Background: TDP-43 pathology and the role of E3 ubiquitin ligases are increasingly recognized in neurodegeneration.
Results: Parkin ubiquitinates TDP-43 and forms a multiprotein complex with HDAC6 to sequester TDP-43 in cytosol.
Conclusion: Parkin E3 ubiquitin ligase activity promotes TDP-43 inclusion formation and nuclear translocation.
Significance: Parkin-TDP-43 interaction may be exploited as a therapeutic strategy in ALS/FTLD pathology.
Keywords: Amyotropic Lateral Sclerosis (Lou Gehrig Disease), E3 Ubiquitin Ligase, Histone Deacetylase, Parkin, Ubiquitin
Abstract
The importance of E3 ubiquitin ligases, involved in the degradation of misfolded proteins or promotion of protein-protein interaction, is increasingly recognized in neurodegeneration. TDP-43 is a predominantly nuclear protein, which regulates the transcription of thousands of genes and binds to mRNA of the E3 ubiquitin ligase Parkin to regulate its expression. Wild type and mutated TDP-43 are detected in ubiquitinated forms within the cytosol in several neurodegenerative diseases. We elucidated the mechanisms of TDP-43 interaction with Parkin using transgenic A315T mutant TDP-43 (TDP43-Tg) mice, lentiviral wild type TDP-43, and Parkin gene transfer rat models. TDP-43 expression increased Parkin mRNA and protein levels. Lentiviral TDP-43 increased the levels of nuclear and cytosolic protein, whereas Parkin co-expression mediated Lys-48 and Lys-63-linked ubiquitin to TDP-43 and led to cytosolic co-localization of Parkin with ubiquitinated TDP-43. Parkin and TDP-43 formed a multiprotein complex with HDAC6, perhaps to mediate TDP-43 translocation. In conclusion, Parkin ubiquitinates TDP-43 and facilitates its cytosolic accumulation through a multiprotein complex with HDAC6.
Introduction
Transactivation response DNA-binding protein 43 (TDP-43) is a 414-amino acid protein with DNA/RNA-binding protein properties (1). TDP-43 has highly conserved RNA recognition motifs (RRM1/RRM2) flanked on either side by N-terminal and glycine-rich C-terminal domains (2). TDP-43 is predominantly nuclear, but under “pathological” conditions, it is translocated to the cytosol where it is ubiquitinated and/or phosphorylated and cleaved into smaller fragments (3–9). Some ubiquitinated inclusions were found in amyotrophic lateral sclerosis (ALS2-TDP) (10) and a subset of fronto-temporal lobar dementia-Tar-DNA-binding protein (7, 11, 12), and TDP-43 was much later discovered as the key protein in these previously uncharacterized ubiquitinated inclusions (7, 11, 12).
Dominantly inherited point mutations in TDP-43 are causal to familial ALS and some cases of FTLD (13–17). However, abnormal TDP-43 staining (TDP-43 with ubiquitin-positive cytoplasmic inclusions) is also detected in the brains of 29% of cognitively normal people over age 65 (18). High frequency of abnormal TDP-43 staining is observed in Huntington disease (19) and dementia-Parkinsonism with ALS complex of Guam (20, 21). TDP-43 pathology is detected throughout the brain in ALS-FTLD and in the limbic system and cortex in dementia-Parkinsonism with the ALS complex of Guam (20, 21) in addition to hippocampus in Alzheimer disease (22–25). However, TDP-43 accumulates in the limbic system in dementia with Lewy body (22, 26), Parkinson disease (PD) (22, 26), cortico-basal degeneration (24, 27, 28), and progressive supranuclear palsy (11, 28). These data suggest disease-specific topographical distribution of TDP-43 pathology in neurodegeneration.
It has been reported that TDP-43 binds to Parkin mRNA and regulates its expression (29), suggesting a potential relationship between TDP-43 and Parkin. More recently, it was demonstrated that TDP-43 depletion results in down-regulation of Park2 mRNA in stem cell-derived human neurons and in motor neurons containing TDP-43 inclusions in sporadic ALS (30). Neurons bearing TDP-43 aggregates showed decreased cytoplasmic Parkin levels (30). Parkin is an E3 ubiquitin ligase involved in degradation of misfolded proteins (31), and Park2 mutations, leading to loss of the E3 ubiquitin ligase function, are linked to autosomal recessive early onset PD (32–34). Parkin functions as part of a number of multiprotein complexes, including PTEN-induced putative kinase 1 (PINK1) (35–37), the Skp1-Cullin-Fbox (SCF)-like complex to facilitate proteasomal degradation (38), the chaperone Hsp70, and the U-box protein C terminus of Hsc70-interacting protein (39). We previously demonstrated that wild type and not mutant loss-of-function Parkin increases proteasome activity (40–42), leading to degradation of ubiquitinated proteins. Therefore, Parkin may be part of a protein complex that regulates, or perhaps is regulated by, TDP-43.
Accumulation of ubiquitinated TDP-43 in the cytosol may change the bioavailability of this predominantly nuclear protein (3–9), either leading to gain of cytosolic or loss of nuclear function. We previously showed that lentiviral expression of wild type TDP-43 can lead to pathological changes, including cleavage, aggregation, and phosphorylation (25, 43). In these studies, we aimed to better understand the mechanisms of TDP-43 interaction with the E3 ubiquitin ligase Parkin. We used transgenic A315T mice, which express TDP-43 exclusively in the nucleus and display early motor symptoms (44), as well as lentiviral gene delivery that leads to expression of nuclear and cytoplasmic TDP-43 and allows examination of nondevelopmental early effects of TDP-43.
EXPERIMENTAL PROCEDURES
Stereotaxic Injection
Stereotaxic surgery was performed to inject the lentiviral (Lv) constructs encoding either LacZ, Parkin, and/or TDP-43 into the primary motor cortex of 2-month-old male Sprague-Dawley rats weighing between 170 and 220 g as described previously (40). Animals were injected into the left side of the motor cortex with 2 × 109 m.o.i. Lv-LacZ and into the right side with 1 × 109 m.o.i. Lv-Parkin + 1 × 109 m.o.i. Lv-LacZ, 1 × 109 m.o.i. Lv-TDP-43 + 1 × 109 m.o.i. Lv-LacZ, or 1 × 109 m.o.i. Lv-Parkin + 1 × 109 m.o.i. Lv-TDP-43. All animals were sacrificed 2 weeks post-injection, and the left cortex was compared with the right cortex. A total of eight animals in each treatment (32 animals) were used for WB, ELISA, and immunoprecipitation and eight animals in each treatment (32 animals) for immunohistochemistry. A total of n = 64 animals were used. Transgenic hemizygous mice harboring human TDP-43 with the A315T mutation under the control of the prion promoter and C57BL6/J mouse controls were used (44). The colony was obtained from The Jackson Laboratory Repository (JAX stock no. 010700) and displayed a life span considerably shorter than previous reports (44), with almost 90% of all pups, including males and females, manifesting motor symptoms around 21–30 days. We bred hemizygous mice via mating of hemizygous with noncarrier wild type C57BL/6, and upon genotyping, half were identified as transgenic and the other half were nontransgenic control mice. All mice used are F1 generation from direct mating between hemizygous and C57BL/6 mice. These studies were approved and conducted according to Georgetown University Animal Care and Use Committee.
Cell Culture and Transfection
Human neuroblastoma M17 cells (seeding density 2 × 105 cells) were grown in 24-well dishes (Falcon) to 70% confluence in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) plus 10% (v/v) heat-inactivated fetal bovine serum (Invitrogen), penicillin/streptomycin, and 2 mm l-glutamine at 37 °C and 5% CO2, washed twice in phosphate-buffered saline (PBS). Transient transfection was performed with 3 μg of Parkin cDNA, 3 μg of TDP-43 cDNA, or 3 μg of LacZ cDNA. Cells were treated with 5 μm tubacin for 24 h and DAPI-stained in 12-well dishes. Cells were harvested 24 h after transfection. Transfection was performed in DMEM without serum using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cells were harvested one time with lysis buffer (20 mm Tris (pH 7.5), 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm sodium orthovanadate, 1 μg/ml leupeptin, and 0.1 mm PMSF) and centrifuged at 10,000 × g for 20 min at 4 °C, and the supernatant was collected. Western blot was performed on NuPAGE 4–12% BisTris gel (Invitrogen). Protein estimation was performed using the microscale Bio-Rad protein assay (Bio-Rad).
Western Blot Analysis
The cortex was dissected out and homogenized in 1× STEN buffer (50 mm Tris (pH 7.6), 150 mm NaCl, 2 mm EDTA, 0.2% Nonidet P-40, 0.2% BSA, 20 mm PMSF, and protease mixture inhibitor). The pellet was then resuspended in 4 m urea and homogenized and centrifuged at 5000 × g, and the supernatant containing the insoluble protein fraction was collected. Total TDP-43 was probed either with (1:1000) mouse monoclonal (2E2-D3) antibody generated against N-terminal 261 amino acids of the full-length protein (Abnova) or (1:1000) rabbit polyclonal (ALS10) antibody (ProteinTech, catalog no. 10782-2-AP). Rabbit polyclonal anti-ubiquitin (Chemicon International) was used (1:1000), and rabbit polyclonal anti-Parkin (Millipore) antibody was used (1:1000) for WB. Rabbit polyclonal anti-actin (Thermo Scientific) was used (1:1000). Rabbit polyclonal anti-SQSTM1/p62 (Cell Signaling Technology) was used (1:500). Rabbit monoclonal (1:1000) HDAC6 (Cell Signaling Technology) was used. SIAH2 was probed (1:400) with mouse monoclonal antibody (Novus Biologicals) and HIF-1α with (1:1000) mouse monoclonal antibody (Novus Biologicals). Immunoprecipitation was performed on a total of 100 mg of protein with (1:100) rabbit polyclonal TDP-43 antibody (ProteinTech) or rabbit monoclonal (1:100) Parkin antibody (Invitrogen) and then compared with the input samples. Western blots were quantified by densitometry using Quantity One 4.6.3 software (Bio Rad). Densitometry was obtained as arbitrary numbers measuring band intensity. Data were analyzed as mean ± S.D., using ANOVA with Neumann Keuls multiple comparison between treatment groups.
Parkin Enzyme-linked Immunosorbent Assay
ELISA was performed on brain-soluble brain lysates (in STEN buffer) or insoluble brain lysates (4 m urea) using a mouse-specific Parkin kit (MyBioSource) in 50 μl (1 μg/μl) of brain lysates detected with 50 μl of primary antibody (3 h) and 100 μl of anti-rabbit antibody (30 min) at RT. Extracts were incubated with stabilized Chromogen for 30 min at RT, and solution was stopped and read at 45 0 nm, according to the manufacturer's protocol.
Parkin E3 Ubiquitin Ligase Activity
To determine the activity of Parkin E3 ligase activity, we used E3LITE customizable ubiquitin ligase kit (Life Sensors, UC 101), which measures the mechanisms of E1-E2-E3 activity in the presence of different ubiquitin chains. To measure Parkin activity in the presence or absence of substrates, we immunoprecipitated Parkin (1:100) with PRK8 antibodies and TDP-43 (1:100) with human TDP-43 (Abnova) from 100 mg of TDP43-Tg brain lysates. We used UbcH7 as an E2 that provides maximum activity with Parkin E3 ligase, and we added E1 and E2 in the presence of recombinant ubiquitin, including wild type containing all seven possible surface lysines, no lysine mutant (Lys-0), or Lys-48 or Lys-63 to determine the lysine-linked type of ubiquitin. We then added E3 as immunoprecipitation Parkin or recombinant Parkin (Novus Biologicals) to an ELISA microplate that captures polyubiquitin chains formed in the E3-dependent reaction, which was initiated with ATP at room temperature for 60 min. We also included an E1-E2-E3 and a polyubiquitin chain control in addition to E1, E2, and TDP-43 without Parkin and assay buffer for background reading. The plates were washed three times and incubated with detection reagent and streptavidin-HRP for 5 min, and the polyubiquitin chains generated by E1-E2-E3 machinery were read on a chemiluminescence plate reader.
Immunoprecipitation and Ubiquitination Assay
Either TDP-43 or Parkin was separately immunoprecipitated in 100 μl (100 μg of proteins) of 1× STEN buffer using (1:100) human-specific anti-TDP-43 monoclonal antibody (Abnova) or (1:100) anti-Parkin mouse monoclonal antibody (PRK8; Signet Labs; Dedham, MA), respectively. Following immunoprecipitation, 300 ng of each substrate protein (Parkin and TDP-43) were mixed in the presence of 1 μg of recombinant human ubiquitin (Boston Biochem, MA), 100 mm ATP, 1 μg of recombinant UbcH7 (Boston Biochem), 40 ng of E1 recombinant enzyme (Boston Biochem) and incubated at 37 °C in an incubator for 20 min. The reaction was heat-inactivated by boiling for 5 min, and the substrates were analyzed by Western blot.
Immunohistology
Immunohistochemistry was performed on 20-μm-thick sections of brain or cervical spinal cord. TDP-43 was probed (1:200) with rabbit polyclonal (ALS10) antibody (ProteinTech, catalog no. 10782-2-AP). Rabbit polyclonal anti-ubiquitin (Chemicon International) was used (1:100), and mouse monoclonal anti-Parkin (Millipore) antibody was used (1:200) for immunohistochemistry. Toluidine blue and DAPI staining were performed according to the manufacturer's instructions (Sigma). Counting of toluidine blue staining of centric axons within 10 random fields of each slide was performed by a blind investigator in n = 8 animals from each treatment. All staining experiments were scored by a blind investigator to the treatments.
20 S Proteasome Activity Assay
Brain extracts of 100 μg were incubated with 250 μm of the fluorescent 20 S proteasome-specific substrate succinyl-LLVY-AMC at 37 °C for 2 h. The medium was discarded, and homogenates were lysed in 50 mm HEPES (pH 7.5), 5 mm EDTA, 150 mm NaCl, and 1% Triton X-100, containing 2 mm ATP. The fluorophore AMC, which was released after cleavage from the labeled substrate succinyl-LLVY-AMC (Chemicon International, Inc.), was detected, and free AMC fluorescence was quantified using a 380/460-nm filter set in a fluorometer (absorption at 351 nm and emission at 430 nm). We measured nonproteasomal side reactivity by adding lactacystin as a specific proteasome inhibitor to the reaction mixture and subtracted these values from the total for an accurate measure of specific proteasome activity.
qRT-PCR in Neuronal Tissues
qRT-PCR was performed on PCR system (Applied Biosystems) with Fast SYBR Green PCR master mix (Applied Biosystems) in triplicate from reverse-transcribed cDNA from control uninjected or lentiviral LacZ, Parkin, TDP-43, and TDP-43 + Parkin injected rat cortical brain tissues. These experiments were repeated in human neuroblastoma M17 cells and A315T-Tg compared with nontransgenic C57BL/6 controls. Human wild type Parkin forward primer CCA TGA TAG TGT TTG TCA GGT TC and a reverse primer GTT GTA CTT TCT CTT CTG CGT AGT GT were used. Gene expression values were normalized using GADPH levels.
RESULTS
TDP-43 Inhibits Proteasome Activity and Alters Parkin Protein Levels
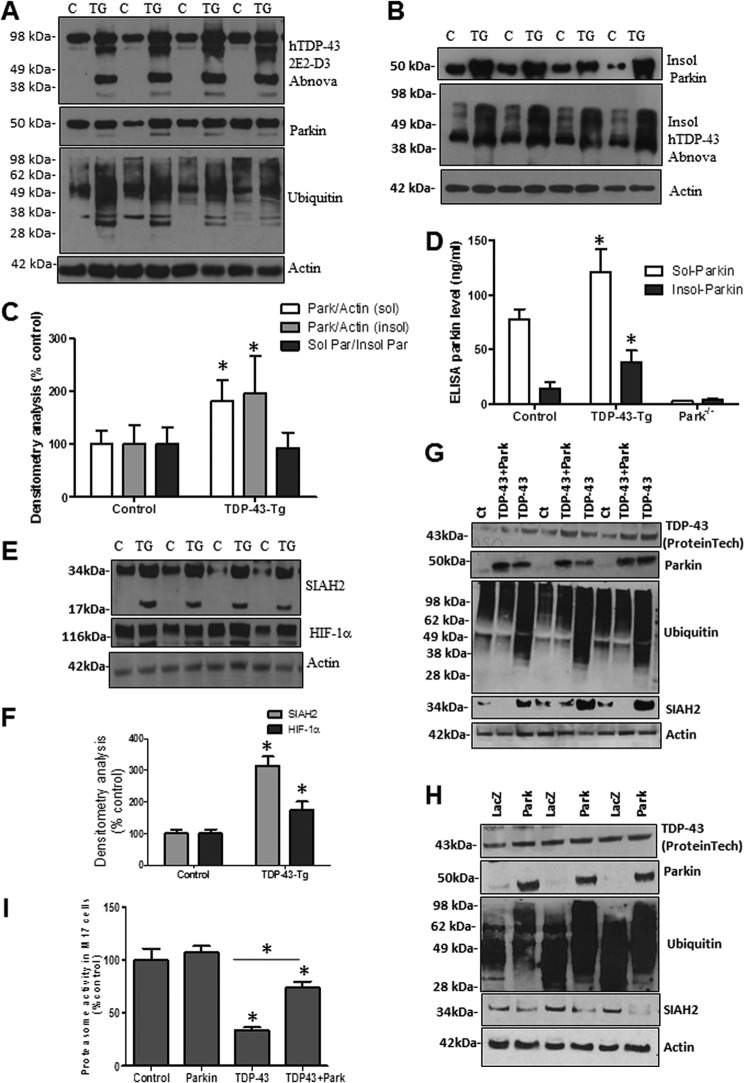
To determine the effects of TDP-43 on Parkin in transgenic animals, we used the A315T mutant TDP-43 transgenic mice (TDP43-Tg), which were reported to have aggregates of ubiquitinated proteins in layer five pyramidal neurons in frontal cortex, as well as spinal motor neurons, without cytoplasmic TDP-43 (44). This model is relevant to our studies because it shows nuclear TDP-43-driven pathology, independent of cytoplasmic TDP-43 inclusions (44). Western blot analysis showed accumulation of full-length and TDP-43 fragments (∼35 kDa) as well as higher molecular weight species with human TDP-43 antibody (Fig. 1A, 1st blot) compared with nontransgenic controls, suggesting TDP-43 pathology. Further analysis of the soluble brain lysate (STEN extract) showed increased Parkin levels by Western blot (Fig. 1A, 2nd blot, 82%, p < 0.05, n = 8) and appearance of a lower molecular weight band, perhaps indicating Parkin cleavage. We also observed increased levels of ubiquitin smears (Fig. 1A, 3rd blot) using anti-ubiquitin antibodies, suggesting accumulation of ubiquitinated proteins. We previously showed that amyloid protein stress and aging alter Parkin solubility (45), so we aimed to determine whether Parkin solubility was altered in TDP43-Tg mice. We resuspended the protein pellet after STEN extraction in 4 m urea to detect the insoluble fraction, and we detected a significant increase (Fig. 1B, 95% by densitometry, p < 0.05, n = 8) in insoluble Parkin in 1-month-old TDP43-Tg mice compared with control (Fig. 1, B and C, p < 0.05, n = 8), suggesting that TDP-43 aggregates are associated with altered Parkin solubility. The ratio of soluble over insoluble Parkin was not significantly changed (Fig. 1C, p < 0.05), suggesting that TDP-43 accumulation increases soluble and insoluble Parkin levels. We also probed for TDP-43 in 4 m urea extracts and detected increased levels of insoluble TDP-43 (Fig. 1B, 2nd blot) in TDP43-Tg compared with control. To verify the changes in Parkin level observed by WB, we performed quantitative Parkin ELISA to determine the levels of both soluble (STEN extract) and insoluble (4 m urea) Parkin, using brain extracts from Parkin−/− mice as control for ELISA specificity (Fig. 1D, n = 8). We detected a significant increase in both soluble (46%, p < 0.05) and insoluble (64%) Parkin in TDP43-Tg mice compared with control levels (Fig. 1D, p < 0.05, n = 8), further suggesting an increase in Parkin level and insolubility in TDP43-Tg mice.
FIGURE 1.
TDP-43 inhibits proteasome activity and alters Parkin levels. A, Western blot analysis of soluble cortical brain lysates from different litters of mixed male and female TDP-43 transgenic mice and nontransgenic control littermates on 4–12% SDS-NuPAGE gel showing human TDP-43 levels probed with 2E2-D3 antibody (1st blot), total Parkin (2nd blot), ubiquitin (3rd blot), and actin (4th blot) levels. B, pellet was resuspended in 4 m urea to extract the insoluble protein fraction, and Western blot was performed showing insoluble Parkin (1st blot) and insoluble TDP-43 (2nd blot) compared with actin loading control (3rd blot). C, densitometry analysis of A and B blots showing soluble and insoluble Parkin protein levels normalized to actin and the ratio of soluble to insoluble Parkin. D, ELISA measurement of Parkin level in soluble (STEN extracts) and insoluble (4 m urea) brain extracts compared with Parkin−/− brain extracts as a specificity control. E, Western blot analysis of cortical brain lysates on 4–12% SDS-NuPAGE gel showing soluble protein levels of the E3 ubiquitin ligase SIAH2 (1st blot) and its target protein HIF-1α (2nd blot) compared with actin loading control. F, densitometry analysis of blots in D normalized to actin control, n = 4, ANOVA with Neumann Keuls, p < 0.05. G, Western blot analysis of M17 cell lysates on 4–12% SDS-NuPAGE gel showing human TDP-43 levels (1st blot), total Parkin (2nd blot), ubiquitin (3rd blot), SIAH2 (4th blot), and actin levels (5th blot) in cells expressing TDP-43 and wild type Parkin. H, Western blot analysis of M17 cell lysates on 4–12% SDS-NuPAGE gel showing human TDP-43 levels (1st blot), total Parkin (2nd blot), ubiquitin (3rd blot), SIAH2 (4th blot), and actin levels (5th blot) in cells expressing LacZ and wild type Parkin. I, histograms represent the chymotrypsin proteasome activity in M17 neuroblastoma cells. * indicates significantly different ANOVA with Neumann Keuls, p < 0.05, n = 6 for cells.
The seven in absentia homolog (SIAH) protein is another E3 ligase involved in ubiquitination and proteasomal degradation of specific proteins (46, 47). SIAH is rapidly degraded via the proteasome (48). The activity of this ubiquitin ligase has been implicated in regulating cellular response to hypoxia, including hypoxia-inducible factor-1 (HIF-1α) (49–51). We used SIAH2 as an E3 ligase control to determine whether TDP-43 decreases Parkin solubility, leading to alteration of its E3 ligase function independent of other E3 ligases. Western blot analysis showed a significant increase (215%) in soluble SIAH2 levels (Fig. 1, E and F, p < 0.05, n = 8) in TDP43-Tg mice compared with control, indicating lack of degradation of SIAH2 perhaps due to proteasomal impairment. However, SIAH2 was not detected in the insoluble fraction. We also observed a lower molecular mass band at 17 kDa (Fig. 1E) in transgenic mice, suggesting possible cleavage of SIAH2 dimeric structure (49–51). Further examination of the level of SIAH2 target molecule HIF-1α showed a significant increase (76%, p < 0.05) in protein level (Fig. 1, E and F), suggesting lack of proteasomal degradation.
To ascertain the effect of TDP-43 on Parkin level and proteasome activity, we expressed wild type TDP-43 (Fig. 1G, 1st blot) in the presence or absence of Parkin (Fig. 1G, 2nd blot) in human M17 neuroblastoma cells. Expression of TDP-43 alone led to the appearance of endogenous Parkin protein (Fig. 1G, 2nd blot), suggesting that TDP-43 regulates Parkin mRNA to induce protein expression (29). Co-expression of exogenous Parkin and TDP-43 led to a slight decrease in TDP-43 levels (Fig. 1G, 1st blot) and a noticeable decrease in ubiquitinated proteins (Fig. 1G, 3rd blot) compared with TDP-43 alone. SIAH2 was difficult to detect in control M17 cells (Fig. 1G, 4th blot), but it accumulated when TDP-43 was expressed despite the increase in endogenous Parkin; however, exogenous Parkin co-expression with TDP-43 led to disappearance of SIAH2 (Fig. 1G, 4th blot). We further compared the effects of Parkin expression alone (Fig. 1G, 2nd blot) compared with LacZ on TDP-43 and SIAH2 levels. No differences were observed between control (Fig. 1F), LacZ-, and Parkin-transfected M17 cells (Fig. 1H) on endogenous TDP-43 expression levels (Fig. 1H, 1st blot). A higher level of ubiquitinated protein smears was observed with Parkin expression (Fig. 1H, 3rd blot), consistent with the role of Parkin as an E3 ubiquitin ligase, but the level of SIAH2 was significantly decreased (Fig. 1H, 4th blot, 74%, p < 0.05) compared with actin control. To determine whether SIAH2 accumulation is due to decreased E3 ligase activity or proteasomal function, we measured proteasome activity (Fig. 1I) and found that TDP-43 significantly decreased (66%) proteasome activity (p < 0.05, n = 12), whereas Parkin co-expression significantly reversed proteasome activity to 74% of control or Parkin levels but remained significantly less (26%) than control. These data suggest that TDP-43 increases Parkin expression levels, whereas proteasomal inhibition leads to decreased degradation of proteins, including the rapidly degrading SIAH2.
Lentiviral Expression of TDP-43 in Rat Motor Cortex Results in Increased Protein Levels in Preganglionic Cervical Spinal Cord Inter-neurons
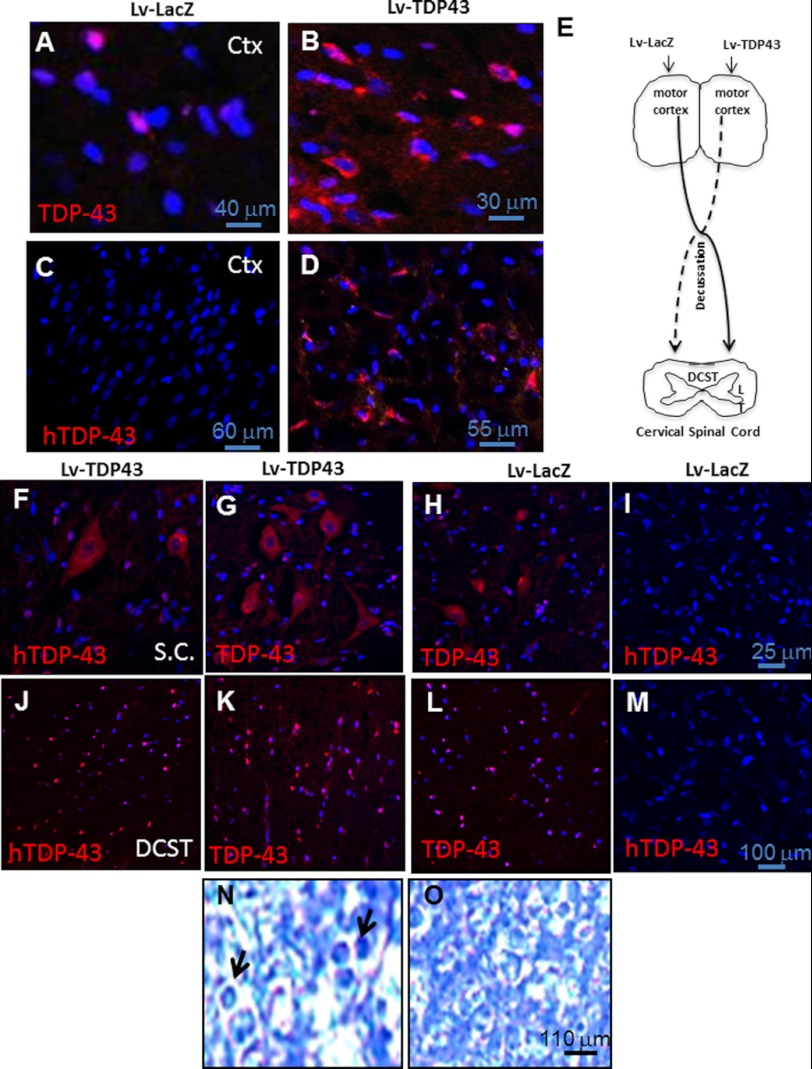
We expressed wild type TDP-43 using lentiviral gene delivery into the motor cortex of 2-month-old Sprague-Dawley rats. We previously showed, using Western blot analysis, significantly increased levels of TDP-43 (41%) 2 weeks after lentiviral gene expression (43). Immunohistochemistry using rabbit polyclonal antibody that recognizes human and rat TDP-43 (ALS10, ProteinTech) showed increased TDP-43 protein levels and cytosolic accumulation 2 weeks post-injection (Fig. 2B) compared with the LacZ-injected contralateral (Fig. 2A) hemisphere. To ascertain specificity of gene expression, we used human-specific (hTDP-43) mouse monoclonal antibody that recognizes amino acids 1–261 (Abcam) and observed positive human TDP-43 staining within a 4-mm radius in 38% (by stereology, n = 8) of cortical neurons (Fig. 2D) compared with a LacZ-injected (Fig. 2C) hemisphere. Further examination of cervical spinal cord revealed a 13% increase in immunoreactivity to hTDP-43 (Fig. 2F) and increased reactivity to TDP-43 antibody (Fig. 2G) in preganglionic inter-neurons, which were morphologically identified in the contralateral side of TDP-43-injected motor cortex (Fig. 2E) compared with the contralateral spinal cord injected with LacZ (Fig. 2, H and I), suggesting that hTDP-43 expression in the motor cortex leads to increased protein levels in the contralateral spinal cord. Furthermore, stereological counting revealed a 46% (by stereology, n = 8) increase in the levels of hTDP-43 (Fig. 2J) and increased immunoreactivity to TDP-43 antibody (Fig. 2K) in the dorso-cortical spinal tract (DCST) of cervical spinal cord contralateral to cortical TDP-43 expression compared with the LacZ-injected side (Fig. 2, L and M). Toluidine blue staining and quantification by a blind investigator of centric axons within 10 random fields of each slide showed an increased number (18%, n = 8) of axons (Fig. 2N, arrows) in enlarged circles, suggesting axonal degeneration compared with the contralateral DCST (Fig. 2O). Some centric axons were detected in all treatments.
FIGURE 2.
Lentiviral expression of TDP-43 in rat motor cortex results in detection of TDP-43 in preganglionic cervical spinal cord inter-neurons. Staining of 20-μm-thick sections from rat brain injected with lentiviral TDP-43 in the right hemisphere and lentiviral LacZ in the left hemisphere show the following: A, neurons in rat motor cortex stained with anti-TDP-43 antibody that detects both human and rat TDP-43 and DAPI-stained nuclei in lentiviral LacZ-injected; B, TDP-43-injected hemisphere; C, neurons in rat motor cortex stained with anti-TDP-43 antibody that detects human TDP-43 and DAPI-stained nuclei in lentiviral LacZ-injected; and D, TDP-43-injected hemispheres. E, schematic representation of injected motor cortex relative to contralateral spinal cord region and DCST. Staining of 20-μm-thick sections showing preganglionic cervical spinal cord inter-neurons stained with hTDP-43 mouse monoclonal antibody (Abnova) that recognizes the following: F, human TDP-43; G, anti-TDP-43 rat polyclonal antibody (ProteinTech) that recognizes both human and rat and DAPI-stained nuclei contralateral to lentiviral TDP-43-injected cortex; H, TDP-43; I, hTDP-43 contralateral to LacZ-injected hemisphere. Staining of 20-μm-thick sections showing fibers in DCST stained with the following: J, mouse monoclonal hTDP-43; K, DAPI and rabbit polyclonal anti-TDP-43 antibody DAPI; L, contralateral to lentiviral TDP-43-injected cortex (TDP-43); M, hTDP-43; N, TDP-43 staining and DAPI in DCST contralateral to LacZ-injected hemisphere. O, toluidine blue-stained DCST contralateral to lentiviral TDP-43-injected cortex compared with Lac-injected hemisphere. Lv, lentivirus.
Lentiviral Parkin Expression Increases Cytosolic Co-localization of TDP-43 with Ubiquitin
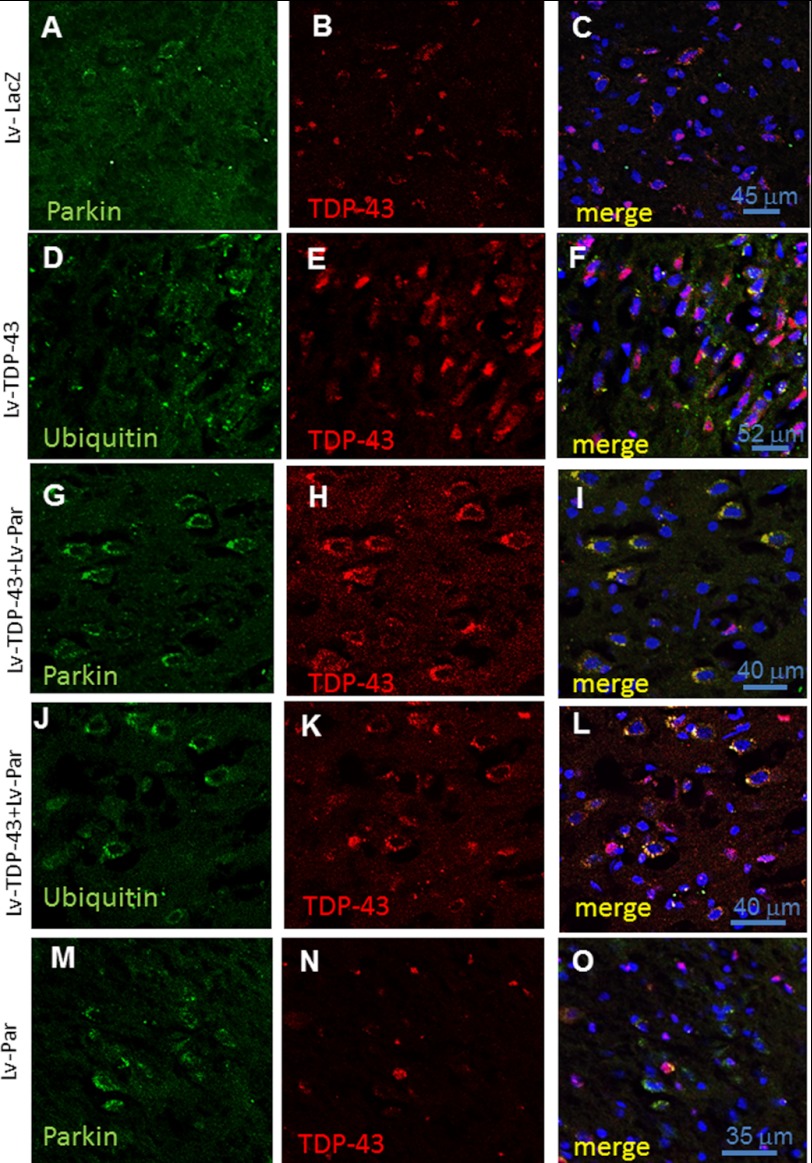
Because TDP-43 is detected in ubiquitinated forms within the cytosol in human disease (3–9), we sought to determine whether ubiquitination is beneficial or detrimental to TDP-43 using Parkin as a ubiquitous E3-ubiquitin ligase in the human brain. We co-expressed TDP-43 with Parkin and sacrificed the animals 2 weeks post-injection. Staining of 20-μm-thick brain sections showed endogenous Parkin expression (Fig. 3A) and TDP-43 (Fig. 3B), which was predominantly localized to DAPI-stained nuclei (Fig. 3C) in the LacZ-injected rat motor cortex. Staining with anti-ubiquitin antibodies (Fig. 3D) in rats expressing TDP-43 in the motor cortex (Fig. 3E) did not result in any noticeable co-localization between TDP-43 and ubiquitin (Fig. 3F). Stereological counting showed 38% increase in hTDP-43-stained cells (Fig. 2D). However, cytosolic TDP-43 was observed in cortical neurons expressing TDP-43 (Fig. 3F) compared with nuclear TDP-43 in LacZ-injected animals (Fig. 3C). We expressed Parkin in the rat motor cortex (Fig. 3G) together with TDP-43 (Fig. 3H) and observed cytosolic co-localization of Parkin and TDP-43 (Fig. 3I, 35% by stereology). We further stained with anti-ubiquitin antibodies and observed increased levels of ubiquitin (Fig. 3J, 35% by stereology) in animals injected with Parkin and TDP-43 (Fig. 3K). Interestingly, enhanced ubiquitin signals co-localized with TDP-43 in the cytosol, suggesting that ubiquitination may result in cytosolic sequestration of TDP-43. To determine whether exogenous Parkin expression affects endogenous TDP-43 protein localization, we stained with Parkin (Fig. 3M, 28% by stereology) and TDP-43 (Fig. 3N) antibodies, but we did not observe any changes in the pattern of TDP-43 staining (Fig. 3O).
FIGURE 3.
Lentiviral Parkin increases cytosolic co-localization of ubiquitin and TDP-43. Staining of 20-μm-thick sections from rat brain injected with lentiviral TDP-43 in the right hemisphere and lentiviral LacZ in the left hemisphere show the following: A, neurons in rat motor cortex stained with mouse monoclonal (Millipore) anti-Parkin; B, rabbit polyclonal anti-TDP-43 antibodies; C, Parkin, TDP-43, and DAPI in lentiviral LacZ-injected hemisphere; D, neurons in rat motor cortex stained with mouse monoclonal anti-ubiquitin; E, rabbit polyclonal anti-TDP-43 antibodies; F, ubiquitin, TDP-43, and DAPI in lentiviral TDP-43-injected hemisphere; G, neurons in rat motor cortex stained with mouse monoclonal anti-Parkin; H, rabbit polyclonal anti-TDP-43 antibodies; I, Parkin, TDP-43, and DAPI in animals co-injected with lentiviral TDP-43 and Parkin; J, neurons in rat motor cortex stained with mouse monoclonal anti-ubiquitin; K, rabbit polyclonal anti-TDP-43 antibodies; L, ubiquitin, TDP-43, and DAPI stained nuclei in animals co-injected with lentiviral TDP-43 and Parkin. Neurons in rat motor cortex stained with the following: M, mouse monoclonal anti-Parkin antibodies; N, rabbit polyclonal anti-TDP-43 antibodies; O, Parkin, TDP-43, and DAPI-stained nuclei in animals injected with lentiviral Parkin alone. Lv, lentiviral.
Parkin Promotes Lys-48- and Lys-63-linked Ubiquitin to TDP-43
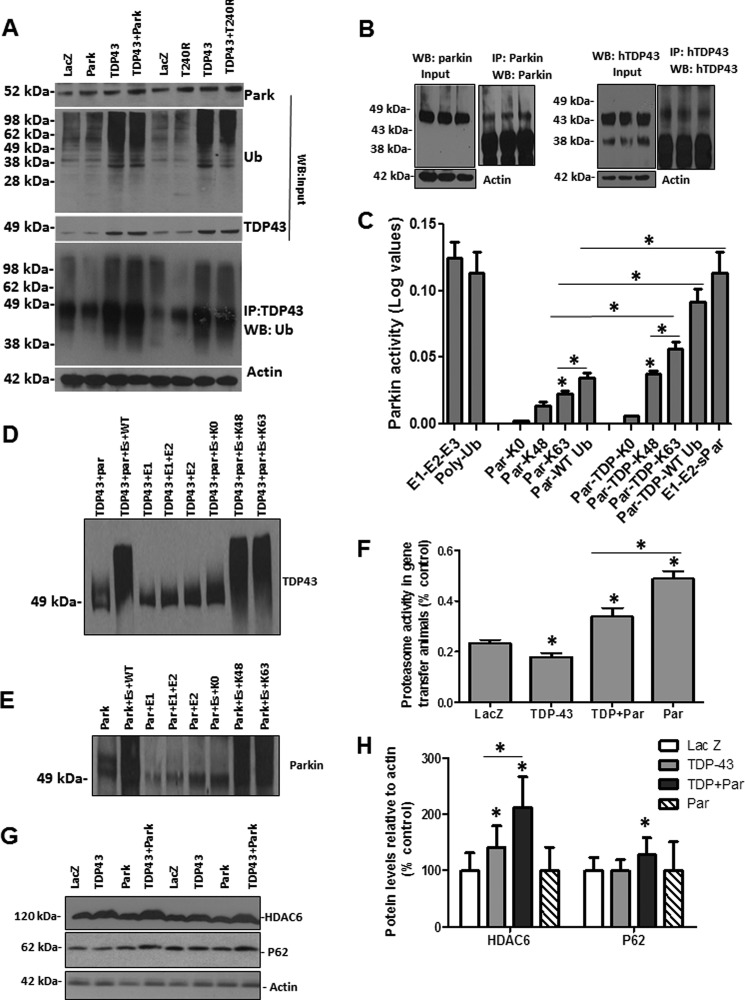
To demonstrate whether Parkin mediates TDP-43 ubiquitination, we performed immunoprecipitation to show ubiquitinated TDP-43 in the presence of Parkin expression. Western blot analysis of the input showed that increased exogenous Parkin (Fig. 4A, 1st blot, n = 8, p < 0.05, 42%) in the rat motor cortex increases the levels of ubiquitinated proteins (Fig. 4A, 2nd blot). Densitometry analysis of TDP-43 blots (Fig. 4A, 3rd blot) showed a significant increase (48%, n = 8) in TDP-43 levels in brains injected with lentiviral TDP-43 (consistent with our previous work (52)) compared with LacZ- or Parkin-injected brains. However, co-injection of TDP-43 and Parkin did not result in any significant changes in TDP-43 levels (p < 0.05, n = 8), suggesting that Parkin mediates TPD-43 ubiquitination, which may not lead to protein degradation. We also used a nonfunctional Parkin mutant (T240R, threonine to arginine mutation), which was co-expressed with TDP-43 (Fig. 4A, top blot) and detected no changes in ubiquitinated proteins (Fig. 4A, 2nd blot) or TDP-43 levels (Fig. 4A, 3rd blot). We then immunoprecipitated TDP-43 and probed with ubiquitin (Fig. 4A, 4th blot) to ascertain that high molecular weight species are ubiquitinated TDP-43 proteins and not some protein aggregates. An increase in protein smear was observed when TDP-43 was co-injected with Parkin, compared with TDP-43, Parkin, or LacZ alone, suggesting increased TDP-43 ubiquitination in the presence of wild type Parkin. However, no differences were observed in the levels of ubiquitinated proteins (Fig. 4A, 4th blot) when TDP-43 was immunoprecipitated with or without expression of T240R mutant Parkin, suggesting that functional Parkin mediates TDP-43 ubiquitination.
FIGURE 4.
Parkin mediates Lys-48 and Lys-63-linked ubiquitination of TDP-43. Western blot of input samples from cortical brain lysates analyzed on 4–12% SDS-NuPAGE gel show the following. A, Parkin expression levels (1st blot); ubiquitin bound protein levels (2nd blot); and TDP-43 levels (3rd blot) compared with actin loading control in rat cortex injected with lentiviral LacZ, TDP-43, Parkin, TDP-43 + Parkin and TDP-43 + T240R mutant. A total of 100 mg of cortical brain samples were immunoprecipitated using rabbit polyclonal anti-TDP-43 and probed (1:1000) with anti-ubiquitin antibody (4th blot) compared with actin-loading control (5th blot) from input samples. B, Western blot of input samples and immunoprecipitated Parkin (top blot) and TDP-43 (bottom blot) from transgenic mice used to measure Parkin E3 ubiquitin ligase activity. C, histograms represent Parkin E3 ubiquitin ligase activity in the presence and absence of human TDP-43 immunoprecipitated from TDP-43 transgenic mice, compared with E3 ubiquitin ligase activity using recombinant Parkin (sPar), polyubiquitin chain as control, and a synthetic E1-E2-E3 control combination. n = 8, p < 0.05, ANOVA with Neumann Keuls. D, Western blot analysis showing ubiquitinated TDP-43 in the presence of Lys-48 and Lys-63. E, Western blot analysis showing ubiquitinated Parkin at Lys-48 and Lys-63. F, histograms represent the chymotrypsin proteasome activity in fresh cortical brain lysates from rats injected with lentiviral LacZ, Parkin, TDP-43, and TDP-43 + Parkin. * indicates significantly different, ANOVA with Neumann Keuls, p < 0.05, n = 8. G, Western blot analysis of cortical brain lysates on 4–12% SDS-NuPAGE gel showing HDAC6 (1st blot) and p62 levels (2nd blot) and actin control (3rd blot). H, densitometry analysis of blots in E from gene transfer animal models. * indicates significantly different, ANOVA with Neumann Keuls, p < 0.05, n = 8.
To determine whether TDP-43 affects Parkin E3 ubiquitin ligase activity, we immunoprecipitated Parkin (Fig. 4B, left blot) and TDP-43 (Fig. 4B, right blot) and performed enzyme activity assay. We used positive controls with E1-E2-E3 or polyubiquitin chains or recombinant Parkin (Novus Biologicals) to measure E3 ubiquitin ligase activity and polyubiquitin chain readings (Fig. 4C). No Parkin activity was detected with the lysine null (Lys-0) ubiquitin, but either mutant Lys-48 or Lys-63-linked ubiquitin showed an increase in Parkin E3 ubiquitin ligase activity compared with control Lys-0 (Fig. 4C, n = 4). Parkin activity with Lys-63 ubiquitin was significantly higher (83%, p < 0.05, n = 4) than Lys-48-linked ubiquitin, suggesting that Parkin undergoes Lys-48 and Lys-63-linked auto-ubiquitination. Parkin was also ubiquitinated using wild type ubiquitin, which contains all seven lysine residues. To determine whether Parkin activity is altered in the presence of TDP-43, we added both Parkin and TDP-43 to the enzyme mixture. As expected, no activity was detected with the lysine null ubiquitin (Lys-0), but Parkin activity was significantly increased compared with Parkin alone (Fig. 4C, p < 0.05, n = 8), with Lys-48 (154%), and Lys-63 (156%) ubiquitin, indicating that Parkin activity is even higher in the presence of a substrate. Parkin also showed a significantly higher level of activity with wild type ubiquitin in the presence of TDP-43 (279%) compared with Parkin alone.
To ascertain that Parkin mediates ubiquitination of TDP-43, we immunoprecipitated Parkin and TDP-43 separately and performed in vitro ubiquitination assays as we reported previously (40). Incubation of both Parkin and TDP-43 in the presence of either wild type (Fig. 4D, 2nd lane), or Lys-48 (7th lane), or Lys-63 (8th lane) ubiquitin (Fig. 4D, n = 3) showed a protein smear upon WB analysis with TDP-43 antibodies compared with lysine null (Lys-0) ubiquitin (6th lane), or in the absence of E1 or E2 or both (all other lanes), suggesting that Parkin mediates Lys-48 and Lys-63-linked ubiquitination of TDP-43. Additionally, Parkin incubation in the presence of either wild type (Fig. 4E, 2nd lane), or Lys-48 (7th lane), or Lys-63 (8th lane) ubiquitin (Fig. 4E, n = 3) showed a protein smear upon WB analysis with Parkin antibodies compared with lysine null (Lys-0) ubiquitin (6th lane), or in the absence of E1 or E2, or both (all other lanes), suggesting that Parkin undergoes Lys-48 and Lys-63-linked auto-ubiquitination.
We then measured the activity of the 20 S proteasome (Fig. 4F), which was significantly decreased (31%, p < 0.05) when TDP-43 was expressed alone (n = 8, p < 0.05), but co-expression of Parkin significantly reversed proteasome activity (48%, p < 0.05) compared with TDP-43 alone. However, proteasome activity in the Parkin-expressing cortex remained significantly higher than LacZ- (73%, p < 0.05) and Parkin + TDP-43 (31%, p < 0.05)-injected animals, indicating that Parkin activity partially reverses proteasome activity.
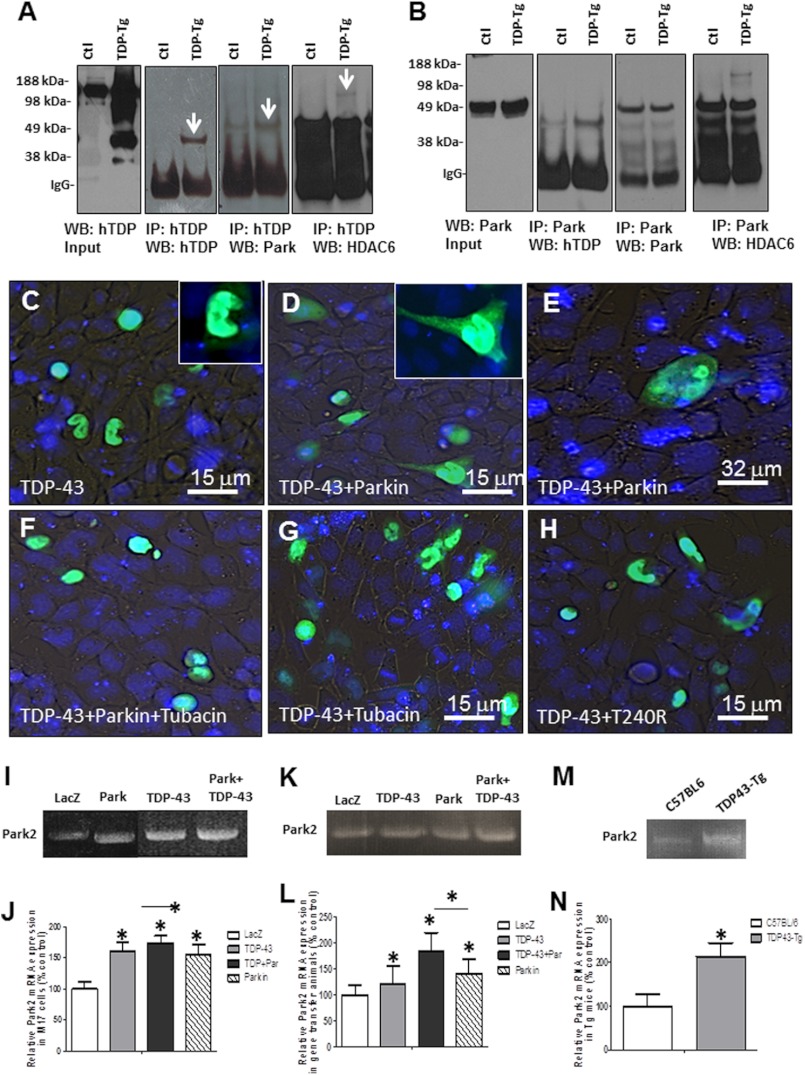
Parkin Forms a Multiprotein Complex with HDAC6 to Mediate TDP-43 Translocation from Nucleus to Cytosol
Lack of degradation of ubiquitinated TDP-43 and cytosolic accumulation of Parkin, TDP-43, and ubiquitin in gene transfer animals led us to examine possible mechanisms to translocate TDP-43 to the cytosol. It was reported that histone deacetylase 6 (HDAC6) directly binds to Parkin and mediates its transport in response to proteasome inhibition (53). Western blot analysis showed a significant increase (41%, p < 0.05) in HDAC6 levels when TDP-43 was expressed compared with LacZ- or Parkin-injected animals (Fig. 4, G and H, 1st blot, p < 0.05, n = 8). However, further increases in HDAC6 levels (Fig. 4, G and H, 112%, p < 0.05) were detected when Parkin was co-expressed with TDP-43, suggesting a possible interaction between these proteins. Examination of molecular markers of autophagy showed a significant increase in p62 (28%, p < 0.05) when Parkin was co-expressed with TDP43 (Fig. 4, G and H, 2nd blot) compared with all other treatments, suggesting accumulation of ubiquitinated proteins. We did not see any changes in other markers of autophagy (LC3, beclin, Atgs) or appearance of autophagic vacuoles by EM (data not shown). We immunoprecipitated human TDP-43 from transgenic mice and verified TDP-43 at 46 kDa using hTDP-43 antibody (Fig. 5A, 1st and 2nd blots). Stripping and re-probing with Parkin antibody showed a slightly higher band around 50 kDa, suggesting the presence of Parkin protein (Fig. 5A, 3rd blot). Further stripping and probing with HDAC6 antibody (Fig. 5A, 4th blot) showed a higher molecular mass band around 120 kDa, indicating a multiprotein complex between Parkin, TDP43, and HDAC6. We then performed a reserve experiment via Parkin immunoprecipitation and verification of human TDP-43 presence (Fig. 5B, 1st and 2nd blot). Stripping and probing with Parkin antibody showed a Parkin band in both transgenic and nontransgenic control mice (Fig. 5B, 3rd blot), indicating that Parkin was successfully immunoprecipitated. We also detected a higher molecular weight band representative of HDAC6 (Fig. 5B, 4th blot) in transgenic but not control mice, further suggesting multiprotein complex formation between TDP43, Parkin, and HDAC6.
FIGURE 5.
TDP-43 forms a multiprotein complex with Parkin and HDAC6. Western blot of input samples from cortical brain lysates in transgenic A315T mice and control littermates analyzed on 4–12% SDS-NuPAGE gel show the following: A, human TDP-43 expression levels (1st blot); immunoprecipitation of TDP-43 showing TDP-43 (2nd blot); Parkin (3rd blot); and HDAC6 (4th blot) forming a protein complex. B represents the reverse immunoprecipitation experiment, where Western blot of input samples from cortical brain lysates in transgenic A315T mice and control littermates analyzed on 4–12% SDS-NuPAGE show Parkin expression levels (1st blot); immunoprecipitation of Parkin showing TDP-43 (2nd blot), Parkin (3rd blot), and HDAC6 (4th blot). GFP fluorescence and nuclear DAPI staining in living human M17 neuroblastoma cells (C) transfected with GFP-TDP-43 alone showing GFP fluorescence within the nucleus (D and E). Cells transfected with GFP-TDP-43 and Parkin showing GFP fluorescence in cytosol and cellular processes. Inset in D shows higher magnification. F, cells transfected with GFP-TDP-43 and Parkin treated with 5 μm HDAC6 inhibitor tubacin for 24 h showing GFP fluorescence within DAPI-stained nuclei. Cells transfected with GFP-TDP-43 for 24 h (G) and treated with tubacin for an additional 24 h (H). Cells transfected with GFP-TDP-43 and T240R show lack of GFP fluorescence with Parkin mutant. I, qRT-PCR showing Park2 mRNA in M17 cells transfected with LacZ TDP-43, Parkin, and TDP-43 + Parkin. J, quantification of qRT-PCR showing relative Park2 mRNA levels normalized to GADPH and expressed as % control. n = 4, p < 0.05, ANOVA with Neumann Keuls. K, qRT-PCR showing Park2 mRNA in rat cortex injected with LacZ (uninjected control), TDP-43, Parkin, and TDP-43 + Parkin. L, quantification of qRT-PCR showing relative Park2 mRNA levels normalized to GADPH and expressed as % control. n = 4, p < 0.05, ANOVA with Neumann Keuls. M, qRT-PCR showing Park2 mRNA in TDP43-Tg and control cortex. N, quantification of qRT-PCR showing relative Park2 mRNA levels normalized to GADPH and expressed as % control. n = 3, p < 0.05, ANOVA with Neumann Keuls.
To ascertain that both Parkin and HDAC6 are required for TDP-43 translocation, we expressed GFP-tagged TDP-43 in M17 neuroblastoma cells in the presence of wild type or loss-of-function mutant (T240R) Parkin and treated with 5 μm selective HDAC6 inhibitor for 24 h. GFP expression was predominantly observed within DAPI-stained nuclei in live M17 cells (Fig. 5C, inset is higher magnification); however, Parkin co-expression led to significant GFP fluorescence within the cytoplasm (Fig. 5, D and E) and neuronal processes (Fig. 5D, inset shows higher magnification of GFP fluorescence). Treatment with the HDAC6 inhibitor, tubacin, did not lead to GFP fluorescence in the cytosol in the presence (Fig. 5F) or absence (Fig. 5G) of Parkin. Loss of Parkin E3 ubiquitin ligase function (T240R) did not lead to TDP-43 accumulation in the cytosol (Fig. 5H), suggesting that the E3 ubiquitin ligase function of Parkin and HDAC6 activity are required to facilitate TDP-43 accumulation within the cytosol.
It was reported that TDP-43 depletion results in down-regulation of Park2 mRNA in stem cell-derived human neurons and in motor neurons containing TDP-43 inclusions in sporadic ALS (30). To verify whether TDP-43 expression increases Parkin mRNA levels, we performed qRT-PCR in samples isolated from rat cortex, human M17 cells, and TDP43-Tg mice. Park2 mRNA levels in M17 cells expressing Parkin was significantly higher (Fig. 5, I and J, 55%, p < 0.05, n = 4) than LacZ but similar to TDP-43-injected brains (61%, p < 0.05). Parkin co-expression with TDP-43 showed significantly higher levels of park2 mRNA (Fig. 5J, 74%, p < 0.05, n = 4) compared with Parkin alone. Similarly, Park2 mRNA levels in rat brains expressing Parkin was significantly higher (Fig. 5, K and L, 41%, p < 0.05, n = 4) than LacZ animals, as well as TDP-43-injected brains (21%, p < 0.05). However, Parkin co-expression with TDP-43 showed significantly higher levels of park2 mRNA (Fig. 5J, 84%, p < 0.05, n = 4) compared with all other treatments. The variation in the data may be due to differences in cDNA transfection in cell culture or efficiency of lentiviral infection in gene transfer animal models. Therefore, we compared park2 mRNA levels between TDP43-Tg and nontransgenic control littermates. A significant increase (Fig. 5, M and N, 114%, n = 4, p < 0.05) in park2 mRNA was observed in TDP43-Tg brains injected compared with C57BL/6 controls, suggesting that Parkin is a transcriptional target for TDP-43 (30).
DISCUSSION
These studies contribute to our knowledge of the poorly understood physiological role of TDP-43 in health and disease. Accumulation of ubiquitinated TDP-43 within the nucleus was shown to cause profound pathological effects in the absence of cytosolic protein (44). Here, we show an important relationship between Parkin E3 ubiquitin ligase and TDP-43. TDP-43 may contribute to increased expression of Parkin mRNA and protein, consistent with other reports that TDP-43 regulates Parkin expression (29, 30). Parkin expression was increased in transgenic TDP-43 mice harboring the A315T mutation as well as M17 neuroblastoma cells, where Parkin protein is difficult to detect, indicating that nuclear TDP-43 may control Parkin expression. The increase in soluble Parkin level failed to protect against TDP-43 pathogenicity in TDP43-Tg mice, perhaps due to Parkin's lack of ability to reduce TDP-43 levels via autophagy or the proteasome. Our data show that Parkin forms a complex with HDAC6 to translocate TDP-43, but it is unable to induce autophagic clearance of this protein (54). HDAC6 directly binds to Parkin and mediates its transport in response to proteasome inhibition, whereas Parkin transport is reversible when proteasome activity is restored (53). Therefore, lack of autophagic clearance and proteasomal inhibition may have led to Parkin accumulation and decreased solubility. We previously demonstrated that wild type and not mutant loss-of-function Parkin increases proteasome activity (40–42), leading to degradation of ubiquitinated proteins, whereas amyloid protein stress and aging alter Parkin solubility (45). Decreased Parkin solubility is associated with alteration of its activity via increased phosphorylation by several kinase activities, including casein kinase 1, protein kinase A, protein kinase C (55), cyclin-dependent kinase 5 (56, 57), c-Abl (58, 59), and potentially PTEN-induced putative kinase 1 (PINK1) (60, 61). It is important to realize that Parkin functions as part of a number of multiprotein complexes, including PINK1 (35–37), the Skp1-Cullin-Fbox (SCF)-like complex to facilitate proteasomal degradation (38), and the chaperone Hsp70, and the U-Box protein C terminus of Hsc70-interacting protein (39). The next phase of this work will be to inject TDP-43 into Parkin−/− mice and determine the role of endogenous Parkin.
It has been reported that accumulation of amyloidogenic proteins can alter Parkin solubility, perhaps leading to changes in its activity (45, 58, 62). It is unlikely that the appearance of Parkin protein is due to proteasome inhibition, leading to lack of degradation as is the case with SIAH2, because the significant reversal of proteasome activity in the presence of exogenous Parkin led to the disappearance of SIAH2 but not Parkin. TDP-43 expression leads to proteasome inhibition and accumulation of ubiquitinated proteins, including the rapidly degrading SIAH2 due to auto-ubiquitination and proteasomal clearance (46–51). Conversely, activation of Parkin E3 ligase function leads to ubiquitination of TDP-43 via Lys-48- and Lys-63-linked ubiquitination, indicating Parkin involvement in TDP-43 metabolism, including its cytosolic accumulation. Parkin led to the cytosolic accumulation of ubiquitinated TDP-43, without any evidence of protein degradation or clearance, suggesting that Parkin-mediated TDP-43 ubiquitination perhaps facilitates nuclear translocation of TDP-43 to the cytosol. Parkin mediated Lys-48 and Lys-63 as well as wild type ubiquitin (which can include any of the five other ubiquitin linkages Lys-6, Lys-11, Lys-27, Lys-29, and Lys-33), indicating several possibilities about the functional role of these ubiquitin linkages (63). Mono-ubiquitination was reported to regulate DNA repair and receptor endocytosis, although Lys-48-linked ubiquitin chains are tags for proteasomal degradation (64). Lys-63-linked ubiquitination was also reported to facilitate DNA repair and target proteins for degradation via interaction between ubiquitinated proteins and p62 (65). However, in a study using autophagy-deficient mice, all ubiquitin-ubiquitin linkages accumulated, in contradiction with the hypothesis that any particular ubiquitin linkage serves as a specific autophagy signal (66). Parkin-mediated TDP-43 ubiquitination led to p62 and cytosolic TDP-43 accumulation. It is therefore possible that the cell contains factors that prevent proteins linked to Lys-63-ubiquitin from degradation (67). For example, Parkin-mediated Lys-63-linked polyubiquitination was reported to link misfolded proteins to the dynein motor complex via the adaptor protein HDAC6, promoting sequestration of misfolded proteins into aggresomes and subsequent clearance by autophagy (54). Other groups showed that HDAC6 directly binds to Parkin and mediates its transport (53). Others reported that Lys-48- and Lys-63-linked polyubiquitination, as well as mono-ubiquitination, contributes to inclusion formation; however, Lys-63-linked polyubiquitin enhances inclusion formation and selectively facilitates clearance via autophagy (68). Our data showed that Parkin forms a multiprotein complex with HDAC6 and TDP-43 without induction of autophagy or proteasomal degradation, suggesting that other factors may determine which type of polyubiquitin chains are specific labels for protein degradation. The multiprotein complex consisting of TDP-43, HDAC6, and Parkin may lead to translocation of nuclear TDP-43 into the cytosol via binding with dynein motors.
Increased detection of TDP-43 in the spinal cord following protein expression in the rat motor cortex is intriguing and may have relevance to human disease, mainly ALS, where affected cortical motor neurons are associated with degenerating spinal tracts (69–72). It is unclear whether TDP-43 protein or mRNA is axonally transported in descending motor neuron fibers or whether increased TDP-43 protein levels in the cortex signals expression in the spinal cord causing axonal degeneration. Detection of TDP-43 in spinal cord and DCST alludes to the susceptibility of this subset of neurons within the descending spinal motor tracts to TDP-43 pathology. More importantly, these studies determine the early effects of TDP-43 accumulation in vivo. It is necessary to mention that no differences were observed in strength or RotaRod behavioral tests 2 weeks post-injection in gene transfer animals, perhaps due to unilateral gene expression (TDP-43 was injected into right hemisphere), but transgenic mice were symptomatic and died between 21 and 30 days. Cytosolic inclusions composed of misfolded accumulating proteins, including ubiquitinated TDP-43, are found in several neurodegenerative diseases (29, 73, 74), whereas TDP-43 knockdown impairs neuronal growth (75) perhaps due to involvement of TDP-43 in RNA/DNA processing of a large number of genes (29). Recent reports show that neurons bearing cytosolic TDP-43 aggregates have decreased Parkin levels (30), suggesting loss of nuclear TDP-43 function to regulate Parkin levels. Conversely, the A315T transgenic mice show an increased Parkin level with decreased solubility, suggesting altered function of de novo synthesized Parkin to facilitate cytosolic accumulation of ubiquitinated TDP-43. Parkin inactivation is described in the nigrostriatal region of patients with sporadic PD (58), and protein aggregates were previously shown to alter Parkin solubility (76). We previously showed that Parkin co-localizes with intraneuronal Aβ(1–42) and has decreased solubility in AD (45). Therefore, TDP-43 stress may alter Parkin solubility and perhaps function. The loss of Parkin function is supported in early onset juvenile PD, where no cytosolic Lewy body inclusions are detected (77), suggesting that functional Parkin mediates cytosolic sequestration of misfolded proteins. Parkin may act alone to increase TDP-43 ubiquitination or collaborate with other ubiquitin ligases to enhance protein ubiquitination. Nuclear TDP-43 regulates a large amount of gene expression and is implicated in many steps of RNA expression and transport, including Parkin (29, 78); therefore, translocation from the nucleus to the cytosol, perhaps via ubiquitination when Parkin is expressed, may be a sequestration strategy.
In conclusion, whether TDP-43 pathology involves loss of nuclear or gain of cytosolic function, it is important to have a functional proteasome to mediate protein clearance and prevent inclusion formation. The type of Lys-linked polyubiquitin chain seems to depend on many factors, perhaps autophagy and the proteasome, to determine the fate of ubiquitinated TDP-43. The activation of Parkin E3 ubiquitin ligase activity and TDP-43 ubiquitination result in cytosolic accumulation, despite the partial reversal of proteasome activity, leading to complex formation with HDAC6 and sequestration of TDP-43. Cytosolic accumulation of TDP-43 may be a coping mechanism to alleviate or temporarily delay the effects of TDP-43 on aberrant mRNA transcription.
This work was supported, in whole or in part, by National Institutes of Health Grant AG30378. This work was also supported by Georgetown University Funding (to C. E.-H. M.).
- ALS
- amyotrophic lateral sclerosis
- ANOVA
- analysis of variance
- DCST
- dorso-cortical spinal tract
- FTLD
- fronto-temporal lobar dementia
- PD
- Parkinson disease
- m.o.i.
- multiplicity of infection
- Lv
- lentiviral
- AMC
- 7-amino-4-methylcoumarin
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- qRT
- quantitative RT
- SIAH
- seven in absentia homolog
- hTDP-43
- human TDP-43
- WB
- Western blot.
REFERENCES
- 1. Ou S. H., Wu F., Harrich D., García-Martínez L. F., Gaynor R. B. (1995) Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J. Virol. 69, 3584–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang H. Y., Wang I. F., Bose J., Shen C. K. (2004) Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics 83, 130–139 [DOI] [PubMed] [Google Scholar]
- 3. Hasegawa M., Arai T., Nonaka T., Kametani F., Yoshida M., Hashizume Y., Beach T. G., Buratti E., Baralle F., Morita M., Nakano I., Oda T., Tsuchiya K., Akiyama H. (2008) Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann. Neurol. 64, 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mackenzie I. R., Bigio E. H., Ince P. G., Geser F., Neumann M., Cairns N. J., Kwong L. K., Forman M. S., Ravits J., Stewart H., Eisen A., McClusky L., Kretzschmar H. A., Monoranu C. M., Highley J. R., Kirby J., Siddique T., Shaw P. J., Lee V. M., Trojanowski J. Q. (2007) Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann. Neurol. 61, 427–434 [DOI] [PubMed] [Google Scholar]
- 5. Neumann M., Kwong L. K., Sampathu D. M., Trojanowski J. Q., Lee V. M. (2007) TDP-43 proteinopathy in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Protein misfolding diseases without amyloidosis. Arch. Neurol. 64, 1388–1394 [DOI] [PubMed] [Google Scholar]
- 6. Neumann M., Mackenzie I. R., Cairns N. J., Boyer P. J., Markesbery W. R., Smith C. D., Taylor J. P., Kretzschmar H. A., Kimonis V. E., Forman M. S. (2007) TDP-43 in the ubiquitin pathology of frontotemporal dementia with VCP gene mutations. J. Neuropathol. Exp. Neurol. 66, 152–157 [DOI] [PubMed] [Google Scholar]
- 7. Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 8. Zhang Y. J., Xu Y. F., Cook C., Gendron T. F., Roettges P., Link C. D., Lin W. L., Tong J., Castanedes-Casey M., Ash P., Gass J., Rangachari V., Buratti E., Baralle F., Golde T. E., Dickson D. W., Petrucelli L. (2009) Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoshiyama Y., Higuchi M., Zhang B., Huang S. M., Iwata N., Saido T. C., Maeda J., Suhara T., Trojanowski J. Q., Lee V. M. (2007) Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 53, 337–351 [DOI] [PubMed] [Google Scholar]
- 10. Leigh P. N., Whitwell H., Garofalo O., Buller J., Swash M., Martin J. E., Gallo J. M., Weller R. O., Anderton B. H. (1991) Ubiquitin-immunoreactive intraneuronal inclusions in amyotrophic lateral sclerosis. Morphology, distribution, and specificity. Brain 114, 775–788 [DOI] [PubMed] [Google Scholar]
- 11. Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., Oda T. (2006) TDP-43 is a component of ubiquitin-positive τ-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611 [DOI] [PubMed] [Google Scholar]
- 12. Mackenzie I. R., Rademakers R., Neumann M. (2010) TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 9, 995–1007 [DOI] [PubMed] [Google Scholar]
- 13. Gitcho M. A., Baloh R. H., Chakraverty S., Mayo K., Norton J. B., Levitch D., Hatanpaa K. J., White C. L., 3rd, Bigio E. H., Caselli R., Baker M., Al-Lozi M. T., Morris J. C., Pestronk A., Rademakers R., Goate A. M., Cairns N. J. (2008) TDP-43 A315T mutation in familial motor neuron disease. Ann. Neurol. 63, 535–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yokoseki A., Shiga A., Tan C. F., Tagawa A., Kaneko H., Koyama A., Eguchi H., Tsujino A., Ikeuchi T., Kakita A., Okamoto K., Nishizawa M., Takahashi H., Onodera O. (2008) TDP-43 mutation in familial amyotrophic lateral sclerosis. Ann. Neurol. 63, 538–542 [DOI] [PubMed] [Google Scholar]
- 15. Kabashi E., Valdmanis P. N., Dion P., Spiegelman D., McConkey B. J., Vande Velde C., Bouchard J. P., Lacomblez L., Pochigaeva K., Salachas F., Pradat P. F., Camu W., Meininger V., Dupre N., Rouleau G. A. (2008) TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 40, 572–574 [DOI] [PubMed] [Google Scholar]
- 16. Van Deerlin V. M., Leverenz J. B., Bekris L. M., Bird T. D., Yuan W., Elman L. B., Clay D., Wood E. M., Chen-Plotkin A. S., Martinez-Lage M., Steinbart E., McCluskey L., Grossman M., Neumann M., Wu I. L., Yang W. S., Kalb R., Galasko D. R., Montine T. J., Trojanowski J. Q., Lee V. M., Schellenberg G. D., Yu C. E. (2008) TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology. A genetic and histopathological analysis. Lancet Neurol. 7, 409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sreedharan J., Blair I. P., Tripathi V. B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J. C., Williams K. L., Buratti E., Baralle F., de Belleroche J., Mitchell J. D., Leigh P. N., Al-Chalabi A., Miller C. C., Nicholson G., Shaw C. E. (2008) TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geser F., Robinson J. L., Malunda J. A., Xie S. X., Clark C. M., Kwong L. K., Moberg P. J., Moore E. M., Van Deerlin V. M., Lee V. M., Arnold S. E., Trojanowski J. Q. (2010) Pathological 43-kDa transactivation response DNA-binding protein in older adults with and without severe mental illness. Arch. Neurol. 67, 1238–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schwab C., Arai T., Hasegawa M., Yu S., McGeer P. L. (2008) Colocalization of transactivation-responsive DNA-binding protein 43 and huntingtin in inclusions of Huntington disease. J. Neuropathol. Exp. Neurol. 67, 1159–1165 [DOI] [PubMed] [Google Scholar]
- 20. Hasegawa M., Arai T., Akiyama H., Nonaka T., Mori H., Hashimoto T., Yamazaki M., Oyanagi K. (2007) TDP-43 is deposited in the Guam parkinsonism-dementia complex brains. Brain 130, 1386–1394 [DOI] [PubMed] [Google Scholar]
- 21. Geser F., Winton M. J., Kwong L. K., Xu Y., Xie S. X., Igaz L. M., Garruto R. M., Perl D. P., Galasko D., Lee V. M., Trojanowski J. Q. (2008) Pathological TDP-43 in parkinsonism-dementia complex and amyotrophic lateral sclerosis of Guam. Acta Neuropathol. 115, 133–145 [DOI] [PubMed] [Google Scholar]
- 22. Higashi S., Iseki E., Yamamoto R., Minegishi M., Hino H., Fujisawa K., Togo T., Katsuse O., Uchikado H., Furukawa Y., Kosaka K., Arai H. (2007) Concurrence of TDP-43, τ, and α-synuclein pathology in brains of Alzheimer's disease and dementia with Lewy bodies. Brain Res. 1184, 284–294 [DOI] [PubMed] [Google Scholar]
- 23. Amador-Ortiz C., Lin W. L., Ahmed Z., Personett D., Davies P., Duara R., Graff-Radford N. R., Hutton M. L., Dickson D. W. (2007) TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer's disease. Ann. Neurol. 61, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. King A., Sweeney F., Bodi I., Troakes C., Maekawa S., Al-Sarraj S. (2010) Abnormal TDP-43 expression is identified in the neocortex in cases of dementia pugilistica, but is mainly confined to the limbic system when identified in high and moderate stages of Alzheimer's disease. Neuropathology 30, 408–419 [DOI] [PubMed] [Google Scholar]
- 25. Herman A. M., Khandelwal P. J., Stanczyk B. B., Rebeck G. W., Moussa C. E. (2011) β-Amyloid triggers ALS-associated TDP-43 pathology in AD models. Brain Res. 1386, 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakashima-Yasuda H., Uryu K., Robinson J., Xie S. X., Hurtig H., Duda J. E., Arnold S. E., Siderowf A., Grossman M., Leverenz J. B., Woltjer R., Lopez O. L., Hamilton R., Tsuang D. W., Galasko D., Masliah E., Kaye J., Clark C. M., Montine T. J., Lee V. M., Trojanowski J. Q. (2007) Co-morbidity of TDP-43 proteinopathy in Lewy body related diseases. Acta Neuropathol. 114, 221–229 [DOI] [PubMed] [Google Scholar]
- 27. Uryu K., Nakashima-Yasuda H., Forman M. S., Kwong L. K., Clark C. M., Grossman M., Miller B. L., Kretzschmar H. A., Lee V. M., Trojanowski J. Q., Neumann M. (2008) Concomitant TAR-DNA-binding protein 43 pathology is present in Alzheimer disease and corticobasal degeneration but not in other tauopathies. J. Neuropathol. Exp. Neurol. 67, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yokota O., Davidson Y., Bigio E. H., Ishizu H., Terada S., Arai T., Hasegawa M., Akiyama H., Sikkink S., Pickering-Brown S., Mann D. M. (2010) Phosphorylated TDP-43 pathology and hippocampal sclerosis in progressive supranuclear palsy. Acta Neuropathol. 120, 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Polymenidou M., Lagier-Tourenne C., Hutt K. R., Huelga S. C., Moran J., Liang T. Y., Ling S. C., Sun E., Wancewicz E., Mazur C., Kordasiewicz H., Sedaghat Y., Donohue J. P., Shiue L., Bennett C. F., Yeo G. W., Cleveland D. W. (2011) Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 14, 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lagier-Tourenne C., Polymenidou M., Hutt K. R., Vu A. Q., Baughn M., Huelga S. C., Clutario K. M., Ling S. C., Liang T. Y., Mazur C., Wancewicz E., Kim A. S., Watt A., Freier S., Hicks G. G., Donohue J. P., Shiue L., Bennett C. F., Ravits J., Cleveland D. W., Yeo G. W. (2012) Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 15, 1488–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shimura H., Hattori N., Kubo Si, Mizuno Y., Asakawa S., Minoshima S., Shimizu N., Iwai K., Chiba T., Tanaka K., Suzuki T. (2000) Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25, 302–305 [DOI] [PubMed] [Google Scholar]
- 32. Cookson M. R., Bandmann O. (2010) Parkinson's disease. Insights from pathways. Hum. Mol. Genet. 19, R21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 [DOI] [PubMed] [Google Scholar]
- 34. Lücking C. B., Dürr A., Bonifati V., Vaughan J., De Michele G., Gasser T., Harhangi B. S., Meco G., Denèfle P., Wood N. W., Agid Y., Brice A., French Parkinson's Disease Genetics Study Group, and European Consortium on Genetic Susceptibility in Parkinson's Disease (2000) Association between early-onset Parkinson's disease and mutations in the parkin gene. N. Engl. J. Med. 342, 1560–1567 [DOI] [PubMed] [Google Scholar]
- 35. Clark I. E., Dodson M. W., Jiang C., Cao J. H., Huh J. R., Seol J. H., Yoo S. J., Hay B. A., Guo M. (2006) Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166 [DOI] [PubMed] [Google Scholar]
- 36. Greene J. C., Whitworth A. J., Kuo I., Andrews L. A., Feany M. B., Pallanck L. J. (2003) Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. U.S.A. 100, 4078–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Park J., Lee S. B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J. M., Chung J. (2006) Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 [DOI] [PubMed] [Google Scholar]
- 38. Staropoli J. F., McDermott C., Martinat C., Schulman B., Demireva E., Abeliovich A. (2003) Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron 37, 735–749 [DOI] [PubMed] [Google Scholar]
- 39. Imai Y., Soda M., Hatakeyama S., Akagi T., Hashikawa T., Nakayama K. I., Takahashi R. (2002) CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol. Cell 10, 55–67 [DOI] [PubMed] [Google Scholar]
- 40. Burns M. P., Zhang L., Rebeck G. W., Querfurth H. W., Moussa C. E. (2009) Parkin promotes intracellular Aβ1–42 clearance. Hum. Mol. Genet. 18, 3206–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Khandelwal P. J., Dumanis S. B., Feng L. R., Maguire-Zeiss K., Rebeck G., Lashuel H. A., Moussa C. E. (2010) Parkinson-related parkin reduces α-synuclein phosphorylation in a gene transfer model. Mol. Neurodegener 5, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rosen K. M., Moussa C. E., Lee H. K., Kumar P., Kitada T., Qin G., Fu Q., Querfurth H. W. (2010) Parkin reverses intracellular β-amyloid accumulation and its negative effects on proteasome function. J. Neurosci. Res. 88, 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Herman A. M., Khandelwal P. J., Rebeck G. W., Moussa C. E. (2012) Wild type TDP-43 induces neuro-inflammation and alters APP metabolism in lentiviral gene transfer models. Exp. Neurol. 235, 297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wegorzewska I., Bell S., Cairns N. J., Miller T. M., Baloh R. H. (2009) TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc. Natl. Acad. Sci. U.S.A. 106, 18809–18814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lonskaya I., Shekoyan A. R., Hebron M. L., Desforges N., Algarzae N. K., Moussa C. E. (2012) Diminished Parkin solubility and co-localization with intraneuronal amyloid-β are associated with autophagic defects in Alzheimer's Disease. J. Alzheimers Dis. 33, 231–247 [DOI] [PubMed] [Google Scholar]
- 46. Hu G., Zhang S., Vidal M., Baer J. L., Xu T., Fearon E. R. (1997) Mammalian homologs of seven in absentia regulate DCC via the ubiquitin-proteasome pathway. Genes Dev. 11, 2701–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dickins R. A., Frew I. J., House C. M., O'Bryan M. K., Holloway A. J., Haviv I., Traficante N., de Kretser D. M., Bowtell D. D. (2002) The ubiquitin ligase component Siah1a is required for completion of meiosis I in male mice. Mol. Cell. Biol. 22, 2294–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hu G., Fearon E. R. (1999) Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol. Cell. Biol. 19, 724–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qi J., Pellecchia M., Ronai Z. A. (2010) The Siah2-HIF-FoxA2 axis in prostate cancer–new markers and therapeutic opportunities. Oncotarget 1, 379–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Qi J., Nakayama K., Cardiff R. D., Borowsky A. D., Kaul K., Williams R., Krajewski S., Mercola D., Carpenter P. M., Bowtell D., Ronai Z. A. (2010) Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell 18, 23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nakayama K., Frew I. J., Hagensen M., Skals M., Habelhah H., Bhoumik A., Kadoya T., Erdjument-Bromage H., Tempst P., Frappell P. B., Bowtell D. D., Ronai Z. (2004) Siah2 regulates stability of prolyl-hydroxylases, controls HIF1α abundance, and modulates physiological responses to hypoxia. Cell 117, 941–952 [DOI] [PubMed] [Google Scholar]
- 52.Deleted in proof
- 53. Jiang Q., Ren Y., Feng J. (2008) Direct binding with histone deacetylase 6 mediates the reversible recruitment of parkin to the centrosome. J. Neurosci. 28, 12993–13002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Olzmann J. A., Chin L. S. (2008) Parkin-mediated K63-linked polyubiquitination. A signal for targeting misfolded proteins to the aggresome-autophagy pathway. Autophagy 4, 85–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yamamoto A., Friedlein A., Imai Y., Takahashi R., Kahle P. J., Haass C. (2005) Parkin phosphorylation and modulation of its E3 ubiquitin ligase activity. J. Biol. Chem. 280, 3390–3399 [DOI] [PubMed] [Google Scholar]
- 56. Avraham E., Rott R., Liani E., Szargel R., Engelender S. (2007) Phosphorylation of Parkin by the cyclin-dependent kinase 5 at the linker region modulates its ubiquitin-ligase activity and aggregation. J. Biol. Chem. 282, 12842–12850 [DOI] [PubMed] [Google Scholar]
- 57. Rubio de la Torre E., Luzón-Toro B., Forte-Lago I., Minguez-Castellanos A., Ferrer I., Hilfiker S. (2009) Combined kinase inhibition modulates parkin inactivation. Hum. Mol. Genet. 18, 809–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ko H. S., Lee Y., Shin J. H., Karuppagounder S. S., Gadad B. S., Koleske A. J., Pletnikova O., Troncoso J. C., Dawson V. L., Dawson T. M. (2010) Phosphorylation by the c-Abl protein tyrosine kinase inhibits parkin's ubiquitination and protective function. Proc. Natl. Acad. Sci. U.S.A. 107, 16691–16696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Imam S. Z., Zhou Q., Yamamoto A., Valente A. J., Ali S. F., Bains M., Roberts J. L., Kahle P. J., Clark R. A., Li S. (2011) Novel regulation of parkin function through c-Abl-mediated tyrosine phosphorylation. Implications for Parkinson's disease. J. Neurosci. 31, 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kim Y., Park J., Kim S., Song S., Kwon S. K., Lee S. H., Kitada T., Kim J. M., Chung J. (2008) PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem. Biophys. Res. Commun. 377, 975–980 [DOI] [PubMed] [Google Scholar]
- 61. Sha D., Chin L. S., Li L. (2010) Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-κB signaling. Hum. Mol. Genet. 19, 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rodríguez-Navarro J. A., Gómez A., Rodal I., Perucho J., Martinez A., Furió V., Ampuero I., Casarejos M. J., Solano R. M., de Yébenes J. G., Mena M. A. (2008) Parkin deletion causes cerebral and systemic amyloidosis in human mutated τ over-expressing mice. Hum. Mol. Genet. 17, 3128–3143 [DOI] [PubMed] [Google Scholar]
- 63. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 64. Ikeda F., Dikic I. (2008) Atypical ubiquitin chains. New molecular signals. Protein modifications. Beyond the usual suspects' review series. EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Seibenhener M. L., Geetha T., Wooten M. W. (2007) Sequestosome 1/p62–more than just a scaffold. FEBS Lett. 581, 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Riley B. E., Kaiser S. E., Shaler T. A., Ng A. C., Hara T., Hipp M. S., Lage K., Xavier R. J., Ryu K. Y., Taguchi K., Yamamoto M., Tanaka K., Mizushima N., Komatsu M., Kopito R. R. (2010) Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J. Cell Biol. 191, 537–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim H. T., Kim K. P., Lledias F., Kisselev A. F., Scaglione K. M., Skowyra D., Gygi S. P., Goldberg A. L. (2007) Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282, 17375–17386 [DOI] [PubMed] [Google Scholar]
- 68. Tan J. M., Wong E. S., Kirkpatrick D. S., Pletnikova O., Ko H. S., Tay S. P., Ho M. W., Troncoso J., Gygi S. P., Lee M. K., Dawson V. L., Dawson T. M., Lim K. L. (2008) Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum. Mol. Genet. 17, 431–439 [DOI] [PubMed] [Google Scholar]
- 69. Dickson D. W., Josephs K. A., Amador-Ortiz C. (2007) TDP-43 in differential diagnosis of motor neuron disorders. Acta Neuropathol. 114, 71–79 [DOI] [PubMed] [Google Scholar]
- 70. Geser F., Brandmeir N. J., Kwong L. K., Martinez-Lage M., Elman L., McCluskey L., Xie S. X., Lee V. M., Trojanowski J. Q. (2008) Evidence of multisystem disorder in whole-brain map of pathological TDP-43 in amyotrophic lateral sclerosis. Arch. Neurol. 65, 636–641 [DOI] [PubMed] [Google Scholar]
- 71. McCluskey L. F., Elman L. B., Martinez-Lage M., Van Deerlin V., Yuan W., Clay D., Siderowf A., Trojanowski J. Q. (2009) Amyotrophic lateral sclerosis-plus syndrome with TAR DNA-binding protein-43 pathology. Arch. Neurol. 66, 121–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mitchell J. D., Borasio G. D. (2007) Amyotrophic lateral sclerosis. Lancet 369, 2031–2041 [DOI] [PubMed] [Google Scholar]
- 73. Polymenidou M., Cleveland D. W. (2011) The seeds of neurodegeneration: prion-like spreading in ALS. Cell 147, 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lagier-Tourenne C., Polymenidou M., Cleveland D. W. (2010) TDP-43 and FUS/TLS. Emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 19, R46–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fiesel F. C., Schurr C., Weber S. S., Kahle P. J. (2011) TDP-43 knockdown impairs neurite outgrowth dependent on its target histone deacetylase 6. Mol. Neurodegener 6, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kawahara K., Hashimoto M., Bar-On P., Ho G. J., Crews L., Mizuno H., Rockenstein E., Imam S. Z., Masliah E. (2008) α-Synuclein aggregates interfere with Parkin solubility and distribution. Role in the pathogenesis of Parkinson disease. J. Biol. Chem. 283, 6979–6987 [DOI] [PubMed] [Google Scholar]
- 77. Mizuno Y., Hattori N., Mori H., Suzuki T., Tanaka K. (2001) Parkin and Parkinson's disease. Curr. Opin. Neurol. 14, 477–482 [DOI] [PubMed] [Google Scholar]
- 78. Tollervey J. R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., König J., Hortobágyi T., Nishimura A. L., Zupunski V., Patani R., Chandran S., Rot G., Zupan B., Shaw C. E., Ule J. (2011) Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 14, 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]