Abstract
Significant advances in genomics underscore the importance of targeted mutagenesis for gene function analysis. Here we have developed a scheme for long-range targeted manipulation of genes in the Drosophila genome. Utilizing an attP attachment site for the phiC31 integrase previously targeted to the nbs gene, we integrated an 80-kb genomic fragment at its endogenous locus to generate a tandem duplication of the region. We achieved reduction to a single copy by inducing recombination via a site-specific DNA break. We report that, despite the large size of the DNA fragment, both plasmid integration and duplication reduction can be accomplished efficiently. Importantly, the integrating genomic fragment can serve as a venue for introducing targeted modifications to the entire region. We successfully introduced a new attachment site 70 kb from the existing attP using this two-step scheme, making a new region susceptible to targeted mutagenesis. By experimenting with different placements of the future DNA break site in the integrating vector, we established a vector configuration that facilitates the recovery of desired modifications. We also show that reduction events can occur efficiently through unequal meiotic crossing over between the large duplications. Based on our results, we suggest that a collection of 1200 lines with attachment sites inserted every 140 kb throughout the genome would render all Drosophila genes amenable to targeted mutagenesis. Excitingly, all of the components involved are likely functional in other eukaryotes, making our scheme for long-range targeted manipulation readily applicable to other systems.
Keywords: I-SceI endunuclease, genome-wide manipulation, homologous recombination, phiC31 integrase, targeted mutagenesis
DROSOPHILA melanogaster is an excellent organism for the genetic analysis of cellular and developmental processes. For the past two decades, methods of gene targeting by homologous recombination have facilitated the engineering of crafted mutations (Gloor et al. 1991; Rong and Golic 2000; Rong et al. 2002; Gong and Golic 2003). The wealth of information provided by bioinformatic analyses has further aided geneticists in the design of mutations. Gene targeting in flies, however, is quite labor intensive, discouraging its use in systematic mutagenesis, which involves more than one targeted allele, although success stories have revealed the benefits of such approaches (Gronke et al. 2010; Spitzweck et al. 2010).
Recently, several laboratories have demonstrated that, by combining homologous recombination with the efficient phiC31-mediated site-specific recombination method, the need for repetitive gene targeting of individual alleles to the endogenous locus can be eliminated (Gao et al. 2008; Huang et al. 2009; Weng et al. 2009; Iampietro et al. 2010). We developed the site-specific integrase-mediated repeated targeting (SIRT) method, in which an attP attachment site for phiC31 integrase is first targeted to the genome in the proximity of the locus of interest using gene targeting by homologous recombination. Further modifications are carried out by integration of a plasmid that carries the modified locus and the “partner” attachment site attB. Recombination between chromosomal attP and plasmid-born attB is irreversible and highly efficient in fly embryos expressing phiC31 (Groth et al. 2004; Bischof et al. 2007). Integration of the plasmid leads to duplication of the chromosomal locus, which is subsequently reduced to one copy by repair of an induced DNA double-strand break (Rong et al. 2002; Gao et al. 2008).
One versatile aspect of SIRT not shared by traditional gene-targeting methods is that a particular attP site can be used to modify not only one’s gene of interest, but all of the genes in its vicinity. In theory, the range amenable to SIRT manipulation from an attP insertion is limited only by the size of the genomic clone in the attB-containing plasmid. Encouragingly, Venken et al. developed the P[acman] system for the introduction of large DNA fragments into the fly genome using phiC31 and achieved integration of genomic fragments on the order of 100 kb with remarkable efficiencies (Venken et al. 2006). We took advantage of this technical innovation and set out to determine whether a preexisting att site could render a surrounding genomic region of similar size amenable to targeted modification. Our study reveals that modifying a genomic region 70 kb away from the attachment site can be quite efficient. In addition, both integration and reduction reactions for our large genomic fragment occurred at a frequency comparable to those previously achieved. Furthermore, reduction events could be recovered from spontaneous events that occur through meiotic recombination. We took the opportunity to introduce a new att site 70 kb downstream from the original one, further expanding the mutable region by at least another 70 kb. We propose to combine our targeting scheme with transposon-delivered att sites genome-wide to render all Drosophila genes amenable to targeted mutagenesis.
Materials and Methods
Primer sequences are listed in Supporting Information, Table S1.
Construction of the pWalkman vector
The unique NheI site in pP[acman] was used to introduce an FRT next to the 3′ inverted repeat of the P element as a product of annealing two oligos (NheFRT48-plus and NheFRT48-minus) with overhangs compatible to NheI. The unique SphI site was used to clone an FRT next to the 5′ inverted repeat of the P element. The FRT oligos used were SphFRT48-plus and SphFRT48-minus. A clone was selected in which the two FRTs are in a directed orientation. The unique PmeI site in pP[acman] was used to clone an I-SceI cut site with the oligos ScBaNhSpMlBsSb+ and ScBaNhSpMlBsSb−. This position corresponds to IS1 in Figure 1. The I-SceI oligos also include restriction sites for BamHI, NheI, SphI, MluI, BsiWI, and SbfI enzymes immediately downstream of the I-SceI cut site but upstream of the existing multiple cloning sites in pP[acman].
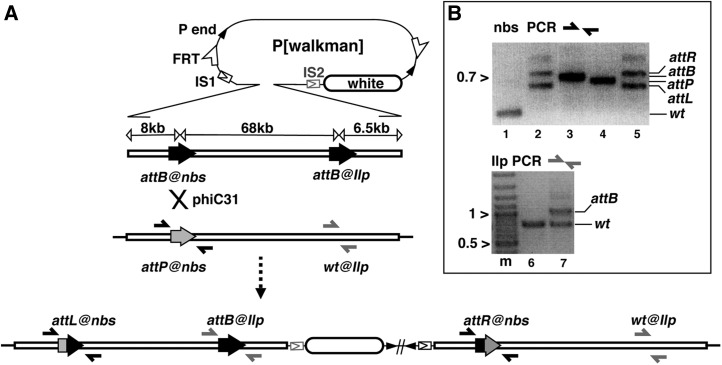
Figure 1 .
Generating an 80-kb duplication by site-specific integration. (A) Integration schematic. At the top is the pWalkman vector built on the pP[acman] backbone, which contains P-element ends (solid arrowheads), a white maker (open oval), FRT sites (half-arrows), and an I-SceI cut site (IS1 in construct pWalkman{nbs-Ilp} IS1 and IS2 in pWalkman{nbs-Ilp} IS2). Below the pWalkman vector is the 82-kb nbs-Ilp insert, which contains two attB sites (solid arrows), one at each locus, separated by 68 kb. Below the 82-kb insert is the chromosomal region with an attP (shaded arrow) targeted to nbs. Note that the chromosomal region of Ilp does not contain any att site (wt). The positions for two sets of PCR primers (solid and shaded half arrows) used in B are indicated. A phiC31-mediated recombination (“X”) between attP and attB at nbs gives rise to the duplication depicted at the bottom. Only the integration of pWalkman{nbs-Ilp}IS1 is shown. The FRTs are not shown but are in close proximity to the P-element ends. (B) Representative results for diagnostic PCR tests. Marker size in kilobases is indicated to the left of the gel images. (Top) PCR analyses with nbs primers. The PCR templates are the following: lane 1, a wild-type line without any attachment site; lane 2, a duplication line from previous plasmid integration at attP@nbs; lane 3, plasmid DNA of pWalkman(nbs-Ilp)IS1; lane 4, the attP@nbs starting line; and lane 5, an integration line with the 80-kb duplication. (Bottom) Results of PCR analyses with Ilp primers. Lane “m” contains the marker. The PCR templates are the following: lane 6, a wild-type line, and lane 7, an integration line from this study.
Construction of pWalkman{nbs-Ilp}
A BAC clone (RP98-7E7) spanning the nbs-Ilp region was obtained from BacPac Resources (http://bacpac.chori.org). The gap repair method based on bacterial recombineering (Venken et al. 2006) was used to subclone the 82-kb nbs-Ilp region into the AscI and EcoRI sites in pWalkman. A detailed description of cloning by recombineering, including the insertion of various DNA elements, can be found in Supporting Information, File S1.
Fly genetics
The pWalkman constructs were injected into embryos with a vasa-driven phiC31 transgene (Bischof et al. 2007) and an attP insertion at nbs (Gao et al. 2008). Crossing schemes are presented in File S2.
Results
Experimental design
We previously used SIRT to create an allelic series for the nbs locus in Drosophila (Gao et al. 2008). To determine if the same attP site could be used to modify a remote region, we set out to target a region ∼70 kb from nbs.
We generated the pWalkman vector, which is based on the P[acman] vector system that was developed to facilitate cloning and large-scale production of plasmids containing large genomic inserts (Venken et al. 2006). pWalkman includes additional features to facilitate SIRT manipulation (Figure 1). First, a cut site for the rare cutting I-SceI enzyme was added to allow for double-strand break (DSB)-induced reduction. I-SceI is more advantageous as a choice of DSB-inducing enzyme than the I-CreI used in previous reduction reactions because overproduction of I-CreI reduces viability and fertility in flies (Rong et al. 2002). Second, two FRT sites were placed in a directed orientation to enable FLP-mediated excision of the plasmid backbone after plasmid integration. Finally, additional restriction sites were added to ease cloning of inserts.
While this targeting system could be used to mutate a specific gene, we chose instead to insert another attachment site at a distant “target region.” We reasoned that this would create a valuable resource for those interested in using SIRT to modify their genes of interest in the vicinity of the new attachment site. We chose the region containing CG8177, CG14168, and a cluster of four Ilp genes as the target region for the new att site (from here on referred to as the Ilp locus). Since the integrating plasmid has an attB site at the nbs locus (attB@nbs), we decided to target an attB instead of an attP to the Ilp locus (attB@Ilp) that is 68 kb away from attB@nbs. Since the phiC31 integrase does not act on identical attachment sites, our design would eliminate any complications due to intramolecular recombination between sites on the same plasmid.
To generate the construct, the 82-kb genomic fragment from the nbs-Ilp region was subcloned into pWalkman, and attB sites were added to nbs and Ilp loci (Figure 1). All three cloning steps were accomplished by using bacterial recombineering (Copeland et al. 2001). In the final construct, which we named pWalkman{nbs-Ilp}, the 8-kb fragment to the left of attB@nbs and the 6.5-kb one to the right of attB@Ilp were included to extend the homology necessary for reduction of the target duplication by homologous recombination (Figures 1 and 2). We have constructed two versions of pWalkman{nbs-Ilp} in which the only difference is the placement of the I-SceI cut site (IS). In pWalkman{nbs-Ilp}IS1, the I-SceI cut site is at position IS1, while in pWalkman{nbs-Ilp}IS2, it is at position IS2 (Figure 1).
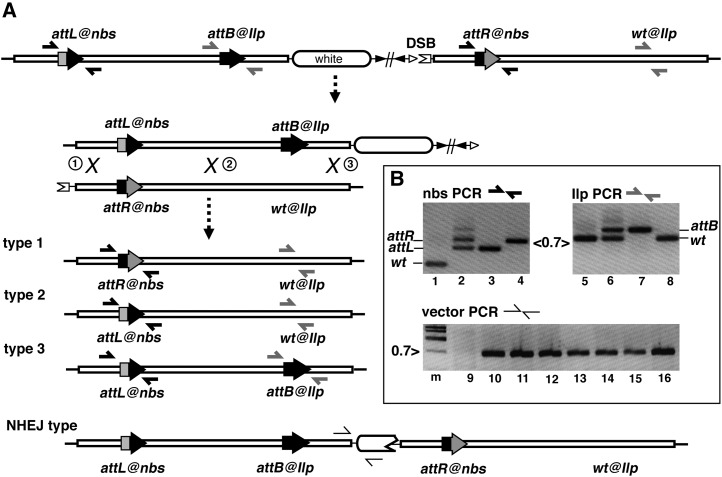
Figure 2 .
I-SceI-induced reduction. (A) Reduction schematic. At the top is the 80-kb duplication resulting from integration of pWalkman{nbs-Ilp}IS1. The I-SceI endonuclease creates a DSB at IS1. The two nbs-Ilp regions align with three possible positions of recombination (“X”). The repair of the DSB gives rise to four types of events diagrammed below, with the first three being the product of recombination between the duplicated copies. The numbers in circles for the potential recombination sites correspond to the resulting types of reduction products. The positions of PCR primer sets are indicated (half arrows). In the NHEJ-type product, the white gene is disrupted (“broken” oval). The primer positions for “vector PCR” in B are indicated as half arrows. (B) Representative results for diagnostic PCR tests. The PCR templates for the top panels are the following: wt for lanes 1 and 5; a duplicated line for lanes 2 and 6; a type 3 reduction for lanes 3 and 7; a type 1 reduction for lane 4; and a type 2 event for lane 8. The 0.7-kb marker is indicated between the gel pictures. The PCR templates for the bottom panel are a reduction line for lane 9 and different NHEJ-type lines for lanes 10–16. Lane “m” represents markers with size indicated to the left in kilobases.
An 80-kb tandem duplication
Embryo injections of the pWalkman{nbs-Ilp}IS1 construct yielded 60 fertile adults, and 3 of these produced red-eyed offspring, which indicates an integration frequency of 5% (3/60, Table 1). All red-eyed progeny (15 in total) were used to establish homozygous lines that were subjected to genetic and molecular characterization. Integration of the plasmid at nbs would place the white marker gene on chromosome 3, a result observed for all 15 lines. It would also generate a duplication of the 80-kb nbs-Ilp region with the left copy having an attL at nbs and the right copy having an attR at nbs (Figure 1). Since all four att elements (attB, attP, attR, attL) differ in size, we used PCR reactions with the same pair of flanking primers to identify each site. We present a sample of the results in Figure 1B. In addition to a wild-type (wt) line (lane 1), we used two additional control lines for the nbs PCR, including the starting line carrying attP@nbs (lane 4) and an integration line from our previous study (Gao et al. 2008) as a reference for the size of attL and attR sites (lane 2). While the starting line produced a PCR product of ∼650 bp for attP, an integration line from this study yielded a doublet consisting of an ∼600-bp product for attL and an ∼750-bp product for attR (lane 5), a result identical to that obtained with a previous integration line (lane 2). At the Ilp locus, the wt line produced an ∼800 bp product (lane 6). An integration line generated a doublet with an additional ∼1050-bp product for attB (lane 7). These results were confirmed for 3 of the 15 lines that originated from two independent parents.
Table 1 . Frequency of reduction events.
| Reduction typee |
||||||
|---|---|---|---|---|---|---|
| Constructa | Source of DSBb | White-loss frequencyc | Reduction frequencyd | 1 | 2 | 3 |
| pWalkman{nbs-Ilp} | ||||||
| IS1 | I-SceI | 10% (24/230) | 29% (6/21) | 5 | 1 | 0 |
| P transposase | 13% (19/150) | 37% (7/19) | 6 | 1 | 0 | |
| Spontaneousf | ||||||
| Female | 0.1% (39/35,000) | 37/37 | 1 | 35 | 1 | |
| Femaleg | 0.1% (22/15,000) | 8/8 | 1 | 7 | 0 | |
| Male | 0.0% (0/8000) | 0 | ||||
| IS2 | I-SceI | 13% (45/350) | 39% (13/33) | 6 | 1 | 6 |
| Spontaneousf female | 0.1% (11/13,000) | 11/11 | 0 | 11 | 0 | |
Constructs used for site-specific integration.
The source of DSB that led to the reported reductions.
For induced events, the frequency was calculated by dividing the number of male parents yielding white-eyed progeny over the total number of males tested. The actual counts are included in parentheses. For spontaneous events, the frequency was calculated by dividing the number of white-eyed offspring over the total number of progeny. The counts are included in parentheses.
Calculated by dividing the number of confirmed reductions over all white-loss events tested by molecular means.
For the molecular structure of different types of reduction events, see Figure 2.
Spontaneous events are categorized according to the gender of the duplication-carrying parents.
Flanking recessive markers were used to demonstrate homolog exchanges (see text).
The rest of the lines did not pass this simple PCR test. Results from some of the PCR tests are shown in Figure S1. We propose that these lines resulted from rearrangement of the nbs-Ilp region. Such rearrangements can be brought about by multiple exchanges between attB and attP at the two loci on different homologous chromosomes or on sister chromatids. For example, once the plasmid was integrated at nbs, attB@Ilp could engage in a second round of phiC-31-mediated exchange with attP@nbs on the homologous chromosome since the plasmid-injected stock is homozygous for that attP insertion. Instead of further characterizing these events, we chose to focus our study on the integration lines that passed the PCR test.
Target reduction induced by an I-SceI-generated DSB
To reduce the duplication to a single copy at the nbs-Ilp region, we followed a reduction procedure (File S2). We initiated reduction by inducing an I-SceI cut in the vector sequences. If the repair of the resulting DSB proceeds through homologous recombination (HR) between the two nbs-Ilp copies, the repair product would be a single-copy locus (Figure 2). We previously observed that the plasmid backbone included in the phiC31-mediated integration decreased efficiency of reduction (Gao et al. 2008). For this reason, we performed a FLP-mediated excision of the vector backbone that is flanked by FRT sites (Figure 1) without deleting any sequence from the white marker, prior to proceeding with the reduction step.
Potential germline reduction events were recovered as white-eyed progeny. These progeny occurred in ∼10% of germlines that carried both the duplication and a source of I-SceI (Table 1). As I-SceI-induced white loss happens in the mitotic component of the germline (Rong and Golic 2000; Rong et al. 2002), we established homozygous lines for only one white progeny from each white-producing germline to ensure that only independent events were being evaluated. We verified the reduction outcomes for these lines using the same PCR strategy employed to verify the duplications (Figure 1). We expected that the single reduced copy would fall into three possible reduction types (Figure 2). If HR occurs within the 8-kb region to the left of nbs, the reduction would carry attR@nbs and be wt at the Ilp locus (reduction type 1). If HR occurs within the 68-kb region between nbs and Ilp, reduction would carry attL@nbs and be wt at Ilp (type 2). Only if HR occurs within the most distal 6.5 kb of the nbs-Ilp homology would it carry the desired attB@Ilp alongside attL@nbs (type 3). Representative products for these PCRs are shown in Figure 2B.
Table 1 summarizes the reduction results obtained from using the pWalkman{nbs-Ilp}IS1 construct. Six of 21 white-loss lines were precise reduction events based on PCR tests. Five of the 6 resulted from a type 1 reduction. The one exception resulted from a type 2 reduction. While we did not obtain any type 3 reduction events in this round, these desirable events were ultimately recovered in experiments described in the next section.
Unexpectedly, we observed a fourth but predominant type from our white-loss lines. For 15 of the 21 lines, the PCR results looked identical to those from the original duplication lines: a doublet in PCR tests for both the nbs and the Ilp loci. We hypothesized that these lines resulted from imprecise nonhomologous end joining (NHEJ) repair of the induced DSB where the white marker had suffered sequence loss before the break was ligated. This hypothesis was confirmed in 14 lines, in which a positive PCR result for the presence of vector sequences was evident (bottom panel in Figure 2B).
We have thus demonstrated two important aspects of this system that suggest that it will be easily applicable for gene targeting. First, we have found that reduction of an 80-kb duplication by DSB-induced HR can be efficient, with 10% of the males tested in our experiment resulting in white-loss events, of which 29% were reduction events. Second, we showed that the types of reduction events can be distinguished by a simple PCR test.
To provide further evidence supporting the accuracy of our PCR-based tests, we genotyped several lines from the reduction screen with regard to two single-nucleotide polymorphisms (SNPs) between the starting nbs-Ilp region on the chromosome and the same region, but originating from the BAC clone. We assayed a C/T SNP at position ∼35 kb (p35) and a single T insertion/deletion SNP at position ∼44 kb (p44) (see Figure 3 for approximate positions). As shown in Figure S2, both SNPs are heterozygous in nonreduced lines (NHEJ type), consistent with their carrying the duplication, while reduced lines are homozygous at both positions.
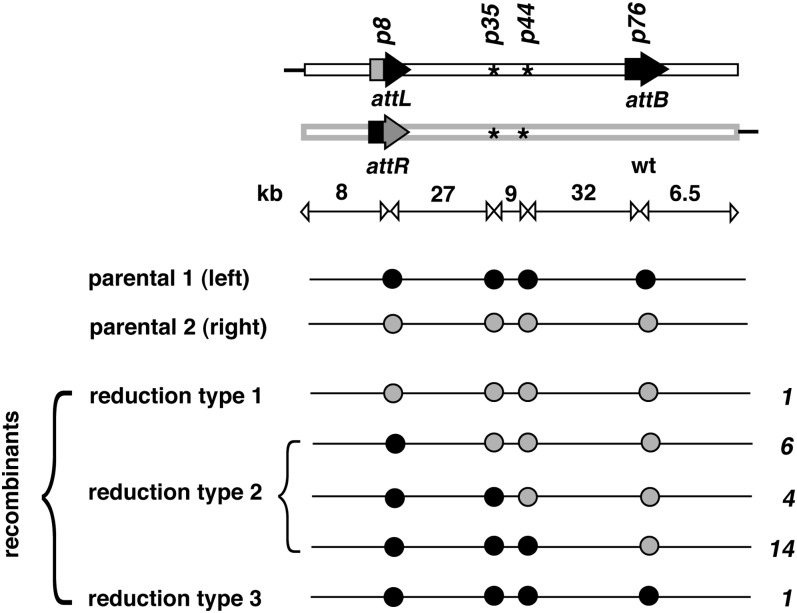
Figure 3 .
Spontaneous products are crossovers between the two duplicated copies. The two parental copies of the nbs-Ilp region are shown on the top with the four pieces of sequence heterology labeled according to their approximate distance from the left end of the nbs-Ilp region. The physical distance between heterologous sites is shown in kilobases below the diagram for the parental copies. (Bottom) Another representation of the nbs-Ilp parental copies and their five crossover products with each heterology depicted as a solid circle. The three types of reduction product have been defined in Figure 2. The number to the right of each product represents the number of lines identified for each outcome.
Effect of I-SceI cut-site placement on the recovery of reduction types
Our inability to recover type 3 reduction events that retain attB@Ilp prompted us to consider whether the IS1 position for the I-SceI cut site in pWalkman{nbs-Ilp}IS1 favors the recovery of type 1 reduction, in which the Ilp locus remains wt. To test this hypothesis, we generated the pWalkman{nbs-Ilp}IS2 construct that carries the I-SceI cut site at position IS2, on the other side of white and closer to attB@Ilp than IS1. This new construct was then taken through the same series of experiments as pWalkman{nbs-Ilp}IS1 to recover attB@Ilp in the reduction events.
The integration frequency for pWalkman{nbs-Ilp}IS2 was 6% (14 independent events of 220 fertile crosses). As before, white mapped to the third chromosome in all 14 lines. Of the 10 lines that we tested by PCR, 3 showed PCR results consistent with an un-rearranged duplication, and one of these lines was taken through the reduction procedure.
Table 1 shows the frequencies for reduction with the new construct. Recovery of white-loss events occurred at a similar frequency to that for the previous construct (13%, 45/350 germlines). As was observed previously, more than half (20/33) of the analyzed white-loss lines remained in a state of duplication (NHEJ type, Figure 2). Remarkably, this time the reaction was not biased against type 3 reduction, resulting in recovery of six lines carrying attB@Ilp out of 13 confirmed reduction events. Therefore, by putting the I-SceI cut site on the side of the vector that is closer to the desired modification (attB@Ilp) with respect to white, we were able to improve its recovery in the final reduced locus from 0 to 46% (6/13).
We also wanted to determine whether an inhibition of NHEJ DSB repair would result in an increase in reduction frequency. For that purpose, we repeated the reduction procedure in a background mutant for DNA ligase 4 (lig4, File S2). Of 330 lig4 mutant males tested, 49 gave white-eyed progeny (15%). Of 10 white-loss events tested by PCR, 2 were reduction events (20%), with both of them carrying the desired attB@Ilp modification. Therefore, the reduction frequencies recovered from the lig4 mutant background are very similar to those obtained in wt males, suggesting that Ligase 4 is not essential for generating the NHEJ-type events in our system.
Reductions induced by P-element transposase
As another way to initiate reduction, we explored the use of the P-element transposase to generate the DSBs. This strategy is possible because pWalkman sequences contain two P-element ends. Although these P-element ends are in an “inside-out” orientation when compared to their natural orientation, they can nevertheless be acted on by the P transposase to generate a DSB (Liang and Sved 2009). To test the efficacy of this approach, we introduced a source of P transposase into the duplicated lines and screened for potential reduction events as before. We observed a white-loss frequency of 13% and a bias toward type 1 reduction (Table 1), similarly to what we obtained using I-SceI.
Recovering reduction by spontaneous recombination
During the course of this study, we observed white-eyed progeny from flies carrying the 80-kb duplication without exposure to I-SceI or the P transposase. We suspected that these events could result from spontaneous recombination between the copies in the nbs-Ilp region. We envisioned three possible modes of crossing over that could lead to the loss of white, including crossovers (1) between the two nbs-Ilp copies on the same chromatid, (2) between copies on sister chromatids but out of register, and (3) between copies on homologous chromosomes also out of register (unequal crossing over). To recover more of these spontaneous events for molecular analyses, we crossed female flies homozygous for the duplication to white males and looked for white-eyed individuals among the heterozygous progeny. Using the pWalkman{nbs-Ilp}IS1 construct, we recovered 39 white individuals from 35,000 offspring (∼0.1%, Table 1). We tested 37 of these by the same PCR scheme as shown in Figure 1, and all were confirmed to be reduction events. One of them contained attB@Ilp. We also observed a strong bias toward one reduction type, specifically type 2 (35/37), which resulted from recombination within the 68-kb region between nbs and Ilp. We confirmed that spontaneous reductions occurred at the same rate of 0.1% in the germlines of females carrying the duplication made from the pWalkman{nbs-Ilp}IS2 construct. All 11 recovered events resulted from a type 2 reduction. In our previous experience with SIRT at the nbs locus, we did not observe any spontaneous white-loss events. However, our prior experimental setup included a smaller 5.5-kb duplication of the region (Gao et al. 2008). Our results here suggest that the presence of a larger duplication can induce another means to recover reduction products via spontaneous recombination.
Spontaneous events are results of meiotic recombination
We hypothesized that the spontaneous recombination events observed in our screen resulted from unequal crossing over between homologous chromosomes in meiosis. We made two observations consistent with this hypothesis. First, most of the events (35/37) were of type 2, which arise from crossovers within the largest portion of homology between the two duplicated copies (68 of 82 kb). Second, we did not observe any presumed NHEJ type events (Table 1), which is consistent with the fact that NHEJ is not a major pathway for the repair of meiotic DSBs (Joyce et al. 2012).
Nonetheless, the nature of the spontaneous events could be accounted for by both mitotic and meiotic events. We set out to further distinguish between these two possibilities. First, a similar screen did not yield any white-eyed events in 8000 progeny from males homozygous for the duplication (Table 1). This is consistent with the fact that meiotic recombination is limited to Drosophila females. Second, we found that spontaneous reduction events are always accompanied by the exchange of flanking markers on homologous chromosomes, a hallmark of meiotic recombination. To track the exchange of flanking markers, we generated females homozygous for the nbs-Ilp duplication and heterozygous for markers on the centromere-distal side of the duplication and on the centromere proximal side. In these females, one duplication carries the hairy1 (h1) distal mutation but is wild type at the proximal locus of approximated (app+), while the other duplication carries h+ and app1. Females of this genotype were used to set up the screen for spontaneous reductions. All resulting reduction events were crossed to flies homozygous for both h1 and app1 to assay the mutation status on the reduced chromosome. All 23 events carried either both or neither of the flanking recessive mutations, indicating that the flanking regions of the homologs were exchanged. Taken collectively, these results serve as strong evidence that the spontaneous events result from unequal crossing over in meiosis.
Mapping points of meiotic crossing over
The large number of recombinant lines that we recovered within the 82-kb interval presented us with an opportunity to investigate whether the crossing-over points are clustered within a relatively small interval. We arbitrarily divided the 82-kb region into five subregions using four pieces of heterology between the duplicate copies (Figure 3). At position 8 kb (p8) from the left end of the nbs-Ilp genomic fragment is the attR/attL heterology at nbs. At positions 35.1 kb (p35) and 43.7 kb (p44) there is a pair of previously described SNPs (Figure S2). At position 75.6 kb (p76) is the attB heterology at Ilp. Reduction homozygotes were genotyped with regard to the four heterologous locations. The results are shown in Figure 3. As expected for the type 1 event, all four sites of heterology correspond to the right copy in the duplication (i.e., parental 2), while for the type 3 event they correspond to the left copy (i.e., parental 1). For type 2 events, we recovered crossovers within all three regions of the 68 kb subdivided by the sites of heterology. Although the crossing-over frequency within this region is not strictly proportional to the extent of homology, we did not observe any region with a crossing-over rate that is greatly disproportional to the extent of homology.
Discussion
We have previously demonstrated that the SIRT method greatly facilitates targeted manipulation of genes situated close to a previously targeted phiC31 integrase attachment site (Gao et al. 2008, 2011). We hypothesized that this principle can be applied to genes farther away from that attachment site with the use of a larger DNA fragment. Here we provided a proof-of-principle study by modifying a region situated almost 70 kb from an existing attachment site. Our results lead us to propose this procedure as a strategy for accessing new genes for repeated mutagenesis.
Long-distance targeted modification
Although phiC31-mediated insertion of large DNA fragments at ectopic positions occurs at a frequency of 2–4% for constructs >50 kb (Venken et al. 2006), we were uncertain of whether such a frequency could be achieved when inserting a large fragment at its endogenous location because of potential instability of the resulting large duplication. This concern was resolved when we recovered integrations at a frequency of 5% or greater and were able to maintain the stocks indefinitely.
Interestingly, we observed infrequent spontaneous reduction events in female germlines homozygous for the duplication. Unequal meiotic crossing over between tandem duplications has been previously observed in many cases (e.g., Green 1962). We provided evidence that spontaneous reduction in our system happens through a similar mechanism. The genetic crosses for the recovery of spontaneous events are quite simple, making it an attractive alternative to the I-SceI-induced mitotic recombination method. In addition, the elimination of the NHEJ-type events simplifies PCR testing. However, this might not be a viable option when dealing with genomic regions with an intrinsically low meiotic recombination frequency.
Our previous experiences in reducing tandem duplications with a site-specific DSB all involved small duplications of a few kilobases in size (Rong et al. 2002; Gao et al. 2008, 2011). Here we faced the question of whether the efficiency of reducing a large duplication by a site-specific DSB would not drop to a prohibitive level. When comparing with our previous results, we observed a substantial drop in white-loss events (from 77–100% to 10–13%) and a substantial increase in NHEJ repair products (from essentially 0% to 60–70%). Nevertheless, our results remain rather encouraging. For the 10–13% of germlines that did experience white loss, about one-third of the white-loss events (29–39%) are reduction events. This translates to about one reduction event for every 30 male germlines tested. More importantly, the goal of our study was to recover not only reduction events of the duplication, but also the specific event with our desired modification, i.e., attB@Ilp. Around 46% of the reduction events from experiments using the pWalkman{nbs-Ilp}IS2 construct retain attB@Ilp. Therefore, a desired reduction event could be recovered for every 65 (30/0.46) males tested.
Recombination mechanisms for the reduction of large duplications
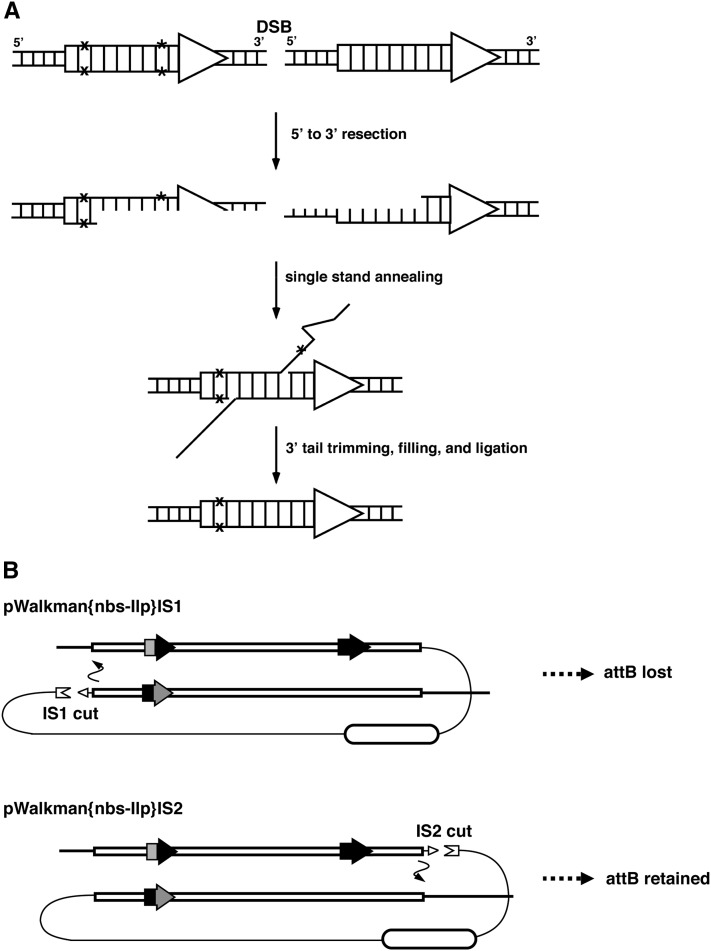
When comparing the efficiency of attB@Ilp retention in the reduction product, we obtained startlingly different results with two different vector configurations depending on which side of the white gene the future DSB site was placed (Figure 1 and Table 1). For construct pWalkman{nbs-Ilp}IS2, in which the I-SceI cut site is physically closer to attB@Ilp with respect to white, nearly half of the reduction events retained attB@Ilp. Remarkably, none of the six events retained attB@Ilp when recovered with pWalkman{nbs-Ilp}IS1, in which the I-SceI cut site is farther away from attB@Ilp. This result is also observed when using P transposase as a means of generating the DSB, most likely due to similar positioning of the DSB to the one generated by I-SceI at position IS1 (see Figures 1 and 2 for the position of P-element ends in the duplication). This preference in regard to the placement of the DSB is opposite to that previously observed when much smaller duplications were tested (Rong et al. 2002; Gao et al. 2008). The previous results are consistent with reduction occurring via the DSB repair mechanism of single strand annealing (SSA), which is the predominant mechanism for repairing a DSB flanked by direct repeats of a few kilobases in size (Rong and Golic 2003; Wei and Rong 2007). In SSA, both ends of the DSB experience single-strand resection at the 5′ to 3′ direction. Resection has to be sufficiently extensive to expose complementary single-stranded regions between the two repeats. Annealing of the two regions followed by trimming of the protruding single-stranded tails complete the repair process, thus reducing the two copies to one. Thus, SSA repair would result in the preferential loss of sequence heterology closer to the DSB (Figure 4A).
Figure 4 .
The effects of different repair mechanisms on reduction outcomes. (A) SSA repair as the mechanism for reduction. (Top) A DSB has been induced between two direct repeats (block arrows) with each horizontal line representing a DNA strand and vertical lines representing base pairing. The two heterologies between the repeats are denoted by an “X” and by an asterisk. The heterology indicated by the asterisk is more likely to become single stranded and thus excluded from the final reduction product due to its being closer to the DSB. (B) The “one-ended invasion crossover” model as the mechanism for reduction. The 80-kb duplication is shown for both pWalkman{nbs-Ilp} constructs. In the top diagram, IS1 is cut and the right end of the DSB in the bottom copy invades the homologous sequences of the top copy, leading to loss of the attB heterology. In the bottom diagram, IS2 is cut and the left end of the DSB in the top copy invades homologous sequences of the bottom copy, leading to retention of the attB. Note that in either case the invaded copy can be located on the sister chromatid instead.
For reducing large duplications, SSA is unlikely to be the major mechanism, simply due to the extent of resection required. In our case, resection of both ends would encompass at least 82 kb of nbs-Ilp plus 5 kb of white sequences. Instead, we propose that a significant portion of the reduction events that we recovered were the result of a “one-ended invasion crossover” mechanism, which has been previously invoked to account for repair products during spontaneous recombination between two direct repeats in yeast (Prado and Aguilera 1995). We envision that resection of the DSB end proximal to one of the repeats would expose single-strand homology leading to that end invading the homologous region in the other repeated copy (Figure 4B). This one-ended invasion ultimately leads to the resolution of a single Holliday junction and the appearance of the reduction product (for a detailed description of the model, see Figure 4 in Prado and Aguilera 1995). This model predicts that sequence heterology that is proximal to the DSB (e.g., attB@Ilp) would be part of the invading end and be more likely to be retained in the reduction product. On the other hand, heterology separated from the DSB end by vector sequences would be less likely to be retained. This is consistent with our inability to recover events that retained attB@Ilp when construct pWalkman{nbs-Ilp}IS1 was used to create the duplication and with the preferred recovery of attR@nbs (type 1 reduction in Figure 2). On the other hand, attB@Ilp was efficiently recovered when pWalkman{nbs-Ilp}IS2 was used. We did recover cases in which attB@Ilp was not retained even with this favorable configuration. Further investigation is needed to shed light on the repair mechanisms responsible for these reduction products.
Based on these results, we suggest that, for reducing a large duplication, the genomic insert within the pWalkman vector should be oriented in such a way as to place the I-SceI site closer to the end of the insert that carries the modification, similar to the IS2 position in Figure 1. In addition, because the FLP-catalyzed vector excision would not be necessary, using P transposase might be a simpler way to induce reduction.
Toward genome-wide targeted manipulations in Drosophila
The goal of knocking out every gene in Drosophila has been a long-lasting crusade for the fly community (Bellen et al. 2011). A significant portion of the 15,000 annotated genes still lack defined mutations after a century of random mutagenesis in Drosophila. The invention of a reliable gene targeting method by Rong and Golic in 2000 opened the possibility for disrupting every fly gene by homologous recombination. However, gene targeting in complex organisms is inherently labor intensive and time-consuming, making its genome-wide application a daunting task. In addition, the return from a genome-wide knock-out collection is limited, as it provides only a single allele for each gene. Yet systematic mutational studies become increasingly important for our complete comprehension of gene function. Thus, even with the availability of a genome-wide knock-out collection, subsequent modifications of a particular gene have to be introduced as transgenes at ectopic loci and might be under the influence of chromosome position effects that are not well understood.
We suggest, as an alternative, the creation of a collection of fly lines carrying att sites distributed throughout the whole genome, which would enable unlimited rounds of targeted manipulations for essentially all genes. If we consider that any att site creates a point of access to ∼140 kb of the surrounding DNA (70 kb in each direction), then the 160 Mb of the fly genome could be covered with 1200 different att stocks. We propose that transposable elements can be utilized to distribute the att sites throughout the genome. Progress in this regard has already been made with the availability of independent collections, such as the MiMIC lines (Venken et al. 2011). Our method will be additionally advantageous in two ways. First, it can be used to modify a region 70 kb or less from an existing att site. Second, it can be used to fill large gaps void of any att site (hence the name “Walkman” for our vector). The estimated number of 1200 new stocks represents a small burden if we consider that, in 2010, the Bloomington Stock Center alone reported holding 38,000 different stocks. The potential utility of these ∼1200 stocks would be unprecedented. Once any locus is equipped with a proximal att site, the possibilities for further modification by SIRT are endless.
Supplementary Material
Acknowledgments
We are grateful to Flavia Amariei for aiding us with screening for spontaneous reductions and to Jemima Barrowman for her assistance in editing the manuscript. Research in our laboratory is supported by the Intramural Program of the National Cancer Institute.
Footnotes
Communicating editor: J. Sekelsky
Literature Cited
- Bellen H. J., Levis R. W., He Y., Carlson J. W., Evans-Holm M., et al. , 2011. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188: 731–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland N. G., Jenkins N. A., Court D. L., 2001. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2: 769–779 [DOI] [PubMed] [Google Scholar]
- Gao G., McMahon C., Chen J., Rong Y. S., 2008. A powerful method combining homologous recombination and site-specific recombination for targeted mutagenesis in Drosophila. Proc. Natl. Acad. Sci. USA 105: 13999–14004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Cheng Y., Wesolowska N., Rong Y. S., 2011. Paternal imprint essential for the inheritance of telomere identity in Drosophila. Proc. Natl. Acad. Sci. USA 108: 4932–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor G. B., Nassif N. A., Johnson-Schlitz D. M., Preston C. R., Engels W. R., 1991. Targeted gene replacement in Drosophila via P element-induced gap repair. Science 253: 1110–1117 [DOI] [PubMed] [Google Scholar]
- Gong W. J., Golic K. G., 2003. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100: 2556–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. M., 1962. The effects of tandem duplications on crossing over in Drosophila melanogaster. Genetica 33: 154–164 [DOI] [PubMed] [Google Scholar]
- Gronke S., Clarke D. F., Broughton S., Andrews T. D., Partridge L., 2010. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 6: e1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R., Calos M. P., 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166: 1775–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhou W., Dong W., Hong Y., 2009. Targeted engineering of the Drosophila genome. Fly (Austin) 3: 274–277 [DOI] [PubMed] [Google Scholar]
- Iampietro C., Gummalla M., Mutero A., Karch F., Maeda R. K., 2010. Initiator elements function to determine the activity state of BX-C enhancers. PLoS Genet. 6: e1001260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce E. F., Paul A., Chen K. E., Tanneti N., McKim K. S., 2012. Multiple barriers to nonhomologous DNA end joining during meiosis in Drosophila. Genetics 191: 739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Sved J. A., 2009. Repair of P element ends following hybrid element excision leads to recombination in Drosophila melanogaster. Heredity (Edinb) 102: 127–132 [DOI] [PubMed] [Google Scholar]
- Prado F., Aguilera A., 1995. Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10 and RAD52 genes. Genetics 139: 109–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018 [DOI] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2003. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics 165: 1831–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., Titen S. W., Xie H. B., Golic M. M., Bastiani M., et al. , 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 16: 1568–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzweck B., Brankatschk M., Dickson B. J., 2010. Distinct protein domains and expression patterns confer divergent axon guidance functions for Drosophila Robo receptors. Cell 140: 409–420 [DOI] [PubMed] [Google Scholar]
- Venken K. J., He Y., Hoskins R. A., Bellen H. J., 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751 [DOI] [PubMed] [Google Scholar]
- Venken K. J., Schulze K. L., Haelterman N. A., Pan H., He Y., et al. , 2011. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8: 737–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D. S., Rong Y. S., 2007. A genetic screen for DNA double-strand break repair mutations in Drosophila. Genetics 177: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng R., Chen Y. W., Bushati N., Cliffe A., Cohen S. M., 2009. Recombinase-mediated cassette exchange provides a versatile platform for gene targeting: knockout of miR-31b. Genetics 183: 399–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.