Abstract
Genomic-enabled prediction is becoming increasingly important in animal and plant breeding and is also receiving attention in human genetics. Deriving accurate predictions of complex traits requires implementing whole-genome regression (WGR) models where phenotypes are regressed on thousands of markers concurrently. Methods exist that allow implementing these large-p with small-n regressions, and genome-enabled selection (GS) is being implemented in several plant and animal breeding programs. The list of available methods is long, and the relationships between them have not been fully addressed. In this article we provide an overview of available methods for implementing parametric WGR models, discuss selected topics that emerge in applications, and present a general discussion of lessons learned from simulation and empirical data analysis in the last decade.
Keywords: genomic selection, prediction, Bayesian methods, GenPred, shared data resources
MODERN animal and plant breeding schemes select individuals based on predictions of genetic merit. Rapid genetic progress requires that such predictions are accurate and that they can be produced early in life. Family-based predictions of genetic values have been used successfully for selection in plants and animals for many decades; however, there is a limit on the annual rate of genetic progress that can be attained with family-based prediction. Molecular markers allow describing the genome of individuals at a large number of loci, and this opens possibilities to derive accurate prediction of genetic values early in life. The first attempts to incorporate marker information into predictions were based on the presumption that one can localize causative mutations underlying genetic variation. This approach, known as QTL mapping (Soller and Plotkin-Hazan 1977; Soller 1978), led to the discovery of a few genes associated to genetic differences of traits of commercial interest. However, the impact on practical breeding programs has been smaller than initially envisaged (Dekkers 2004; Bernardo 2008). Several factors contributed to this: first, with a few exceptions, the proportion of the variance accounted by mapped QTL has commonly been small. Second, the financial resources required to develop the populations needed to map QTL were considerable, limiting the adoption of this technology (see Dekkers 2004, Bernardo 2008, Collard and Mackill 2008, and Hospital 2009 for insightful discussions of lessons learned from QTL studies in animal and plant breeding).
There is a general consensus that most traits are affected by large numbers of small-effect genes (see Buckler et al. 2009, for examples of traits in maize affected by large numbers of small-effect loci) and that the prediction of complex traits requires considering large numbers of variants concurrently. The continued advancement of high-throughput genotyping and sequencing technologies allowed the discovery of hundreds of thousands of genetic markers (e.g., single-nucleotide polymorphisms, SNPs) in the genomes of humans and several plant and animal species. Such dense panels of molecular markers allow exploiting multilocus linkage disequilibrium (LD) between QTL and genome-wide markers (e.g., SNPs) to predict genetic values. Although earlier contributions exist (Nejati-Javaremi et al. 1997; Haley and Visscher 1998; Whittaker et al. 2000), the foundations of genome-enabled selection (GS) were largely defined in the ground-breaking article by Meuwissen et al. (2001), who proposed to incorporate dense molecular markers into models using a simple, but powerful idea: regress phenotypes on all available markers using a linear model. And in recent years this approach has gained ground both in animal (VanRaden et al. 2009) and plant breeding (Bernardo and Yu 2007; Crossa et al. 2010).
With high-density SNP panels the number of markers (p) can vastly exceed the number of records (n), and fitting this large-p with-small-n regression requires using some type of variable selection or shrinkage estimation procedure. Owing to developments of penalized and Bayesian estimation procedures, as well as advances in the field of nonparametric regressions, several shrinkage estimation methods have been proposed and used for whole-genome regression (WGR) and prediction (WGP) of phenotypes or breeding values. However, the relationships between these methods have not been fully addressed and many important topics emerging in empirical applications have been often overlooked. In this article we provide an overview of parametric Bayesian methods as applied to GS (Methods), a discussion of selected topics that emerge when these models are used for empirical analysis (Selected Topics Emerging in Empirical Applications), and a discussion of lessons learned in the past years based on a literature review of simulation and empirical studies (Lessons Learned from Simulation and Empirical Data Analysis).
Methods
Early proposals for implementing GS (Meuwissen et al. 2001) used linear regression methods. More generally, one can regress phenotypes on marker covariates using a regression function, that may be parametric or not so that . Here, the regression function, , should be viewed as an approximation to the true unknown genetic values, , which can be a complex function involving the genotype of the ith individual at a large number of genes as well as cryptic interactions between genes and between genes and environmental conditions. Therefore, in a WGR model residuals represent random variables capturing nongenetic effects, plus approximation errors, , which can emerge due to imperfect LD between markers and QTL or because of model misspecification (e.g., unaccounted interactions).
The linear model appears as a special case with , where μ is an intercept, is the genotype of the ith individual at the jth marker (j = 1, . . . , p), and is the corresponding marker effect. Alternatively the regression function could be represented using semiparametric approaches (Gianola et al. 2006; de los Campos et al. 2010a) such as reproducing kernel Hilbert spaces (RKHS) regressions or neural networks (NN). Therefore, a first element of model specification in WGR is whether genetic values are approximated by using linear regression procedures or using semiparametric methods. In this article we focus on linear regression models; a review and a discussion about nonparametric procedures can be found in Gianola et al. (2010).
With modern genotyping technologies the number of markers, and therefore the number of parameters to be estimated, can vastly exceed the number of records. To confront the problems emerging in these large-p with small-n regressions, estimation procedures performing variable selection, shrinkage of estimates, or a combination of both are commonly used. Therefore, a second element of model choice pertains to the type of shrinkage estimation procedure used. Next, we discuss briefly the effects of shrinkage on the statistical properties of estimates and subsequently review some of the most commonly used penalized and Bayesian variable selection and shrinkage estimation procedures.
Effects of shrinkage on the mean-squared error of estimates
The accuracy of an estimator can be measured with the squared Euclidean distance between the estimated, , and the true value of the parameter, θ. In the case of scalars this is simply the squared deviation: . Here, we write to stress that the estimator is a function of the sampled data. The mean-squared error (MSE) is the expected value (over possible realizations of the data) of the squared Euclidean distance, . This can be decomposed into two terms: the variance of the estimator plus the square of its bias, . The variance of the estimator (and in some cases its bias) decreases with sample size. With standard estimation procedures, such as ordinary least squares (OLS) or maximum likelihood (ML), with fixed sample size, the variance of estimates increases rapidly as p does, yielding high MSE of estimates. One way of confronting the MSE problem emerging in large-p with small-n regressions is by shrinking estimates toward a fixed point (e.g., 0); this may increase bias but reduces the variance of the estimator. To illustrate the effects of shrinkage on MSE of estimates, consider a simple shrinkage estimator obtained by multiplying an unbiased estimator by a constant so that . The new estimator shrinks the original one toward 0. If , is biased; however, the variance of the new estimator, is guaranteed to be lower for any α< 1. Penalized and Bayesian methods are the two most commonly used shrinkage estimation procedures, an overview of these methods is given next.
Penalized methods
In penalized regressions estimates are derived as the solution to an optimization problem that balances model goodness of fit to the training data and model complexity. For continuous outcomes, lack of fit to the training data is usually measured by the residual sum of squares, (alternatively, one can use the negative of the logarithm of the likelihood or some other loss function) and model complexity is commonly defined as a function of model unknowns, ; therefore, penalized estimates are commonly derived as the solution to an optimization problem of the form
| (1) |
where is a regularization parameter that controls the trade-offs between lack of fit and model complexity. Ordinary least squares appear as a special case of (1) with . Usually, not all model unknowns are penalized; for instance, in (1) the intercept is not included in the penalty function. The features of the regression function that are not penalized [the overall mean in (1)] are then perfectly fitted.
Several penalized estimation procedures have been proposed, and they differ on the choice of penalty function, . In ridge regression (RR) (Hoerl and Kennard 1970), the penalty is proportional to the sum of squares of the regression coefficients or L2 norm, . A more general formulation, known as bridge regression (Frank and Friedman 1993), uses with . RR is a particular case with yielding the L2 norm, . Subset selection occurs as a limiting case with , which penalizes the number of nonzero effects regardless of their magnitude, . Another special case, known as least absolute angle and selection operator (LASSO) (Tibshirani 1996), occurs with , yielding the L1 penalty: . Using this penalty induces a solution that may involve zeroing out some regression coefficients and shrinkage estimates of the remaining effects; therefore LASSO combines variable selection and shrinkage of estimates. LASSO has become very popular in several fields of applications. However, LASSO and subset selection approaches have two important limitations. First, by construction, in these methods the solution admits at most n nonzero estimates of regression coefficients (Park and Casella 2008). In WGR of complex traits, there is no reason to restrict the number of markers with nonzero effect to be limited by n (the number of observations). Second, when predictors are correlated, something that occurs when LD span over long regions, methods performing variable selection such as the LASSO are usually outperformed by RR (Hastie et al. 2009). Therefore, in an attempt to combine the good features of RR and of LASSO in a single estimation framework, Zou and Hastie (2005) proposed to use as a penalty a weighted average of the L1 and L2 norm, that is, for , , and termed the method the elastic net (EN). This model involves two tuning parameters that need to be specified, the regularization parameter (λ) and α.
Bayesian shrinkage estimation
Bayesian methods can also be used for variable selection and shrinkage of estimates. Most penalized estimates are equivalent to posterior modes of certain types of Bayesian models (Kimeldorf and Wahba 1970; Tibshirani 1996). An illustration of the equivalence between some penalized and some Bayesian estimates is given in supporting information, File S1.
The general structure of the standard Bayesian linear models used in GS is
| (2) |
where is the posterior density of model unknowns given the data and hyperparameters , is the conditional density of the data given the unknowns, which for continuous traits are commonly independent normal densities with mean and with variance , and is the joint prior density of model unknowns, including the intercept , which is commonly assigned a flat prior, marker effects , which are commonly assigned IID informative priors, and the residual variance , which is commonly assigned a scaled-inverse chi-square prior with degree of freedom and scale parameter S, which is ; here we use a parameterization .
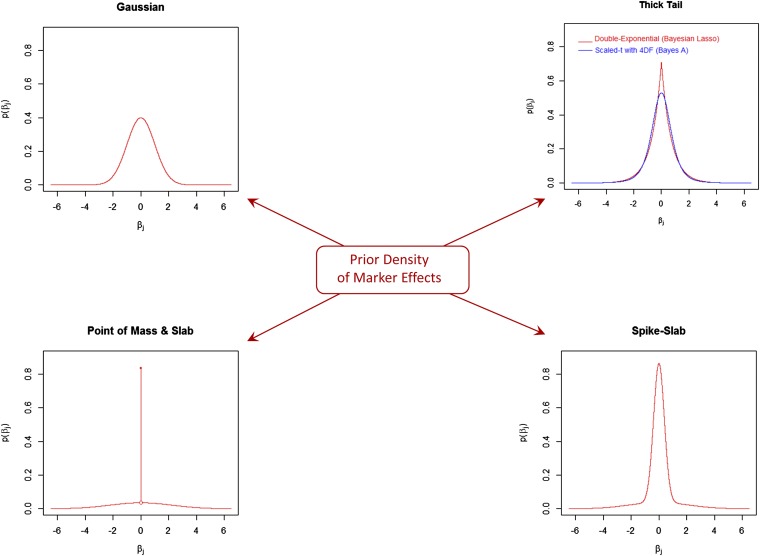
In these Bayesian models, the prior density of marker effects, , defines whether the model will induce variable selection and shrinkage or shrinkage only. Also, the choice of prior will define the extent and type of shrinkage induced. Two important features of these priors are how much mass they have in the neighborhood of zero and how thick or flat the tails of the density are. Based on these two features we classified the most commonly used priors into four big categories and in Figure 1 we have arranged them in a way that, starting from the Gaussian prior located in the top left corner, as one moves clockwise there is an increase in the peak of mass at zero and the tails are allowed to become thicker.
Figure 1 .
Commonly used prior densities of marker effects (all with zero mean and unit variance). The densities are organized in a way that, starting from the Gaussian in the top left corner, as one moves clockwise, the amount of mass at zero increases and tails become thicker and flatter.
Gaussian prior:
This density (depicted in the top left corner of Figure 1) has two hyperparameters: the mean (commonly set to zero) and the variance ; therefore, in this model, . If the intercept and the variance parameters are known, the posterior distribution of marker effects, , can be shown to be multivariate normal, with posterior mean given by , where is a matrix of marker genotypes and is a vector of (centered) phenotypes. This is exactly the RR estimate with . Because of this characteristic, this model is sometimes referred to as Bayesian ridge regression (BRR). Also, can be shown to be the best linear unbiased predictor (BLUP) of marker effects; therefore, this model is also sometimes referred to as RR-BLUP (standing for ridge regression BLUP).
Ridge regression and the BRR both perform an extent of shrinkage that is homogenous across markers; this approach may not be optimal if some markers are linked to QTL while others are in regions that do not harbor QTL. To overcome this potential limitation, other prior densities can be used.
Thick-tailed priors:
The two most commonly used thick-tailed densities, the scaled t and the double exponential, are represented in the top right corner of Figure 1. The scaled-t density is the prior used in model BayesA (Meuwissen et al. 2001) and the double-exponential or Laplace prior is the density used in the Bayesian LASSO (BL) (Park and Casella 2008). Relative to the Gaussian, these densities have higher mass at zero (inducing strong shrinkage toward zero of estimates of effects of markers with small effects) and thicker tails (inducing, relative to the BRR, less shrinkage of estimates of markers with sizable effects).
For computational convenience, the thick-tail densities are commonly represented as infinite mixtures of scaled-normal densities (Andrews and Mallows 1974) of the form , where is a prior density assigned to marker-specific variance parameters and ω denotes hyperparameters indexing this density. When is a scaled inverse chi-square density, the resulting marginal prior of marker effects is scaled t, and this is the approach used in model BayesA of Meuwissen et al. (2001). When is an exponential density, the resulting marginal prior of marker effects is double exponential, and this is the approach followed in the Bayesian LASSO of Park and Casella (2008). The double-exponential density is indexed by a single parameter (rate) and the scaled t is indexed by two parameters (scale and degree of freedom); this gives the scaled t more flexibility for controlling how thick the tails may be. An even higher degree of flexibility to control the shape of the prior can be obtained by using priors that are finite mixtures.
Spike–slab priors:
These models use priors that are mixtures of two densities: one with small variance (the spike) and one with large variance (the slab) (e.g., George and McCulloch 1993; see bottom left corner in Figure 1). Commonly, the spike and the slab are both zero-mean normal densities. A graphical representation of one of such mixtures is given in the bottom right corner of Figure 1. The general form of these mixtures is where is a mixture proportion and and are variance parameters. To prevent the so-called label-switching problem a common approach is to restrict so that π can be interpreted as the proportion of effects coming from the “small” variance component. Another approach is to reparameterize the prior so that the variance of one of the components is a scaled version of the variance of the other component; for instance, with . Model Bayes “stochastic search variable selection” (SSVS) (Calus et al. 2008; Verbyla et al. 2009) follows this approach with τ commonly fixed at a certain value (e.g., ). Another possibility is to link the proportion of effects coming from the “small-variance” component with the proportion of variance accounted for. For instance, following Yu and Meuwissen (2011), one could assume that of the markers account for of genetic variance. The so-called Pareto principle represents a specific case of the more general principle with . This reduces the number of hyperparameters that need to be chosen, at the expense of imposing restrictions that may or may not hold.
Although spike–slab models are usually formed by mixing two Gaussian components, similar models may be obtained by mixing other densities such as scaled t (Zou et al. 2010) or double exponential (DE). There are two limiting cases of the spike–slab model that are of special interest. The first one occurs when or , and this case corresponds to the standard Gaussian prior (see above); the second one occurs when , and in this case, the small-variance component of the mixture collapses to a point of mass at zero, giving rise to a prior that consists of a mixture of a point of mass at zero and a slab.
Point of mass at zero and slab priors:
These are used to induce a combination of variable selection and shrinkage (see bottom left corner in Figure 1). These priors are used, for example, in models BayesB (Meuwissen et al. 2001) and BayesC (Habier et al. 2011). In Bayes B the slab is a scaled-t density, while in BayesC the slab is a normal density.
Genome-enabled BLUP
An alternative parameterization of the BRR can be obtained by replacing with or in matrix notation . In the BRR, marker effects are IID normal random variables. From properties of the multivariate normal density it follows that , where for some k. For instance, a common choice is to use , where is (an estimate of) the frequency of the allele coded as one at the jth marker. Indeed, can be regarded as an estimate of the realized matrix of additive relationships (Habier et al. 2007; VanRaden 2007). Therefore, an equivalent representation of the BRR is given by the following model [genome-enabled BLUP, G-BLUP]:
| (3) |
The posterior mode of this model can be shown to be
| (4) |
with . This is also the best linear unbiased predictor of u, and therefore this model is usually referred as to G-BLUP. Above, we have motivated G-BLUP by exploiting its equivalence with the BRR; however, these methods can be also motivated simply as an additive infinitesimal model in which we replace the standard pedigree-based numerator relationship matrix with a marker-based estimate of additive relationships. Indeed, these methods have existed long before GS emerged (Ritland 1996, 2002; Nejati-Javaremi et al. 1997; Lynch and Ritland 1999; Eding and Meuwissen 2001).
Computing genomic relationships for G-BLUP:
Several proposals exist as to how to map from pairs of marker genotypes onto estimates of genetic relationships, and no one is considered superior. A first distinction is between methods that aim at estimating realized genomic relationships [or proportion of identical by state (IBS) (VanRaden 2007; Yang et al. 2010)] and those that attempt to estimate probability of sharing alleles due to inheritance from a known common ancestor [or probability of identical by descent (IBD)]. The IBD methods (Pong-Wong et al. 2001; Villanueva et al. 2005) are essentially multilocus extensions of the single-locus IBD approach of Fernando and Grossman (1989) with IBD coefficients averaged across multiple putative QTL.
Within the IBS framework, the most common approach, at least in GS, is to estimate genomic relationships using moment-based estimators, which in general take the form of cross-products of marker genotypes: . Here p is the number of loci and are the genotypes of individuals i and i′ at the jth locus. In matrix notation we have . As with any other regression procedure, marker genotypes can be centered by subtracting the mean of the marker genotype or centered and standardized to a unit variance; that is, , where are estimates of the frequency of the allele coded as one. Therefore, another common estimator is . Centering implies that variances and covariances between genetic values are measured as deviations with respect to a center defined by the average genotype. Following the tradition of pedigree-based infinitesimal models, one can define the “center” to be the average genotype in a “base” population. In such case allele frequencies should be estimated in that population. Alternatively, allele frequencies could be estimated directly from the sample without further consideration about an ancestral base population of nominally unrelated individuals. In this case the “origin” is defined as the average genotype in the sample. When this approach is used, some entries of G may become negative, some diagonal elements become <1, and the average diagonal value has an expected value equal to 1. Therefore, we cannot interpret the entries of G as proportion of allele sharing or as probabilities. Nevertheless, from the point of view of the Gaussian process simply defines a covariance function and nothing precludes assigning negative prior covariances between pairs of genetic values. For G to define a proper Gaussian process, it must be positive semidefinite; this is guaranteed when or . However, other methods do not guarantee that this condition will hold. Therefore, a good practice is to check that this condition is satisfied, by, for example, checking that the associated eigenvalues of G are all nonnegatives.
Relative to estimates of genomic relationship based on unstandardized markers, , standardization, , increases the “weight” given to markers with extreme allele frequency on the computation of genomic relationships and this occurs because the denominator used in the standardization, , is maximum at intermediate allele frequencies and minimum at extreme allele frequencies. Yang et al. (2010) proposed a modified version of where a different formula is used to compute the diagonal elements of G. The proposed formula has a sampling variability that does not depend on allele frequency and equals one in absence of inbreeding; however, to the best of our knowledge, the proposed method is not guaranteed to yield a positive semidefinite matrix.
Relationships between Bayesian methods
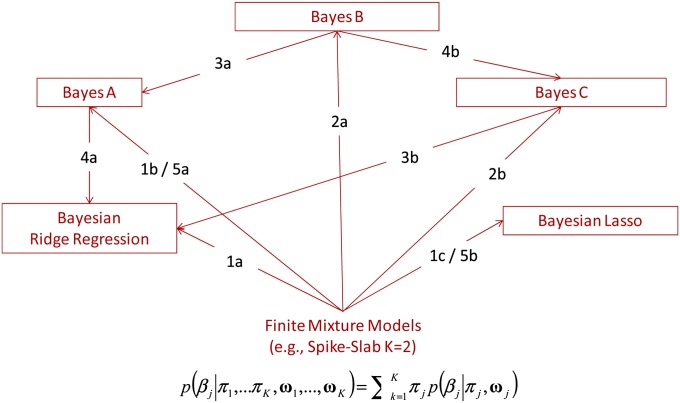
The categorization of priors given in Figure 1 is somehow arbitrary and some models can be considered special or limiting cases of others. In Figure 2 we represent some of these relationships. (Figure 2 is partially inspired by a presentation given by Robert Tempelman at University of Alabama at Birmingham, who discussed connections between G-BLUP, BayesA, and BayesB.):
Figure 2 .
Relationships between some prior densities commonly assigned to marker effects.
In finite mixture models we mix K densities; therefore, models using two or a single density component can be seen as special cases of the finite mixture model with K = 2 and K = 1, respectively (see paths 1a–1c in Figure 2).
Starting with a two-component mixture, such as the spike–slab, we can obtain models with a point of mass at zero and a slab, such as BayesB or –C, by fixing the variance of one of the components at zero (see paths 2a and 2b in Figure 2).
Models BayesA and BRR can be obtained as special cases of models BayesB and BayesC, respectively. This is done by setting in either BayesB or BayesC the proportion of markers with no effect (π) equal to zero (see paths 3a and 3b in Figure 2).
The scaled-t density has two parameters, the scale and the d.f.; as d.f. increases, the scaled-t density becomes increasingly similar to the Gaussian density. Therefore, starting from BayesA (BayesB) one can obtain the BRR (BayesC) by simply setting the d.f. to a very large value (see 4a and 4b in Figure 2).
The spike–slab prior is commonly formed by mixing two normal densities. The flexibility of such mixtures can be increased by increasing the number of components; eventually, we could consider an infinite number of components, each of which will have its own variance. However, there is a limit on the number of variance parameters that we can estimate. To confront this, a common approach is to regard these variances as random variables that are drawn from a common process. This is precisely what the scaled-t or DE densities are: these are infinite mixtures of scaled-normal densities (see paths 5a and 5b in Figure 2).
In view of the fact that many models (BRR, BayesA, and BayesC) appear as special cases of BayesB (for some values of parameters π and d.f.), a reasonable strategy would be to use a modified version of BayesB with π, scale and d.f. estimated from data (Nadaf et al. 2012). However, usually, with long-range LD and with p ≫ n, different configurations of marker effects can yield very similar values at the likelihood. Therefore, estimating π, the scale and d.f. parameters jointly from the data may not be possible.
Dealing with hyperparameters
The hyperparameters indexing the prior density of marker effects (ω) control the extent and strength of shrinkage of estimates of marker effects and they can have important impacts on inferences; therefore, dealing with these hyperparameters appropriately is crucial. These unknowns can be dealt with in different ways.
Heritability-based rules:
One possibility is to choose these hyperparameters based on prior expectation about the genetic variance of the trait. This approach was used, for instance, by Meuwissen et al. (2001), who derived hyperparameter values of the models by solving for them as a function of genetic variance. In their derivation they assume that genetic variance emerges due to uncertainty both about genotypes and about marker effects; however, this is not entirely consistent with the Bayesian models used in GS where genotypes are regarded as fixed and marker effects as random. Therefore, here we present a simple derivation that is consistent with models used in GS (Equation 2) where marker genotypes are regarded as fixed and marker effects are viewed as random IID variables. In linear models we have ; therefore the prior variance of the ith genomic value is where is the prior variance of marker effects that is a function of ω. Therefore, the average prior variance of genetic values in the sample is where is the average sum of squares of marker genotypes. Commonly, the model includes an intercept and variance is defined as deviations of genomic values from the center of the sample; therefore, should be computed using centered genotypes; that is, where is the average genotype at the jth marker. Moreover, when marker genotypes are centered and standardized to a null mean and unit variance equals the number of markers (p). A natural approach is to replace with the product of (an estimate of) heritability and of the variance of phenotypes, yielding
| (5) |
Equation 5 can be used to solve for values of ω. We have only one equation; therefore, if ω involves more than one hyperparameter, others need to be fixed. Table 1 shows examples of the use of this formula for BRR, BayesA, BayesB, BayesC, Bayesian LASSO, and Bayes SSVS.
Table 1 . Prior density of marker effects, prior variance of marker effects, and suggested formulas for choosing hyperparameter values by model.
| Model | Prior variance | Solution for scale/variance parameter | |
|---|---|---|---|
| Hyperparameters | |||
| Bayesian ridge regression | |||
| Bayesian LASSO | |||
| BayesA | |||
| Spike–slab | |||
| BayesC | |||
| BayesB | |||
where represents number of copies of the allele coded as one at the jth (j = 1,…,p) locus of the ith (i = 1,…,n) individual, and is the average genotype at the jth marker.
Validation methods:
Another possibility is to fit models over a grid of values of ω and then retain the value that maximizes predictive performance. To that end some type of internal validation (e.g., using a tuning data set) needs to be carried out. However, this approach can be computationally demanding. This happens because the grid of values of ω may involve a large number of cells (especially when ω involves several parameters) and because standard validation schemes usually involve fitting the model several times (e.g., across different folds of a cross-validation) for each possible value of ω in the grid. Other alternatives such as leave-one-out cross-validation or generalized cross-validation could be used; however, unlike ordinary least squares, in many of the models of interest, the leave-one-out residual sum of squares does not have a closed form. Because of these reasons this approach has not been a very popular one in GS [although examples of the use of validation methods for choosing regularization parameters exist (Usai et al., 2009)].
Fully Bayesian treatment:
The fully Bayesian approach regards ω as unknown. This is done by assigning a prior to ω; the model in expression (2) becomes
| (6) |
where is a prior density assigned to ω and H denotes a set of hyperparameters of higher order. The above-mentioned heritability rule can be seen as a limiting case of (6), where is set to be simply a point of mass at some value, say , which was chosen using prior knowledge (e.g., using the formulas in Table 1). In the fully Bayesian treatment, we may choose H so that has a prior mean or prior mode in the neighborhood of . This incorporates prior information into the model but in a more flexible way than heritability-based rules. The choice of prior, depends on the nature of the hyperparameters. For BRR ; therefore, it is natural to choose ; for BL Park and Casella (2008) suggested using a Gamma prior for , ; therefore, . For model BayesA , a common choice is to assign a Gamma prior to the scale parameter and either fix the degree of freedom parameter (usually to some small value >4 to guarantee a finite prior variance) or assign a prior to with support on the positive real numbers.
Empirical Bayes methods:
The empirical-based approach involves replacing in (3) with a point of mass located and an estimate of ω; that is, . In this respect this approach is similar to heritability-based rules; however, in the empirical Bayes method (EB) is a data-derived estimate. This approach is also commonly used in pedigree-based models where first, variance components are estimated from the data using restricted maximum likelihood and then, BLUPs of breeding values are derived with variance parameters replaced with those estimates. Ideally, we want to be the posterior mean of ω; however, in most cases it is difficult to derive a closed-form formula for the marginal posterior mean of ω. An alternative is to use the empirical Bayes principle within a Gibbs sampler (Casella 2001); however, the convergence of the algorithm may be too slow, and evidence does not suggest superiority of this approach relative to the fully Bayesian treatment.
Relaxing IID assumptions
All of the above-mentioned Bayesian models use IID priors for marker effects; that is, is the same for all markers; therefore, the prior mean and variances are the same for all marker effects. This assumption can be justified based on “ignorance”; however, in many instances we may have additional prior information about markers and we may want to incorporate such information into the prior assigned to marker effects. Examples of such information include, but are not limited to, (a) location of the marker in the genome; (b) whether the marker is located in a coding or a noncoding region; (c) whether the marker is in a region that harbors genes that we may believe affect the trait of interest; and (d) any prior information about the marker from an independent study, such as P-values or estimates of effects derived from a genome-wide association study. One of the great advantages of the Bayesian framework is that we can potentially include all these different types of information into the model via prior specification. With increasing volumes of information coming from multiple studies, the topic of how prior information can be incorporated into models is becoming increasingly important.
One possibility is to assign different priors for different sets of markers, an approach referred to as variance decomposition or variance partition. For instance, in BRR or in G-BLUP one can estimate variance parameters peculiar to chromosomes (Calus et al. 2010). Such an approach could also be used with any other prior information that allows grouping the markers such as information about gene function or ontology. Another possibility is to structure the prior to induce borrowing of information across marker effects. For instance, Yang and Tempelman (2012) propose using an antedependence covariance function to specify prior covariances between marker effects.
Algorithms
In Bayesian analysis inferences are based on the posterior distribution of the unknowns given the data with the general form of the posterior density is given in (3). In most cases, especially when prior hyperparameters are treated as random, the posterior distribution does not have a closed form. However, features of the posterior distribution (e.g., the posterior mean or standard deviation of marker effects) can be approximated using Monte Carlo Markov chain (MCMC) methods (Gelman et al. 2003).
Gibbs sampler:
Among the many MCMC algorithms the Gibbs sampler (Geman and Geman 1984; Casella and George 1992) is the most commonly used. In a Gibbs sampler draws from the joint posterior density are obtained by sampling from fully conditional densities; therefore, this algorithm is convenient when the fully conditional densities have closed form and are easy to sample from. This occurs, for example, in BRR, where all fully conditionals have closed form. However, this does not directly occur when the priors assigned to marker effects are from the thick-tailed family. To circumvent this problem, the most common approach consists of representing the thick-tailed densities as mixtures of scaled-normal densities (see Thick-tailed priors section above). With this approach, used in models BayesA, BayesB, and BL, the fully conditional densities of marker effects as well as those of the conditional variances of marker effects have closed forms. The typical iterations of the Gibbs sampler are illustrated next, using model BayesA as an example.
Update the intercept with a sample drawn from a normal density with mean equal to and variance , where .
For j in {1, … , p} update marker effects with a draw from a normal density with mean and variance , where and is the prior conditional variance of the jth marker effect.
For j in {1, … , p} update the variance of marker effects with a draw from a scaled-inverse chi-square density with scale and degree of freedom parameters and , respectively, where and are the prior scale and prior degree of freedom assigned to the variances of marker effects.
Update the residual variance, with a draw from a scaled-inverse chi-square density with degree of freedom and scale . Here, and are the prior scale and prior degree of freedom assigned to the residual variance and are the current model residuals.
Updating location parameters (intercept and marker effects) requires forming offsets obtained by subtracting from the phenotypes all the regression terms except the one that is being updated. In practice it is computationally less demanding to form the same offset by adding to the current residuals the current sample of the effect that will be updated. For instance, the offset required for sampling the jth marker effect can be also formed by adding to the current residual the contribution to the regression of the marker whose effect will be updated; that is, . Once location parameters are updated, residuals are updated by subtracting from the offset the contribution to the conditional expectation of the effect just updated (e.g., after drawing the jth marker effect, the updated residuals are ). Steps 3 and 4 require updating dispersion parameters. In most models the residual variance is updated from an inverse chi-square density. The fully conditional density of the variances of marker effects changes across models. In BayesA these are also inverse chi square. Note that these densities depart from the prior density by only 1 d.f. (see step 3, above), suggesting that data contain little information about these unknowns and that the influence of the prior on inferences about these unknowns can be substantial (Gianola et al. 2009).
The structure of the Gibbs sampler for BRR and BL is very similar to that described in steps 1–4, above. For BL the structure is very similar with two main differences: and has an exponential prior. Therefore, in step 3 the are updated from inverse Gaussian densities, and in step 4, and . For BRR, is the same for all markers; therefore: (a) in step 2 (see above), the term is also the same for all markers and (b) in step 3 only one variance parameter is updated, in this case from a scaled-inverse chi-square density with scale and degree of freedom parameters and , respectively. Therefore, in this case the fully conditional density departs from the prior by p d.f.
Although the Gibbs sampler is extremely flexible and in general easy to implement, the computational burden increases linearly with the number of records (due to computation of offsets) and with the number of markers (see steps 2 and 3 above). Many future applications will be using large data sets with hundreds of thousands of markers. In this context the Gibbs sampler can be extremely computationally demanding. To circumvent this problem, a few “fast” methods have been developed; these are briefly discussed next.
Fast methods attempt to estimate the posterior mode by maximizing the posterior density using expectation-maximization (Dempster et al. 1977) type algorithms (Yi and Banerjee 2009; Hayashi and Iwata 2010; Shepherd et al. 2010). Other proposals attempt to approximate the posterior mean using iterative-conditional expectation procedures (Meuwissen et al. 2009). Finally, a third approach consists of using two-step procedures where first, parameters other than marker effects are estimated from their marginal posterior density, and subsequently marker effects are estimated conditional on the other parameters (Cai et al. 2011). Thus far, only few studies have compared any of those methods with their MCMC-based counterparts and generally concluded that accuracies of the fast implementations were close to those of the MCMC counterparts; however, it is important to note that many of the algorithms are heuristic and the convergence properties are not well known. This is particularly relevant because in many of the models used in GS the posterior density is not guaranteed to be unimodal; therefore, there is great risk for the algorithm to arrive at, and not move from, local maxima. Moreover, unlike MCMC algorithms, the fast methods usually do not provide estimates of uncertainty about the estimated marker effects or predicted breeding values.
Two-step approaches for G-BLUP:
Above we discussed algorithms that estimate variance parameters and marker effects simultaneously. In G-BLUP there are only two variance parameters to be estimated, and . All data points contribute information about these unknowns; therefore with moderate to large sample size and with genetically related individuals the posterior densities of these unknowns are reasonably sharp. Consequently, a common approach consists of first estimating these variance parameters using a non-Bayesian algorithm, usually restricted maximum likelihood, and subsequently computing BLUP of genetic effects from standard mixed-model equations (see Equation 4). This is computationally much more convenient than the MCMC algorithms described above. The form of the restricted maximum-likelihood (REML) objective function and that of the mixed-model equations of G-BLUP are the same as those used in standard pedigree-based models with G replacing A. However, unlike G, which is usually a dense matrix, A and its inverse are sparse and most available software developed for pedigree models use sparse-matrix algorithms. Therefore, despite the similarity between G-BLUP and standard BLUP using A, some of the existing BLUP software cannot be readily used for G-BLUP. However, several packages such as ASReml (Gilmour et al. 2009) and DMU (Madsen and Jensen 2010) have options that allow providing a dense matrix, G, or its inverse, instead of pedigree data.
Selected Topics Emerging in Empirical Applications
The application of models for GS to real data usually involves several preprocessing steps. These steps can, in some cases, have great impact on model performance. Here we discuss selected topics that emerge in application of GS with real data.
Coding of marker genotypes
Centering and standardization of covariates is common practice in regression. When regression coefficients are estimated using OLS, centering and standardizing predictors have no effect on predictions. However, when shrinkage estimation procedures are used, transformations of the predictors can potentially impact estimates of effects and predictions. Centering is less relevant because models include an intercept that is usually not penalized; therefore, the effects of centering are “absorbed” in the intercept (Strandén and Christensen 2011). However, rescaling genotypes does have an effect. We explain this from a Bayesian perspective. Let be the contribution to the genomic value of the jth marker when genotypes are standardized to a unit variance. Here are genotype codes in original scale, is the frequency of the allele coded as one, and is the effect of the jth marker when genotypes are standardized. Further, following standard assumptions (Equation 2) let denote the prior variance of the effects of standardized markers. It follows that the effects of unstandardized markers are (j = 1, … , p). From here we observe that . The denominator in the expression is maximum at intermediate allele frequencies and approaches zero as θj approaches either zero or one; therefore, assigning equal prior variances for the effects of standardized markers implies smaller prior variance (i.e., strongest shrinkage toward zero) for the effects of markers with intermediate allele frequencies and less informative priors (i.e., larger prior variance) for the effects of markers with extreme allele frequencies. In short, other things being equal, standardization induces less (more) shrinkage of estimates of markers with extreme (intermediate) allele frequency.
Preadjusting phenotypes with estimates of systematic effects
The models discussed so far ignore effects other than those of markers and the intercept. In practice, phenotypes are affected by nongenetic effects such as those of contemporary groups (e.g., heard–year–season, sex, or age in animals). Theoretically, one can extend the model described in expression (2) by adding effects other than the intercept and markers. This allows joint estimation of all effects and when this is feasible, joint estimation of effects is the preferred option. However, in practice the joint estimation of effects may not be feasible because the software available for GS does not allow that or because of high computational requirements or because raw data cannot be shared. In such instances, a common approach is to preadjust data with estimates of nongenetic effects. However, caution must be exercised because, for reasons discussed below, precorrection of data can have undesirable consequences.
Bias:
In practice, marker effects and nonmarker covariates are not orthogonal to each other; therefore, estimation in two steps is likely to induce bias (and inconsistency) of estimates of marker effects. For instance, this may occur if genetically superior individuals are overrepresented in some contemporary groups. Similar problems emerge if selection takes place and data are preadjusted with estimates of year effects. All these problems are mitigated if the size of the contemporary groups is large and when the assignment of individuals to groups is at random. In an attempt to reduce biases induced by precorrection, a common practice is to add into the model for precorrection a genetic effect, commonly a pedigree regression. Preadjusted records are then computed by summing predictions of genetic and residual effects. This approach may reduce some of the confounding between genetic and nongenetic effects but it can also bring other types of biases. For instance, in comparing pedigree vs. marker-based models, precorrections based on a pedigree model may “favor” the pedigree model in the second step.
Induced heterogeneous residual variances and residual correlation:
In most cases, preadjustments are carried out using a linear model; therefore predictions and fitted residuals are linear functions of data of the form and , respectively, where: and are predicted phenotypes and model residuals, and H is the so-called hat matrix. For example, if the model for precorrection has the form and b is estimated by OLS, then . The variance–covariance matrix of the adjusted residuals is . This matrix is likely to have heterogeneous entries in the diagonal and nonzero off diagonals, implying that predicted residuals may have heterogeneous variances and correlations induced by the preprocessing. A common practice is to “weight” the second-step regression by dividing data with square roots of the diagonal elements of Σ. Such scaling makes the residual variance homoskedastic; however, this does not account for correlations between residuals that may have been induced by preadjustment of the data.
Dealing with unphenotyped and ungenotyped individuals
In many instances the set of individuals for which genotypic records are available may be different from that of the individuals with genotypes; for example, in dairy cattle many productive and reproductive traits are measured in females and, most commonly, a large proportion of genotypes are from sires. Schematically, we have two data sets: one comprising phenotypic records and pedigree and one comprising genotypic records. These two sets may be completely disjoint or may partially overlap. Roughly speaking, there are two main strategies that can be used to deal with unphenotyped and ungenotyped individuals.
Two-step genomic evaluations:
A common approach is to preprocess the phenotypic data in a way that a phenotypic score is produced for each of the genotyped individuals. Examples of these are the use of so-called daughter-yield deviations (DYD), predicted transmitting abilities (PTA), or deregressed proofs (DP) as “phenotypes” in genomic models (VanRaden and Wiggans 1991; Garrick et al. 2009). For reasons similar to those discussed in the previous section, these procedures are likely to induce not only heterogeneous residual variances but also correlations between the phenotypic scores, , unless special care is taken in computing these scores (Garrick et al. 2009). Heterogeneous residual variances can easily be accounted for in the following genomic evaluation by weighing the residual variance for each individual with an appropriate weight (Fikse and Banos 2001). And Garrick et al. (2009) discuss alternatives for computing family means in ways that avoid inducing residual correlations.
Single-step evaluations:
The single-step evaluations aim at combining information from genotyped and ungenotyped individuals in a single analysis. This requires deriving the joint distribution of the genetic effects of ungenotyped () and genotyped () animals, and most proposals attempt to do this in G-BLUP type models. We show in File S1 that if the genotypes of some individuals are unobserved, the joint density of is not multivariate normal; rather, it is a mixture of multivariate normal densities (see File S1). Therefore, existing proposals for combining data from genotyped and ungenotyped individuals (Legarra et al. 2009; Christensen and Lund 2010) that assume multivariate normality should be regarded as linear approximations to a nonlinear problem.
In the proposed methods (Aguilar et al. 2010; Christensen and Lund 2010) the standard genomic relationship matrix, G, is replaced with a matrix, , computed using observed genotypes (from the subset of genotyped individuals) and pedigree information (which is assumed to include all individuals with phenotypes or genotypes or both sources of information). In , genotypic information propagates into the relationships of ungenotyped individuals, using a linear regression procedure that implicitly predicts unobserved genotypes as linear combinations of observed genotypes with regression coefficients derived from pedigree relationships. The inverse of , which can be used to compute G-BLUP (see Equation 4), has a relatively simple form (Aguilar et al. 2010; Christensen and Lund 2010); however, computing requires inverting the matrix of genomic relationships of individuals with genotypes that may be singular. To circumvent this problem several procedures have been proposed (see Aguilar et al. 2010).
Lessons Learned from Simulation and Empirical Data Analysis
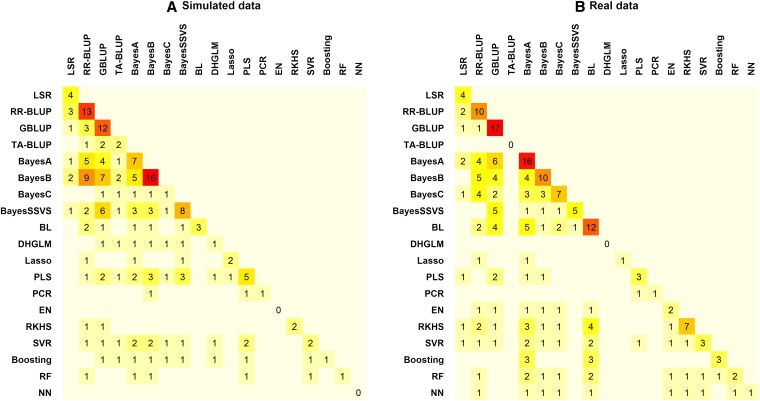
In the past few years, many studies have evaluated the performance of various WGP methods. In this section, we review this literature with a focus on extracting what we consider are the lessons learned after almost a decade of research in GS. Early publications were mainly based on simulated data but in recent years empirical evaluations have become more important. Based on a review of published articles we present in Table 2 a list of methods whose predictive performance has been compared to at least one other method for GS. In addition to the method’s name and abbreviation we indicate whether the estimation procedure is a penalized or a Bayesian regression. Most of the methods are parametric, in the sense that genomic values are represented as parametric functions of marker genotypes (see Equation 2), but some are nonparametric in nature and this is indicated in the last column of Table 2 .
Table 2 . Classification and abbreviations of the models included in Figure 3, A and B.
| Name (abbreviation) | Bayesian | Penalized | Nonparametric |
|---|---|---|---|
| Least-squares regression (LSR) | |||
| Bayesian ridge regression (BRR) or RR-BLUP | X | X | |
| BLUP using a genomic relationship matrix (G-BLUP) | X | X | |
| Trait-specific BLUP (TA-BLUP) | X | X | |
| BayesA | X | ||
| BayesB | X | ||
| BayesC | X | ||
| Bayes SSVS | X | ||
| Bayesian LASSO (BL) | X | ||
| Double hierarchical generalized linear models (DHGLM) | |||
| Least absolute shrinkage and selection operator (LASSO) | X | ||
| Partial least-squares regression (PLS) | X | ||
| Principal component regression (PCR) | X | ||
| Elastic net (EN) | X | ||
| Reproducing kernel Hilbert spaces regressions (RKHS) | X | X | X |
| Support vector regression (SVR) | X | X | |
| Boostinga | NA | NA | NA |
| Random forests (RF) | X | ||
| Neural networks (NN)b | X | X | X |
The following are early references of the use of the above methods for genomic prediction (references with the original description of some of the methods are also given in earlier sections of this article and in the references given here). LSR, BRR, BayesA, and BayesB, Meuwissen et al. (2001); G-BLUP, VanRaden (2008); TA-BLUP, Zhang et al. (2010); BayesC, Habier et al. (2011); Bayes SSVS, Calus et al. (2008); BL, de los Campos et al. (2009); DHGLM, Shen et al. (2011); LASSO, Usai et al. (2009); PLS and SVR, Moser et al. (2009); PCR, Solberg et al. (2009); EN, croiseau et al. (2011); RKHS, gianola et al. (2006); Boosting, González-Recio et al. (2010); RF, González-Recio and Forni (2011); and NN, Okut et al. (2011).
Boosting as an estimation technique could be applied to any method, Bayesian or penalized, parametric or nonparametric.
NN could be implemented in a nonpenalized, penalized, or Bayesian framework.
Using the methods listed in Table 2 and a sample of articles we reviewed (references provided at the bottom of Table 2), we provide in Figure 3 the number of times each pair of methods was compared, with the diagonal entries giving the number of times a particular method was used in a comparison study. Figure 3A counts simulation-based studies and Figure 3B summarizes those based on real data. The great majority of the studies included in our review evaluated linear regressions. Nonparametric procedures have the potential of capturing nonadditive effects as well; however, the use of nonparametric procedures remains limited thus far.
Figure 3 .
(A and B) Number of articles reviewed comparing one or more methods using simulated (A) or real (B) data. The abbreviations used for the methods are given in Table 2. The following references were used: (Meuwissen et al. 2001; Habier et al. 2007; Piyasatian et al. 2007; González-Recio et al. 2008; Lee et al. 2008; Bennewitz et al. 2009; de los Campos et al. 2009; Gonzalez-Recio et al. 2009; Hayes et al. 2009a,b; Lorenzana and Bernardo 2009; Luan et al. 2009; Lund et al. 2009; Meuwissen 2009; Meuwissen et al. 2009; Moser et al. 2009; Solberg et al. 2009; Usai et al. 2009; Verbyla et al. 2009; Zhong et al. 2009; Andreescu et al. 2010; Bastiaansen et al. 2010; Coster et al. 2010; Crossa et al. 2010; Daetwyler et al. 2010a,b; de los Campos et al. 2010a,b; Gonzalez-Recio et al. 2010; Gredler et al. 2010; Guo et al. 2010; Habier et al. 2010; Konstantinov and Hayes 2010; Meuwissen and Goddard 2010; Mrode et al. 2010; Pérez et al. 2010; Shepherd et al. 2010; Zhang et al. 2010; Calus and Veerkamp 2011; Clark et al. 2011; Croiseau et al. 2011; de Roos et al. 2011; Gonzalez-Recio and Forni 2011; Habier et al. 2011; Heffner et al. 2011; Iwata and Jannink 2011; Legarra et al. 2011; Long et al. 2011a,b; Makowsky et al. 2011; Mujibi et al. 2011; Ober et al. 2011; Ostersen et al. 2011; Pryce et al. 2011; Pszczola et al. 2011; Wiggans et al. 2011; Wittenburg et al. 2011; Wolc et al. 2011a,b; Yu and Meuwissen 2011; Bastiaansen et al. 2012; Heslot et al. 2012).
Among the additive models, the Bayesian regressions and G-BLUP were the most commonly used. G-BLUP is attractive because its implementation is straightforward using existing REML+BLUP software. The Bayesian methods have been discussed and described widely and are generally chosen because many of them (e.g., BayesA, -B, and -C and BL) allow departures from the infinitesimal model. Many of the penalized regressions (e.g., LASSO and EN) also allow this, but their use in GS is much more limited. This may appear to be surprising considering the diversity of available methods [RR, partial least-squares regression (PLS), principal component regression (PCR), LASSO, EN, and support vector regression (SVR)] and the fact that most of these methods are implemented in relatively efficient packages. However, although limited, empirical evidence suggests somewhat lower prediction accuracy of these methods relative to the more frequently used Bayesian regressions (Solberg et al. 2009; Coster et al. 2010; Gredler et al. 2010; Pszczola et al. 2011; Heslot et al. 2012).
Simulation studies
Simulation studies (Meuwissen et al. 2001; Habier et al. 2007) have systematically shown higher prediction accuracy of GS relative to standard pedigree-based predictions regardless of trait architecture or model of choice. However, many studies have shown that factors such as size of the reference data set (Meuwissen et al. 2001; VanRaden and Sullivan 2010), trait heritability, the number of loci affecting the trait (Daetwyler et al. 2008), and the degree of genetic relationships between training and validation samples (Habier et al. 2007) can greatly affect the prediction accuracy of WGP.
Effects of genetic architecture, marker density, and model on prediction accuracy:
The choice of model has also been shown to affect predictive performance, but simulation studies suggest that the effect of model choice on prediction accuracy depends on genetic architecture.
Daetwyler et al. (2010b) showed in a simulation study that the accuracy of G-BLUP is not affected by the number of QTL; however, the predictive performance of BayesC was greatly affected by that factor and its accuracy was higher when the number of QTL was low and decreased with increasing number of QTL. Similar trends were observed by Coster et al. (2010) and Clark et al. (2011). In the study of Coster et al. (2010), variable selection methods (e.g., some Bayesian regressions and LASSO) showed an increase in accuracy when the number of QTL decreased, while a method that includes all SNPs (PLS) was shown to be unaffected by the number of QTL. In the study by Clark et al. (2011) it was also shown that with BayesB greater accuracies were obtained than with G-BLUP in scenarios involving different distributions of allele frequencies (from rare to common variants) at casual loci. For a scenario that closely resembled the infinitesimal model there was no difference in accuracy between BayesB and GBLUP, while Daetwyler et al. (2010b) reported a lower accuracy for BayesC in such a scenario. Other comparisons based on simulated data also show that there are differences in accuracies across models and generally confirm that, as one would expect, accuracy is greater for the model that better fits the genetic architecture of the trait (Lund et al. 2009; Bastiaansen et al. 2010; Pszczola et al. 2011).
Apart from the number of QTL and the distribution of their effects, there are several other factors that theoretically are expected to result in differences in accuracy for variable selection methods vs. G-BLUP type models. With low marker density markers are unlikely to be tightly linked to QTL and each marker may track signal from different QTL, inducing a less extreme distribution of effects than obtained at higher density. Meuwissen et al. (2009) showed that the superiority of BayesB over G-BLUP increased with increasing marker density. This result is also clearly confirmed in the study by Meuwissen and Goddard (2010), where genomic prediction based on the whole-genome sequence including the causal loci was substantially more accurate for BayesB compared to G-BLUP. However, both studies simulated very few QTL, giving variable selection methods an advantage. On the other hand, for any given marker density, the ability of variable selection methods to detect variants tightly linked to QTL increases as the span of LD decreases.
Real data analysis
A number of empirical evaluations of GS have been published in recent years. These studies have confirmed some but not all of the findings anticipated by simulation studies.
Pedigree vs. marker-enabled prediction:
In general, empirical studies have confirmed the superiority of GS relative to family-based predictions that was anticipated by simulations. The most clear case occurs in Holstein dairy cattle where several studies (Hayes et al. 2009a; VanRaden et al. 2009) have confirmed that GS can attain higher prediction accuracy than standard family-based prediction (e.g., parental average). The potential of GS has also been confirmed in several breeds of beef cattle (Garrick 2011; Saatchi et al. 2011), sheep (Daetwyler et al. 2010a), broilers (Gonzalez-Recio et al. 2008), layer chickens (Wolc et al. 2011a,b), and several plant species (de los Campos et al. 2009; Crossa et al. 2010; Heslot et al. 2012). For applications in plants, it has been shown that GS outperforms conventional marker-assisted selection (Heffner et al. 2010, 2011) and that it has the potential to be substantially more efficient per unit of time than phenotypic selection (Grattapaglia and Resende 2011; Zhao et al. 2012). However, the superiority of GS, relative to pedigree-based predictions, has not always been as high as anticipated by simulations. This is particularly clear in applications for sheep (Daetwyler et al. 2010a) and in beef cattle (Saatchi et al. 2011). Many reasons may contribute to this: in general in these cases the size of the training data set is limited and the phenotypes used may have been noisy, in some cases (e.g., some breeds in sheep) population structure (e.g., a mixture of breeds) may be a reason, and in others the relatively small family size may limit the potential accuracy that can be attained with GS.
Some studies have shown benefits of extending the standard models of GS by adding a random effect representing a regression on pedigree information (de los Campos et al. 2009; Crossa et al. 2010); however, the benefits of jointly modeling pedigree and marker data relative to a markers-only model seem to vanish as marker density increases (Vazquez et al. 2010).
Effects of marker density:
Several empirical studies have evaluated the effects of marker density on prediction accuracy (Weigel et al. 2009; Vazquez et al. 2010; Makowsky et al. 2011). When shrinkage estimation procedures are used, prediction accuracy increases monotonically with marker density, but it does at diminishing marginal rates of response. Consequently, prediction accuracy reaches a plateau and does not increase beyond certain marker density. The level at which this plateau takes place depends on two factors mainly: the span of LD in the genome and sample size. For instance, Vazquez et al. (2010) found little increase on prediction accuracy beyond 10,000 SNPs for the prediction of several traits in U.S. Holsteins, a population where LD span over long regions. However, in a study involving WGP of human data, which exhibit much shorter span of LD than found in cattle data, Makowsky et al. (2011) found response to increased marker density even beyond 100,000 SNPs; more importantly the rates of response to increases in marker density were greatly affected by the number of close relatives used in the training data set, suggesting that “local sample size” also affects the effects of marker density on prediction accuracy.
Genetic architecture, sample size, and model:
For traits involving a limited number of large-effect QTL, simulation studies have consistently predicted superiority of methods using variable selection and differential shrinkage of estimates of effects such as BayesB. However, this has not been fully confirmed by real data analysis. Indeed, in most studies comparing different genomic prediction models based on real data, there are only small differences observed in accuracies between models. An important question is whether the genetic architecture of real data is perhaps less extreme than suggested by QTL-mapping studies (Kearsey and Farquhar 1998; Hayes and Goddard 2001) and generally assumed in simulation studies or whether other characteristics of the data (number of markers, span of LD) used prohibit greater distinction between models. Variable selection is most effective with short span of LD, high marker density, and large sample size; however, these conditions are not always met in applications in plant and animal breeding.
In empirical studies genetic architecture is less well known and grouping traits based on their architecture is not straightforward. In animal breeding, there are only a few examples of traits where one or a few major genes explain a sizable proportion of genetic variance. One of such examples is the DGAT1 gene that has a large effect on fat percentage in dairy cattle (Grisart et al. 2002; Winter et al. 2002). For this particular case, it is shown in several studies that models with a thick-tailed prior distribution of marker effects such as BayesA and variable selection methods such as Bayes SSVS yield higher accuracy than G-BLUP.
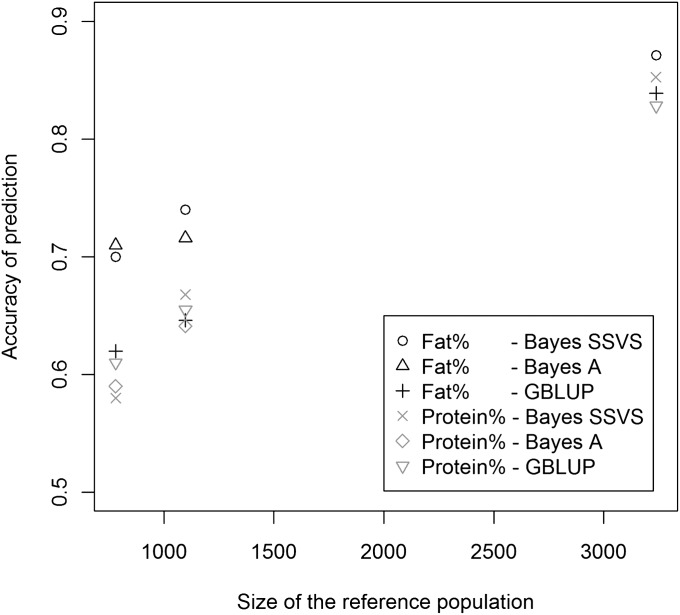
In Figure 4 we summarize results from three studies (Hayes et al. 2009b; Verbyla et al. 2009; de Roos et al. 2011) where prediction accuracy of G-BLUP and of a Bayesian regression using either a thick-tailed (BayesA) or a spike–slab (Bayes SSVS) density was evaluated for fat and protein percentage at varying sizes of the training data set. The results of these studies illustrate important concepts. First, prediction accuracy increases markedly as the size of the training data set does. This has been anticipated by simulation studies (VanRaden and Sullivan 2010) and consistently confirmed in empirical analyses (Lorenzana and Bernardo 2009; Bastiaansen et al. 2010). Second, in general, for fat percentage there is a clear superiority of the models performing differential shrinkage of estimates of effects (e.g., Bayes SSVS or BayesA) relative to G-BLUP. All traits were analyzed with model BayesA and the authors concluded that prediction accuracy was higher for traits with simpler genetic architecture. This also represents a confirmation of results from simulations that anticipated superiority of methods performing variable selection and differential shrinkage of estimates, especially with traits having some large-effects QTL (Meuwissen et al. 2001; Daetwyler et al. 2010b). However, the differences detected in empirical evaluations are not as large as those usually reported in simulations. More importantly, differences in predictive performance between methods decreased when the sample size increased (see Figure 4), reflecting that, as expected, the influence of the prior (the choice of prior density of marker effects in this case) decreases as sample size increases. This is in agreement with findings from simulation studies such as that of Daetwyler et al. (2010b).
Figure 4 .
Accuracies of G-BLUP, BayesA, and Bayes SSVS models for fat and protein percentage, estimated using three different Holstein–Friesian reference populations (Hayes et al. 2009b; Verbyla et al. 2009; de Roos et al. 2011). Note that the data used by Hayes et al. (2009b) are a subset of the data used by Verbyla et al. (2009).
A recent study on genomic prediction for different traits in loblolly pine indicated generally small differences in prediction accuracy between the models RR-BLUP, BL, BayesA, and BayesC (Resende et al. 2012). Nevertheless, for traits related to fusiform rust, known to be controlled by a few genes of large effects, BayesA and BayesC outperformed RR-BLUP and BL.
Theoretically, it can be expected that a method that is flexible enough to model any type of genetic architecture (e.g., model BayesB or BayesC when all hyperparameters are estimated from data) will perform relatively well across a wide range of traits with different characteristics; however, empirical evidence has not always confirmed this. For instance, the study of Heslot et al. (2012) compared 10 different methods for 18 different traits measured on barley, wheat, and maize, and RKHS was the only method that clearly outperformed others across traits and data sets. Among the linear regressions, the Bayesian methods outperformed the penalized regressions, and the differences among the Bayesian linear regressions were very small. However, marker density in this study was very low and this clearly limits the possibility of some methods to express their potential.
The effects of sample size on prediction accuracy are clear. First, the accuracy of estimates of marker effects increases with sample size. This occurs because bias and variance of estimates of marker effects decrease with sample size. Additionally, in some designs an increase in sample size may also increase the extent of genetic relationships between subjects in the training and validation data sets.
Liu et al. (2011) evaluated the effect of the size of the training data set on the dispersion of estimates of marker effects and on prediction accuracy using BRR-BLUP. The authors reported an increase in dispersion among estimates of SNP effects by more than a factor of 5, when the reference population was increased from 735 to 5025 bulls (Liu et al. 2011). This essentially reflects that the extent of shrinkage of estimates of effects decreases as simple size increases. Moreover, they showed that the correlation between estimated SNP effects was as low as 0.42 between the two extreme scenarios, but correlations were >0.9 when ∼500 bulls were added to the reference population that already contained >3500 bulls. However, they reported that the correlation between genomic predictions with different numbers in the reference population was much closer to 1.0, compared to the correlation between estimated SNP effects. This illustrates two important points: (a) when p ≫ n, one can arrive at similar predictions of total genomic merit with very different estimates of marker effects and (b), because of the same reason, with small sample size, one needs to be cautious about interpretation of estimates of marker effects because these may be highly influenced by the choice of prior.
Concluding remarks
Both simulation and empirical studies have systematically shown higher prediction accuracy of GS relative to standard pedigree-based predictions and it is now clear that GS offers great opportunities to further increase the rate of genetic progress achieved in plant and animal breeding. Most of the benefits of GS arise from the possibility of obtaining accurate predictions early in the breeding cycle; therefore, getting the most out of GS may require changes in breeding programs.
Implementing GS involves making important decisions regarding the choice of model, the size of the training data, and marker density, just to mention a few. In recent years simulation and empirical studies have produced valuable information that can be used to guide researchers in making those decisions. Several factors, including span of LD, trait heritability, genetic architecture, marker density, size of the training data set, and the model used can affect the prediction accuracy of GS.
Some of the factors affecting prediction accuracy, such as trait heritability, genetic architecture, and to a large extent LD, cannot be controlled; however, we can have control of the design of reference data sets, including size and relationships, marker density, and the model used for estimation of effects. Among the factors that are under control of the researcher, the size of the training data set and the strength of genetic relationships between training and validation samples are by far the most important factors affecting prediction accuracy. The model of choice is also important; however, the differences between models reported by simulation studies have not always been confirmed by real data analysis. Empirical analyses have shown only small differences between methods, with a slight advantage of models performing “selection and shrinkage” such as BayesB for traits with “large-effect QTL.” But in general thick-tailed models such as BayesA or Bayesian LASSO perform well across traits and G-BLUP performs well for most traits. An important reason is that, due to the fact that p ≫ n, there are a multitude of different prediction equations that yield about the same likelihood and minimum prediction error rate (Breiman 2001): often we encounter multiple equally-good models.
The unexpected generally good performance of G-BLUP in real data is connected to several issues that have been revealed to some extent. First, the real genetic architecture of traits appears less extreme than expected based on QTL-mapping results. Second, most of the gain in accuracy due to using markers in current applications arises from explaining the Mendelian sampling term, rather than from tracing signals generated at individual QTL. Overall, it seems that with a long span of LD and relatively sparse platforms (e.g., 50,000 SNPs) variable selection may not be needed. However, the relative performance of G-BLUP and variable selection methods may change with denser coverage (e.g., with genotyping by sequencing) and in populations with short LD span.
Supplementary Material
Acknowledgments
The authors thank D. J. de Koning and Lauren McIntyre for encouraging us to write this review article and for insightful comments provided on earlier versions of the manuscript. Mario Calus acknowledges financial support from the Dutch Ministry of Economic Affairs, Agriculture, and Innovation (Programme “Kennisbasis Research,” code KB-17-003.02-006). H.D.D. was funded by the Cooperative Research Centre for Sheep Industry Innovation. J.M.H. was funded by the Australian Research Council project LP100100880 of which Genus Plc, Aviagen LTD, and Pfizer are cofunders.
Note added in proof: See Daetwyler et al. 2013 (pp. 347–365) for a related work.
Footnotes
Edited by D. J. de Koning and Lauren M. McIntyre
Literature Cited
- Aguilar I., Misztal I., Johnson D. L., Legarra A., Tsuruta S., et al. , 2010. Hot topic: a unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 93: 743–752 [DOI] [PubMed] [Google Scholar]
- Andreescu C., Habier D., Fernando R. L., Kranis A., Watson K., et al. , 2010. Accuracy of genomic predictions across breeding lines of chickens. 9th World Congress on Genetics Applied to Livestock Production, Leipzig, Germany. CD-ROM Communication 0956
- Andrews D. F., Mallows C. L., 1974. Scale mixtures of normal distributions. J. R. Stat. Soc., B 36: 99–102 [Google Scholar]
- Bastiaansen, J. W. M., M. C. A. M. Bink, A. Coster, C. Maliepaard, and M. P. L. Calus, 2010 Comparison of analyses of the QTLMAS XIII common dataset. I: Genomic selection. BMC Proc. 4(Suppl. 1): S1. [DOI] [PMC free article] [PubMed]
- Bastiaansen J. W. M., Coster A., Calus M. P. L., van Arendonk J. A. M., Bovenhuis H., 2012. Long-term response to genomic selection: effects of estimation method and reference population structure for different genetic architectures. Genet. Sel. Evol. 44: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennewitz J., Solberg T., Meuwissen T., 2009. Genomic breeding value estimation using nonparametric additive regression models. Genet. Sel. Evol. 41: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo R., 2008. Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci. 48: 1649–1664 [Google Scholar]
- Bernardo R., Yu J., 2007. Prospects for genomewide selection for quantitative traits in maize. Crop Sci. 47: 1082–1090 [Google Scholar]
- Breiman L., 2001. Statistical modeling: the two cultures. Stat. Sci. 16: 199–215 [Google Scholar]
- Buckler E. S., Holland J. B., Bradbury P. J., Acharya C. B., Brown P. J., et al. , 2009. The genetic architecture of maize flowering time. Science 325: 714–718 [DOI] [PubMed] [Google Scholar]
- Cai X., Huang A., Xu S., 2011. Fast empirical Bayesian LASSO for multiple quantitative trait locus mapping. BMC Bioinformatics 12: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calus M. P. L., Veerkamp R. F., 2011. Accuracy of multi-trait genomic selection using different methods. Genet. Sel. Evol. 43: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calus M. P. L., Meuwissen T. H. E., de Roos A. P. W., Veerkamp R. F., 2008. Accuracy of genomic selection using different methods to define haplotypes. Genetics 178: 553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calus, M. P. L., H. A. Mulder, and R. F. Veerkamp, 2010 Estimation of breeding values for haploid chromosomes. 9th World Congress on Genetics Applied to Livestock Production, Leipzig, Germany. CD-ROM Communication 0269.
- Casella G., 2001. Empirical Bayes Gibbs sampling. Biostatistics 2: 485–500 [DOI] [PubMed] [Google Scholar]
- Casella G., George E. I., 1992. Explaining the Gibbs sampler. Am. Stat. 46: 167–174 [Google Scholar]
- Christensen O. F., Lund M. S., 2010. Genomic prediction when some animals are not genotyped. Genet. Sel. Evol. 42: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. A., Hickey J. M., van der Werf J. H. J., 2011. Different models of genetic variation and their effect on genomic evaluation. Genet. Sel. Evol. 43: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collard B. C. Y., Mackill D. J., 2008. Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B 363: 557–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coster A., Bastiaansen J. W., Calus M. P., van Arendonk J. A., Bovenhuis H., 2010. Sensitivity of methods for estimating breeding values using genetic markers to the number of QTL and distribution of QTL variance. Genet. Sel. Evol. 42: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croiseau P., Legarra A., Guillaume F., Fritz S., Baur A., et al. , 2011. Fine tuning genomic evaluations in dairy cattle through SNP pre-selection with the Elastic-Net algorithm. Genet. Res. 93: 409–417 [DOI] [PubMed] [Google Scholar]
- Crossa J., de los Campos G., Perez P., Gianola D., Burgueño J., et al. , 2010. Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics 186: 713–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Villanueva B., Woolliams J. A., 2008. Accuracy of predicting the genetic risk of disease using a genome-wide approach. PLoS ONE 3: e3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Hickey J. M., Henshall J. M., Dominik S., Gredler B., et al. , 2010a Accuracy of estimated genomic breeding values for wool and meat traits in a multi-breed sheep population. Anim. Prod. Sci. 50: 1004–1010 [Google Scholar]
- Daetwyler H. D., Pong-Wong R., Villanueva B., Woolliams J. A., 2010b The impact of genetic architecture on genome-wide evaluation methods. Genetics 185: 1021–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Calus M. P. L., Pong-Wong R., de los Campos G., Hickey J. M., 2013. Genomic prediction in animals and plants: simulation of data, validation, reporting, and benchmarking. Genetics 193: 347–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers J. C., 2004. Commercial application of marker-and gene-assisted selection in livestock: strategies and lessons. J. Anim. Sci. 82: E313–E328 [DOI] [PubMed] [Google Scholar]
- de los Campos G., Naya H., Gianola D., Crossa J., Legarra A., et al. , 2009. Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 182: 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G., Gianola D., Rosa G. J. M., Weigel K. A., Crossa J., 2010a. Semi-parametric genomic-enabled prediction of genetic values using reproducing kernel Hilbert spaces methods. Genet. Res. 92: 295–308 [DOI] [PubMed] [Google Scholar]
- de los Campos G., Gianola D., Rosa G. J. M., Weigel K., Vazquez A. I., et al. , 2010b. Semi-parametric marker-enabled prediction of genetic values using reproducing kernel Hilbert spaces regressions. 9th World Congress on Genetics Applied to Livestock Production, Leipzig, Germany. CD-ROM Communication 0520
- Dempster A. P., Laird N. M., Rubin D. B., 1977. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. B 39: 1–38 [Google Scholar]
- de Roos A. P. W., Schrooten C., Druet T., 2011. Genomic breeding value estimation using genetic markers, inferred ancestral haplotypes, and the genomic relationship matrix. J. Dairy Sci. 94: 4708–4714 [DOI] [PubMed] [Google Scholar]
- Eding H., Meuwissen T. H. E., 2001. Marker based estimates of between and within population kinships for the conservation of genetic diversity. J. Anim. Breed. Genet. 118: 141–159 [Google Scholar]
- Fernando R., Grossman M., 1989. Marker assisted selection using best linear unbiased prediction. Genet. Sel. Evol. 21: 467 [Google Scholar]
- Fikse W., Banos G., 2001. Weighting factors of sire daughter information in international genetic evaluations. J. Dairy Sci. 84: 1759–1767 [DOI] [PubMed] [Google Scholar]
- Frank I. E., Friedman J. H., 1993. A statistical view of some chemometrics regression tools. Technometrics 35: 109–135 [Google Scholar]
- Garrick D., 2011. The nature, scope and impact of genomic prediction in beef cattle in the United States. Genet. Sel. Evol. 43: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick D. J., Taylor J. F., Fernando R. L., 2009. Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet. Sel. Evol. 41: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A., Carlin J. B., Stern H. S., Rubin D. B., 2003. Bayesian Data Analysis, Ed. 2. Chapman & Hall/CRC, London/New York/Washington, D.C./Boca Raton, FL [Google Scholar]
- Geman S., Geman D., 1984. Stochastic relaxation, Gibbs distributions, and the Bayesian restoration of images. IEEE Trans. Patt. Anal. Machine Intell. 6: 721–741
- George E. I., McCulloch R. E., 1993. Variable selection via Gibbs sampling. J. Am. Stat. Assoc. 88: 881–889 [Google Scholar]
- Gianola D., Fernando R. L., Stella A., 2006. Genomic-assisted prediction of genetic value with semiparametric procedures. Genetics 173: 1761–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianola D., de los Campos G., Hill W. G., Manfredi E., Fernando R., 2009. Additive genetic variability and the Bayesian alphabet. Genetics 183: 347–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianola, D., G. de los Campos, O. González-Recio, N. Long, H. Okut et al., 2010 Statistical learning methods for genome-based analysis of quantitative traits. World Genetic Congress Applied to Livestock Production, Leipzig, Germany, CD-ROM Communication 0014.
- Gilmour A., Gogel B., Cullis B., Thompson R., 2009. ASReml User Guide. VSN International, Hemel Hempstead, UK [Google Scholar]
- González-Recio O., Forni S., 2011. Genome-wide prediction of discrete traits using Bayesian regressions and machine learning. Genet. Sel. Evol. 43: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Recio O., Gianola D., Long N., Weigel K. A., Rosa G. J., et al. , 2008. Nonparametric methods for incorporating genomic information into genetic evaluations: an application to mortality in broilers. Genetics 178: 2305–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Recio O., Gianola D., Rosa G. J. M., Weigel K. A., Kranis A., 2009. Genome-assisted prediction of a quantitative trait measured in parents and progeny: application to food conversion rate in chickens. Genet. Sel. Evol. 41: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Recio O., Weigel K. A., Gianola D., Naya H., Rosa G. J. M., 2010. L2-Boosting algorithm applied to high-dimensional problems in genomic selection. Genet. Res. 92: 227–237 [DOI] [PubMed] [Google Scholar]
- Grattapaglia D., Resende M. D. V., 2011. Genomic selection in forest tree breeding. Tree Genet. Genomes 7: 241–255 [Google Scholar]
- Gredler, B., H. Schwarzenbacher, C. Egger-Danner, C. Fuerst, and R. Emmerling, 2010 Accuracy of genomic selection in dual purpose Fleckvieh cattle using three types of methods and phenotypes. 9th World Congress on Genetics Applied to Livestock Production, Leipzig, Germany. CD-ROM Communication 0907.
- Grisart B., Coppieters W., Farnir F., Karim L., Ford C., et al. , 2002. Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res. 12: 222–231 [DOI] [PubMed] [Google Scholar]
- Guo G., Lund M. S., Zhang Y., Su G., 2010. Comparison between genomic predictions using daughter yield deviation and conventional estimated breeding value as response variables. J. Anim. Breed. Genet. 127: 423–432 [DOI] [PubMed] [Google Scholar]
- Habier D., Fernando R. L., Dekkers J. M., 2007. The impact of genetic relationship information on genome-assisted breeding values. Genetics 177: 2389–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Tetens J., Seefried F. R., Lichtner P., Thaller G., 2010. The impact of genetic relationship information on genomic breeding values in German Holstein cattle. Genet. Sel. Evol. 42: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Fernando R., Kizilkaya K., Garrick D., 2011. Extension of the Bayesian alphabet for genomic selection. BMC Bioinformatics 12: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley C. S., Visscher P. M., 1998. Strategies to utilize marker-quantitative trait loci associations. J. Dairy Sci. 81(Suppl. 2): 85–97 [DOI] [PubMed] [Google Scholar]
- Hastie, T., R. Tibshirani, and J. Friedman, 2009 The Elements of Statistical Learning: Data Mining, Inference, and Prediction, Ed. 2. Springer-Verlag, Berlin/Heidelberg, Germany/New York.
- Hayashi T., Iwata H., 2010. EM algorithm for Bayesian estimation of genomic breeding values. BMC Genet. 11: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B., Goddard M. E., 2001. The distribution of the effects of genes affecting quantitative traits in livestock. Genet. Sel. Evol. 33: 209–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B. J., Bowman P. J., Chamberlain A. J., Goddard M. E., 2009a Invited review: genomic selection in dairy cattle: progress and challenges. J. Dairy Sci. 92: 433–443 [DOI] [PubMed] [Google Scholar]
- Hayes B., Bowman P., Chamberlain A., Verbyla K., Goddard M., et al. , 2009b Accuracy of genomic breeding values in multi-breed dairy cattle populations. Genet. Sel. Evol. 41: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffner E. L., Lorenz A. J., Jannink J.-L., Sorrells M. E., 2010. Plant breeding with genomic selection: gain per unit time and cost. Crop Sci. 50: 1681–1690 [Google Scholar]
- Heffner E. L., Jannink J.-L., Iwata H., Souza E., Sorrells M. E., 2011. Genomic selection accuracy for grain quality traits in biparental wheat populations. Crop Sci. 51: 2597–2606 [Google Scholar]
- Heslot N., Sorrells M. E., Jannink J. L., Yang H. P., 2012. Genomic selection in plant breeding: a comparison of models. Crop Sci. 52: 146–160 [Google Scholar]
- Hoerl A. E., Kennard R. W., 1970. Ridge regression: biased estimation for nonorthogonal problems. Technometrics 12: 55–67 [Google Scholar]
- Hospital F., 2009. Challenges for effective marker-assisted selection in plants. Genetica 136: 303–310 [DOI] [PubMed] [Google Scholar]
- Iwata H., Jannink J. L., 2011. Accuracy of genomic selection prediction in barley breeding programs: a simulation study based on the real single nucleotide polymorphism data of barley breeding lines. Crop Sci. 51: 1915–1927 [Google Scholar]
- Kearsey M. J., Farquhar A. G. L., 1998. QTL analysis in plants; where are we now? Heredity 80: 137–142 [DOI] [PubMed] [Google Scholar]
- Kimeldorf G. S., Wahba G., 1970. A correspondence between Bayesian estimation on stochastic processes and smoothing by splines. Ann. Math. Stat. 41: 495–502 [Google Scholar]
- Konstantinov K. V., Hayes B. J., 2010. Comparison of BLUP and Reproducing kernel Hilbert spaces methods for genomic prediction of breeding values in Australian Holstein Friesian cattle. Proceedings of the 9th World Congress on Genetics Applied to Livestock Production, Leipzig, Germany. CD-ROM Communication 0224
- Lee S. H., van der Werf J. H. J., Hayes B. J., Goddard M. E., Visscher P. M., 2008. Predicting unobserved phenotypes for complex traits from whole-genome SNP data. PLoS Genet. 4: e1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legarra A., Aguilar I., Misztal I., 2009. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 92: 4656–4663 [DOI] [PubMed] [Google Scholar]
- Legarra A., Robert-Granie C., Croiseau P., Guillaume F., Fritz S., 2011. Improved Lasso for genomic selection. Genet. Res. 93: 77–87 [DOI] [PubMed] [Google Scholar]
- Liu Z., Seefried F. R., Reinhardt F., Rensing S., Thaller G., et al. , 2011. Impacts of both reference population size and inclusion of a residual polygenic effect on the accuracy of genomic prediction. Genet. Sel. Evol. 43: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long N., Gianola D., Rosa G. J. M., Weigel K. A., 2011a. Dimension reduction and variable selection for genomic selection: application to predicting milk yield in Holsteins. J. Anim. Breed. Genet. 128: 247–257 [DOI] [PubMed] [Google Scholar]
- Long N., Gianola D., Rosa G. J. M., Weigel K. A., 2011b. Application of support vector regression to genome-assisted prediction of quantitative traits. Theor. Appl. Genet. 123: 1065–1074 [DOI] [PubMed] [Google Scholar]
- Lorenzana R. E., Bernardo R., 2009. Accuracy of genotypic value predictions for marker-based selection in biparental plant populations. Theor. Appl. Genet. 120: 151–161 [DOI] [PubMed] [Google Scholar]
- Luan T., Woolliams J. A., Lien S., Kent M., Svendsen M., et al. , 2009. The accuracy of genomic selection in Norwegian Red cattle assessed by cross-validation. Genetics 183: 1119–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, M. S., G. Sahana, D. J. De Koning, G. Su, and Ö. Carlborg, 2009 Comparison of analyses of the QTLMAS XII common dataset. I: Genomic selection. BMC Proc. 3(Suppl. 1): S1. [DOI] [PMC free article] [PubMed]
- Lynch M., Ritland K., 1999. Estimation of pairwise relatedness with molecular markers. Genetics 152: 1753–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen, P. A., and J. Jensen, 2010 A User’s Guide to DMU. A Package for Analysing Multivariate Mixed Models. Version 6, release 5.0. University of Aarhus, Denmark. http://www.dmu.agrsci.dk/dmuv6_guide-R4-6-7.pdf.
- Makowsky R., Pajewski N. M., Klimentidis Y. C., Vazquez A. I., Duarte C. W., et al. , 2011. Beyond missing heritability: prediction of complex traits. PLoS Genet. 7: e1002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H. E., 2009. Accuracy of breeding values of ‘unrelated’ individuals predicted by dense SNP genotyping. Genet. Sel. Evol. 41: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T., Goddard M., 2010. Accurate prediction of genetic values for complex traits by whole-genome resequencing. Genetics 185: 623–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H., Hayes B. J., Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T., Solberg T. R., Shepherd R., Woolliams J. A., 2009. A fast algorithm for BayesB type of prediction of genome-wide estimates of genetic value. Genet. Sel. Evol. 41: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser G., Tier B., Crump R. E., Khatkar M. S., Raadsma H. W., et al. , 2009. A comparison of five methods to predict genomic breeding values of dairy bulls from genome-wide SNP markers. Genet. Sel. Evol. 41: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrode R., Coffey M. P., Stranden I., Meuwissen T. H. E., Van Kaam J. B. C. H. M., et al. , 2010. A comparison of various methods for the computation of genomic breeding values of dairy bulls using software at Genomicselection.net. Proceedings of the 9th World Congress on Genetics Applied to Livestock Production, Leipzig, Germany. CD-ROM Communication 0518
- Mujibi F. D. N., Nkrumah J. D., Durunna O. N., Grant J. R., Mah J., et al. , 2011. Associations of marker panel scores with feed intake and efficiency traits in beef cattle using preselected single nucleotide polymorphisms. J. Anim. Sci. 89: 3362–3371 [DOI] [PubMed] [Google Scholar]
- Nadaf, J., V. Riggio, T.-P. Yu, and R. Pong-Wong, 2012 Effect of the prior distribution of SNP effects on the estimation of total breeding value. BMC Proc. 6(Suppl. 2): S6. [DOI] [PMC free article] [PubMed]
- Nejati-Javaremi A., Smith C., Gibson J. P., 1997. Effect of total allelic relationship on accuracy of evaluation and response to selection. J. Anim. Sci. 75: 1738–1745 [DOI] [PubMed] [Google Scholar]
- Ober U., Erbe M., Long N. Y., Porcu E., Schlather M., et al. , 2011. Predicting genetic values: a kernel-based best linear unbiased prediction with genomic data. Genetics 188: 695–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okut H., Gianola D., Rosa G. J. M., Weigel K. A., 2011. Prediction of body mass index in mice using dense molecular markers and a regularized neural network. Genet. Res. 93: 189–201 [DOI] [PubMed] [Google Scholar]
- Ostersen T., Christensen O., Henryon M., Nielsen B., Su G., et al. , 2011. Deregressed EBV as the response variable yield more reliable genomic predictions than traditional EBV in pure-bred pigs. Genet. Sel. Evol. 43: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T., Casella G., 2008. The Bayesian Lasso. J. Am. Stat. Assoc. 103: 681–686 [Google Scholar]
- Pérez P., de los Campos G., Crossa J., Gianola D., 2010. Genomic-enabled prediction based on molecular markers and pedigree using the Bayesian Linear Regression package in R. Plant Gen. 3: 106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piyasatian N., Fernando R. L., Dekkers J. C. M., 2007. Genomic selection for marker-assisted improvement in line crosses. Theor. Appl. Genet. 115: 665–674 [DOI] [PubMed] [Google Scholar]
- Pong-Wong R., George A. W., Woolliams J. A., Haley C. S., 2001. A simple and rapid method for calculating identity-by-descent matrices using multiple markers. Genet. Sel. Evol. 33: 453–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce J. E., Gredler B., Bolormaa S., Bowman P. J., Egger-Danner C., et al. , 2011. Short communication: genomic selection using a multi-breed, across-country reference population. J. Dairy Sci. 94: 2625–2630 [DOI] [PubMed] [Google Scholar]
- Pszczola, M., T. Strabel, A. Wolc, S. Mucha, and M. Szydlowski, 2011 Comparison of analyses of the QTLMAS XIV common dataset. I: Genomic selection. BMC Proc. 5(Suppl. 3): S1. [DOI] [PMC free article] [PubMed]
- Resende M. F. R., Jr, Muñoz P., Resende M. D. V., Garrick D. J., Fernando R. L., et al. , 2012. Accuracy of genomic selection methods in a standard dataset of Loblolly pine (Pinus taeda L.). Genetics 190: 1503–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritland K., 1996. A marker-based method for inferences about quantitative inheritance in natural populations. Evolution 50: 1062–1073 [DOI] [PubMed] [Google Scholar]
- Ritland K., 2002. Extensions of models for the estimation of mating systems using n independent loci. Heredity 88: 221–228 [DOI] [PubMed] [Google Scholar]
- Saatchi M., McClure M. C., McKay S. D., Rolf M. M., Kim J. W., et al. , 2011. Accuracies of genomic breeding values in American Angus beef cattle using k-means clustering for cross-validation. Genet. Sel. Evol. 43: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, X., L. Rönnegård, and Ö. Carlborg, 2011 Hierarchical likelihood opens a new way of estimating genetic values using genome-wide dense marker maps. BMC Proc. 5(Suppl. 3): S14. [DOI] [PMC free article] [PubMed]
- Shepherd R., Meuwissen T., Woolliams J., 2010. Genomic selection and complex trait prediction using a fast EM algorithm applied to genome-wide markers. BMC Bioinformatics 11: 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg T. R., Sonesson A. K., Woolliams J. A., Meuwissen T. H. E., 2009. Reducing dimensionality for prediction of genome-wide breeding values. Genet. Sel. Evol. 41: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller M., 1978. The use of loci associated with quantitative effects in dairy cattle improvement. Anim. Prod. 27: 133–139 [Google Scholar]
- Soller M., Plotkin-Hazan J., 1977. The use of marker alleles for the introgression of linked quantitative alleles. Theor. Appl. Genet. 51: 133–137 [DOI] [PubMed] [Google Scholar]
- Strandén I., Christensen O. F., 2011. Allele coding in genomic evaluation. Genet. Sel. Evol. 43: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R., 1996. Regression shrinkage and selection via the lasso. J. R. Stat. Soc., B 58: 267–288 [Google Scholar]
- Usai M. G., Goddard M. E., Hayes B. J., 2009. LASSO with cross-validation for genomic selection. Genet Res. Camb 91: 427–436 [DOI] [PubMed] [Google Scholar]
- VanRaden P., 2007. Genomic measures of relationship and inbreeding. Interbull Bull. 37: 33–36. [Google Scholar]
- VanRaden P., 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91: 4414–4423 [DOI] [PubMed] [Google Scholar]
- VanRaden P. M., Sullivan P. G., 2010. International genomic evaluation methods for dairy cattle. Genet. Sel. Evol. 42: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P. M., Wiggans G. R., 1991. Derivation, calculation, and use of national animal model information. J. Dairy Sci. 74: 2737–2746 [DOI] [PubMed] [Google Scholar]
- VanRaden P. M., Van Tassell C. P., Wiggans G. R., Sonstegard T. S., Schnabel R. D., et al. , 2009. Invited review: reliability of genomic predictions for North American Holstein bulls. J. Dairy Sci. 92: 16–24 [DOI] [PubMed] [Google Scholar]
- Vazquez A. I., Rosa G. J. M., Weigel K. A., de los Campos G., Gianola D., et al. , 2010. Predictive ability of subsets of single nucleotide polymorphisms with and without parent average in US Holsteins. J. Dairy Sci. 93: 5942–5949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbyla K. L., Hayes B., Bowman P. J., Goddard M. E., 2009. Short note: accuracy of genomic selection using stochastic search variable selection in Australian Holstein Friesian dairy cattle. Genet. Res. 91: 307–311 [DOI] [PubMed] [Google Scholar]
- Villanueva B., Pong-Wong R., Fernández J., Toro M., 2005. Benefits from marker-assisted selection under an additive polygenic genetic model. J. Anim. Sci. 83: 1747–1752 [DOI] [PubMed] [Google Scholar]
- Weigel K. A., de los Campos G., González-Recio O., Naya H., Wu X. L., et al. , 2009. Predictive ability of direct genomic values for lifetime net merit of Holstein sires using selected subsets of single nucleotide polymorphism markers. J. Dairy Sci. 92: 5248–5257 [DOI] [PubMed] [Google Scholar]
- Whittaker J. C., Thompson R., Denham M. C., 2000. Marker-assisted selection using ridge regression. Genet. Res. 75: 249–252 [DOI] [PubMed] [Google Scholar]
- Wiggans G. R., VanRaden P. M., Cooper T. A., 2011. The genomic evaluation system in the United States: past, present, future. J. Dairy Sci. 94: 3202–3211 [DOI] [PubMed] [Google Scholar]
- Winter A., Krämer W., Werner F. A. O., Kollers S., Kata S., et al. , 2002. Association of a lysine-232/alanine polymorphism in a bovine gene encoding acyl-CoA:diacylglycerol acyltransferase (DGAT1) with variation at a quantitative trait locus for milk fat content. Proc. Natl. Acad. Sci. USA 99: 9300–9305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenburg D., Melzer N., Reinsch N., 2011. Including non-additive genetic effects in Bayesian methods for the prediction of genetic values based on genome-wide markers. BMC Genet. 12: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolc A., Arango J., Settar P., Fulton J. E., O’Sullivan N. P., et al. , 2011a Persistence of accuracy of genomic estimated breeding values over generations in layer chickens. Genet. Sel. Evol. 43: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolc A., Stricker C., Arango J., Settar P., Fulton J. E., et al. , 2011b Breeding value prediction for production traits in layer chickens using pedigree or genomic relationships in a reduced animal model. Genet. Sel. Evol. 43: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Tempelman R. J., 2012. A Bayesian antedependence model for whole genome prediction. Genetics 190: 1491–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi N., Banerjee S., 2009. Hierarchical generalized linear models for multiple quantitative trait locus mapping. Genetics 181: 1101–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Meuwissen T. H. E., 2011. Using the Pareto principle in genome-wide breeding value estimation. Genet. Sel. Evol. 43: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Liu J., Ding X., Bijma P., De Koning D. J., et al. , 2010. Best linear unbiased prediction of genomic breeding values using a trait-specific marker-derived relationship matrix. PLoS ONE 5: e12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Gowda M., Longin F., Würschum T., Ranc N., et al. , 2012. Impact of selective genotyping in the training population on accuracy and bias of genomic selection. Theor. Appl. Genet. DOI: 10.1007/s00122-012-1862-2 [DOI] [PubMed] [Google Scholar]
- Zhong S., Dekkers J. C. M., Fernando R. L., Jannink J.-L., 2009. Factors affecting accuracy from genomic selection in populations derived from multiple inbred lines: a barley case study. Genetics 182: 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F., Huang H., Lee S., Hoeschele I., 2010. Nonparametric Bayesian variable selection with applications to multiple quantitative trait loci mapping with epistasis and gene–environment interaction. Genetics 186: 385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., Hastie T., 2005. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B Stat. Methodol. 67: 301–320 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.