Abstract
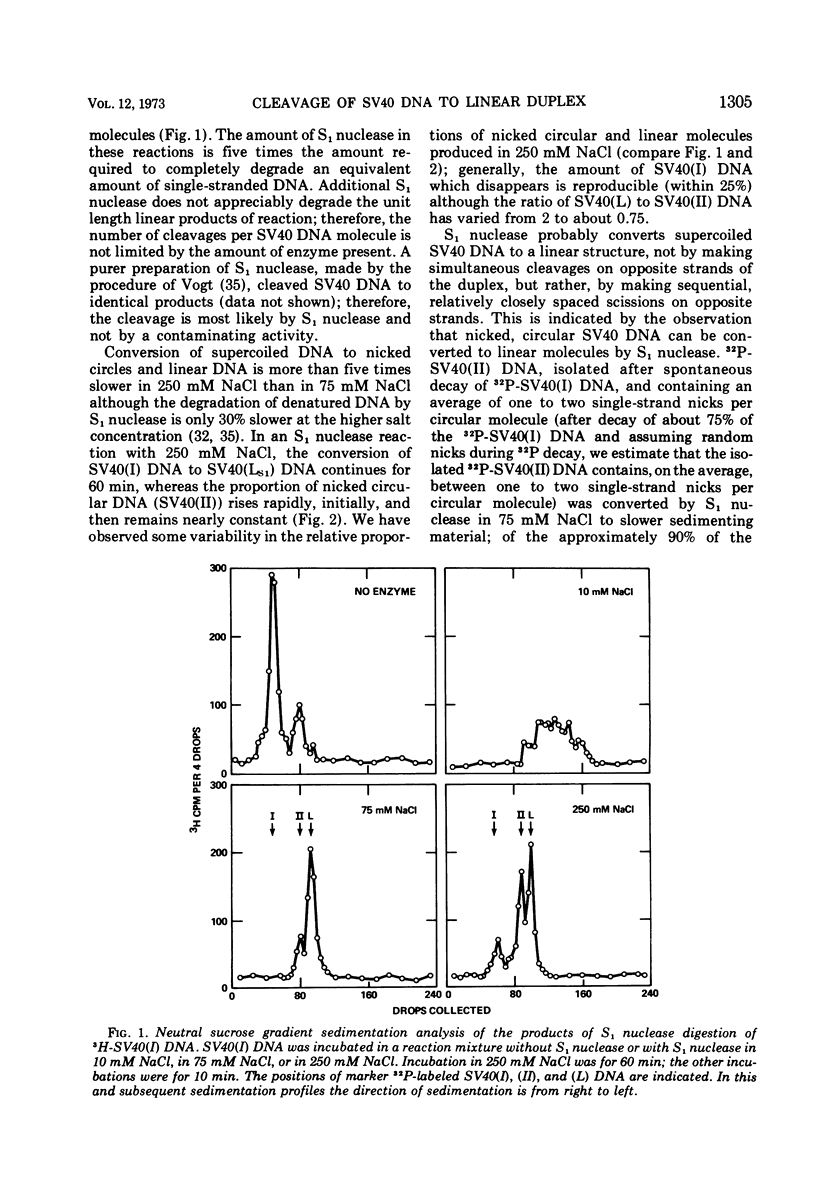
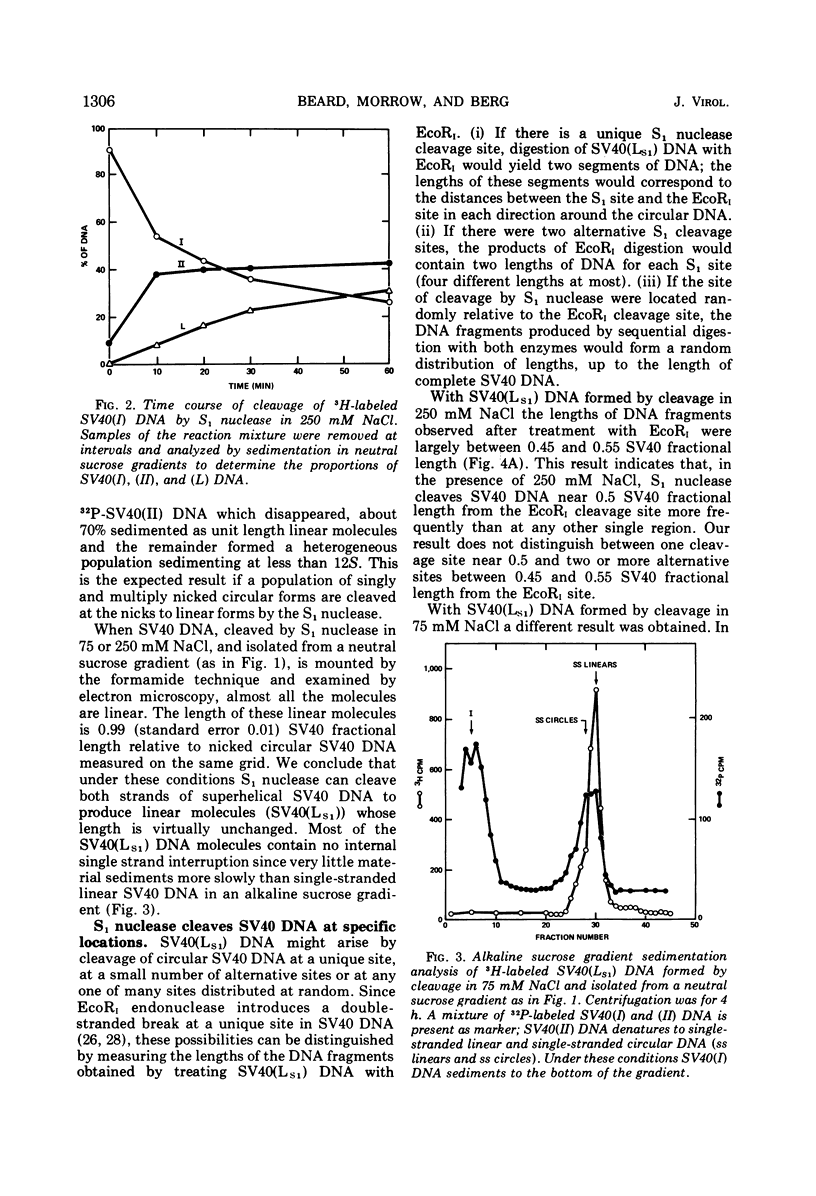
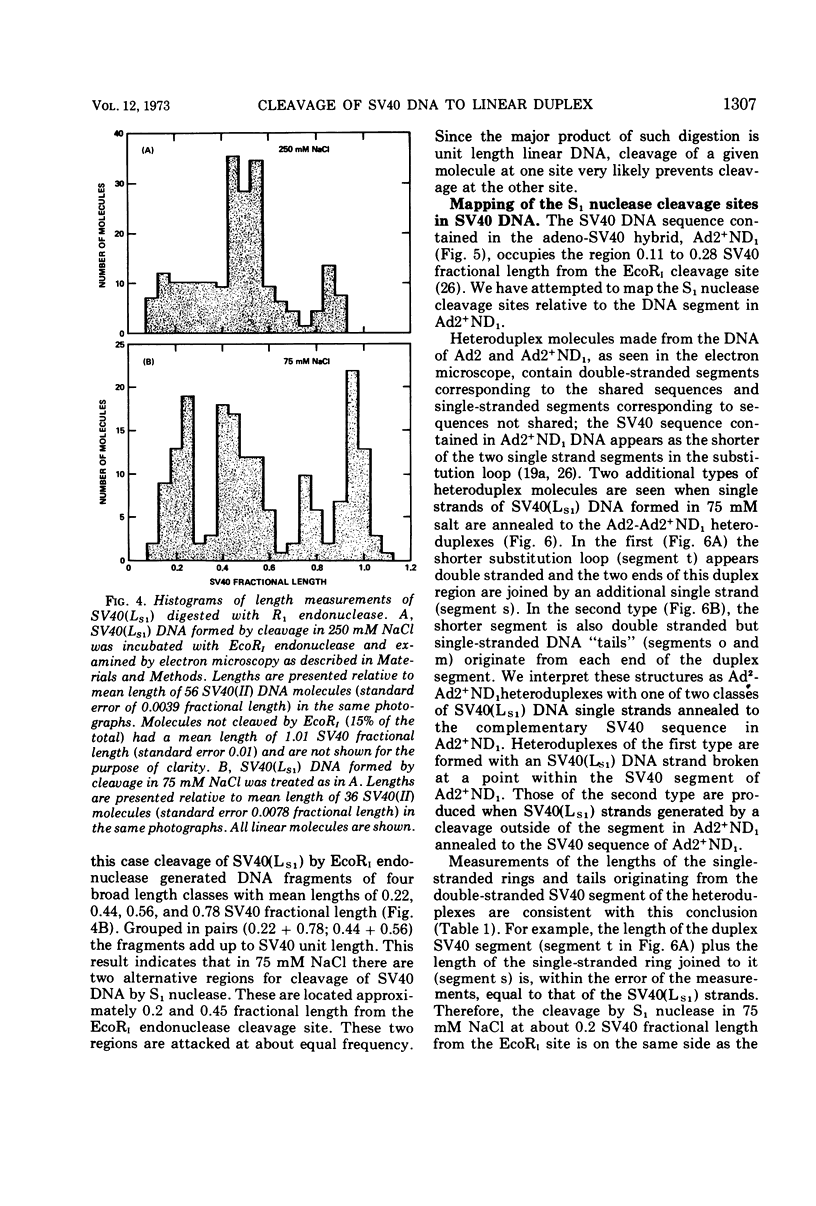
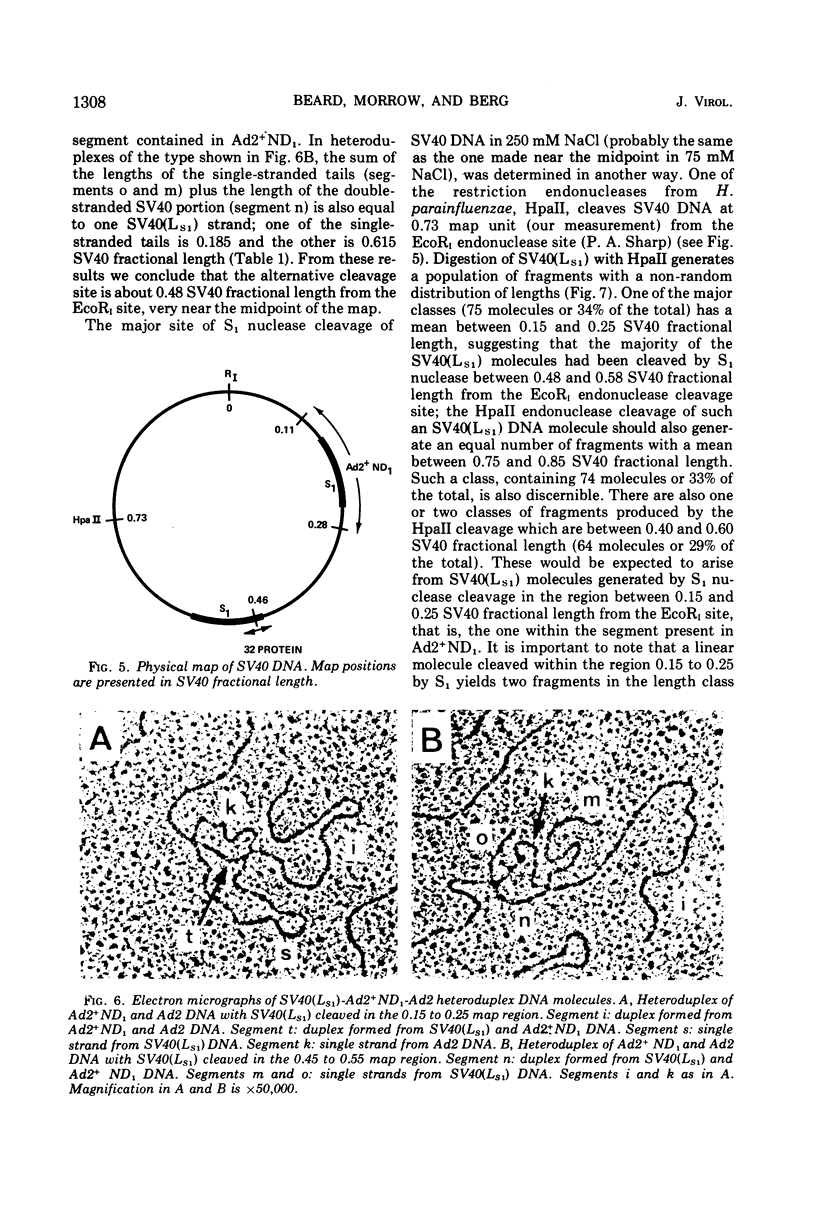
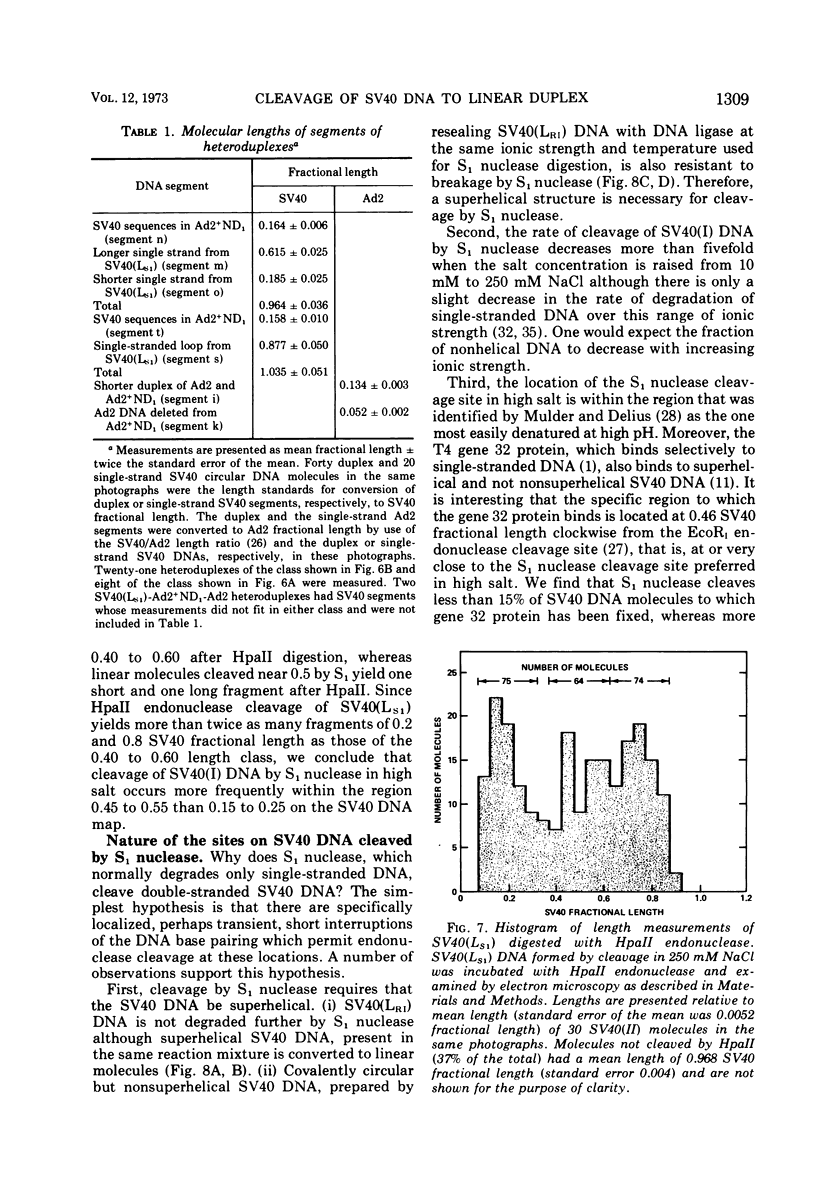
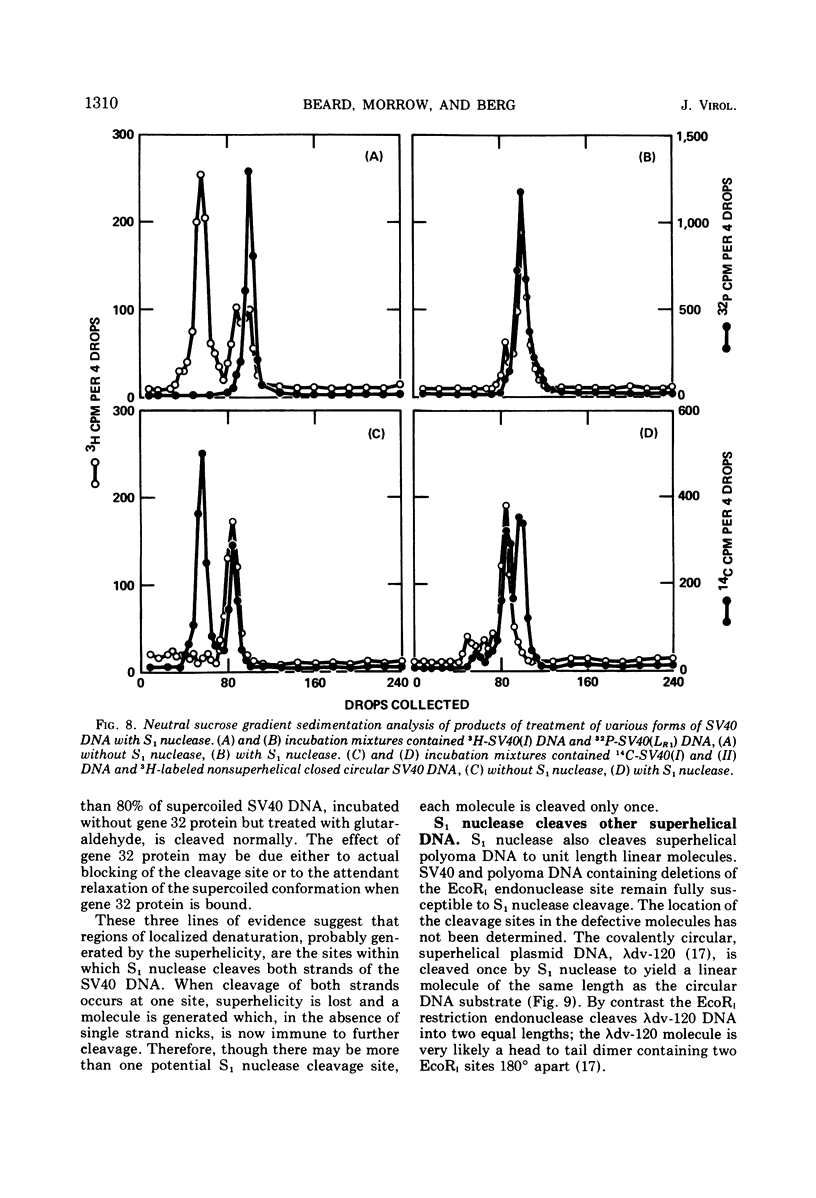
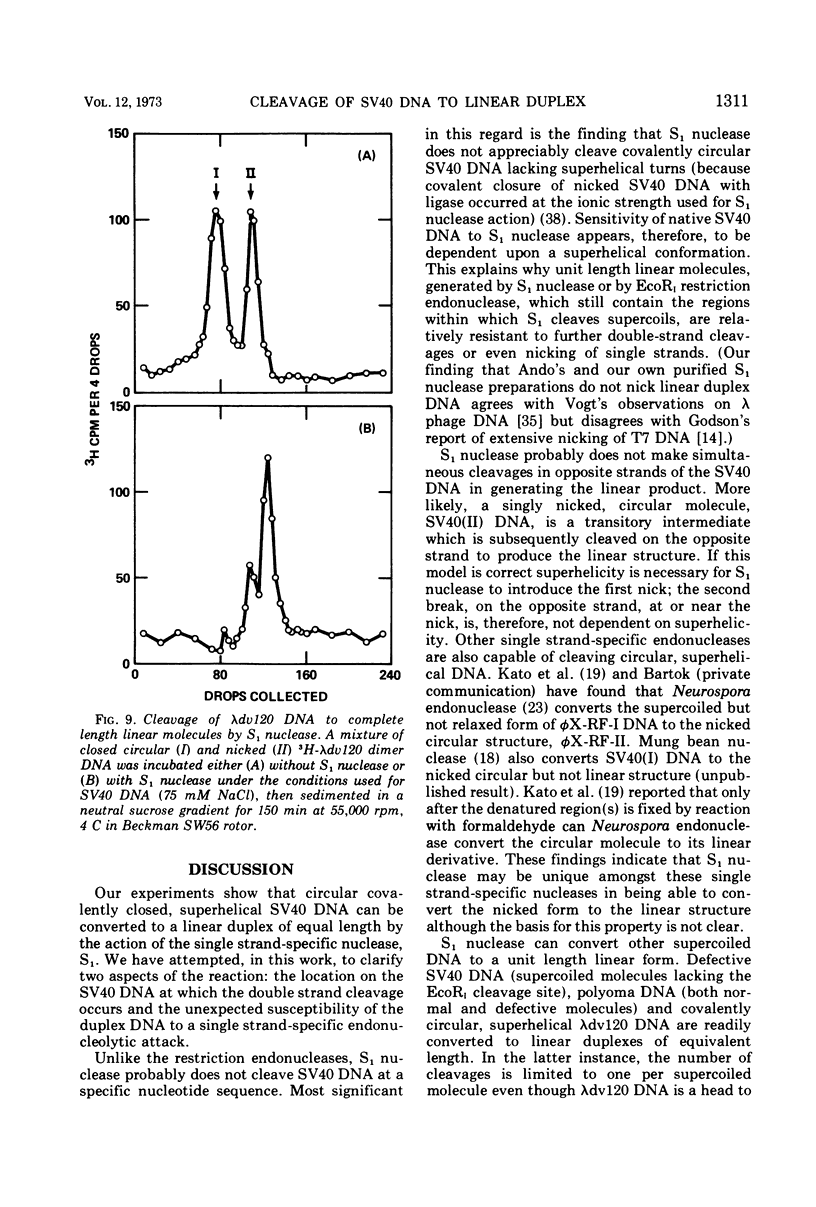
S1 nuclease, the single-strand specific nuclease from Aspergillus oryzae can cleave both strands of circular covalently closed, superhelical simian virus 40 (SV40) DNA to generate unit length linear duplex molecules with intact single strands. But circular, covalently closed, nonsuperhelical DNA, as well as linear duplex molecules, are relatively resistant to attack by the enzyme. These findings indicate that unpaired or weakly hydrogen-bonded regions, sensitive to the single strand-specific nuclease, occur or can be induced in superhelical DNA. Nicked, circular SV40 DNA can be cleaved on the opposite strand at or near the nick to yield linear molecules. S1 nuclease may be a useful reagent for cleaving DNAs at regions containing single-strand nicks. Unlike the restriction endonucleases, S1 nuclease probably does not cleave SV40 DNA at a specific nucleotide sequence. Rather, the sites of cleavage occur within regions that are readily denaturable in a topologically constrained superhelical molecule. At moderate salt concentrations (75 mM) SV40 DNA is cleaved once, most often within either one of the two following regions: the segments defined as 0.15 to 0.25 and 0.45 to 0.55 SV40 fractional length, clockwise, from the EcoRI restriction endonuclease cleavage site (defined as the zero position on the SV40 DNA map). In higher salt (250 mM) cleavage occurs preferentially within the 0.45 to 0.55 segment of the map.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M., Frey L. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970 Sep 26;227(5265):1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- Ando T. A nuclease specific for heat-denatured DNA in isolated from a product of Aspergillus oryzae. Biochim Biophys Acta. 1966 Jan 18;114(1):158–168. doi: 10.1016/0005-2787(66)90263-2. [DOI] [PubMed] [Google Scholar]
- Bauer W., Vinograd J. Interaction of closed circular DNA with intercalative dyes. II. The free energy of superhelix formation in SV40 DNA. J Mol Biol. 1970 Feb 14;47(3):419–435. doi: 10.1016/0022-2836(70)90312-8. [DOI] [PubMed] [Google Scholar]
- Bauer W., Vinograd J. The interaction of closed circular DNA with intercalative dyes. I. The superhelix density of SV40 DNA in the presence and absence of dye. J Mol Biol. 1968 Apr 14;33(1):141–171. doi: 10.1016/0022-2836(68)90286-6. [DOI] [PubMed] [Google Scholar]
- CRAWFORD L. V., BLACK P. H. THE NUCLEIC ACID OF SIMIAN VIRUS 40. Virology. 1964 Nov;24:388–392. doi: 10.1016/0042-6822(64)90176-x. [DOI] [PubMed] [Google Scholar]
- Danna K. J., Sack G. H., Jr, Nathans D. Studies of simian virus 40 DNA. VII. A cleavage map of the SV40 genome. J Mol Biol. 1973 Aug 5;78(2):363–376. doi: 10.1016/0022-2836(73)90122-8. [DOI] [PubMed] [Google Scholar]
- Danna K., Nathans D. Specific cleavage of simian virus 40 DNA by restriction endonuclease of Hemophilus influenzae. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2913–2917. doi: 10.1073/pnas.68.12.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean W. W., Lebowitz J. Partial alteration of secondary structure in native superhelical DNA. Nat New Biol. 1971 May 5;231(18):5–8. [PubMed] [Google Scholar]
- Delius H., Mantell N. J., Alberts B. Characterization by electron microscopy of the complex formed between T4 bacteriophage gene 32-protein and DNA. J Mol Biol. 1972 Jun 28;67(3):341–350. doi: 10.1016/0022-2836(72)90454-8. [DOI] [PubMed] [Google Scholar]
- Doerfler W. Nonproductive infection of baby hamster kidney cells (BHK21) with adenovirus type 12. Virology. 1969 Aug;38(4):587–606. doi: 10.1016/0042-6822(69)90179-2. [DOI] [PubMed] [Google Scholar]
- Fareed G. C., Garon G. F., Salzman N. P. Origin and direction of simian virus 40 deoxyribonucleic acid replication. J Virol. 1972 Sep;10(3):484–491. doi: 10.1128/jvi.10.3.484-491.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godson G. N. Action of the single-stranded DNA specific nuclease S1 on double-stranded DNA. Biochim Biophys Acta. 1973 Apr 21;308(7):59–67. doi: 10.1016/0005-2787(73)90122-6. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Huang E. S., Newbold J. E., Pagano J. S. Analysis of simian virus 40 DNA with the restriction enzyme of Haemophilus aegyptius, endonuclease Z. J Virol. 1973 Apr;11(4):508–514. doi: 10.1128/jvi.11.4.508-514.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. A., Symons R. H., Berg P. Biochemical method for inserting new genetic information into DNA of Simian Virus 40: circular SV40 DNA molecules containing lambda phage genes and the galactose operon of Escherichia coli. Proc Natl Acad Sci U S A. 1972 Oct;69(10):2904–2909. doi: 10.1073/pnas.69.10.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. H., Laskowski M., Sr Sugar-unspecific mung bean nuclease I. J Biol Chem. 1968 Jun 25;243(12):3421–3424. [PubMed] [Google Scholar]
- Kato A. C., Bartok K., Fraser M. J., Denhardt D. T. Sensitivity of superhelical DNA to a single-strand specific endonuclease. Biochim Biophys Acta. 1973 Apr 21;308(7):68–78. doi: 10.1016/0005-2787(73)90123-8. [DOI] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Lewis A. M., Jr Use of nondefective adenovirus-simian virus 40 hybrids for mapping the simian virus 40 genome. J Virol. 1973 Sep;12(3):643–652. doi: 10.1128/jvi.12.3.643-652.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Rose J. A. Simian virus 40 integration site in an adenovirus 7-simian virus 40 hybrid DNA molecule. Proc Natl Acad Sci U S A. 1971 May;68(5):1037–1041. doi: 10.1073/pnas.68.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klump H., Ackermann T. Experimental thermodynamics of the helix--random coil transition. IV. Influence of the base composition of DNA on the transition enthalpy. Biopolymers. 1971;10(3):513–522. doi: 10.1002/bip.360100307. [DOI] [PubMed] [Google Scholar]
- LINN S., LEHMAN I. R. AN ENDONUCLEASE FROM NEUROSPORA CRASSA SPECIFIC FOR POLYNUCLEOTIDES LACKING AN ORDERED STRUCTURE. II. STUDIES OF ENZYME SPECIFICITY. J Biol Chem. 1965 Mar;240:1294–1304. [PubMed] [Google Scholar]
- Levin M. J., Crumpacker C. S., Lewis A. M., Jr, Oxman M. N., Henry P. H., Rowe W. P. Studies of nondefective adenovirus 2-simian virus 40 hybrid viruses. II. Relationship of adenovirus 2 deoxyribonucleic acid and simian virus 40 deoxyribonucleic acid in the Ad2+ND genome. J Virol. 1971 Mar;7(3):343–351. doi: 10.1128/jvi.7.3.343-351.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz J. E., Davis R. W. Cleavage of DNA by R 1 restriction endonuclease generates cohesive ends. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3370–3374. doi: 10.1073/pnas.69.11.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow J. F., Berg P. Cleavage of Simian virus 40 DNA at a unique site by a bacterial restriction enzyme. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3365–3369. doi: 10.1073/pnas.69.11.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow J. F., Berg P. Location of the T4 gene 32 protein binding site on simian virus 40 DNA. J Virol. 1973 Dec;12(6):1631–1632. doi: 10.1128/jvi.12.6.1631-1632.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder C., Delius H. Specificity of the break produced by restricting endonuclease R 1 in Simian virus 40 DNA, as revealed by partial denaturation mapping. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3215–3219. doi: 10.1073/pnas.69.11.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffler I. E., Elson E. L., Baldwin R. L. Helix formation by d(TA) oligomers. II. Analysis of the helix-coli transitions of linear and circular oligomers. J Mol Biol. 1970 Feb 28;48(1):145–171. doi: 10.1016/0022-2836(70)90225-1. [DOI] [PubMed] [Google Scholar]
- Sobell H. M. Molecular mechanism for genetic recombination. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2483–2487. doi: 10.1073/pnas.69.9.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton W. D. A crude nuclease preparation suitable for use in DNA reassociation experiments. Biochim Biophys Acta. 1971 Jul 29;240(4):522–531. doi: 10.1016/0005-2787(71)90709-x. [DOI] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Radloff R., Watson R., Laipis P. The twisted circular form of polyoma viral DNA. Proc Natl Acad Sci U S A. 1965 May;53(5):1104–1111. doi: 10.1073/pnas.53.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Watson R. Early and late helix-coil transitions in closed circular DNA. The number of superhelical turns in polyoma DNA. J Mol Biol. 1968 Apr 14;33(1):173–197. doi: 10.1016/0022-2836(68)90287-8. [DOI] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Baumgarten D., Olivera B. M. On the origin of tertiary turns in covalently closed double-stranded cyclic DNA. Proc Natl Acad Sci U S A. 1967 Nov;58(5):1852–1858. doi: 10.1073/pnas.58.5.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. Degree of superhelicity of covalently closed cyclic DNA's from Escherichia coli. J Mol Biol. 1969 Jul 28;43(2):263–272. doi: 10.1016/0022-2836(69)90266-6. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Variation of the average rotation angle of the DNA helix and the superhelical turns of covalently closed cyclic lambda DNA. J Mol Biol. 1969 Jul 14;43(1):25–39. doi: 10.1016/0022-2836(69)90076-x. [DOI] [PubMed] [Google Scholar]
- Zain B. S., Dhar R., Weissman S. M., Lebowitz P., Lewis A. M., Jr Preferred site for initiation of RNA transcription by Escherichia coli RNA polymerase within the simian virus 40 DNA segment of the nondefective adenovirus-simian virus 40 hybrid viruses Ad2 + ND 1 and Ad2 + ND 3 . J Virol. 1973 May;11(5):682–693. doi: 10.1128/jvi.11.5.682-693.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]



