Background: Glycosyltransferases (GTs) have important functions in plant secondary metabolism.
Results: A gene encoding an N-methylanthranilic acid O-glucosyltransferase forms part of a biosynthetic cluster for the synthesis of acylated defense compounds in oat.
Conclusion: This GT synthesizes the activated acyl donor required for triterpene acylation.
Significance: These findings open up new opportunities for metabolic engineering for disease control.
Keywords: Antibiotics, Glycosyltransferases, Metabolic Engineering, Plant, Terpenoids, Cereals, Plant Defense, Saponins
Abstract
Plants produce a huge array of specialized metabolites that have important functions in defense against biotic and abiotic stresses. Many of these compounds are glycosylated by family 1 glycosyltransferases (GTs). Oats (Avena spp.) make root-derived antimicrobial triterpenes (avenacins) that provide protection against soil-borne diseases. The ability to synthesize avenacins has evolved since the divergence of oats from other cereals and grasses. The major avenacin, A-1, is acylated with N-methylanthranilic acid. Previously, we have cloned and characterized three genes for avenacin synthesis (for the triterpene synthase SAD1, a triterpene-modifying cytochrome P450 SAD2, and the serine carboxypeptidase-like acyl transferase SAD7), which form part of a biosynthetic gene cluster. Here, we identify a fourth member of this gene cluster encoding a GT belonging to clade L of family 1 (UGT74H5), and show that this enzyme is an N-methylanthranilic acid O-glucosyltransferase implicated in the synthesis of avenacin A-1. Two other closely related family 1 GTs (UGT74H6 and UGT74H7) are also expressed in oat roots. One of these (UGT74H6) is able to glucosylate both N-methylanthranilic acid and benzoic acid, whereas the function of the other (UGT74H7) remains unknown. Our investigations indicate that UGT74H5 is likely to be key for the generation of the activated acyl donor used by SAD7 in the synthesis of the major avenacin, A-1, whereas UGT74H6 may contribute to the synthesis of other forms of avenacin that are acylated with benzoic acid.
Introduction
Glycosylation of small molecules is essential for a wide range of biological processes in plants, including regulation of the activity of hormones, neutralization of xenobiotics, and synthesis and storage of secondary metabolites (1, 2). The glycosyltransferases (GTs)7 that catalyze these modifications belong to carbohydrate-active enzyme (CAZY) family 1, as defined by Cantarel et al. (3). These enzymes transfer sugars from nucleotide diphosphate-activated sugar moieties to small hydrophobic acceptor molecules. Alterations in the properties of small molecules as a consequence of glycosylation include changes in water solubility, stability, bioactivity, and pharmacological properties (2).
The family 1 GTs are one of the largest groups of secondary metabolic tailoring enzymes in higher plants. The expansion of this family is likely to be a reflection of chemical diversification during the adaptation of plants to life on land (4–6). The activated sugar donor used by these enzymes is usually UDP-d-glucose, although some family 1 GTs have been reported to use other sugar donors such as UDP-d-galactose, UDP-l-rhamnose, UDP-d-glucuronic acid, or UDP-l-arabinose (2). Family 1 GTs catalyze the transfer of these UDP-activated sugars onto either a small lipophilic acceptor or onto the sugar moiety of a glycoside (2). Glycosylation of the acceptor is regiospecific and can occur at a variety of different functional groups (7). Although family 1 GTs show specificity for sugar donors, they may produce diverse glycosides due to the permissiveness of their acceptor binding sites (8, 9).
Mining of the complete genome sequence of thalecress (Arabidopsis thaliana) has identified 107 predicted family 1 GT genes (2, 6). The recent release of complete genome sequences for several cereal species (e.g. rice, sorghum, and maize) and the growing resource of transcriptomics data emerging from high-throughput sequencing have similarly revealed numerous predicted family 1 GT sequences in the genomes of monocots. For example, there are 213 predicted family 1 GT genes in rice, 201 in Sorghum bicolor, 168 in Zea mays, and 143 in purple false brome (Brachypodium distachyon) (5). The considerable expansion of the family 1 GTs in higher plants suggests that these enzymes have evolved to have diverse and important functions. However, despite the overwhelming body of sequence data that is emerging, information about the biochemical properties and biological significance of these enzymes is limited.
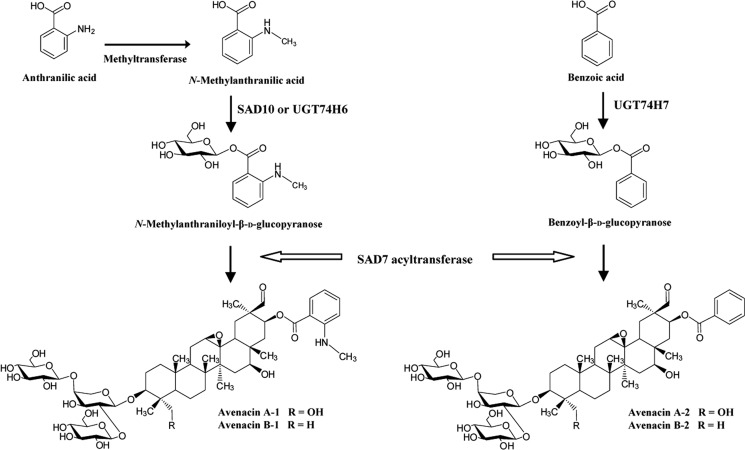
Oats (Avena spp.) produce antimicrobial triterpenes known as avenacins that are synthesized in the roots and that provide protection against soil-borne pathogens (10). Avenacins are acylated molecules that are conjugated with either N-methylanthranilic acid or benzoic acid (Fig. 1). The major avenacin found in oat roots is avenacin A-1. Acylation of avenacins is carried out by the serine carboxypeptidase-like acyl transferase SAD7, which requires an acyl glucose donor (11). Here, we identify a gene predicted to encode a family 1 GT gene belonging to the UGT74 subfamily that forms part of a metabolic gene cluster for avenacin synthesis (UGT74H5). This gene is co-expressed with other previously characterized genes in the avenacin cluster, suggesting that UGT74H5 is required for avenacin synthesis. UGT74H5 was expressed in Escherichia coli and shown to be an N-methylanthranilic acid O-glucosyltransferase implicated in the synthesis of avenacin A-1. A second related enzyme UGT74H6 was able to glucosylate both N-methylanthranilic acid and benzoic acid. Our biochemical analysis, coupled with investigation of the distribution of these enzymes in oat roots, indicates that UGT74H5 is likely to be key for the synthesis of the major avenacin, A-1, whereas UGT74H6 may be required for the synthesis of other avenacins that are acylated with benzoic acid.
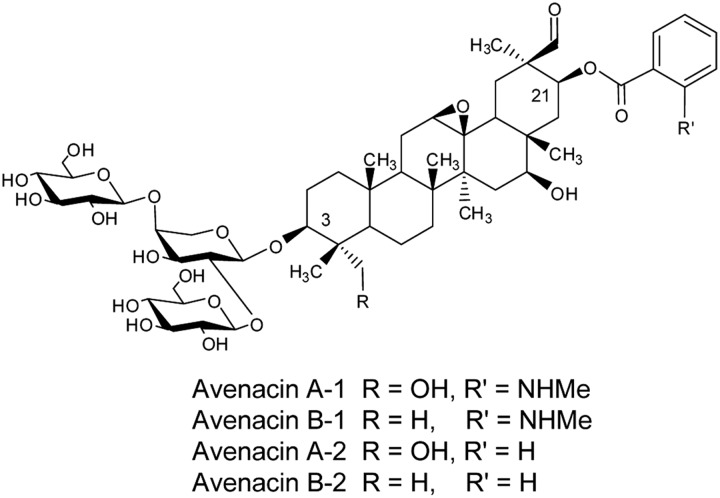
FIGURE 1.
Structures of the four avenacins. Avenacins A-1 and B-1 are esterified at the C-21 position with N-methylanthranilate, and avenacins A-2 and B-2 are esterified with benzoic acid.
EXPERIMENTAL PROCEDURES
Materials
Chemicals and reagents were obtained from Sigma-Aldrich unless stated otherwise. Glucose esters of anthranilic acid, N-methylanthranilic acid, and benzoic acid were produced by chemical synthesis (full details to be published in due course). Protein quantification was performed using the Bio-Rad protein assay kit (Dye Reagent Concentrate, Bio-Rad) and calibrated with bovine serum albumin (BSA). Proteins were analyzed by SDS-PAGE on 10% cross-linked gels and detected with Coomassie Brilliant Blue (12).
Bacterial Artificial Chromosome (BAC) and Expressed Sequence Tag (EST) Resources
Construction of the Avena strigosa BAC library is described in Qi et al. (13). Sad1 and Sad2 both reside on BAC 460D15 (13). An overlapping BAC containing Sad7 (BAC 341P21) was reported in Mugford et al. (11). The full sequence of BAC 341P21 was determined by standard shotgun sequencing. An EST resource from a root cDNA library derived from diploid oat (A. strigosa) was generated previously (14). Sequence similarity searches were carried out using the tFASTA algorithm (15).
Phylogenetic Analysis
Family 1 glycosyltransferase amino acid sequences from a variety of different plant species were aligned using the MAFFT program (version 6). The phylogenetic tree was built with the neighbor joining method, using Mega program (version 4.01) (16) with 500 bootstrap replicates. Details of the sequences used to build the tree are provided in supplemental Table 1.
Gene Expression Analysis
Gene expression analysis was carried out by RT-PCR as described previously (13, 17).
Recombinant Glycosyltransferases
Specific oligonucleotides were designed to amplify the complete open reading frames of UGT74H5, UGT74H6, and UGT74H7 from oat root tip cDNA (supplemental Table 2). The PCR products of the three full-length cDNAs were amplified using Phusion DNA polymerase (Finnzymes). Full-length coding sequences were cloned into the pET-19b expression vector, which includes an N-terminal His6 tag sequence (Novagen) and transformed into Escherichia coli BL21(DE3).
For expression of the histidine-tagged proteins, transformed E. coli strains containing the recombinant constructs were grown in LB medium containing 34 μg/ml of chloramphenicol, 50 μg/ml ampicillin, 2.5 mm betaine, and 0.6 m sorbitol. Cultures were grown at 37 °C with shaking to an A600 of 0.6. They were then incubated at 16 °C for 30 min prior to the addition of isopropyl 1-thio-β-d-galactopyranoside to 0.1 mm final concentration and left overnight at 16 °C. Bacterial cells were harvested by centrifugation at 7000 × g for 10 min at 4 °C. The pellet was then resuspended in 5–10 ml of lysis buffer (300 mm NaCl, 50 mm sodium phosphate, 20 mm imidazole, 5% glycerol, pH 7.8) with one tablet of EDTA-free protease inhibitor per 50 ml (Roche Applied Science). Cells were lysed twice using a French press (1000 psi), keeping ice-cold throughout. The lysate was centrifuged at 10,000 × g for 15 min at 4 °C, and the supernatant was filtered using a 0.22-μm filter (Minisart) to generate the crude soluble protein preparation for purification.
Recombinant enzymes were purified using a cobalt immobilized metal ion affinity column (5-ml Hi-Trap chelating HP column (GE Healthcare)), followed by size exclusion chromatography (Superdex 200 16/60 prep grade column (GE Healthcare)). The peak fractions corresponding to GT monomers were concentrated using an Amicon Ultra 30000 molecular weight cut-off membrane (Millipore), snap frozen in aliquots (50 μl) in liquid nitrogen, and stored at −80 °C. Polyclonal antisera to UGT74H5 were raised in rabbit using purified histidine-tagged protein as the antigen (BioGenes GmbH, Berlin, Germany).
Glucosyltransferase Activity Assays
For kinetic analysis, assay mixtures (200 μl) consisted of recombinant enzyme (1 μg/ml) in 50 mm Tris-HCl, pH 7.8, containing 5 mm UDP-d-glucose, 3 mm MnCl2, and 1 mm substrate (benzoic acid, salicylic acid, anthranilic acid, or N-methylanthranilic acid). The reactions were carried out at 30 °C for 30 min and stopped by the addition of 20 μl of trichloroacetic acid (240 mg/ml), snap-frozen in liquid nitrogen, and stored at −20 °C before HPLC analysis. The reaction products were analyzed by reverse phase HPLC on a Dionex Ultimate 3000 instrument equipped with a LUNA 5-μm C18 column (150 × 4.6 mm; Phenomenex) eluted at 0.5 ml/min. Compounds were detected using a photodiode array detector: N-methylanthranilic acid (320 nm); anthranilic acid (320 nm); benzoic acid (272 nm); salicylic acid (296 nm). Compounds were separated using a linear gradient of acetonitrile in water acidified with 0.1% trifluoroacetic acid (10–40% for anthranilic acid and N-methylanthranilic acid; 10–60% for salicylic acid and benzoic acid). The retention times (Rt) of the glucose conjugates were determined as follows: salicyloyl-β-d-glucopyranose (Rt, 12.1 min); benzoyl-β-d-glucopyranose (Rt, 11.6 min); anthraniloyl-β-d-glucopyranose (Rt, 9.2 min); N-methylanthraniloyl-β-d-glucopyranose (Rt, 11.8 min) (supplemental Fig. 1). Kinetic parameters, Km and kcat, for the acceptor substrates in the presence of saturating donor substrate, UDP-d-glucose (5 mm), were determined by fitting the initial velocity data to the Michaelis-Menten equation using SigmaPlot software (Sigma-Aldrich).
Protein Extraction and Western Blot Analysis
For expression of native GTs in E. coli, full-length cDNAs were cloned into the pET-24a E. coli expression vector (Novagen). Recombinant plasmids were transformed into E. coli BL21(DE3). Crude protein preparations were generated as described above. For analysis of oat root proteins, soluble protein was extracted from the terminal 0.5 cm of roots of 3-day-old Avena strigosa seedlings (accession no. S75) (10) in protein extraction buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 10% glycerol, 1% Triton X-100, 1% polyvinylpyrrolidone, and protease inhibitor mixture (Roche Applied Science)). Root material was ground, the extract centrifuged at 4 °C for 30 min, and the supernatant was collected and kept at −20 °C. Proteins were denatured in the presence of Nupage reducing agent (Invitrogen) and separated and blotted onto nitrocellulose using Nupage gels (4 to 12% acrylamide gradient) according to the manufacturer's protocol. Membranes were blocked with 5% skimmed milk in 1× TBS (0.15 m NaCl, 10 mm Tris-HCl, pH 7.5) for 1 h then incubated with UGT74H5 antisera (1:3000), followed by detection with secondary antibody (anti-rabbit IgG conjugated with horseradish peroxidise, 1:8000) according to the manufacturer's protocol (Sigma-Aldrich).
For mass spectrometry proteomics by LC-MS/MS, gel slices were excised from SDS-PAGE gels run as described above. Each gel slice was washed, reduced, and alkylated and treated with trypsin according to standard procedures (18). Peptides were extracted with 5% (v/v) formic acid/50% acetonitrile (v/v), dried down, and redissolved in 0.1% (v/v) TFA. Nano-LC-MS/MS experiments were performed on an LTQ-OrbitrapTM mass spectrometer (Thermo Fisher Scientific, Inc.) coupled to an EASY-nLC HPLC via an ion source (Proxeon). Aliquots of the extracted peptides were loaded onto a C18 PepMapTM trap column (Dionex), which was then switched in-line to an analytical column (BEH C18, 1.7 μm, Waters, 75 μm × 120 mm, self-packed) for separation. The LC system was run at a flow rate of 250 nl min−1 with a gradient of 5–40% acetonitrile in water, 0.1% formic acid at a rate of 1% min−1. The mass spectrometer was operated in positive ion mode at a capillary temperature of 200 °C.
Raw files were processes with MaxQuant (version 1.3.0.5) (19). Database searches were carried out using an in-house Mascot® 2.4 server (Matrix Science Ltd., London, UK). Mascot searches were performed on a custom database containing all available root-expressed A. strigosa GT protein sequences (extracted from European Bioinformatics Institute database no. ERA148431) in a background of 1000 random E. coli sequences downloaded from Uniprot using 7 ppm precursor tolerance, 0.7 Da fragment tolerance, two missed cleavages (trypsin), and carbamidomethylation (C) as fixed and oxidation (M) as variable modifications. Mascot search results were imported and evaluated in Scaffold (version 3.6.3, Portland, OR).
RESULTS
The Avenacin Gene Cluster Contains a Gene for a Predicted Family 1 GT
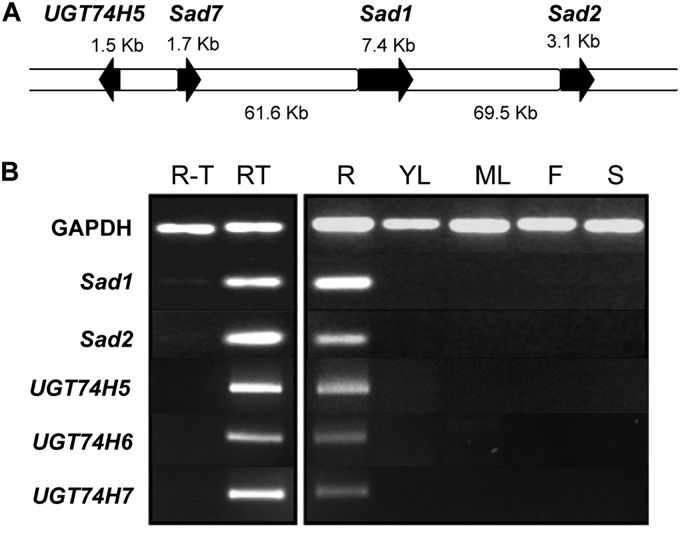
In our earlier investigations of avenacin synthesis, we cloned and characterized three genes that are required for different steps in the pathway. These include the gene for β-amyrin synthase, which catalyzes the first committed step in the pathway (Sad1) (14), and a cytochrome P450 that carries out subsequent modifications to the β-amyrin skeleton (Sad2) (13). A third gene (Sad7) encodes a serine carboxypeptidase-like acyltransferase that is required for triterpene acylation (11). Rather surprisingly, these three genes are contiguous, lying within a 150-kb region of the genome (11, 13). Although secondary metabolic gene clusters are a common feature of microbial genomes, they are a relatively new and emerging theme in plant biology (20). We then extended the region of the genome encompassing Sad1, Sad2, and Sad7 by BAC sequencing and discovered a fourth gene that is predicted to encode a family 1 GT adjacent to Sad7 (Fig. 2A). Similar to the three other previously characterized avenacin biosynthetic genes, this gene (UGT74H5) is expressed specifically in the root tips (Fig. 2B). It is therefore implicated in avenacin synthesis by virtue of its location within the gene cluster and its expression pattern.
FIGURE 2.
The gene for UGT74H5 forms part of the avenacin gene cluster and is co-expressed with other characterized avenacin biosynthetic genes. A, BAC contig showing the locations of the Sad1, Sad2, Sad7, and UGT74H5 genes. B, RT-PCR analysis of transcript levels of Sad1, Sad2, Sad7, UGT74H5, UGT74H6, and UGT74H7 in different oat tissues. R-T, roots without tips; RT, root tips; R, root; YL, young leaf; ML, mature leaf; F, flowers; S, stem. RNA loading was monitored with methylene blue. The oat GAPDH was used as a control.
Analysis of Root-expressed Sequences in Oat
Previously, we generated an EST resource (>16,000 sequences) from roots of diploid oat (A. strigosa) (14). Sequence similarity searches revealed that UGT74H5 is represented in this EST database. We also found two other closely related EST sequences that are expressed in roots (UGT74H6 and UGT74H7). Sequence similarity comparisons of the predicted products of the full-length coding sequences revealed that UGT74H6 and UGT74H7 share 78 and 79% amino acid similarity with UGT74H5, respectively (supplemental Fig. 2). Similar to UGT74H5, the corresponding genes also have similar expression patterns to previously characterized avenacin biosynthetic genes (Fig. 2B) and so may also contribute to avenacin synthesis.
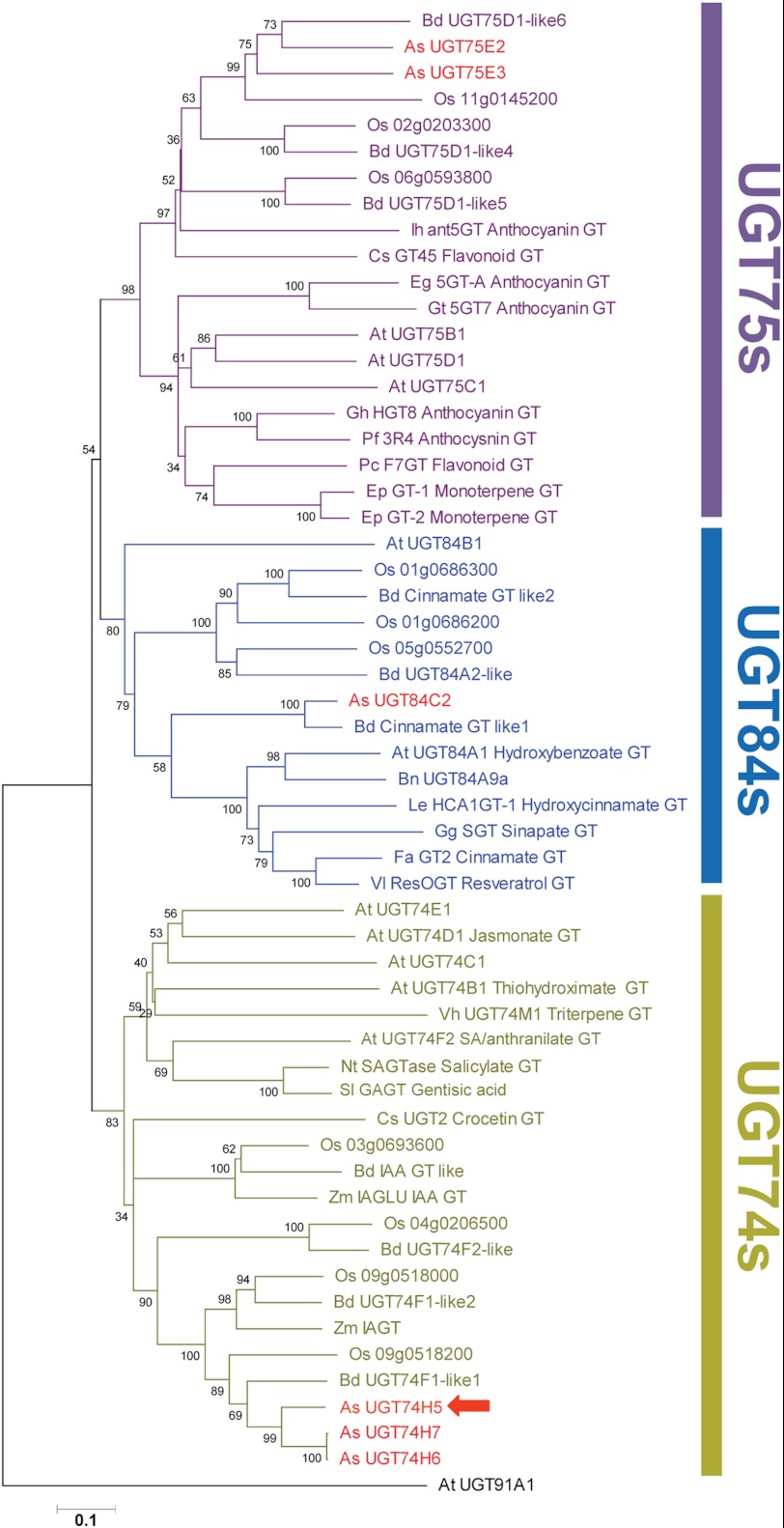
UGT74H5, UGT74H6, and UGT74H7 belong to clade L of GT family 1 (6). Group L GTs have been shown to glucosylate small molecule acceptors with various functional groups, for example carboxyl, hydroxyl, and thiol groups (4). UGT74H5, UGT74H6, and UGT74H7 belong to the UGT74 subfamily of clade L, as shown in Fig. 3. Other characterized UGT74 enzymes include an indole-3-acetic acid GT (ZmIAGT) from Z. mays (21), a crocetin GT (CsUGT2) from Crocus sativus (22), a gentisic acid xylosyltransferase from Solanum lycopersicum (SlGAGT) (23), a salicylic acid GT from Nicotiana tabacum (NtSAGTase) (24), a salicylic acid/anthranilic acid GT (UGT74F2), a thiohydroximate GT (AtUGT74B1) and a jasmonate GT (AtUGT74D1) from A. thaliana (25–27), and a triterpene GT from Saponaria vaccaria (synonym Vaccaria hispanica) (SvUGT74M1) that catalyzes the addition of glucose onto gypsogenic acid (28). A. thaliana UGT74E2 is thought to be an auxin indole-3-butyric acid glucosyltransferase (29) and UGT74C1 is implicated in aliphatic glucosinolate biosynthesis (30). Other predicted monocot UGT74 sequences from B. distachyon and O. sativa are also included in the tree shown in Fig. 3 for reference.
FIGURE 3.
Comparison of amino acid sequences of plant UGTs belonging to group L. Predicted sequences from diploid oat (indicated in red) were aligned with biochemically characterized enzymes from subfamilies UGT74, UGT75, and UGT84 along with predicted GTs from rice (O. sativa), thalecress (A. thaliana), and purple false brome (Brachypodium distachyon) (see supplemental Table 1 for GenBank accession numbers and further information). UGT74H5 is indicated with a red arrow. The phylogenetic tree was drawn by neighbor joining with 500 bootstrap replicates (percentage values shown at branch points). The scale bar represents 10% divergence. At, Arabidopsis thaliana; As, Avena strigosa; Bd, Brachypodium distachyon; Bn, Brassica napus; Cs, Crocus sativus; Ep, Eucalyptus perriniana; Eg, Eustoma grandiflorum; Fa, Fragaria x ananassa; Gt, Gentiana triflora; Gh, Glandularia x hybrida; Gg, Gomphrena globosa; Ih, Iris hollandica; Le, Lobelia erinus; Nt, Nicotiana tabacum; Os, Oryza sativa; Pf, Perilla frutescens; Pc, Pyrus communis; Sl, Solanum lycopersicon; Vh, Vaccaria hispanica; Vl, Vitis labrusca; Zm, Zea mays.
Oats produce four structurally related avenacins A-1, B-1, A-2, and B-2 (Fig. 1), of which avenacin A-1 is the most abundant (31). These molecules are pentacyclic triterpenes with a trisaccharide moiety consisting of one molecule of l-arabinose and two molecules of d-glucose. Avenacins A-1 and B-1 are acylated with N-methylanthranilic acid, and avenacins A-2 and B-2 are acylated with benzoic acid. Serine carboxypeptidase-like proteins have recently emerged as a new class of acyltransferase proteins. Unlike other characterized plant acyltransferases, these enzymes utilize O-glucose ester acyl donors instead of CoA-thioester acyl donor substrates (32–34). Oat lines with mutations in the gene encoding the serine carboxypeptidase-like acyltransferase SAD7 accumulate unacylated avenacins with a hydroxyl at the carbon 21 position instead of an acyl group (11). They also accumulate N-methylanthraniloyl-β-d-glucopyranose, the fluorescent activated donor required by SAD7 for synthesis of avenacins A-1 and B-1 (11). Because these mutants are unable to synthesize any of the four avenacins, this suggests that SAD7 is able to transfer both N-methylanthranilic acid (for synthesis of avenacins A-1 and B-1) and benzoic acid (for synthesis of avenacins A-2 and B-2) onto the triterpene acceptor. Thus, although it is possible that UGT74H5 and possibly also the two other related GTs may be involved in the addition of sugars to the trisaccharide chain of the avenacins, the close relatedness of these enzymes to GTs that glucosylate carboxylic acids suggests a role in the synthesis of the acyl glucose donors used by SAD7.
Biochemical Analysis of the Properties of UGT74H5 and Related GTs
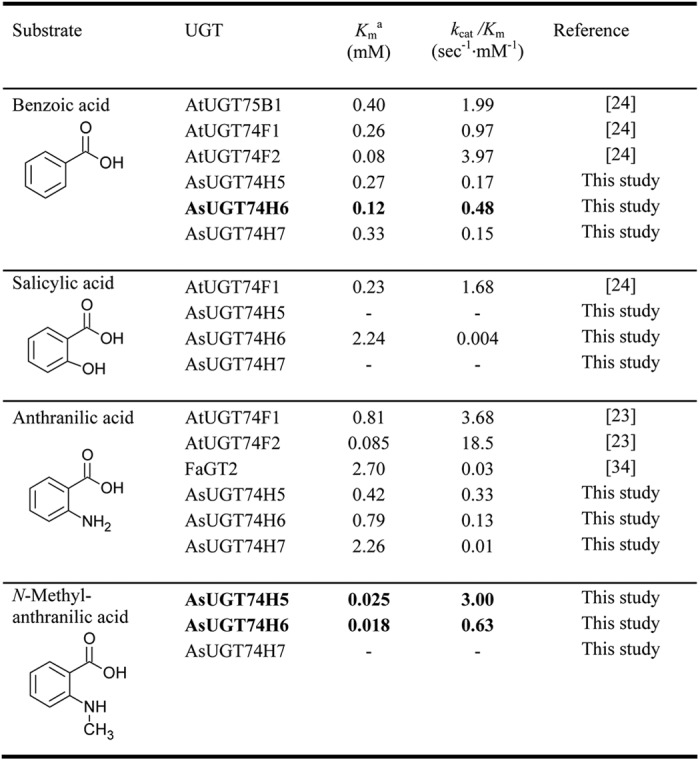
UGT74H5 and the two other closely related GTs, UGT74H6 and UGT74H7, were expressed in E. coli as N-terminal histidine-tagged fusion proteins (supplemental Fig. 3). Recombinant enzymes were purified using immobilized metal ion affinity column followed by size exclusion chromatography (Hiload 16/60 Superdex 200). Protein purity was confirmed by SDS-PAGE. The yields of purified recombinant UGT74H5, UGT74H6, and UGT74H7 protein obtained were 37, 24, and 12 mg/l of E. coli culture, respectively. The substrate specificity of the three recombinant GTs was assessed with respect to four potential acyl acceptor substrates: anthranilic acid, N-methylanthranilic acid, salicylic acid, and benzoic acid. All three GTs displayed typical Michaelis-Menten behavior for the various acceptor substrates assessed (supplemental Fig. 4). In contrast, activity toward the simple triterpenes β-amyrin and oleanolic acid was not observed. The kinetic parameters for the acyl acceptors are summarized in Table 1. Of the four prospective acceptor substrates assessed, UGT74H5 showed a strong preference for N-methylanthranilic acid (kcat/Km, 3.00 mm−1·s−1) over anthranilic acid (9-fold lower) and benzoic acid (18-fold lower) and did not have detectable activity toward salicylic acid. Of the three GTs, UGT74H5 also showed the greatest ability to utilize N-methylanthranilic acid as a substrate (kcat/Km, 5-fold higher than for UGT74H6, with UGT74H7 showing no turnover). UGT74H6 showed moderate and comparable selectivity for benzoic acid and N-methylanthranilic acid (kcat/Km, 0.42 and 0.61 mm−1·s−1, respectively). With a kcat/Km for N-methylanthranilic acid within 5 fold of that of UGT74H5, and within 3-fold for benzoic acid, it is possible that UGT74H5 and UGT74H6 may both be capable of effecting the synthesis of acyl donor substrates for the incorporation of N-methylanthranilate or benzoate into avenacins in planta. UGT74H7 was, at best, only able to turn over benzoic acid and anthranilic acid rather weakly (kcat/Km, 0.15 and 0.01 mm−1·sec−1, respectively) and did not have detectable activity toward N-methylanthranilic acid or salicylic acid. This suggests that the physiological substrate for UGT74H7 is not one of the four compounds assessed in this study.
TABLE 1.
Summary of kinetic data for the three oat GTs towards different acyl substrates
| UGT/Substrates | Kma | Vmax | kcat | kcat/Km |
|---|---|---|---|---|
| mm | mm·s−1 | s−1 | s−1·mm−1 | |
| UGT74H5 | ||||
| Salicylic acid | −b | − | − | − |
| Benzoic acid | 0.269 ± 0.046 | 6.70 × 10−6 | 0.045 | 0.167 |
| Anthranilic acid | 0.421 ± 0.101 | 2.17 × 10−5 | 0.140 | 0.333 |
| N-Methylanthranilic acid | 0.025 ± 0.005c | 1.16 × 10−5 | 0.075 | 3.000 |
| UGT74H6 | ||||
| Salicylic acid | 2.240 ± 0.450 | 1.34 × 10−6 | 0.009 | 0.004 |
| Benzoic acid | 0.119 ± 0.011 | 8.20 × 10−6 | 0.050 | 0.420 |
| Anthranilic acid | 0.794 ± 0.250 | 1.62 × 10−5 | 0.105 | 0.132 |
| N-Methylanthranilic acid | 0.018 ± 0.003 | 1.80 × 10−5 | 0.011 | 0.611 |
| UGT74H7 | ||||
| Salicylic acid | − | − | − | − |
| Benzoic acid | 0.330 ± 0.096 | 8.00 × 10−6 | 0.051 | 0.154 |
| Anthranilic acid | 1.857 ± 0.380 | 1.80 × 10−6 | 0.014 | 0.006 |
| N-Methylanthranilic acid | − | − | − | − |
a Values are the means ± S.D. (three replicates).
b −, no activity detected.
c The parameters of preferential substrate activities are highlighted in boldface type.
Detection of UGT74H5 and Related GTs in Oat Root Tips
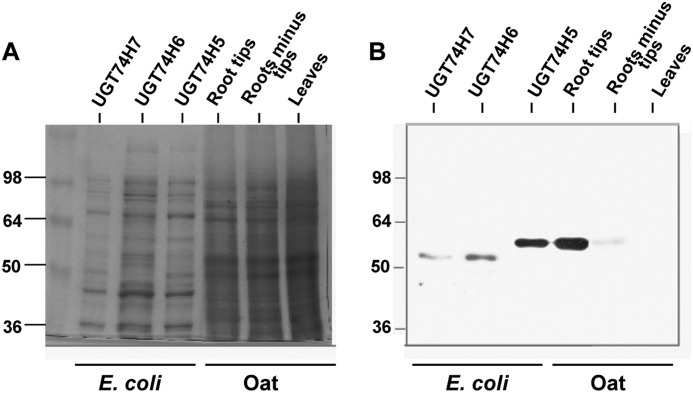
The UGT74H5, UGT74H6, and UGT74H7 transcripts are all expressed in oat root tips (Fig. 2B). The predicted molecular masses of UGT74H5 and the other two closely related oat GTs are very similar (between 51 and 52 kDa). Western blot analysis was carried out using antisera raised against the purified UGT74H5 protein to detect the three GTs in oat roots. Crude protein extracts from E. coli expressing the recombinant GTs (without His6 tags) were prepared, along with protein extracts from the root tips, upper parts of roots, and leaves of A. strigosa seedlings. These samples were subjected to SDS-PAGE and Western blot analysis. The UGT74H5 antisera recognize all three proteins. However, UGT74H5 could clearly be separated from the two other oat GTs under appropriate SDS-PAGE conditions. The UGT74H6 and UGT74H7 proteins both had lower apparent molecular masses compared with UGT74H5 when expressed in E. coli (Fig. 4). A single band of the same size as UGT74H5 was detected in protein preparations from oat root tips, with trace amounts detectable in extracts from the upper parts of the roots and no signal with leaf extracts. The UGT74H6 and UGT74H7 proteins were not detected in any of the protein preparations from oat. These results suggest that of the three enzymes, UGT74H5 is the major GT present in protein preparations derived from oat root tips. The apparent molecular mass of UGT74H5 as assessed by SDS-PAGE gel was larger than the predicted mass (∼55 kDa compared with 51 kDa). Because this difference is also evident when the three proteins are expressed in E. coli, it is unlikely to be due to glycosylation or phosphorylation, rather some intrinsic property of UGT74H5 that affects migration under the gel running conditions. LC-MS/MS analysis confirmed that the protein band detected by Western blot analysis in protein preparations derived from oat root tips was indeed UGT74H5 (supplemental Fig. 5). Amino acid sequences derived from UGT74H6 and UGT74H7 were not detected by LC-MS/MS in oat root tip-derived protein preparations.
FIGURE 4.
A, SDS-PAGE gel of protein extracts from E. coli expressing each of the three oat GTs (UGT74H7, UGT74H6, and UGT74H5) and from root tips, roots minus tips, and leaves of seedlings of diploid oat (A. strigosa). Proteins were detected by staining with Coomassie Brilliant Blue. B, Western blot analysis of a comparable SDS-PAGE gel probed with polyclonal antisera raised against UGT74H5.
DISCUSSION
Previously, three avenacin pathway enzymes have been identified. These include those encoded by Sad1 (β-amyrin synthase) (14) and Sad2 (cytochrome P450) (13). Recently a third gene, Sad7, was cloned and shown to encode an SCPL acyltransferase that catalyzes the addition of either N-methylanthranilic acid or benzoic acid to the C-21 hydroxyl group of the triterpene precursor (11). UGT74H5 lies adjacent to Sad7 in the gene cluster (Fig. 2A) and is predicted to encode a family 1 GT. Because UGT74H5, similar to the other cloned avenacin genes, is expressed specifically in roots (Fig. 2B), this gene is also implicated in avenacin synthesis. UGT74H5 is closely related to two other GTs that are expressed in oat roots (UGT74H6 and UGT74H7). In this study, the properties of these three oat GTs were characterized.
Phylogenetic analysis (Fig. 3) revealed that UGT74H5, UGT74H6, and UGT74H7 were most closely related to GTs that glucosylate carboxylic acids, suggesting a role for UGT74H5, and possibly the two other oat GTs, in the synthesis of the acyl glucose donor substrates used by the acyltransferase SAD7. Because avenacins are acylated with N-methylanthranilic acid (avenacin A-1 and B-1) or benzoic acid (avenacin A-2 and B-2), these two acids represented potential substrates for UGT74H5. Previously, we have shown that sad7 mutants accumulate N-methylanthraniloyl-β-d-glucopyranose, indicating that N-methylation of anthranilate and subsequent glucosylation occurs before transfer of the acyl group to the triterpene backbone (11). However, it is also possible that anthranilate may serve as a substrate for UGT74H5 in planta. The three GT enzymes studied here displayed different levels of specificity for the prospective acceptor substrates benzoic acid, anthranilic acid, N-methylanthranilic acid, and salicylic acid. UGT74H5 had a clear preference for N-methylanthranilic acid, whereas UGT74H6 showed greater specificity for benzoic acid. In contrast, salicylic acid was a poor substrate for all three GTs. UGT74H7 only showed weak activity on benzoic acid and very weak activity with anthranilic acid.
The efficiency and selectivity of UGT74H5 acting on N-methylanthranilic acid was very good (kcat/Km, 3.00 mm−1·s−1) compared with benzoic acid and anthranilic acid (0.167 and 0.333 mm−1·s−1, respectively); UGT74H5 did not turn over salicylic acid. This suggests that the nature of the functional groups on the aromatic ring affects substrate binding/turnover. The presence and position of hydroxyl groups around aromatic rings has previously been shown to impact on the activity of A. thaliana UGTs (27, 35). The Km values of UGT74H5 and UGT74H6 for N-methylanthranilic acid are very similar (0.025 and 0.018 mm, respectively). This may be important when considering the function of these enzymes in vivo because UGTs with overlapping substrate specificity that are co-expressed in the same cell types may show functional redundancy. For example, in A. thaliana UGT72E2 has been shown to glycosylate monolignol in the phenylpropanoid pathway, yet a ugt72E2 T-DNA insertion line shows only a 50% reduction in monolignol glucoside due to functional redundancy (36). Two other related GTs (UGT72E1 and UGT72E3) are also able to glucosylate monolignol. The triple knock-out line showed a 90% reduction in monolignol glucoside, consistent with functional redundancy resulting from overlapping, but not necessarily identical, substrate specificity. Functional redundancy has also been demonstrated for two closely related A. thaliana anthranilate glucosyltransferases, UGT74F1 and UGT74F2, because increasing the level of UGT74F1 is sufficient to rescue anthranilate glucoside levels in the ugt74f2 mutant (26). Western blot and LC-MS/MS analysis indicate that UGT74H5 is likely to be the key player in the synthesis of the acyl glucose donor N-methylanthraniloyl-β-d-glucopyranose (which serves as the acyl donor for avenacins A-1 and B-1), although UGT74H6 may also contribute to avenacin synthesis. It seems likely that UGT74H5 and UGT74H6 are required for the synthesis of activated acyl donors N-methylanthraniloyl-β-d-glucopyranose (which serves as the acyl donor for avenacin A-1 and B-1) and benzoyl-β-d-glucopyranose (for avenacin A-2 and avenacin B-2), as proposed in Fig. 5.
FIGURE 5.
Predicted functions of UGT74H5 and UGT74H6 in avenacin biosynthesis.
A comparison of the kinetic properties of the three oat GTs with those of other plant GTs with activity against related compounds is shown in Table 2. These data highlight the important of functional groups on the aromatic ring in determining the substrate specificity of different GTs. UGT74F1 is predicted to work as a salicylic acid glucosyltransferase in planta (27). It is interesting to note that UGT74H5 and UGT74H6 are either inactive (UGT74H5) or far less active (UGT74H6) toward salicylic acid than is UGT74F1 (kcat/Km of 0.062, 0.004, and 1.68 mm−1·s−1, respectively) but have higher activity toward N-methylanthranilic acid. Salicylic acid and N-methylanthranilic acid differ in having either a hydroxyl or a methylamine, respectively, adjacent to the carboxyl sugar acceptor group. The influence of the functional group on the aromatic ring on activity is also clearly illustrated when using anthranilate as an acceptor. In addition to glucosylating salicylic acid, the A. thaliana GTs UGT74F1 and UGT74F2 are known to form glucose esters of anthranilate in planta (27). In vitro, the multifunctional enzyme FaGT2 from Fragaria x ananassa is also able to form anthraniloyl glucose, but with a lower efficiency (37). Although UGT74H5 and UGT74H6 show good recognition of anthranilate (Km of 0.42 and 0.79 mm, respectively) compared with that of A. thaliana anthranilate glucosyltransferases UGT74F1 (Km of 0.81 mm), their specificity for anthranilate is 10 to 30 times lower than that of UGT74F2 ((kcat/Km of UGT74H5, 0.33 mm−1·s−1; kcat/Km of UGT74H6, 0.13 mm−1·s−1; kcat/Km of UGT74F2, 18.5 mm−1·s−1). In the current study, UGT74H7 proved to have poor activity against anthranilate (kcat/Km of 0.01 mm−1·s−1), similar to FaGT2 (kcat/Km of 0.03 mm−1·s−1). Similarly, recognition of benzoic acid by UGT74H6 (Km of 0.12 mm) and A. thaliana GTs (Km of 0.08–0.40 mm) is good, but the specificity of UGT74H6 for benzoate (kcat/Km of 0.48 mm−1·s−1) is lower than that of the A. thaliana GTs shown in Table 2 (0.97–3.97 mm−1·s−1) (27).
TABLE 2.
Comparison of the kinetic properties of the three oat GTs with those of other plant UGTs with activity towards related compounds
Enzymes beginning with At are from A. thaliana; FaGT2 is from Fragaria x ananassa (see supplemental Table 1 for further information). Enzymes beginning with As are from diploid oat (A. strigosa). −, this study; -, no activity detected. The parameters of preferential substrate activites are highlighted in boldface type.
In summary, our kinetic analysis indicates that the oat root-expressed GT UGT74H5 preferentially glucosylates N-methylanthranilic acid to generate the corresponding glucose ester. Our data provide support for a scenario in which anthranilate is methylated prior to glucosylation by UGT74H5, consistent with our previous results indicating that sad7 mutants accumulate N-methylanthraniloyl-β-d-glucopyranose (11). N-Methylanthraniloyl-β-d-glucopyranose then serves as an activated acyl donor for the SAD7 acyltransferase, which transfers N-methylanthranilic acid onto the triterpene backbone (Fig. 5). SAD7 is also required for transfer of the benzoate group to avenacin A-2 and B-2 (11). In this case, UGT74H6 may be important for synthesis of the benzoyl glucose donor. With a kcat/Km for N-methylanthranilic acid within 5-fold of that of UGT74H5, and within 3-fold for benzoic acid, UGT74H6 may also contribute to avenacin synthesis in vivo. However analysis of protein extracts from oat roots indicates that UGT74H5 is likely to the most important of the three GTs in oat root tips, the site of avenacin synthesis (14).
Several family 1 GTs have been shown to have roles in the synthesis of protective compounds in other cereals, for example cyanogenic glycosides in sorghum (38) and benzoxazinones in maize (39). The development of a TILLING platform (40) for diploid oat based on the mutagenised populations that we have developed for A. strigosa is now underway. This will open up opportunities to carry out functional analysis in planta using reverse genetics, with the aim of identifying mutations in UGT74H5, UGT74H6, and other target genes of interest and assessing the role of these genes in specialized metabolism, plant defense, and other important crop traits. UGT74H5 and UGT74H6 now form part of the growing tool kit of characterized genes and enzymes available from plants that can be deployed for triterpene metabolic engineering for disease control and synthetic biology applications (41–43).
Supplementary Material
This work was supported by a Thai Royal Government Studentship award (to A. O.), Biotechnology and Biological Sciences Research Council Grants BBSRC 83/P13269 and BB/C504435/1 (to X. Q., S. B., and B. T.) and DuPont Agricultural Products (to H. J.).

This article contains supplemental Tables 1 and 2, Figs. 1–5, and additional references.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (43) with the data set identifier PXD000083 and DOI 10.6019/PXD000083.
- GT
- glycosyltransferases
- BAC
- bacterial artificial chromosome
- EST
- expressed sequence tag.
REFERENCES
- 1. Gachon C. M., Langlois-Meurinne M., Saindrenan P. (2005) Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci. 10, 542–549 [DOI] [PubMed] [Google Scholar]
- 2. Bowles D., Lim E. K., Poppenberger B., Vaistij F. E. (2006) Glycosyltransferases of lipophilic small molecules. Annu Rev. Plant Biol. 57, 567–597 [DOI] [PubMed] [Google Scholar]
- 3. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yonekura-Sakakibara K., Hanada K. (2011) An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 66, 182–193 [DOI] [PubMed] [Google Scholar]
- 5. Caputi L., Malnoy M., Goremykin V., Nikiforova S., Martens S. (2012) A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 69, 1030–1042 [DOI] [PubMed] [Google Scholar]
- 6. Ross J., Li Y., Lim E., Bowles D. J. (2001) Higher plant glycosyltransferases. Genome Biol. 2, reviews 3004.1–3004.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang X. (2009) Structure, mechanism and engineering of plant natural product glycosyltransferases. FEBS Lett. 583, 3303–3309 [DOI] [PubMed] [Google Scholar]
- 8. Brazier-Hicks M., Offen W. A., Gershater M. C., Revett T. J., Lim E. K., Bowles D. J., Davies G. J., Edwards R. (2007) Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc. Natl. Acad. Sci. U.S.A. 104, 20238–20243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shao H., He X., Achnine L., Blount J. W., Dixon R. A., Wang X. (2005) Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 17, 3141–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Papadopoulou K., Melton R. E., Leggett M., Daniels M. J., Osbourn A. E. (1999) Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. U.S.A. 96, 12923–12928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mugford S. T., Qi X., Bakht S., Hill L., Wegel E., Hughes R. K., Papadopoulou K., Melton R., Philo M., Sainsbury F., Lomonossoff G. P., Roy A. D., Goss R. J., Osbourn A. (2009) A serine carboxypeptidase-like acyltransferase is required for synthesis of antimicrobial compounds and disease resistance in oats. Plant Cell 21, 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, pp. 18.47–18.59, Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 13. Qi X., Bakht S., Qin B., Leggett M., Hemmings A., Mellon F., Eagles J., Werck-Reichhart D., Schaller H., Lesot A., Melton R., Osbourn A. (2006) A different function for a member of an ancient and highly conserved cytochrome P450 family: from essential sterols to plant defense. Proc. Natl. Acad. Sci. U.S.A. 103, 18848–18853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haralampidis K., Bryan G., Qi X., Papadopoulou K., Bakht S., Melton R., Osbourn A. (2001) A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proc. Natl. Acad. Sci. U.S.A. 98, 13431–13436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pearson W. R., Lipman D. J. (1988) Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U.S.A. 85, 2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tamura K., Dudley J., Nei M., Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 17. Qi X., Bakht S., Leggett M., Maxwell C., Melton R., Osbourn A. (2004) A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. U.S.A. 101, 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 19. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 20. Kliebenstein D. J., Osbourn A. (2012) Making new molecules - evolution of pathways for novel metabolites in plants. Curr. Opin. Plant Biol. 15, 415–423 [DOI] [PubMed] [Google Scholar]
- 21. Szerszen J. B., Szczyglowski K., Bandurski R. S. (1994) iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science 265, 1699–1701 [DOI] [PubMed] [Google Scholar]
- 22. Moraga A. R., Nohales P. F., Pérez J. A., Gómez-Gómez L. (2004) Glucosylation of the saffron apocarotenoid crocetin by a glucosyltransferase isolated from Crocus sativus stigmas. Planta 219, 955–966 [DOI] [PubMed] [Google Scholar]
- 23. Tárraga S., Lisón P., López-Gresa M.P., Torres C., Rodrigo I., Bellés J.M., Conejero V. (2010) Molecular cloning and characterization of a novel tomato xylosyltransferase specific for gentisic acid. J. Exp. Bot. 61, 4325–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee H.I., Raskin I. (1999) Purification, cloning, and expression of a pathogen inducible UDP-glucose:salicylic acid glucosyltransferase from tobacco. J. Biol. Chem. 274, 36637–36642 [DOI] [PubMed] [Google Scholar]
- 25. Dean J. V., Delaney S. P. (2008) Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol. Plant. 132, 417–425 [DOI] [PubMed] [Google Scholar]
- 26. Quiel J. A., Bender J. (2003) Glucose conjugation of anthranilate by the Arabidopsis UGT74F2 glucosyltransferase is required for tryptophan mutant blue fluorescence. J. Biol. Chem. 278, 6275–6281 [DOI] [PubMed] [Google Scholar]
- 27. Lim E. K., Doucet C. J., Li Y., Elias L., Worrall D., Spencer S. P., Ross J., Bowles D. J. (2002) The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J. Biol. Chem. 277, 586–592 [DOI] [PubMed] [Google Scholar]
- 28. Meesapyodsuk D., Balsevich J., Reed D. W., Covello P. S. (2007) Saponin biosynthesis in Saponaria vaccaria. cDNAs encoding β-amyrin synthase and a triterpene carboxylic acid glucosyltransferase. Plant Physiol. 143, 959–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tognetti V. B., Van Aken O., Morreel K., Vandenbroucke K., van de Cotte B., De Clercq I., Chiwocha S., Fenske R., Prinsen E., Boerjan W., Genty B., Stubbs K. A., Inzé D., Van Breusegem F. (2010) Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22, 2660–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gachon C. M., Langlois-Meurinne M., Henry Y., Saindrenan P. (2005) Transcriptional co-regulation of secondary metabolism enzymes in Arabidopsis: functional and evolutionary implications. Plant Mol. Biol. 58, 229–245 [DOI] [PubMed] [Google Scholar]
- 31. Crombie W. M., Crombie L. (1986) Distribution of avenacins A-1, A-2, B-1 and B-2 in oat roots: Their fungicidal activity towards ‘take-all’ fungus. Phytochemistry 25, 2069–2073 [Google Scholar]
- 32. Lehfeldt C., Shirley A. M., Meyer K., Ruegger M. O., Cusumano J. C., Viitanen P. V., Strack D., Chapple C. (2000) Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell 12, 1295–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shirley A. M., McMichael C. M., Chapple C. (2001) The sng2 mutant of Arabidopsis is defective in the gene encoding the serine carboxypeptidase-like protein sinapoylglucose:choline sinapoyltransferase. Plant J 28, 83–94 [DOI] [PubMed] [Google Scholar]
- 34. Fraser C. M., Thompson M. G., Shirley A. M., Ralph J., Schoenherr J. A., Sinlapadech T., Hall M. C., Chapple C. (2007) Related Arabidopsis serine carboxypeptidase-like sinapoylglucose acyltransferases display distinct but overlapping substrate specificities. Plant Physiol. 144, 1986–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lim E. K., Baldauf S., Li Y., Elias L., Worrall D., Spencer S. P., Jackson R. G., Taguchi G., Ross J., Bowles D. J. (2003) Evolution of substrate recognition across a multigene family of glycosyltransferases in Arabidopsis. Glycobiology 13, 139–145 [DOI] [PubMed] [Google Scholar]
- 36. Lanot A., Hodge D., Jackson R. G., George G. L., Elias L., Lim E. K., Vaistij F. E., Bowles D. J. (2006) The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J. 48, 286–295 [DOI] [PubMed] [Google Scholar]
- 37. Landmann C., Fink B., Schwab W. (2007) FaGT2: a multifunctional enzyme from strawberry (Fragaria x ananassa) fruits involved in the metabolism of natural and xenobiotic compounds. Planta 226, 417–428 [DOI] [PubMed] [Google Scholar]
- 38. Hansen K. S., Kristensen C., Tattersall D. B., Jones P. R., Olsen C. E., Bak S., Møller B. L. (2003) The in vitro substrate regiospecificity of recombinant UGT85B1, the cyanohydrin glucosyltransferase from Sorghum bicolor. Phytochemistry 64, 143–151 [DOI] [PubMed] [Google Scholar]
- 39. von Rad U., Hüttl R., Lottspeich F., Gierl A., Frey M. (2001) Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize. Plant J. 28, 633–642 [DOI] [PubMed] [Google Scholar]
- 40. McCallum C.M., Comai L., Greene E.A., Henikoff S. (2000) Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 123, 439–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xue Z., Duan L., Liu D., Guo J., Ge S., Dicks J., ÓMáille P., Osbourn A., Qi X. (2012) Divergent evolution of oxidosqualene cyclases in plants. New Phytol. 193, 1022–1038 [DOI] [PubMed] [Google Scholar]
- 42. Augustin J. M., Kuzina V., Andersen S.B., Bak S. (2011) Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry 72, 435–457 [DOI] [PubMed] [Google Scholar]
- 43. Vizcaíno J.A., Côté R., Reisinger F., Barsnes H., Foster J.M., Rameseder J., Hermjakob H., Martens L. (2010) The Proteomics Identifications database: 2010 update. Nucleic Acids Res. 38, D736–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.