Abstract
Viral nanoparticles (VNPs) are structurally regular, highly stable, tunable nanomaterials that can be conveniently produced in high yields. Unmodified VNPs from plants and bacteria generally do not show tissue specificity or high selectivity in binding to or entry into mammalian cells. They are, however, malleable by both genetic and chemical means, making them useful scaffolds for the display of large numbers of cell- and tissue-targeting ligands, imaging moieties, and/or therapeutic agents in a well-defined manner. Capitalizing on this attribute, we modified the genetic sequence of the Cowpea mosaic virus (CPMV) coat protein to display an RGD oligopeptide sequence derived from human adenovirus type 2 (HAdV-2) Concurrently, wild-type CPMV was modified via NHS acylation and Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) chemistry to attach an integrin-binding cyclic RGD peptide. Both types of particles showed strong and selective affinity for several different cancer cell lines that express RGD-binding integrin receptors.
Introduction
The development of therapeutic agents that selectively target and destroy diseased cells while avoiding healthy cells1 is an important goal for modern clinical medicine. Toward this end, nanoparticles bearing targeting, imaging, and/or cytotoxic functional units have been of intense interest. The most popular of these platforms include liposomes, polymer vesicles, quantum dots, nanotubes, and viral nanoparticles (VNPs). VNPs offer several advantages over their synthetic counterparts: they are structurally well-defined, monodisperse, can be produced in high yields, and can be modified both chemically and genetically. Their structural uniformity allows for precise control over functional unit attachment via selective bioconjugation reactions to either their exterior or interior surfaces.2, 3
Taking advantage of their natural mechanisms of infectivity, mammalian VNPs have long been studied as platforms for gene-delivery4 and vaccine development.5–9 We and others, however, have focused on VNPs derived from plants and bacteria, which have no natural ability to infect or propagate in mammalian cells and are therefore regarded as benign materials for biomedical applications.10, 11 Although there are occasional exceptions,12 VNP derivation from non-mammalian viruses means that specific biologically functional elements are not intrinsic to the particles and must be added. One such example is the grafting of poly(ethylene glycol) (PEG) onto the VNP surface in order to decrease immunogenicity,13 reduce undesirable non-specific tissue interactions, prolong plasma circulation, and increase stability of the VNP.14–18 Another example is the chemical attachment of fluorescent labels, allowing tracking of VNPs in vitro and in vivo.19–21 Additionally, specific tissue destinations can be programmed into the particle by the attachment of targeting ligands such as folic acid.22, 23
An attractive receptor class for targeting is the integrins, a large group of heterodimeric transmembrane glycoproteins that attach cells to extracellular matrix molecules. Out of 24 reported integrin receptors, eight recognize the RGD (arginine-glycine-aspartic acid) sequence.24 These eight receptors are responsible for various functions, including cell adhesion, cell survival, binding of blood platelets, cell migration, and angiogenesis.25 Several viruses that use αv integrins to gain entry into host cells26, 27 feature the RGD sequence as the primary integrin binding motif. These viruses include adenovirus (Ad), a nonenveloped DNA virus associated with numerous human respiratory and gastrointestinal illnesses,28 and foot-and-mouth-disease virus (FMDV), a nonenveloped RNA virus that is highly contagious in livestock.29 These two evolutionarily unrelated particles share a remarkable similarity in the spatial distribution of RGD-containing peptides on their surfaces: in each case the RGD sequences are located within a flexible loop positioned at the five-fold symmetry axes with approximately 60 Å separating nearest-neighbor RGD sites.27 This similarity of display pattern suggests that such an arrangement facilitates integrin clustering, thereby promoting an efficient receptor-mediated process of cell entry.30
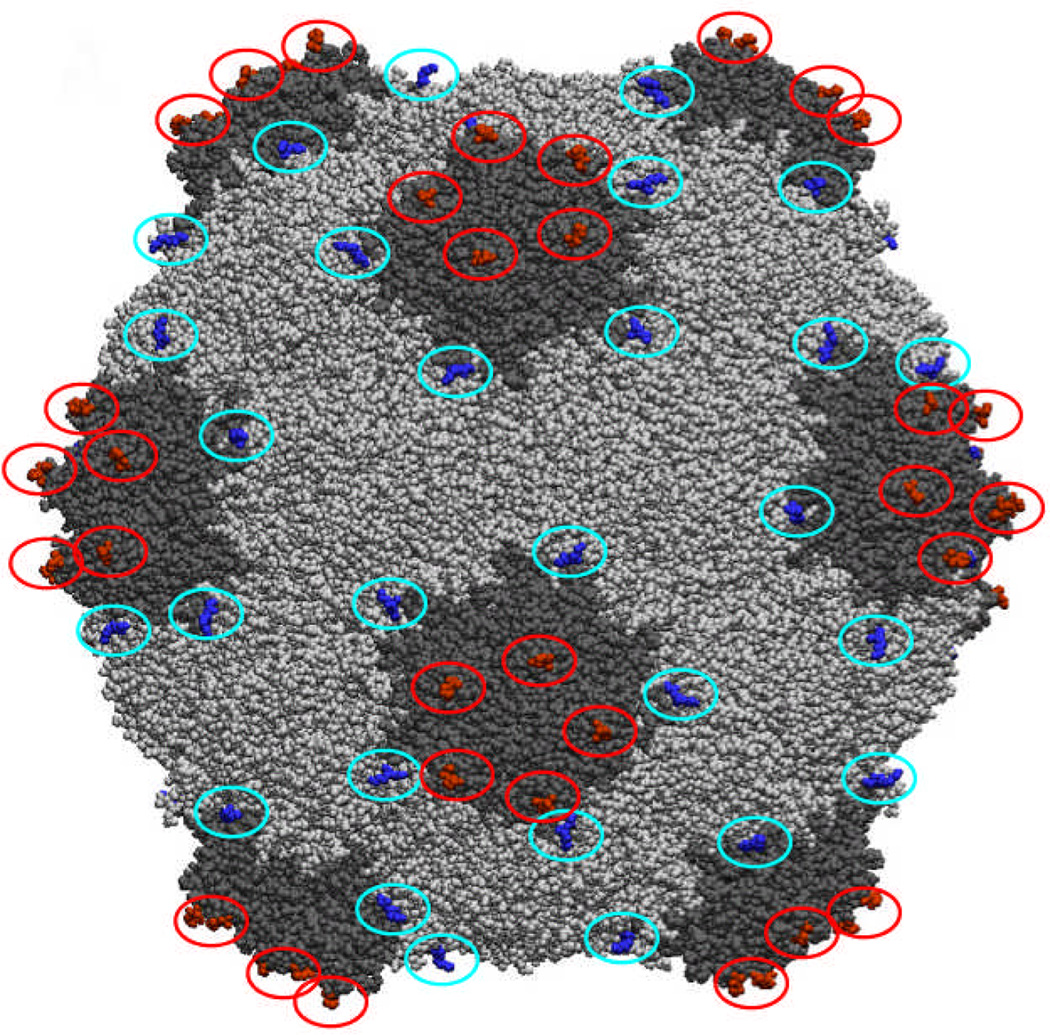
We focus here on the nanoparticles derived from Cowpea mosaic virus (CPMV), a ~30 nm-sized icosahedral plant virus. Its multivalent capsid is composed of 60 copies each of a large (42 kDa) and a small (24 kDa) coat protein, and the structure of the capsid is known to near-atomic resolution (Figure 1).31, 32 CPMV propagates efficiently in black-eyed pea plants and can be obtained in yields of about one gram per kilogram of infected leaf tissue.32 Previous work employing CPMV as a platform for the polyvalent display of foreign peptide sequences revealed that additional amino acids were best installed in one of two solvent exposed sites, the βB-βC loop of the small subunit or the βE-βF loop of the large subunit (Figure 1A).33–35 Genetic inserts in the βB-βC loop are positioned around five-fold symmetry axes with adjacent peptides being separated by ~28 Å, whereas sequences inserted in the βE-βF loop afford peptide spacings of ~55 Å around the five-fold axes and ~65 Å between the nearest neighbors in adjacent five-fold clusters. In addition to this type of genetic modification, CPMV has also been chemically modified by virtue of nucleophilic reactivity of exposed lysine36, 37 and cysteine38 residues. The former provides a high surface loading, with 300 addressable lysine residues on the exterior surface.10, 39 CPMV was also recently addressed by hydrazone ligation and azide-alkyne cycloaddition (click) chemistries to attach a peptidic VEGFR-1 ligand or folic acid for targeted imaging.22, 40 Related work has recently been published by Wang and colleagues, using click chemistry to attach linear RGD-containing peptides to the surface of turnip yellow mosaic virus.41
Figure 1.
Space-filling structure of CPMV, showing the positions of the βB-βC (red) and βE-βF (blue) loops.
Since many cancers overexpress integrin receptors to enable growth of new vasculature toward the tumor tissue,42 RGD-containing compounds have attracted attention for anticancer therapy. To inhibit tumor growth, selective integrin ligands are necessary that bind only to the integrin receptors responsible for angiogenesis, while not interfering with essential biological functions, such as the binding of blood platelets. Natural or linear RGD ligands generally do not possess this level of selectivity, which has led to the development of a large number of optimized peptides and peptidomimetics.43–45 The cellular trafficking of integrin-binding ligands varies depending on their nature and size. For example, small RGD-containing peptides can enter cells through non-receptor-mediated endocytic pathways,25 and can have biochemical effects that depend on,46 or are independent of,47, 48 integrin receptor clustering. Monoclonal antibodies can bind and be internalized by integrin receptors, blocking access to other ligands and slowing recycling of the receptors back to the cell surface.25 Certain cyclic RGD structures target a variety of solid tumors and distribute throughout tumor parenchyma.49, 50
The cyclic pentapeptide Cilengitide [cyclo(RGDfMeV)]51, 52 selectively binds integrin receptors53, 54 αvβ3, αvβ5 and α5β1, which are known cancer-related integrins52, and is currently in phase III clinical trials for the treatment of glioblastoma. Since the behavior of RGD-decorated nanoparticles cannot be assumed to be the same as the peptides alone, we wished to explore RGD decoration by both genetic and chemical means, using CPMV as a prototypical protein nanoparticle.
Experimental Section
Synthesis of the cyclic peptides and PEG linker
cyclo(RGDfMeK) and cyclo(RβADfK) were prepared on solid support using standard Fmoc chemistry.55 Details are given in Supporting Information, along with those for the synthesis of the PEG linker and its connection to the cyclic peptides.
Virus Growth and Purification
CPMV virions were propagated in black-eyed pea plants and purified by ultracentrifugation through sucrose gradients followed by ultrapelleting, as previously described.56 Purified VNPs were stored at 4°C in 0.1 M potassium phosphate buffer at pH 7.0 (referred to as buffer in the following text). The concentration of purified VNPs was determined by Bradford assay or photometrically; at 1 cm path length, a buffered solution of 0.1 mg/mL CPMV gives an absorbance of 0.81 at a wavelength of λ = 260 nm.20
Chemical Modification of Viral Nanoparticles
CPMV particles were functionalized on solvent exposed lysine side chains with azide moieties. To this purpose, 5-(3-azidopropylamino)-5-oxopentanoic acid NHS ester (1.34 µmol, 1500 equiv with respect to CPMV particle) was dissolved in 40 µL DMSO and added to the CPMV solution (final concentration 5 mg/mL, 0.89 µM in particles) to give a final DMSO concentration of less than 20%. After incubation for 4 hours at room temperature, excess reagents were removed using 10 kDa molecular weight cutoff centrifugal filters (Amicon Ultra, Millipore). The virus particles were then derivatized with either RGD or RβAD cyclic peptides (200 equiv per particle, 3.3 equiv per asymmetric unit) using a standard copper-catalyzed azide-alkyne cycloaddition procedure.57 Excess reagents were again removed using 10 kDa molecular weight cutoff filters. Finally, the particles were labeled with NHS ester derivatives of Oregon Green (O488), AlexaFluor 568, or AlexaFluor 555 dyes (Supporting Information). For this purpose, dye conjugated NHS ester (0.5 µmol, 2500 equiv with respect to CPMV VNPs) was dissolved in 50 µL DMSO and added to the CPMV solution (final volume 1.1 mL; final concentration 1 mg/mL = 0.18 µM in particles) to give a final DMSO concentration of less than 20%. The final particles were purified by size-exclusion chromatography (Superose 6 resin). In all cases, the isolated particles were discrete, intact, and of the same diameter as the wild-type structures.
Propagation and Purification of CPMV Chimeras
The installation of the 12-mer RGD sequence into the βB-βC and βE-βF loops of CPMV was achieved by site-directed mutagenesis of the cDNA encoding for the viral coat protein as described elsewhere.38 The primers utilized for the mutagenesis were as follows: BC (forward), 5’-ct agc act cct cca gct gat cac gca ata aga ggt gat aca ttt gca act aga cca ttt tca gac gt-3’; BC (reverse), 5’-c tga aaa tgg tct agt tgc aaa tgt atc acc tct tat tgc gtg atc agc tgg agg agt g-3’; EF (forward), 5’-aga gga gat cat gct att aga gga gat aca ttt gca acc aga aag tat agt ac-3’; EF (reverse), 5’-t ata ctt tct ggt tgc aaa tgt atc tcc tct aat agc atg atc tcc tct-3’. In all cases, the isolated particles were discrete, intact, and of the same diameter as the wild-type structures.
Cell culture
HeLa, HT-29, SW480, A549, 293, and MCF-7 cells were purchased from ATCC (Manassas, VA). All were cultured at 37°C in a humidified 5% CO2/95% air atmosphere, in the following media: HeLa, DMEM or MEM with 7% fetal bovine serum (FBS), 1% l-glutamine, and 1% penicillin-streptomycin (PS); MCF-7, MEM with 10% FBS, 1% l-glutamine, and 1% PS; HT-29, RPMI with 10% FBS, 1% l-glutamine, and 1% PS; SW480, A549, and 293, DMEM with 10% FBS, 1% l-glutamine, and 1% PS. No indications of cellular toxicity were observed for any of the particles at the indicated concentrations with any of the cell lines tested.
Flow cytometry
Cells were collected using enzyme-free cell dissociation buffer (Gibco/Invitrogen) and distributed in 200 µL portions at a concentration of 5 × 106 cells mL−1 in 96-well V-bottom shaped plates. VNP formulations were added to live cells at a concentration of 0.83–8.3 nM (1 × 105−1 × 106 particles per cell) and incubated in media at 37°C for 1 hour prior to fixing with 2% formaldehyde and permeabilizing with 0.2 % (w/v) saponin in FACS buffer (PBS pH 7.4 containing 0.2 % (v/v) 0.5 M EDTA, 2.5% (v/v) 1 M HEPES, and 1% (v/v) FBS). CPMV was labeled with fluorescent dye for detection or stained using a polyclonal anti-CPMV antibody (1:1000 in FACS buffer) followed by a goat anti-rabbit secondary antibody conjugated to AlexaFluor 647 or AlexaFluor 488 (1:2000 in FACS buffer). All steps were carried out at 4°C. Cells were resuspended for FACS analysis, acquiring at least 10,000 gated events. Experiments were repeated at least twice with triplicates of each sample. Data were analyzed using FlowJo 8.6.3 software (Tree Star, Inc.).
ELISA Protocol for Determination of Relative RGD Exposure on CPMV Chimeras
96-well Immulon 2 plates were coated overnight at room temperature with CPMV particles (100 µL, 1 µg/well) in NaHCO3 buffer (0.1 M, pH 8.5) in triplicate. The coating solution was removed and the wells were incubated for 1 h with blocking buffer (PBS, 1% dry skimmed milk), followed by a 5 min wash with PBS. To remove non-specific reactivity between WT CPMV and anti-Ad2 penton base antibodies, the virus was incubated overnight at 4 °C with the probing antibody (blocking buffer, 0.05% Tween-20, 16 mg WT CPMV, 1:200 diluted anti-Ad2 penton base antibodies, total volume of stock solution = 1.5 mL) prior to its use in the assay. Immobilized virus particles were probed by incubating the wells at room temperature with the WT CPMV/anti-Ad2 penton base solution for 1 h, washed 3 × 5 min with PBS containing 0.05% Tween-20, and then treated with HRP-conjugated goat anti-rabbit IgG (1:500 dilution in blocking buffer) for 1 h. The wells were thoroughly washed 4 × 5 min with PBS containing 0.05% Tween-20. After a final 5 min wash using PBS, a 100 µL solution of ABTS peroxidase substrate was added to each well. The absorbance at 405 nm was recorded 0 – 90 min later using a microplate reader.
Confocal microscopy
Approximately 1 × 105 HeLa cells were plated in glass bottom dishes (MatTek) and allowed to adhere overnight at 37°C in a humidified 5% CO2/95% air atmosphere. CPMV particle solutions were prepared in complete growth media directly before addition to the cells. Cells were rinsed once with PBS before CPMV solutions were added to a final concentration of 9 or 18 nM (3.4 and 6.8 × 107 particles/cell). After incubation for 1, 3, or 6 h at 37°C, the cells were washed with PBS at room temperature. The cells were then fixed with 2% paraformaldehyde for 10 min and permeabilized with 0.2% Triton X-100 for 10 min. Staining of endolysosomes and CPMV particles was carried out sequentially, and all antibody dilutions were made in 1% of the serum used for blocking. The first blocking step was performed with 10% goat serum (Omega Scientific) for 1 h, before incubation with anti-human CD107a (Lamp-1) primary antibody (BioLegend) overnight at 4°C. The following morning, staining was performed with goat anti-mouse AlexaFluor 488 conjugated secondary antibody (Cell Signaling Technologies). The second blocking step was done in 10% donkey serum (Jackson ImmunoResearch), and CPMV particles were stained with rabbit polyclonal anti-CPMV antibody, followed by DyLight 549 conjugated donkey anti-rabbit secondary antibody (Biolegend). Nuclei were stained with DAPI (Biotium, Inc.), cell membranes were stained with AlexaFluor 555 conjugated wheat-germ agglutinin (Invitrogen) and glass cover slips were mounted over cells with ImmunO-Fluore mounting media (MP Biomedicals).
Results and Discussion
Genetic incorporation of RGD sequences
Incorporation of the RGD peptide sequence derived from HAdV-2 penton base into the protein capsid of CPMV was achieved by site-directed mutagenesis of the cDNA encoding for the viral coat protein of the plant virus. Initial screening studies included insert lengths of 8, 12, or 16 amino acids in the βB-βC or βE-βF loops. The 8-mer sequences proved too short for sufficient recognition by integrin expressing cells, whereas the 16-mer sequences produced virus particles that were difficult to purify and underwent extensive proteolytic cleavage of the peptide insert. This phenomenon is routinely observed during the insertion of a foreign peptide sequence into the CPMV capsid and the extent of proteolytic cleavage is often proportional to the insert length.33, 59 We therefore focused on the 12 amino acid insert as this peptide length afforded good recognition by integrin binding cells while minimizing proteolytic cleavage of the peptide insert. Two chimeras were constructed, inserting the 12-mer RGD sequence DHAIRGDTFATR60 in the βB-βC loop of the small subunit (BC mutant) or the βE-βF loop of the large subunit (EF mutant). Analysis of the X-ray crystal structure of CPMV31 reveals the spacing between adjacent βB-βC loops to be ~28 Å, whereas the separation of adjacent βE-βF loops is ~60 Å (Figure 1). The resulting particles were isolated in approximately 25% yield relative to wild type (WT) CPMV. Particles were stored at 2 mg/mL, a concentration at which aggregation was not observed after extended storage at 4°C.
Preparation and Characterization of BC and EF Mutants
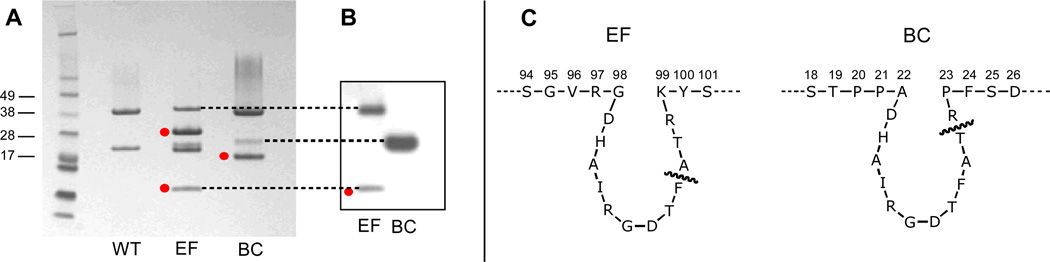
Isolated RNA from purified virions was analyzed by reverse transcription (RT) PCR to confirm the genetic modifications, and insertion of the RGD peptide sequence was determined to be stable over multiple successive passages in plants, as revealed by sequence analysis of RNA isolated from the BC and EF mutants. Size-exclusion FPLC chromatography verified that isolated particles were intact with little or no aggregation. As expected, analysis of the two chimeras by SDS-PAGE revealed that the peptide inserts are located in the small (~24 kDa) and large (~42 kDa) subunits of the BC and EF mutants, respectively, as indicated by a shift in molecular weight of the protein bands corresponding to these subunits (Figure 2A). Furthermore, a western blot using antibodies directed against the RGD sequence of HAdV-2 (anti-HAdV-2 penton base) confirmed binding of the antibodies to the genetically modified subunits of the denatured particles (Figure 2B).
Figure 2.
(A) SDS-PAGE of CPMV particles. Cleaved subunits are marked by red dots; the doubled band for the EF mutant small subunit is sometimes observed in standard preparations.58 (B) Western blot of EF and BC mutants using anti-Ad2 penton base antibodies. (C) Peptide sequences around the sites of insertion for EF and BC chimeras, with the site of cleavage marked in each case.
The presence of additional protein bands of lower molecular weight indicates cleavage of the peptide insert, presumably by plant proteases present during either virus propagation or isolation.33, 59 N-terminal protein sequencing of the cleavage products revealed that the inserted peptides were cleaved between Thr and Arg in the BC mutant, and between Phe and Ala in the EF mutant. Proteolytic processing between the last two residues of inserted peptides in the βB-βC loop is well-established and occurs irrespective of sequence or length,59 whereas the cleavage event in the βE-βF loop has received considerably less attention.61 In neither case is the stability of the particle substantially compromised. Because of these cleavage events, approximately 90% of the inserted sequences on the BC particle were presented as dangling chains rather than loops, containing the RGD sequence and four amino acids on its C-terminal side. For the EF particle, approximately 30% of the loops were uncleaved, but the linearized sequence contained only two amino acids downstream of the RGD motif. This was still enough to be recognized by the anti-HAdV-2 penton base antibody, along with the full-length large subunit of the uncleaved portion of the capsid (Figure 2B). The corresponding Western blot of the BC mutant showed anti-HAdV-2 penton base binding only to the full-length small subunit, since the cleavage product containing the major portion of the inserted sequence was too small to be retained on the gel.
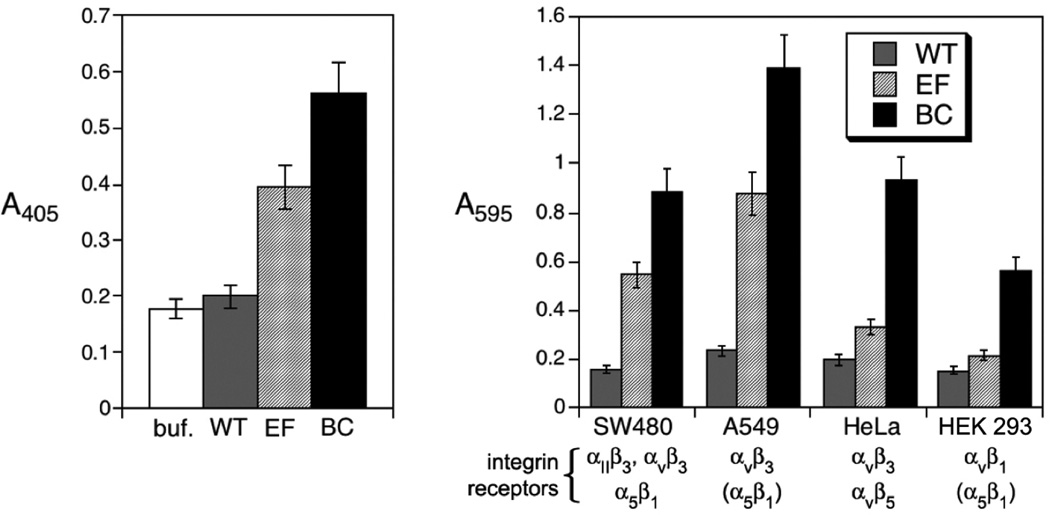
The relative exposure of the inserted RGD sequences in the BC and EF mutants was determined by ELISA using anti-HAdV-2penton base antibodies. Wild-type CPMV, used as a negative control, exhibited substantial interaction and necessitated pre-incubation of the antibodies with WT particles in order to remove non-specific binding (Figure 3A). Under these conditions, the antibodies bound significantly better to the BC mutant than to the EF mutant, consistent with the more solvent-exposed position of the βB-βC loop relative to the βE-βF loop (Figure 1). Cell adhesion aassays were performed to assess the ability of the two CPMV chimeras to bind four human cell lines (SW480, A549, and HeLa cancer cells; HEK293 normal cells) that over-express αv integrins, with the results shown in Figure 3B. In no case did WT CPMV particles bind the cells to a significant extent, whereas the EF mutant bound to two cell lines (SW480 and A549), and the BC insertion promoted strong adhesion to all four cell lines. The superiority of the BC chimera in cell adhesion is consistent with the ELISA results.
Figure 3.
(A) ELISA of CPMV particles using anti-Ad2 penton base antibodies; “buf.” refers to the control wells using only buffer. (B) Cell adhesion assay of CPMV particles using four cell types which overexpress surface integrins.62–64 Integrin receptors in parentheses are reported to not be involved in ligand internalization.
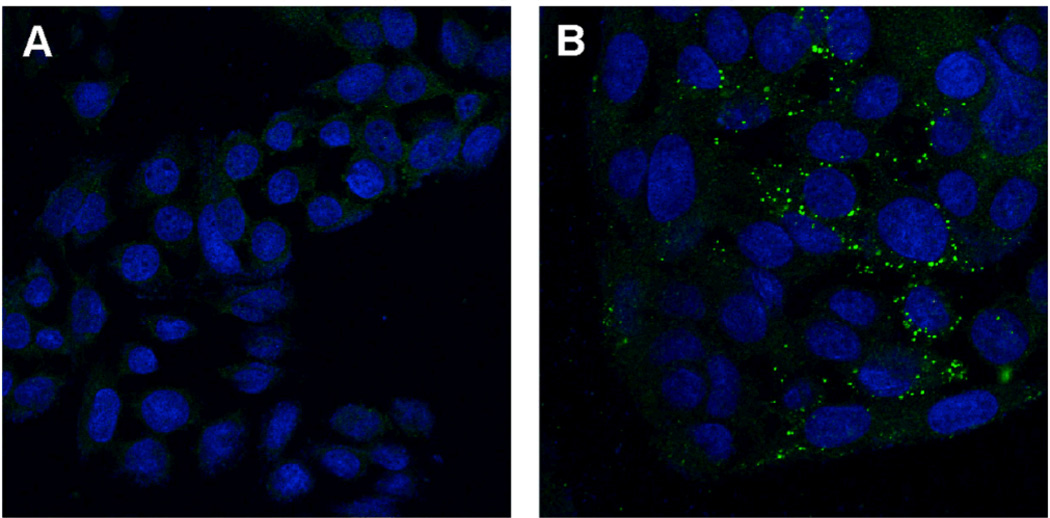
The ability of the inserted Ad-related peptide sequences to stimulate cell entry was probed using confocal microscopy using the superior BC-mutated particles. When the candidate particles were labeled on their exterior surfaces with fluorescein,37 wild-type virions were found to remain associated with cells to a significant degree, either by virtue of a capsid interaction with a cell surface receptor,12 or nonspecific interaction of the hydrophobic dye with one or more membrane components. To distinguish between these possibilities, unlabeled particles were detected with anti-CPMV primary and AlexaFluor 488-conjugated secondary antibodies. Figure 4 shows the microscopy images obtained in the study of SW480 cells with WT CPMV and the BC mutant. After incubation for 2 h at 37°C, the interaction of WT CPMV with the cells was minimal, whereas uptake of the RGD-bearing particles was clearly evident, in a punctate pattern consistent with endosomal localization. Very little signal was observed when incubation was conducted at 4 °C (data not shown), suggesting that uptake is an energy-dependent process as expected for receptor-mediated endocytosis.
Figure 4.
Immunofluorescence analysis of (A) WT CPMV and (B) the BC mutant incubated with SW480 cells at 37°C, imaged using confocal microscopy. Virus particles were probed using rabbit anti-CPMV and AlexFluor 488-conjugated goat anti-rabbit antibodies (green); cell nuclei are stained blue (ToPro-3).
Chemical attachment of an optimized cyclic RGD ligand
To test the feasibility of uptake using a chemically-attached RGD motif, and to attempt to direct particles to the αvβ5 integrin receptor which is used efficiently by adenovirus,65 a variant of the Cilengitide structure was employed. Replacement of MeVal by MeLys retains the activity profile of the cyclic peptide66 while providing an amino functionality that can be used for attachment to the scaffold protein. We chose to install a poly(ethylene glycol) (PEG) linker to improve aqueous solubility of the molecule and inhibit aggregation of the resulting polyvalent CPMV conjugates, as well as to inhibit non-selective internalization of the particles into cells.13, 18, 21 Alkyne units were incorporated at the end of the PEG spacer for use in the Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) reaction.57 The resulting conjugates were evaluated for their interactions in vitro with three human cancer cell lines using flow cytometry and confocal microscopy.
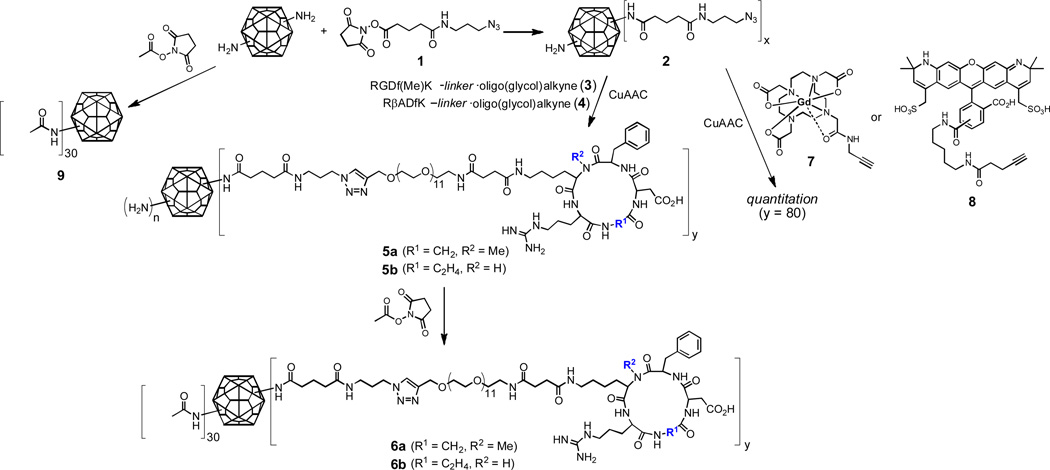
To covalently decorate CPMV with fluorescent dyes and PEG-peptide ligands, sequential lysine acylation and click chemistry steps were performed, as shown in Figure 5. Thus, NHS ester 1 at moderate concentration was used to provide azide groups on the capsid surface while leaving some accessible amino groups for later attachment. The resulting particles 2 were addressed by CuAAC57 with cyclic RGD-alkyne reagent 3 or negative control 4, to give particles 5. The remaining amino groups were then capped in a final labeling step, which added approximately 30 dyes per particle (after 2 h reaction time) or 60 dyes per particle (4–5 h reaction time), as determined by UV-Vis absorbance spectroscopy of the purified particles and the known molar extinction coefficient of the dye. The final particles in each case were purified by size exclusion chromatography and additionally characterized using dynamic light scattering and native gel electrophoresis. To assess the number of click chemistry attachments to the particle, parallel reactions using a Gd(DOTA)-alkyne reagent (7)67 or dye-alkyne (8) were performed under the same conditions used for the PEG-peptide click reactions. The resulting particles were analyzed either with inductively coupled plasma optical emission spectroscopy for Gd or by UV-Vis absorbance for dyes, as has been done previously.68 Both model reactions resulted in the formation of 80 ± 8 triazoles per particle.
Figure 5.
Derivatization of CPMV with dyes (represented by colored stars) and PEG-derivatized cyclic RGD ligands using NHS ester and CuAAC chemistry. The same reactions were performed with the analogous PEG-cyclic RβAD reagent to provide negative control particles.
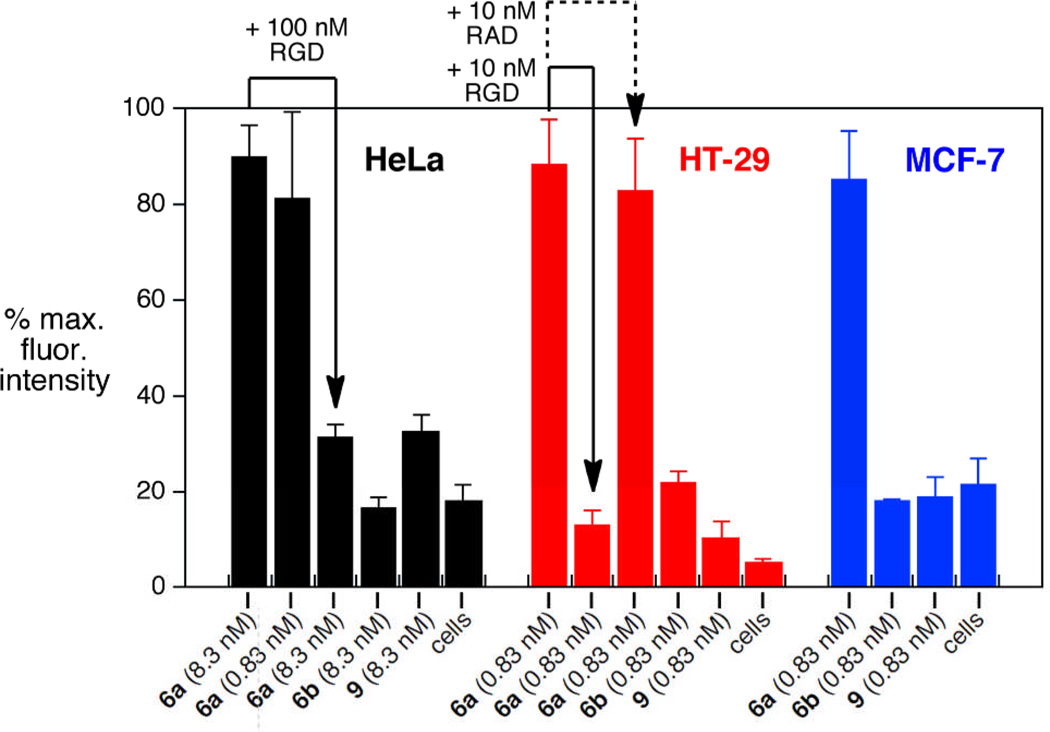
The binding of derivatized particles to integrin receptor-expressing cells was evaluated by flow cytometry using established cell lines HeLa (cervical, expressing integrins αvβ3 and αvβ5),69 HT-29 (colon, expressing αvβ5),70 and MCF-7 (breast, expressing αvβ5 and potentially αvβ3 or α5β1 as well).71, 72 The CPMV(cyclic-RGD)80 particle 6a was found to interact strongly with each cell line, as expected given the biological activity of the parent peptide Cilengitide. The effect of concentration was explored on HeLa cells, with a 10-fold higher dose of particle (8.3 nmol particle, 46 µg/mL) not producing significantly higher internalization than the standard concentration (0.83 nmol particle, 4.6 µg/mL). Native CPMV derivatized with AlexaFluor dye (particle 9) showed much weaker association,21 and CPMV(cyclic-RβAD)80 (6b) showed no detectable binding (Figure 6). A 12-fold excess of free cyclic RGD peptide (but not the analogous cyclic RβAD peptide), added shortly before the addition of the functionalized particle was found to abolish binding of 6a to both HeLa and HT-29 cells, supporting the conclusion that the association is integrin-dependent.
Figure 6.
Flow cytometry of mixtures of the indicated cells (5 × 106 cells per mL) with various CPMV formulations, 37°C, 1 h, with detection by immunostaining for CPMV particles. Statistical analysis of data was performed with FlowJo software; error bars show standard deviation in averaged mean fluorescence intensity for three independent experiments. 100% values for fluorescence intensity: HeLa = 620, HT-29 = 1800, MCF-7 = 900. Added peptides: “RGD” = cyclo(RGDfMeK); “RAD” = cyclo(RβADfK).
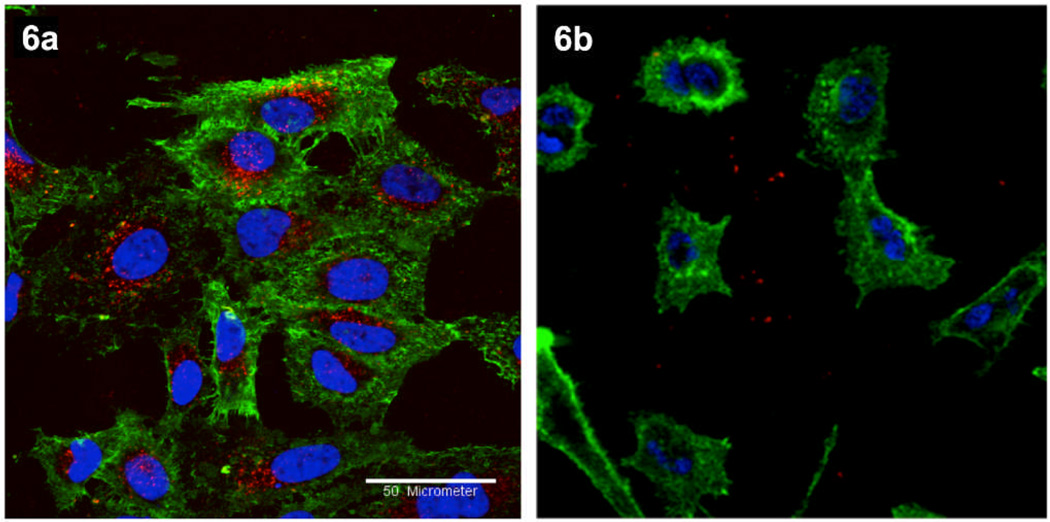
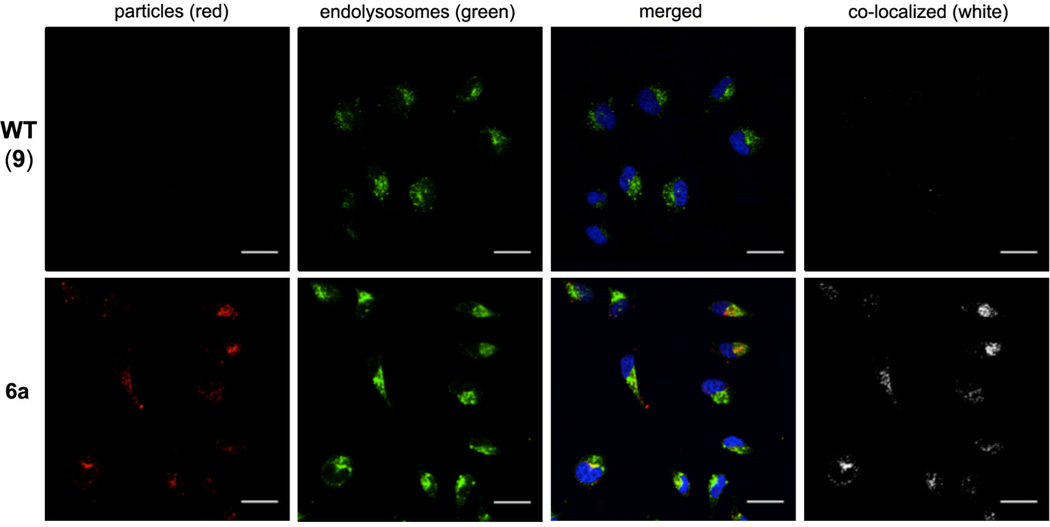
Representative confocal fluorescence microscopy images of HeLa cells treated with 6a and 6b particles are shown in Figure 7A,B. Internalization was observed only for the cyclic-RGD-coated CPMV particles; cyclic-RβAD-coated particles neither bound nor internalized. Native CPMV is known to bind to surface-expressed vimentin and internalize to endolysosomes20 of various cell types, including endothelial cells and cancer cells.12, 21 The interactions of dye-labeled CPMV particles were therefore examined relative to those of 6a (Figure 8). Even after 6 hours incubation, the non-targeted particles were not detected on or in HeLa cells, in contrast to the efficient uptake of 6a into endolysosomal compartments, as indicated by co-localization of the CPMV-PEG-RGD particles with endolysosome-specific Lamp-1 antibodies. Similar results showing increasing uptake of 6a with time are included in Supporting Information.
Figure 7.
Confocal microscopy of HeLa cells treated with the indicated particles (6a or 6b, 100 µg/mL = 18 nM), 37°C, 1 hr. Red = AlexaFluor 568 conjugated CPMV particles, green = cell membrane stain (Oregon Green 488 conjugated wheat-germ agglutinin), blue = cell nuclei stain (DAPI). Scale bar = 50 µm.
Figure 8.
Confocal microscopy of HeLa cells after 6 h incubation with the indicated particles (50 µg/mL = 9 nM) at 37°C, followed by washing, cell fixation, and staining with the following reagents: red CPMV particles, blue = cell nuclei stain (DAPI), green = endolysosomes. Co-localization of particles and endolysosomes was determined using ImageJ software (white). Scale bars = 30 µm.
Conclusions
We report here the genetic and chemical engineering of RGD-bearing CPMV particles to bind and internalize into cancer cells in vitro. A 12-mer loop derived from adenovirus was encoded into two positions of the CPMV structure, providing RGD sites approximately 28 and 60 Å apart, respectively. Both chimeras showed significant interaction with the target cells, but differences in binding domain exposure and proteolytic cleavage sites ruled out a comparative study of spacing vs. activity. CPMV particles were also chemically modified by the attachment of shielding oligo(ethylene glycol) chains tipped with a powerful integrin-binding cyclic peptide, made convenient by CuAAC bioconjugation chemistry.
While likely differences in cell-surface integrin receptor density and other properties makes comparisons between cells difficult, it is notable that a fairly light loading of RGD motifs (60–80 per particle, corresponding to an average density of one ligand for every 35–50 nm2 of surface area) was sufficient to stimulate binding and internalization of the attached nanoparticles. The particles were taken up at low concentrations into endosomal compartments, as expected for integrin receptor mediation. The fact that integrin-dependent binding and internalization was accessed by the addition of different RGD motifs shows that this function is a robust one in the modular design of multifunctional entities. These results provide a basis for the development of more sophisticated particles by the addition of other useful functions such as endosomal escape and triggered release of cargo for gene or drug delivery.
Supplementary Material
Acknowledgements
This work was supported by the NIH (CA112075, HL054352, K99-EB009105), a NSF predoctoral fellowship to M.L.H., and postdoctoral fellowships from the American Heart Association (to N.F.S.) and the Deutsche Akademische Austausch Dienst (to B.L.). We thank Dr. Vu Hong for assistance with CuAAC reactions.
Footnotes
Supporting Information
Synthetic procedures and compound characterization data; assay procedures; additional confocal micrographs at different time points. This material is available free of charge via the Internet at http://pubs.rsc.org.
References
- 1.Strebhardt K, Ullrich A. Nat. Rev. Cancer. 2008;8:473–480. doi: 10.1038/nrc2394. [DOI] [PubMed] [Google Scholar]
- 2.Strable E, Finn MG. Curr Top Microbiol. 2009;327:1–21. doi: 10.1007/978-3-540-69379-6_1. [DOI] [PubMed] [Google Scholar]
- 3.Pokorski JK, Steinmetz NF. Mol Pharmaceut. 2011;8:29–43. doi: 10.1021/mp100225y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seow Y, Wood MJ. Mol Ther. 2009;17:767–777. doi: 10.1038/mt.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang JH, Faust SM, Rabinowitz JE. J. Mol. Cell. Cardiol. 2011;50:793–802. doi: 10.1016/j.yjmcc.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 6.Peek LJ, Middaugh CR, Berkland C. Adv Drug Deliver Rev. 2008;60:915–928. doi: 10.1016/j.addr.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jennings GT, Bachmann MF. Biol. Chem. 2008;389:521–536. doi: 10.1515/bc.2008.064. [DOI] [PubMed] [Google Scholar]
- 8.Hinton HJ, Jegerlehner A, Bachmann MR. Curr. Top. Microbiol. Immunol. 2008;319:1–15. doi: 10.1007/978-3-540-73900-5_1. [DOI] [PubMed] [Google Scholar]
- 9.Chackerian B. Exp. Rev. Vaccines. 2007;6:381–390. doi: 10.1586/14760584.6.3.381. [DOI] [PubMed] [Google Scholar]
- 10.Singh P, Prasuhn D, Yeh RM, Destito G, Rae CS, Osborn K, Finn MG, Manchester M. J. Control. Release. 2007;120:41–50. doi: 10.1016/j.jconrel.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaiser CR, Flenniken ML, Gillitzer E, Harmsen AL, Harmsen AG, Jutila MA, Douglas T, Young MJ. Int. J. Nanomed. 2007;2:715–733. [PMC free article] [PubMed] [Google Scholar]
- 12.Koudelka KJ, Destito G, Plummer EM, Trauger SA, Siuzdak G, Manchester M. PLoS Pathogen. 2009;5:e1000417. doi: 10.1371/journal.ppat.1000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raja KS, Wang Q, Gonzalez MJ, Manchester M, Johnson JE, Finn MG. Biomacromolecules. 2003;3:472–476. doi: 10.1021/bm025740+. [DOI] [PubMed] [Google Scholar]
- 14.Yoo JW, Chambers E, Mitragotri S. Curr. Pharm. Des. 2010;16:2298–2307. doi: 10.2174/138161210791920496. [DOI] [PubMed] [Google Scholar]
- 15.Schipper ML, Iyer G, Koh AL, Cheng Z, Ebenstein Y, Aharoni A, Keren S, Bentolila LA, Li JQ, Rao JH, Chen XY, Banin U, Wu AM, Sinclair R, Weiss S, Gambhir SS. Small. 2009;5:126–134. doi: 10.1002/smll.200800003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Mol Pharmaceut. 2009;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otsuka H, Nagasaki Y, Kataoka K. Adv Drug Deliver Rev. 2003;55:403–419. doi: 10.1016/s0169-409x(02)00226-0. [DOI] [PubMed] [Google Scholar]
- 18.Steinmetz NF, Manchester M. Biomacromolecules. 2009;10:784–792. doi: 10.1021/bm8012742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leong HS, Steinmetz NF, Ablack A, Destito G, Zijlstra A, Stuhlmann H, Manchester M, Lewis JD. Nat. Protocols. 2010;5:1406–1417. doi: 10.1038/nprot.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis JD, Destito G, Zijlstra A, Gonzalez MJ, Quigley JP, Manchester M, Stuhlmann H. Nat. Med. 2006;12:354–360. doi: 10.1038/nm1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinmetz NF, Cho CF, Ablack A, Lewis JD, Manchester M. Nanomedicine. 2011;6:351–364. doi: 10.2217/nnm.10.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Destito G, Yeh R, Rae CS, Finn MG, Manchester M. Chem. Biol. 2007;10:1152–1162. doi: 10.1016/j.chembiol.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren Y, Wong SM, Lim L-Y. Bioconjugate Chem. 2007;18:836–843. doi: 10.1021/bc060361p. [DOI] [PubMed] [Google Scholar]
- 24.Hynes RO. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 25.Castel S, Pagan R, Mitjans F, Piulats J, Goodman S, Jonczyk A, Huber F, Vilaro S, Reina M. Lab. Invest. 2001;81:1615–1626. doi: 10.1038/labinvest.3780375. [DOI] [PubMed] [Google Scholar]
- 26.Nemerow GR, Cheresh DA. Nature Cell Biol. 2002;4:E69–E71. doi: 10.1038/ncb0402-e69. [DOI] [PubMed] [Google Scholar]
- 27.Nemerow GR, Stewart PL. Virology. 2001;288:189–191. doi: 10.1006/viro.2001.1095. [DOI] [PubMed] [Google Scholar]
- 28.Nemerow GR. Virology. 2000;274:1–4. doi: 10.1006/viro.2000.0468. [DOI] [PubMed] [Google Scholar]
- 29.Arzt J, Baxt B, Grubman MJ, Jackson T, Juleff N, Rhyan J, Rieder E, Waters R, Rodriguez LL. Transboundary and Emerging Diseases. 2011;58:305–326. doi: 10.1111/j.1865-1682.2011.01236.x. [DOI] [PubMed] [Google Scholar]
- 30.Arnold M, Cavalcanti-Adam EA, Glass R, Blümmel J, Eck W, Kantlehner M, Kessler H, Spatz JP. ChemPhysChem. 2004;5:383–388. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- 31.Lin T, Chen Z, Usha R, Stauffacher CV, Dai J-B, Schmidt T, Johnson JE. Virology. 1999;265:20–34. doi: 10.1006/viro.1999.0038. [DOI] [PubMed] [Google Scholar]
- 32.Lomonossoff GP, Johnson JE. Prog Biophys Mol Bio. 1991;55:107–137. doi: 10.1016/0079-6107(91)90003-b. [DOI] [PubMed] [Google Scholar]
- 33.Taylor KM, Lin T, Porta C, Mosser A, Giesing H, Lomonossoff GP, Johnson JE. J. Mol. Recog. 2000;13:71–82. doi: 10.1002/(SICI)1099-1352(200003/04)13:2<71::AID-JMR489>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 34.Porta C, Spall VE, Lin T, Johnson JE, Lomonossoff GP. Intervirology. 1996;39:79–84. doi: 10.1159/000150478. [DOI] [PubMed] [Google Scholar]
- 35.Lomonossoff GP, Hamilton WDO. Curr. Top. Microbiol. Immun. 1999;240:177–189. doi: 10.1007/978-3-642-60234-4_9. [DOI] [PubMed] [Google Scholar]
- 36.Chatterji A, Ochoa WF, Paine M, Ratna BR, Johnson JE, Lin T. Chem. Biol. 2004;11:855–863. doi: 10.1016/j.chembiol.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Kaltgrad E, Lin T, Johnson JE, Finn MG. Chem. Biol. 2002;9:805–811. doi: 10.1016/s1074-5521(02)00165-5. [DOI] [PubMed] [Google Scholar]
- 38.Wang Q, Lin T, Johnson JE, Finn MG. Chem. Biol. 2002;9:813–819. doi: 10.1016/s1074-5521(02)00166-7. [DOI] [PubMed] [Google Scholar]
- 39.Steinmetz NF, Lin T, Lomonossoff GP, Johnson JE. Curr Top Microbiol. 2009;327:23–58. doi: 10.1007/978-3-540-69379-6_2. [DOI] [PubMed] [Google Scholar]
- 40.Brunel FM, Lewis JD, Destito G, Steinmetz NF, Manchester M, Stuhlmann H, Dawson PE. Nano Lett. 2010;10:1093–1097. doi: 10.1021/nl1002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng Q, Saha S, Lee LA, Barnhill H, Oxsher J, Dreher T, Wang Q. Bioconjugate Chem. 2011;22:58–66. doi: 10.1021/bc100351n. [DOI] [PubMed] [Google Scholar]
- 42.Schottelius M, Laufer B, Kessler H, Wester HJ. Accounts Chem Res. 2009;42:969–980. doi: 10.1021/ar800243b. [DOI] [PubMed] [Google Scholar]
- 43.Ruoslahti E. Annu. Rev. Cell Develop. Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 44.Meyer A, Auemheimer J, Modlinger A, Kessler H. Curr. Pharm. Des. 2006;12:2723–2747. doi: 10.2174/138161206777947740. [DOI] [PubMed] [Google Scholar]
- 45.Weide T, Modlinger A, Kessler H. Top. Curr. Chem. 2007;272:1–50. [Google Scholar]
- 46.Hersel U, Dahmen C, Kessler H. Biomaterials. 2003;24:4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 47.Buckley CD, Pilling D, Henriquez NV, Parsonage G, Threlfall K, Scheel-Toellner D, Simmons DL, Albar AN, Lord JM, Salmon M. Nature. 1999;397:534–539. doi: 10.1038/17409. [DOI] [PubMed] [Google Scholar]
- 48.Ruoslahti E, Reed J. Nature. 1999;397:479–480. doi: 10.1038/17229. [DOI] [PubMed] [Google Scholar]
- 49.Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Girard OM, Hanahan D, Mattrey RF, Ruoslahti E. Cancer Cell. 2009;16:510–520. doi: 10.1016/j.ccr.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang HT, Li HC, Li ZW, Guo CH. Anti-Cancer Drugs. 2011;22:409–415. [Google Scholar]
- 51.Dechantsreiter MA, Planker E, Mathä B, Lohof E, Hölzemann G, Jonczyk A, Goodman SL, Kessler H. J. Med. Chem. 1999;42:3033–3040. doi: 10.1021/jm970832g. [DOI] [PubMed] [Google Scholar]
- 52.Mas-Moruno C, Rechenmacher F, Kessler H. Anti-Cancer Agent Me. 2010;10:753–768. doi: 10.2174/187152010794728639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hynes RO. Nat. Med. 2002;8:918–921. doi: 10.1038/nm0902-918. [DOI] [PubMed] [Google Scholar]
- 54.Hynes RO. J. Thromb. Haemost. 2007;5 Suppl 1:32–40. doi: 10.1111/j.1538-7836.2007.02569.x. [DOI] [PubMed] [Google Scholar]
- 55.Carpino LA, Han GY. J. Org. Chem. 1972;37:3404–3409. [Google Scholar]
- 56.Wellink J. Methods Mol. Biol. 1998;81:205–209. doi: 10.1385/0-89603-385-6:205. [DOI] [PubMed] [Google Scholar]
- 57.Hong V, Presolski SI, Ma C, Finn MG. Angew Chem Int Edit. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia JA, Schrijvers L, Tan A, Vos P, Wellink J, Goldbach R. Virology. 1987;159:67–75. doi: 10.1016/0042-6822(87)90348-5. [DOI] [PubMed] [Google Scholar]
- 59.Taylor KM, Porta C, Lin T, Johnson JE, Barker PJ, Lomonossoff GP. Biol. Chem. 1999;280:387–392. doi: 10.1515/BC.1999.051. [DOI] [PubMed] [Google Scholar]
- 60.Zubieta C, Schoehn G, Chroboczek J, Cusack S. Mol. Cell. 2005;17:121–135. doi: 10.1016/j.molcel.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 61.Phelps JP, Dang N, Rasochova L. J. Virol. Meth. 2007;141:146–153. doi: 10.1016/j.jviromet.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chiang H-S, Peng H-C, Huang T-F. Biochim. Biophys. Acta. 1994;1224:506–516. doi: 10.1016/0167-4889(94)90288-7. [DOI] [PubMed] [Google Scholar]
- 63.Odrljin TM, Haidaris CG, Lerner NB, Simpson-Haidaris PJ. Am. J. Respir. Cell Mol. Biol. 2001;24:12–21. doi: 10.1165/ajrcmb.24.1.3992. [DOI] [PubMed] [Google Scholar]
- 64.Li E, Brown SL, Stupack DG, Puente XS, Cheresh DA, Nemerow GR. J. Virol. 2001;75:5405–5409. doi: 10.1128/JVI.75.11.5405-5409.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wickham TJ, Filardo EJ, Cheresh DA, Nemerow GR. J. Cell Biol. 1994;127:257–264. doi: 10.1083/jcb.127.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Laufer B, Frank AO, Chatterjee J, Neubauer T, Mas-Moruno C, Kummerlowe G, Kessler H. Chem-Eur J. 2010;16:5385–5390. doi: 10.1002/chem.201000545. [DOI] [PubMed] [Google Scholar]
- 67.Prasuhn, D.E. J, Yeh RM, Obenaus A, Manchester M, Finn MG. Chem. Commun. 2007:1269–1271. doi: 10.1039/b615084e. [DOI] [PubMed] [Google Scholar]
- 68.Prasuhn DE, Singh P, Strable E, Brown S, Manchester M, Finn MG. J Am Chem Soc. 2008;130:1328–1334. doi: 10.1021/ja075937f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Werner J, Decarlo CA, Escott N, Zehbe I, Ulanova M. Innate Immun. 2012;18:55–69. doi: 10.1177/1753425910392934. [DOI] [PubMed] [Google Scholar]
- 70.Burvenich I, Schoonooghe S, Vervoort L, Dumolyn C, Coene E, Vanwalleghem L, Van Huysse J, Praet M, Cuvelier C, Mertens N, De Vos F, Slegers G. Mol Cancer Ther. 2008;7:3771–3779. doi: 10.1158/1535-7163.MCT-08-0600. [DOI] [PubMed] [Google Scholar]
- 71.Wong NC, Mueller BM, Barbas CF, Ruminski P, Quaranta V, Lin ECK, Smith JW. Clin. Exp. Metastasis. 1998;16:50–61. doi: 10.1023/a:1006512018609. [DOI] [PubMed] [Google Scholar]
- 72.Santis G, Lord R, Parsons M, Kirby I, Beavil A, Hunt J, Sutton B. J. Gen. Virol. 2006;87:2497–2505. doi: 10.1099/vir.0.81620-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.