. J Biol Chem. 1997;272:17899–17902. doi: 10.1074/jbc.272.29.17899. [DOI] [PubMed] [Google Scholar]
- 13.Burnham MR, Bruce-Staskal PJ, Harte MT, Weidow CL, Ma A, Weed SA, Bouton AH. Regulation of c-SRC activity and function by the adaptor protein CAS. Mol Cell Biol. 2000;20:5865–5878. doi: 10.1128/mcb.20.16.5865-5878.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas JW, Ellis B, Boerner RJ, Knight WB, White GC, Schaller MD. SH2- and SH3-mediated Interactions between Focal Adhesion Kinase and Src. J Biol Chem. 1998;273:577–583. doi: 10.1074/jbc.273.1.577. [DOI] [PubMed] [Google Scholar]
- 15.Okada M, Nakagawa H. A protein tyrosine kinase involved in regulation of pp60c-src function. J Biol Chem. 1989;264:20886–20893. [PubMed] [Google Scholar]
- 16.Cole PA, Shen K, Qiao Y, Wang D. Protein tyrosine kinases Src and Csk: A tail’s tale. Curr Opin Chem Biol. 2003;7:580–585. doi: 10.1016/j.cbpa.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Miller MA, Malik IA, Shenk MA, Steele RE. The Src/Csk regulatory circuit arose early in metazoan evolution. Oncogene. 2000;19:3925–3930. doi: 10.1038/sj.onc.1203714. [DOI] [PubMed] [Google Scholar]
- 18.Suga H, Katoh K, Miyata T. Sponge homologs of vertebrate protein tyrosine kinases and frequent domain shufflings in the early evolution of animals before the parazoan-eumetazoan split. Gene. 2001;280:195–201. doi: 10.1016/s0378-1119(01)00784-3. [DOI] [PubMed] [Google Scholar]
- 19.Nada S, Yagi T, Takeda H, Tokunaga T, Nakagawa H, Ikawa Y, Okada M, Aizawa S. Constitutive activation of Src family kinases in mouse embryos that lack Csk. Cell. 1993;73:1125–1135. doi: 10.1016/0092-8674(93)90642-4. [DOI] [PubMed] [Google Scholar]
- 20.Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. A Phylogenomic Investigation into the Origin of Metazoa. Mol Biol Evol. 2008;25:664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- 21.King N. The unicellular ancestry of animal development. Dev Cell. 2004;7:313–325. doi: 10.1016/j.devcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Rokas A. The molecular origins of multicellular transitions. Curr Opin Genet Dev. 2008;18:472–478. doi: 10.1016/j.gde.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Lim WA, Pawson T. Phosphotyrosine signaling: Evolving a new cellular communication system. Cell. 2010;142:661–667. doi: 10.1016/j.cell.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–363. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- 25.King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JGI, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pincus D, Letunic I, Bork P, Lim WA. Evolution of the phosphotyrosine signaling machinery in premetazoan lineages. Proc Natl Acad Sci USA. 2008;105:9680–9684. doi: 10.1073/pnas.0803161105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manning G, Young SL, Miller WT, Zhai Y. The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan. Proc Natl Acad Sci USA. 2008;105:9674–9679. doi: 10.1073/pnas.0801314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller WT. Tyrosine kinase signaling and the emergence of multicellularity. Biochim Biophys Acta. 2012;1823:1053–1057. doi: 10.1016/j.bbamcr.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hertel LA, Bayne CJ, Loker ES. The symbiont Capsaspora owczarzaki, nov. gen nov sp., isolated from three strains of the pulmonate snail Biomphalaria glabrata is related to members of the Mesomycetozoea. Int J Parasitol. 2002;32:1183–1191. doi: 10.1016/s0020-7519(02)00066-8. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz-Trillo I, Inagaki Y, Davis LA, Sperstad S, Landfald B, Roger AJ. Capsaspora owczarzaki is an independent opisthokont lineage. Curr Biol. 2004;14:R946–R947. doi: 10.1016/j.cub.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 31.Owczarzak A, Stibbs HH, Bayne CJ. The destruction of Schistosoma mansoni mother sporocysts in vitro by amoebae isolated from Biomphalaria glabrata: An ultrastructural study. J Invertebr Pathol. 1980;35:26–33. doi: 10.1016/0022-2011(80)90079-8. [DOI] [PubMed] [Google Scholar]
- 32.Torruella G, Derelle R, Paps J, Lang BF, Roger AJ, Shalchian-Tabrizi K, Ruiz-Trillo I. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single-copy protein domains. Mol Biol Evol. 2012;29:531–544. doi: 10.1093/molbev/msr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suga H, Dacre M, de Mendoza A, Shalchian-Tabrizi K, Manning G, Ruiz-Trillo I. Genomic survey of premetazoans shows deep conservation of cytoplasmic tyrosine kinases and multiple radiations of receptor tyrosine kinases. Sci Signaling. 2012;5:ra35. doi: 10.1126/scisignal.2002733. [DOI] [PubMed] [Google Scholar]
- 34.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(Suppl 2):215–225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 35.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 36.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 37.Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 38.Casnellie JE. Assay of protein kinases using peptides with basic residues for phosphocellulose binding. Methods Enzymol. 1991;200:115–120. doi: 10.1016/0076-6879(91)00133-h. [DOI] [PubMed] [Google Scholar]
- 39.Pellicena P, Stowell KR, Miller WT. Enhanced phosphorylation of Src family kinase substrates containing SH2 domain binding sites. J Biol Chem. 1998;273:15325–15328. doi: 10.1074/jbc.273.25.15325. [DOI] [PubMed] [Google Scholar]
- 40.Scott MP, Miller WT. A peptide model system for processive phosphorylation by Src family kinases. Biochemistry. 2000;39:14531–14537. doi: 10.1021/bi001850u. [DOI] [PubMed] [Google Scholar]
- 41.Patwardhan P, Shen Y, Goldberg GS, Miller WT. Individual Cas Phosphorylation Sites Are Dispensable for Processive Phosphorylation by Src and Anchorage-independent Cell Growth. J Biol Chem. 2006;281:20689–20697. doi: 10.1074/jbc.M602311200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W, Young SL, King N, Miller WT. Signaling Properties of a Non-metazoan Src Kinase and the Evolutionary History of Src Negative Regulation. J Biol Chem. 2008;283:15491–15501. doi: 10.1074/jbc.M800002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Segawa Y. Functional development of Src tyrosine kinases during evolution from a unicellular ancestor to multicellular animals. Proc Natl Acad Sci USA. 2006;103:12021–12026. doi: 10.1073/pnas.0600021103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klinghoffer RA, Sachsenmaier C, Cooper JA, Soriano P. Src family kinases are required for integrin but not PDGFR signal transduction. EMBO J. 1999;18:2459–2471. doi: 10.1093/emboj/18.9.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S, Lin X, Nam NH, Parang K, Sun G. Determination of the substrate-docking site of protein tyrosine kinase C-terminal Src kinase. Proc Natl Acad Sci USA. 2003;100:14707–14712. doi: 10.1073/pnas.2534493100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sondhi D, Cole PA. Domain interactions in protein tyrosine kinase Csk. Biochemistry. 1999;38:11147–11155. doi: 10.1021/bi990827+. [DOI] [PubMed] [Google Scholar]
- 47.Mikkola ET, Gahmberg CG. Hydrophobic Interaction between the SH2 Domain and the Kinase Domain Is Required for the Activation of Csk. J Mol Biol. 2010;399:618–627. doi: 10.1016/j.jmb.2010.04.045. [DOI] [PubMed] [Google Scholar]
- 48.Lin X, Wang Y, Ahmadibeni Y, Parang K, Sun G. Structural Basis for Domain-Domain Communication in a Protein Tyrosine Kinase, the C-terminal Src Kinase. J Mol Biol. 2006;357:1263–1273. doi: 10.1016/j.jmb.2006.01.046. [DOI] [PubMed] [Google Scholar]
- 49.Li W, Scarlata S, Miller WT. Evidence for Convergent Evolution in the Signaling Properties of a Choano-flagellate Tyrosine Kinase. Biochemistry. 2009;48:5180–5186. doi: 10.1021/bi9000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sebé-Pedrós A, Roger AJ, Lang FB, King N, Ruiz-Trillo I. Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc Natl Acad Sci USA. 2010;107:10142–10147. doi: 10.1073/pnas.1002257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Songyang Z, Carraway KL, III, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, Lorenzo MJ, Poner BAJ, Mayer BJ, Cantley LC. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.