Abstract
Site-specific recombinases (SSRs) are valuable tools for manipulating genomes. In Drosophila, thousands of transgenic insertions carrying SSR recognition sites have been distributed throughout the genome by several large-scale projects. Here we describe a method with the potential to use these insertions to make custom alterations to the Drosophila genome in vivo. Specifically, by employing recombineering techniques and a dual recombinase-mediated cassette exchange strategy based on the phiC31 integrase and FLP recombinase, we show that a large genomic segment that lies between two SSR recognition-site insertions can be “captured” as a target cassette and exchanged for a sequence that was engineered in bacterial cells. We demonstrate this approach by targeting a 50-kb segment spanning the tsh gene, replacing the existing segment with corresponding recombineered sequences through simple and efficient manipulations. Given the high density of SSR recognition-site insertions in Drosophila, our method affords a straightforward and highly efficient approach to explore gene function in situ for a substantial portion of the Drosophila genome.
Keywords: recombinase-mediated cassette exchange (RMCE), ΦC31, targeted mutagenesis, genomic engineering
MANY approaches to genetic research rely on an ability to alter DNA sequences in vivo. To this end, site-specific recombinases (SSRs) such as Cre, FLP, and phiC31 integrase have become invaluable tools for manipulating genes and genomes on small and large scales (reviewed by Branda and Dymecki 2004; Venken and Bellen 2005). The introduction of recognition sequences for SSRs into a genome can permit the generation of insertions, deletions, and/or inversions at precisely mapped genomic locations, facilitating analyses of genome structure and gene function.
In the model organism Drosophila melanogaster, SSRs play a pivotal role in several methods that make predictable changes to the genome sequence (reviewed by Venken and Bellen 2005; Venken and Bellen 2007). For example, recombination between two FRT sites located at different positions on homologous chromosomes can generate deletions, duplications, and inversions spanning several megabases depending on the relative positions and orientations of the FRTs (Golic and Golic 1996; Parks et al. 2004; Ryder et al. 2004). Several multi-step schemes have used SSRs in conjunction with the endogenous homologous recombination repair machinery to generate more precisely defined changes to the genome sequence, including small deletions, point mutations, and protein fusions (Gao et al. 2008; Choi et al. 2009; Huang et al. 2009; Weng et al. 2009). While these latter methods are of great utility to the Drosophila community, their efficiency and ease of use are limited by their reliance on the endogenous homologous recombination machinery during at least one step in each method. Thus, an approach to generate similarly precise changes to the genome that could circumvent the use of homologous recombination would be of great benefit.
In addition to methods that alter existing sequences in the Drosophila genome, several strategies that rely on SSRs have been used to improve methods of transgenesis. Much recent attention has been focused on site-specific integration of transgenes using phiC31 integrase, which catalyzes recombination between two distinct recognition sequences, attP and attB (reviewed by Venken and Bellen 2007 and Smith et al. 2010). Recombination between these sites creates two new sequences, attL and attR, that are not substrates for the integrase, resulting in a directional recombination event that is not reversible under normal conditions. Site-specific integration of a transgene can be achieved via recombination between an attP in the genome and an attB-bearing plasmid that carries the transgene of interest, resulting in a stable integrant of the plasmid at the site of the original attP (Groth et al. 2004; Bischof et al. 2007).
In an alternative strategy called recombinase-mediated cassette exchange (RMCE; reviewed by Baer and Bode 2001), a transgene of interest can be integrated into a defined site in a genome without incorporating the associated plasmid sequences. In this case, a “target cassette” carrying a selectable marker flanked by two SSR recognition sites is first inserted into the genome by other means. In the presence of the relevant SSR, introduction of a plasmid carrying a “donor” cassette flanked by compatible recognition sites can result in recombination at both ends of the cassettes, effectively exchanging the target and donor cassettes and thereby integrating the donor sequence into the genome. Methods for RMCE in Drosophila and other organisms have been demonstrated using Cre, FLP, or phiC31 integrase (reviewed by Baer and Bode 2001; Venken and Bellen 2007). Furthermore, “dual” RMCE strategies have been developed wherein the donor and target cassettes carry loxP recognition sites for Cre at one end and FRT sites for FLP at the other end (Lauth et al. 2002; Osterwalder et al. 2010; Anderson et al. 2012). In these schemes, cassette exchange relies on the combined activity of both enzymes, either simultaneously or in successive steps of plasmid insertion and subsequent deletion of the target cassette and plasmid backbone. A variant of dual RMCE has been applied to transgenesis in Drosophila, wherein a plasmid carrying a transgene is first integrated by recombination between phiC31 attP and attB sites, and the plasmid backbone is then deleted in a second step using Cre-mediated recombination between loxP sites (Bischof et al. 2007; Huang et al. 2009).
In parallel with the development of technologies that rely on SSRs, several large-scale projects have incorporated thousands of SSR recognition sequences throughout the Drosophila genome. In particular, >23,000 PiggyBac insertions carrying FRT sequences were generated as part of the Exelixis collection (Thibault et al. 2004), and an additional ∼3000 FRT-bearing P-element insertions were created for the DrosDel collection (Ryder et al. 2004). In addition, the Drosophila Gene Disruption Project recently reported >1200 insertions of MiMIC, a Minos-based transposon carrying phiC31 attP sites, and plans to generate >6000 insertions by 2015 (Venken et al. 2011). These large collections are an important asset to the Drosophila research community and provide an invaluable resource for the development of new genetic technologies in Drosophila.
Here we describe a method aimed at using publicly available insertions of SSR recognition sites to generate custom alterations in the Drosophila genome. Our strategy is made possible by advances in recombineering, a system for manipulating large DNA fragments (e.g., BACs) using homologous recombination in bacteria (reviewed by Sharan et al. 2009). Recombineering facilitates the introduction of insertions, deletions, and/or point mutations into DNA molecules carried by bacteria, allowing countless modifications to be applied to a cloned genomic fragment of interest in bacterial cells. In our scheme, a region of the Drosophila genome that is flanked on one side by an attP insertion and on the other by an FRT insertion can be exchanged for a corresponding recombineered DNA sequence from a donor BAC using a dual RMCE strategy. We demonstrate this approach by replacing a 50-kb chromosomal segment encompassing the teashirt (tsh) gene with a corresponding 50-kb BAC-derived genomic fragment that we engineered in bacteria. Furthermore, we show that this method can be used to incorporate large- and small-scale alterations into the captured segment through recombineering of the donor BAC sequence. With the vast number of attP and FRT insertions that are available to the Drosophila community, we believe that this method will be applicable to a substantial proportion of the genome.
Materials and Methods
Stocks and fly husbandry
A stock carrying pBac(WH)f06252 was obtained from the Exelixis Collection at Harvard Medical School, and the stocks y1 w1118 P(70FLP)3F, y1 w1118; PBac(y+-attP-3B)VK00003a PBac(y+-attP-3B)VK00003b, and tsh04319 cn/CyO; ry and w1118; Df(2L)BSC151/CyO were obtained from the Bloomington Drosophila Stock Center. PBac(y+-attP-3B)VK00003a was removed from the chromosome carrying PBac(y+-attP-3B)VK00003b by meiotic recombination prior to the analyses described here. Stocks carrying the insertions attP52 and attP64 were provided by Michele Markstein. All flies were maintained at 25° on standard Drosophila cornmeal, yeast, sugar, and agar medium with p-hydroxybenzoic acid methyl ester as a mold inhibitor (Morris et al. 1998).
Cloning, gap repair, and recombineering
P[acman]-ApR (Venken et al. 2006) was obtained from the Drosophila Genomics Resource Center. To enhance eye pigmentation of transformants, a fragment encoding the eye enhancer GMR (Moses and Rubin 1991) was digested from pcr2.1-GMR (Bateman et al. 2012) and inserted into a unique EcoRI site upstream of the mini-white coding region of P[acman]-ApR. We then removed BamHI and NotI sites that were carried on the GMR fragment by digesting the vector with BamHI and ligating in overlapping oligonucleotides PacBK_5 and PacBK_3 (see Supporting Information, Table S1, for sequences of primers used in this study) to create GMR-P[acman].
An ∼50-kb fragment of Drosophila genomic DNA spanning from the insertion point of PBac(y+-attP-3B)VK00003b to that of pBac(WH)f06252 was inserted into GMR-P[acman] by gap repair using existing methods (Venken et al. 2006; Sharan et al. 2009) with modifications. Briefly, 0.5-kb left and right homology arms were amplified from the BAC RP98-9H20 (BACPAC Resource Center, Children’s Hospital Oakland Research Institute) using primer pairs VK3b_5_3/VK3b_3_1 and f06252_5_2/f06252_3_1; the former pair includes a 40-bp attB sequence on the forward primer, and the latter pair includes 20 bp of sequence complementary to the left homology arm on the forward primer and a 35-bp FRT sequence on the reverse primer. The two 0.5-kb arms were combined into a single 1-kb fragment via Splicing by Overlap Extension (SOEing; Horton et al. 1990) using primers VK3b_5_3 and f06252_3_1, and the 1-kb fragment was subcloned into pcr2.1 using a TOPO-TA cloning kit (Invitrogen). Next, the 1-kb fragment was excised from pcr2.1 and inserted into GMR-P[acman] using AscI and PacI. The resulting plasmid, GMR-P[acman]-tsh1, was linearized at a BamHI site between the left and right homology arms and electroporated into heat-shocked SW102 bacteria (Warming et al. 2005) carrying RP98-9H20. Following selection on LB plates containing 100 µg/ml ampicillin, candidate gap-repaired plasmids were screened using the primer pairs PacmanMCS-F/tsh-5-vk3b-check-R and tsh-3-06252-check-F/PacmanMCS-R and then confirmed by fingerprinting digests with EcoRI. The resulting plasmid, GMR-P[acman]-tsh50, was electroporated into EPI300 bacterial cells (Epicentre Biotechnologies) for large-scale plasmid isolation.
To delete noncoding sequences downstream of the tsh-coding region in GMR-P[acman]-tsh50, we used a galK-based recombineering strategy as previously described (Warming et al. 2005; Sharan et al. 2009). Briefly, we targeted a 312-bp sequence (tshNC1) and an adjacent 550-bp sequence (tshNC2) ∼10 kb downstream of the tsh coding region by designing galK-specific PCR primers that carried 50-bp sequences complementary to the regions flanking the intended deletions. These primers were used to amplify the galK gene from pgalK (Warming et al. 2005), and the resulting PCR products were electroporated into heat-shocked SW102 cells carrying GMR-P[acman]-tsh50. Cells were then washed in 1× M9 salts and plated on M63 minimal media plates with galactose, leucine, biotin, and ampicillin to select for GMR-P[acman]-tsh50 clones that had incorporated the galK cassette. Replacement of the deleted genomic DNA by galK was verified using the primer pairs NC1check1for/NC1check1rev and NC1check1for/NC2check1rev for tshNC1 and tshNC2 deletions, respectively, and the structure of each BAC was assessed by fingerprinting digests with EcoRI.
Drosophila transformation and captured segment exchange
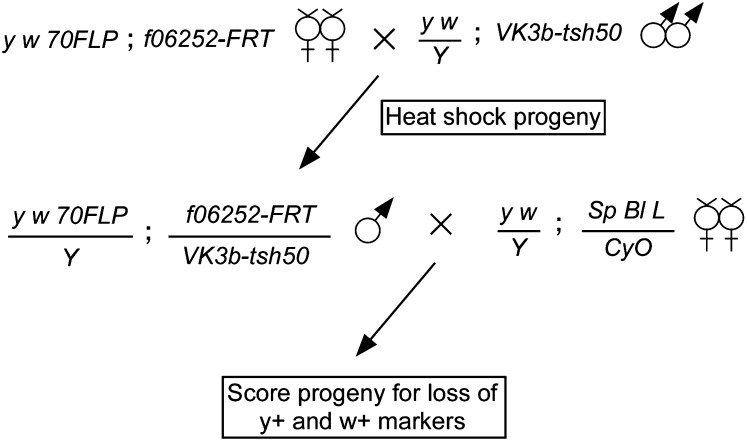
GMR-P[acman] clones were injected into embryos carrying PBac(y+-attP-3B)VK00003b and an endogenous source of the phiC31 integrase (Bischof et al. 2007), and transformants were identified by screening for red eye pigmentation as previously described (Venken et al. 2006). Injections, screening, and PCR-based confirmation of correctly targeted insertions were performed by BestGene Drosophila Embryo Injection Services (see Table S2 for integration rates). To exchange the captured genomic segment for the engineered sequence, we crossed y1 w1118 P(70FLP)3F; pBac(WH)f06252 virgin females with males carrying a GMR-P[acman] construct inserted at VK00003b and heat-shocked the progeny on day 3 for 90 min at 37°. The adult males arising from this cross were mated individually to y w; Sp Bl Lrm/CyO virgin females, and the progeny were screened for y− and w− phenotypes.
Molecular analysis
Candidate flies in which the endogenous captured segment was exchanged using the donor GMR-P[acman]-tsh50 were verified by PCR-amplifying each end of the captured segment using primer pairs tsh_RRF3/tsh_RRR3 for the distal end and tsh_RLF/tsh_RLR for the proximal end. Genomic DNA from flies that had undergone captured segment exchange was used as a template, and amplified products were sequenced to confirm the expected structures. To identify single nucleotide polymorphisms (SNPs) in the captured segment, PCR was used to amplify several ∼500-bp regions of noncoding DNA from BAC RP98-9H20, genomic DNA from flies carrying pBac(WH)f06252, and genomic DNA from flies carrying PBac(y+-attP-3B)VK00003b. Sequences were then aligned, and for the amplified regions defined by primer pairs SNP2F/SNP2R and SNP5F/SNP5R, several SNPs were detected that differentiate BAC DNA from both genomic templates. Genomic DNA from flies that had undergone captured segment exchange was then analyzed by the same method.
Candidate flies in which the endogenous captured segment was deleted via exchange using the donor GMR-P[acman]-tsh1 were verified by PCR amplification using the primer pair SOE_eF/SOE_eR and genomic DNA from flies heterozygous for the exchanged chromosome as template. These primers are complementary to genomic sequences flanking the captured segment and are predicted to generate an ∼3-kb product if the captured segment is deleted, and no product if the captured segment is unaltered (due to their >50 kb spacing on an unaltered chromosome). Amplified products were sequenced to confirm the expected structure.
Candidate flies in which the endogenous captured segment was exchanged using the donors GMR-P[acman]-tsh50ΔNC1 and GMR-P[acman]-tsh50ΔNC2 were verified by PCR amplification from genomic DNA of flies homozygous for the exchanged chromosome using the primer pairs NC1check1for/NC1check1rev and NC2check1for/NC2check1rev, respectively. Genomic DNA from flies carrying pBac(WH)f06252 or PBac(y+-attP-3B)VK00003b were each used as controls.
Captured segment exchange using two attP sites
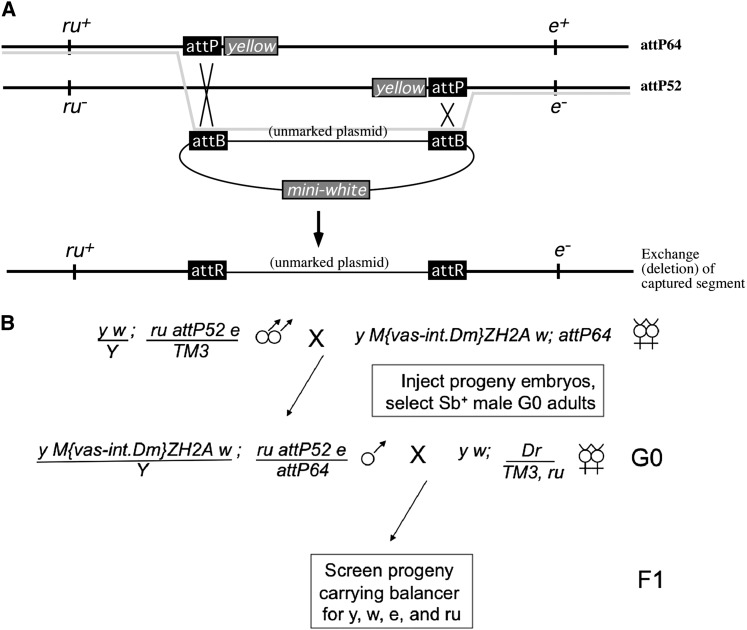
The visible markers e1 and ru1 were recombined onto a chromosome carrying the insertion attP52 to create flies of genotype y1 w1118; ru1 attP52 e1/TM3. Embryos from the cross depicted in Figure 6B were injected with piB-miniwhite, constructed by ligating a 4-kb PCR fragment carrying mini-white into the vector backbone of Xba-digested piB-GFP (Bateman et al. 2006) at a concentration of ∼400 ng/µl.
Figure 6 .
Captured segment exchange via single-step RMCE. (A) Schematic showing homologous chromosomes carrying attP64 (top) and attP52 (bottom), which are separated by ∼40 kb. The chromosome carrying attP52 is marked by mutations in ru to the left of the insert and in e to the right. Following injection of a plasmid carrying attB sites, captured segment exchange will result in a recombinant chromosome that is ru+, e−, lacks yellow markers, and carries unmarked plasmid sequence in place of the deleted captured segment (the faded line traces the regions of the parental chromosomes and the injected plasmid that are found in the final recombinant chromosome). (B) Cross scheme for accomplishing the exchange depicted in A (see text for details).
Results
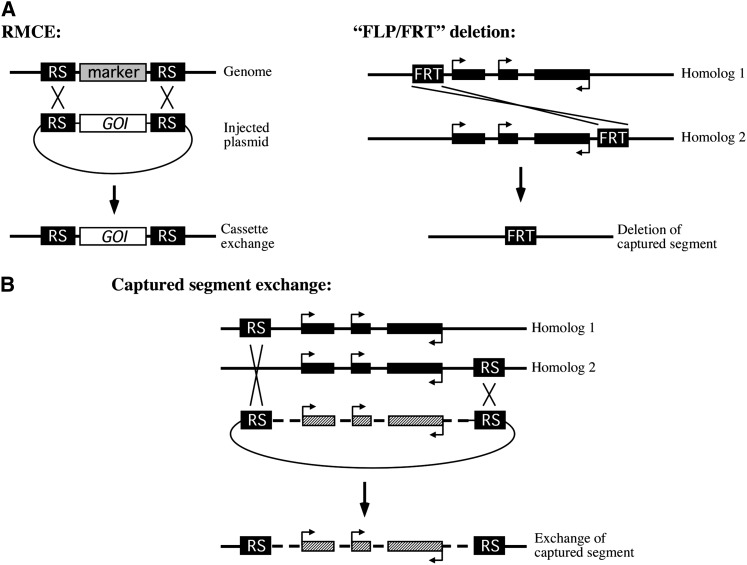
Our goal was to establish a method for altering Drosophila genomic sequences that could take advantage of the large number of publicly available SSR recognition-site insertions. We developed a strategy that combined elements of two existing technologies for genome manipulation in Drosophila: RMCE, where a genomic DNA sequence that is flanked by SSR recognition sites can be exchanged for a donor cassette flanked by compatible recognition sequences, and the FLP/FRT method for generating deletions, where SSR recognition sites can interact between homologous chromosomes to produce a recombinant chromosome carrying a defined deficiency (Figure 1A). We imagined that the latter method could be adapted so that, rather than deleting a sequence, a genomic segment between two SSR recognition sites could instead be “captured” as a target cassette and exchanged with a donor sequence (Figure 1B). As with the FLP/FRT deletion approach, we reasoned that the SSR recognition sites could be carried on homologous chromosomes, allowing the direct use of existing Drosophila lines carrying SSR recognition sites without the need to recombine elements onto the same chromosome.
Figure 1 .
Conceptual view of captured segment exchange. (A) Existing technologies that form the basis of captured segment exchange. (Left) In RMCE, a marker flanked by SSR recognition sequences (RS) in the genome can be exchanged for a gene of interest (GOI) that is flanked by compatible RS on an injected plasmid. (Right) Crossovers between FRTs at nearby positions on homologous chromosomes can create a deletion of the intervening sequence on a resulting recombinant chromosome. (B) Captured segment exchange combines principles of the two technologies in A. RS at nearby positions on homologous chromosomes can effectively capture the intervening sequence such that a corresponding sequence flanked by compatible RS on an injected plasmid can replace the captured segment on a recombinant chromosome. Hatched region indicates the engineered sequence. Diagrams are not to scale.
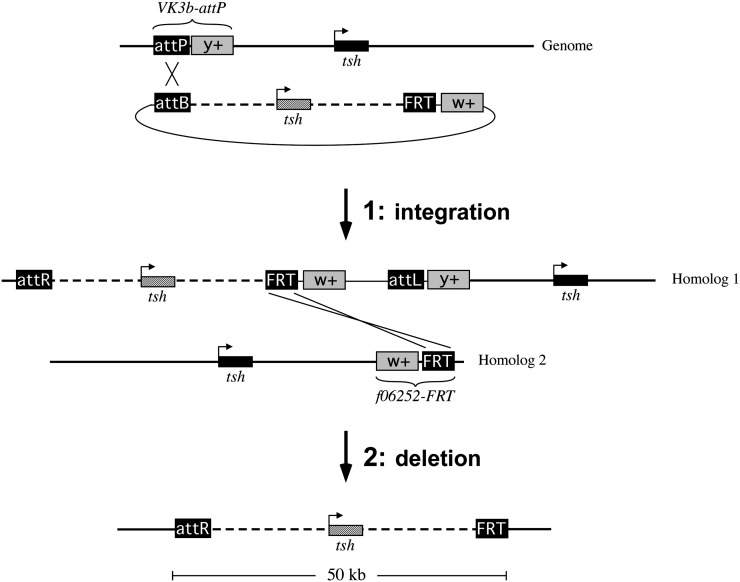
To demonstrate the feasibility of this method, we employed a dual RMCE strategy using a phiC31 attP site at one end of a captured segment and an FRT at the other. We reasoned that the use of an attP would maximize the efficiency of integration, while the inclusion of an FRT would provide the greatest flexibility in adapting the method to different genomic regions, given the large number of FRT insertions available. Specifically, we targeted a 50-kb genomic region defined by the attP insertion PBac(y+-attP-3B)VK00003b (VK3b-attP; Venken et al. 2006) upstream of tsh and the FRT-bearing insertion pBac(WH)f06252 (f06252-FRT; Thibault et al. 2004) downstream of tsh. Our strategy accomplishes exchange in two steps (Figure 2): first, integration of an engineered donor BAC into the VK3b-attP site via phiC31-mediated transgenesis, and second, deletion of the “endogenous” tsh segment via recombination between an engineered FRT in the donor BAC on one homolog and f06252-FRT on the other, leaving only the engineered tsh segment on a recombinant chromosome. Importantly, VK3b-attP, f06252-FRT, and the donor BAC are each marked with either a mini-white gene (BAC and f06252-FRT) or a yellow gene (VK3b-attP), allowing the two steps of the exchange to be monitored simply by scoring eye color and cuticle pigmentation in adult flies (Figure 2); given the positions of the yellow and mini-white markers relative to the att and FRT sites, individuals in which the endogenous segment is deleted can be identified based on their loss of y+ and w+ phenotypes. Accordingly, all of our crosses were carried out in a y− w− background.
Figure 2 .
Two-step dual RMCE strategy for captured segment exchange at the tsh locus. In step 1, a P[acman] clone carrying an engineered tsh sequence is integrated upstream of the endogenous tsh gene via phiC31-mediated transgenesis using the attP insertion PBac(y+-attP-3B)VK00003b (VK3b-attP). The mini-white carried by P[acman] and yellow carried by VK3b-attP serve as markers such that successful transformants are y+ w+ in an otherwise y− w− background. In step 2, the endogenous tsh locus is deleted from a recombinant chromosome resulting from FLP-mediated crossing over between FRTs on the two homologs, leaving only the engineered sequence; this event also deletes all mini-white and yellow markers from the recombinant chromosome, allowing candidates to be identified by a y− w− phenotype. Not shown is a reciprocal chromosome resulting from the FRT exchange that will carry a tandem duplication of the captured sequence along with all yellow and mini-white markers. Diagrams are not to scale.
To generate a compatible donor vector, we used the P[acman] BAC system that allows sequence modification by recombineering and carries a mini-white gene for identification of Drosophila transformants (Venken et al. 2006). In our preliminary experiments, we found that the eye pigmentation caused by mini-white expression was difficult to detect when P[acman] clones were integrated into the tsh locus, so we modified the vector to include the strong eye enhancer GMR (Moses and Rubin 1991) upstream of mini-white. The modified vector, GMR-P[acman], produces robust eye pigmentation when integrated at the tsh locus (data not shown) and was used for all subsequent experiments.
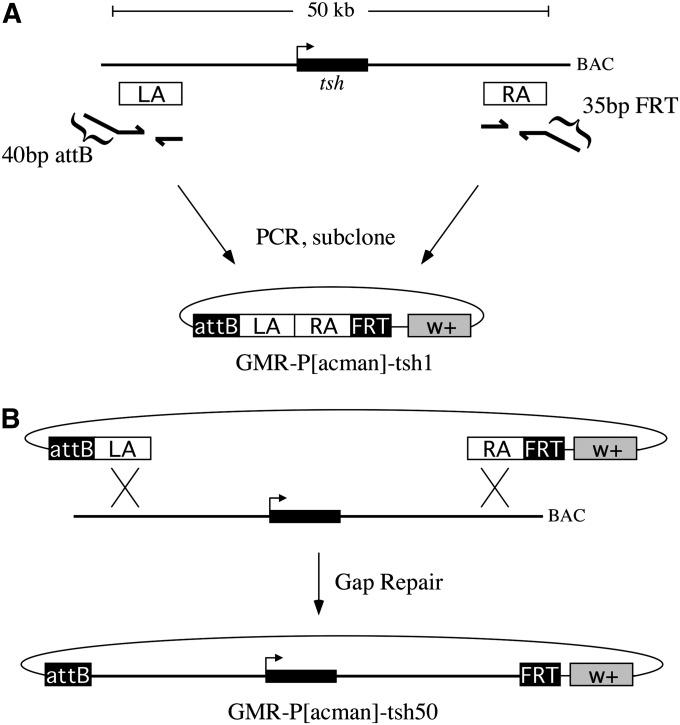
We used recombineering-mediated gap repair to copy a 50-kb sequence spanning the captured tsh segment from a publicly available genomic clone into GMR-P[acman]. In this strategy, two 0.5 kb homology arms corresponding to the ends of the captured segment were first amplified by PCR and ligated into GMR-P[acman] to create GMR-P[acman]-tsh1 (Figure 3A). We designed our primers so that the outer ends of these homology arms corresponded to the precise nucleotides of the VK3b and f06252 insertions that define the boundaries of the captured segment. In addition, we incorporated a 40-bp attB sequence and a 35-bp FRT sequence as “tails” on the outermost primers (Bateman and Wu 2008) so that the homology arms would be flanked by attB and FRT sites as required for the exchange reaction. We then linearized GMR-P[acman]-tsh1 at a site between the homology arms and transformed it into bacteria carrying the corresponding tsh genomic region and Red recombination functions (Lee et al. 2001; Court et al. 2003) to create our donor BAC, GMR-P[acman]-tsh50 (Figure 3B; see Materials and Methods), which carries 50 kb of genomic DNA corresponding to the captured segment and is flanked by attB and FRT sites.
Figure 3 .
Recombineering strategy to generate a donor BAC for captured segment exchange of tsh. (A) Two “homology arms” (LA and RA) are generated by PCR and subcloned into GMR-P[acman] to create GMR-P[acman]-tsh1. The forward primer for LA incorporates a 40-bp attB sequence at its 5′ end, whereas the reverse primer for RA incorporates a 35-bp FRT sequence at its 5′ end such that the final PCR product is flanked by attP and FRT sites. Not shown is an additional 20 bp at the 5′ end of the forward RA primer that is complementary to the reverse LA primer, which was used to splice by overlap extension the two PCR fragments together prior to subcloning (Horton et al. 1990), and a recognition site for BamHI, which was used to linearize the plasmid for gap repair. (B) GMR-P[acman]-tsh1 is linearized between the homology arms and transformed into bacteria that carry a tsh genomic clone (BAC) and Red recombination functions. The homology arms direct repair of the linearized plasmid from the BAC, creating GMR-P[acman]-tsh50, which carries a 50-kb sequence corresponding to the captured segment flanked by attP and FRT sites. Diagrams are not to scale.
GMR-P[acman]-tsh50 was targeted to VK3b-attP via existing methods for phiC31-mediated transgenesis (Groth et al. 2004; Bischof et al. 2007; Venken and Bellen 2007) using a commercial injection service, and successful VK3b-tsh50 integrants were identified by the eye pigmentation produced by mini-white expression. To carry out the second step of the exchange reaction, we crossed VK3b-tsh50 males to virgin females carrying f06252-FRT and an X-chromosomal insertion of FLP driven by a heat-shock promoter (hs-FLP) (Figure 4). Heat-shocked male progeny that carried f06252-FRT on one homolog and VK3b-tsh50 on the other homolog were then individually mated to virgin females carrying a second chromosome balancer, and their progeny were analyzed for evidence of a FLP-mediated crossover between homologs. As anticipated, we observed a y− w− phenotype, indicative of a completed exchange of the captured segment, at a high rate, with ∼20–30% of the progeny scored as putative exchange candidates (Table 1). Notably, we also observed a class of y+ w+ progeny with darker eye pigmentation than expected (data not shown), which is consistent with a prediction for the reciprocal recombinant chromosome resulting from an FRT crossover; this chromosome would carry tandem copies of the captured segment in addition to two mini-white genes and the yellow marker. We did not analyze these candidate reciprocal chromosomes further.
Figure 4 .
Cross scheme for FLP-mediated deletion of the endogenous tsh locus. Virgin females carrying a heat-shock-inducible FLP gene (70FLP) and f06252-FRT are crossed to males in which GMR-P[acman]-tsh50 was integrated into VK3b-attP (VK3b-tsh50). The resulting larvae are heat-shocked to activate FLP expression, and male progeny are mated individually to virgin females carrying a second chromosome balancer (CyO) and arbitrary dominant markers (Sp Bl L). In the F2, male progeny carrying CyO are scored for y and w phenotypes, with y− w− indicative of completed exchange. All crosses were carried out in a y− w− background.
Table 1 . Captured segment exchange at the tsh locus.
| P[acman] constructa | Insert size (kb) | Engineered mutation | y− w− (exchange)b | Total scored | % exchangedc |
|---|---|---|---|---|---|
| tsh50 #1 | 50 | None | 27 | 132 | 20.5 |
| tsh50 #2 | 50 | None | 10 | 31 | 32.3 |
| tsh1 #1 | 1 | ∼49-kb deletion | 21 | 151 | 13.9 |
| tsh1 #2 | 1 | ∼49-kb deletion | 11 | 132 | 8.3 |
| tsh50ΔNC1 | ∼50 | 312-bp deletion | 8 | 99 | 8.1 |
| tsh50ΔNC2 | ∼50 | 550-bp deletion | 8 | 45 | 17.8 |
Flies were subjected to the schemes outlined in Figures 2 and 4 using the indicated GMR-P[acman] construct as a donor for captured segment exchange. Only male progeny carrying the balancer CyO were scored for y and w phenotypes.
Two independent insertions of GMR-P[acman]-tsh50 and of GMR-P[acman]-tsh1 were tested.
Number of individual progeny in the F2 generation of the cross scheme in Figure 4 that had y− cuticle and w− eyes, indicative of a completed exchange.
Percentage of all F2 flies scored that had y− cuticle and w− eyes.
To verify that the endogenous captured segment had been exchanged for the engineered sequence, we amplified and sequenced genomic DNA surrounding each end of the captured segment in one of our candidate lines and confirmed that the sequence matched that predicted for the exchange reaction. In addition, we identified SNPs within the captured segment that could differentiate the BAC-derived sequence from that of the chromosomes carrying VK3b-attP or f06252-FRT. When we analyzed these SNPs in our candidate line, we saw only the alleles corresponding to the BAC and not those from either parent chromosome (Figure S1). Thus, our data are consistent with a clean exchange of the endogenous captured segment for the engineered BAC sequence.
Flies homozygous for the engineered chromosome are viable and fertile, with no evidence of abnormal adult structures. In addition, staining of embryos with an anti-Tsh antibody shows that the overall pattern of tsh expression is not altered by incorporating the engineered sequence (data not shown). We conclude that captured segment exchange of tsh has no adverse effect on development.
Captured segment exchange can be used to custom-engineer genomic sequences
Thus far, our demonstration of captured segment exchange has used a donor sequence from a genomic clone carrying wild-type sequences. However, we imagined that our method would be most useful for making changes to genomic sequences in situ. Thus, we tested whether donor BACs carrying defined sequence changes could be easily exchanged for the captured segment.
We first tested whether the captured segment could be replaced with a large deletion. To do so, we completed the steps for segment exchange outlined above using the donor BAC GMR-P[acman]-tsh1, which carries only the 0.5-kb homology arms and therefore represents a deletion of ∼49 kb of the tsh locus. Following dual RMCE, we once again observed high rates of candidate exchange events as indicated by progeny with a y− w− phenotype (Table 1), implying that the size of the donor DNA molecule does not significantly impact the rate of crossover between FRTs on homologous chromosomes. We verified that these candidate deletion-bearing flies carried the expected sequence by amplifying and sequencing a ∼3-kb fragment spanning the engineered sequence using primers complementary to genomic DNA flanking the VK3b-attP and f06252-FRT insertions (data not shown). Notably, the resulting engineered chromosome caused lethality when homozygous, as expected for a deletion of the essential tsh gene. In addition, the chromosome failed to complement the independent tsh deficiency Df(2L)BSC151 (Parks et al. 2004) and the lethal allele tsh04319 (Bellen et al. 2004) (data not shown). Thus, our method can be used to make large deletions within the captured segment.
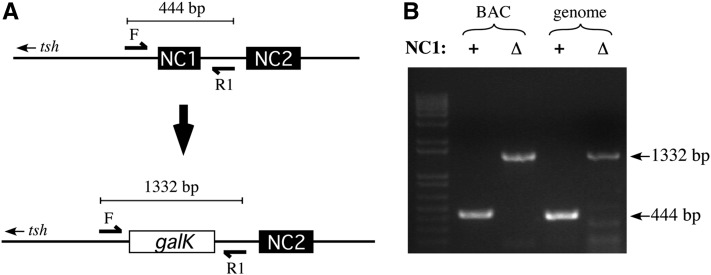
We next used our method to make more fine-scale changes to the tsh locus. The tsh gene is situated in a small gene desert, with regions of noncoding DNA extending 33.1 kb upstream and 59.6 kb downstream from the transcription unit (Crosby et al. 2007). Notably, analyses from the ModEncode consortium predict that these noncoding sequences are rich in binding sites for transcription factors and other DNA-binding proteins, suggesting that much of the region plays a regulatory function (Roy et al. 2010; Negre et al. 2011). We used recombineering schemes to delete two small regions of feature-rich noncoding DNA ∼10 kb downstream of tsh in GMR-P[acman]-tsh50, replacing each with the small bacterial gene galK (Warming et al. 2005). We then used the resulting altered BACs, GMR-P[acman]-tsh50ΔNC1 and GMR-P[acman]-tsh50ΔNC2, as donor vectors in the captured segment exchange procedure above. For both types of deletion, flies carrying the resulting engineered chromosomes were homozygous viable (the effects of these deletions on Drosophila development will be described in a later publication). We confirmed that the deletions were incorporated into the genome via PCR using primers flanking the altered sequences (Figure 5). In both cases, flies homozygous for chromosomes carrying the candidate-exchanged segments showed only the PCR product corresponding to the deletion, and not that of the endogenous unaltered sequence (Figure 5B; Figure S2). Thus, captured segment exchange represents a simple and efficient method to engineer custom alterations into the Drosophila genome.
Figure 5 .
Confirmation of tshNC1 deletion following captured segment exchange. (A) Schematic showing relative positions of tshNC1, tshNC2, and confirmation primers NC1check1for (F) and NC1check1rev (R1) ∼10 kb downstream of the tsh transcription unit. Captured segment exchange using GMR-P[acman]-tsh50ΔNC1 as a donor effectively results in deletion of tshNC1 from the chromosome, leaving the ∼1-kb bacterial galK gene in its place. Diagrams are not to scale. (B) Ethidium-stained gel showing PCR products from templates where tshNC1 is unaltered (+) or deleted (Δ) and replaced with galK using purified BAC DNA or Drosophila genomic DNA as templates. Candidate flies homozygous for the recombinant chromosome carrying the deletion (right-most lane) show the predicted PCR product. A similar strategy was used to confirm deletion of tshNC2 (Figure S2).
Captured segment exchange via single-step RMCE using two attP insertions
We designed our strategy for captured segment exchange to take advantage of the large number of FRT- and attP-bearing insertions that are available to the Drosophila community. However, other configurations of SSR recognition sequences could also be employed to accomplish captured segment exchange. In one alternative configuration, a captured segment could be flanked at both ends by attP sites such that exchange can be accomplished by the action of phiC31 integrase alone. To explore this possibility, we used the attP insertions attP52 and attP64 (Markstein et al. 2008), which are separated by ∼40 kb and are oriented such that the yellow marker of each insertion is within the captured segment defined by the attP sites (Figure 6A). We reasoned that exchange of the endogenous segment for a donor sequence would create a recombinant chromosome lacking a yellow marker and could therefore be detected by screening for a y− phenotype in a y w mutant background. To add further evidence that a candidate exchange event is accompanied by recombination between homologs, we used the visible mutations roughoid (ru) and ebony (e), which flank the captured segment, as markers to detect recombination. According to our model, captured segment exchange will result in a recombinant chromosome with a mutant allele of e and a wild-type allele of ru (Figure 6A). Finally, for a donor plasmid, we used a construct carrying two attB sites and a mini-white marker gene. In this case, the orientations of the attB sites are such that captured segment exchange will integrate the plasmid backbone rather than the mini-white gene, whereas “single” integration events resulting from crossovers between one of the attB sites of the donor and one of the parental attP sites will incorporate the entire plasmid and produce red eye pigmentation in the resulting progeny; this serves as a useful control, as these insertions should not be accompanied by a recombination event and should therefore be either e+ ru+ (insertion into attP64) or e− ru− (insertion into attP52). In sum, captured segment exchange using this strategy will result in deletion of the endogenous segment, replacing it with bacterial plasmid sequence, and is predicted to result in flies with a y− w− e− ru+ phenotype.
We injected 390 embryos from a cross of y M[vas-int.Dm]ZH2A w; attP64 virgin females to y w; ru attP52 e/TM3 males, and selected male G0 adult progeny of the genotype y M[vas-int.Dm]ZH2A w/Y; ru attP52 e/attP64 to mate singly to virgin females with the third chromosome balancer TM3 carrying e and ru markers (Figure 6B). We then scored the F1 progeny for y, w, e, and ru phenotypes. Of the 31 vials analyzed, 10 (32%) produced y− w− e− ru+ progeny consistent with the occurrence of segment exchange. Importantly, 46/46 y− flies that we identified among the 10 vials were also w− e− and ru+, indicating that spurious loss of the yellow marker genes did not occur. Furthermore, of the 10 vials in which we identified candidate flies carrying a deletion (y− w− e− ru+), 8 vials also produced flies of the predicted reciprocal chromosome resulting from segment exchange with y+ w+ e+ ru− phenotypes. Finally, as predicted, 9 of the 10 vials also yielded flies consistent with a single attP/attB exchange with the chromosome carrying either attP64 (yielding y+ w+ e+ ru+ phenotypes) or attP52 (y+ w+ e− ru−). In sum, our genetic data are consistent with the occurrence of captured segment exchange when using insertions of attP sites on homologous chromosomes to define the captured segment.
Discussion
Captured segment exchange joins a suite of useful approaches for custom genome manipulation in Drosophila. Importantly, existing methods for making precise changes to Drosophila genomic sequences each employ steps that rely on the endogenous homologous recombination repair machinery and/or require pretreatment of a locus for subsequent manipulation, both of which limit efficiency and throughput (Gloor et al. 1991; Rong and Golic 2000; Gong and Golic 2003; Gao et al. 2008; Choi et al. 2009; Huang et al. 2009; Weng et al. 2009). In contrast, captured segment exchange relies only on highly efficient SSR activity via a dual RMCE reaction and aims to make direct use of existing stocks carrying SSR recognition-site insertions without the need for pretreatment. Furthermore, we have shown that a large segment of 50 kb is efficiently exchanged with diverse donor sequences and that necessary DNA sequence manipulations, including the incorporation of attB and FRT sites, are easily accomplished using the P[acman] recombineering system (Venken et al. 2006). Thus, by taking advantage of several existing resources, captured segment exchange further improves the genetic toolkit of Drosophila researchers.
In addition to demonstrating captured segment exchange using a wild-type donor BAC, we used two strategies to engineer changes into the genome. In the first, we used a version of our donor BAC that carried only 1 kb of the 50-kb segment to create a large deletion, while in the second, we used galK-based recombineering to delete two small regions of noncoding DNA. Although our ΔNC1 and ΔNC2 donor BACs carried the galK gene in place of the deleted Drosophila sequence, the bacterial gene can easily be removed via a simple recombineering strategy prior to the dual RMCE reaction (Warming et al. 2005), making it possible to incorporate “clean” deletions, point mutations, and/or gene fusions into the captured segment. The similar efficiency of exchange achieved with donor BACs carrying large vs. small deletions implies that captured segment exchange will be capable of incorporating diverse sequence alterations into the genome.
Our method should be easily adaptable to other genomic segments that are flanked by existing insertions of an attP and an FRT. As of this writing, the Gene Disruption Project has generated 4071 mapped insertions of the attP-bearing MiMIC transposon (Venken et al. 2011), which corresponds to one insertion every ∼30 kb on average. In addition, a modest number of attP insertions, including the PBac(y+-attP-3B) element used in our study, have been generated by other projects (Groth et al. 2004; Venken et al. 2006; Bischof et al. 2007; Markstein et al. 2008). Similarly, ∼15,000 mapped FRT insertions are publicly available from stock collections at Harvard Medical School, Bloomington, and Kyoto (Ryder et al. 2004; Thibault et al. 2004; Crosby et al. 2007), making it likely that the majority of attP insertions will be within tens of kilobases of the nearest FRT insertion. The upper limit on the potential size of a captured segment will depend on whether a corresponding donor BAC can be recombineered and incorporated into the genome in the first step of the exchange protocol; notably, a 133-kb fragment of the ten-m gene was previously recombineered and inserted into an attP site using P[acman] (Venken et al. 2006), making it reasonable to assume that a captured segment of >100 kb could be easily exchanged for a recombineered sequence. Thus, our method should be applicable to much of the genome, with the number of potential captured segments increasing as new insertions of MiMIC continue to be generated. Furthermore, although we designed captured segment exchange with existing stock collections in mind, it is also possible to incorporate attP and/or FRT sequences into specific sites in the genome via homologous recombination or other methods, which may facilitate establishing a captured segment exchange strategy for those loci where appropriate insertions are unavailable.
In our demonstration at the tsh locus, the yellow and mini-white genes were located within the boundaries of the captured segment and were therefore lost from the recombinant chromosome, making them convenient markers to monitor the exchange reaction. In some cases, it is possible that a potentially useful insertion carrying an FRT or attP would be oriented such that the marker lies outside the captured segment. In this and other cases (e.g., using an unmarked insertion), the deletion of the endogenous segment may not be identifiable by eye and body phenotypes. However, given the high efficiency of FRT-mediated recombination following step 2 of our procedure, it should be straightforward to screen for these events using a simple molecular approach such as PCR. In addition, the use of MiMIC rather than the PBac(y+-attP-3B) element used here should alleviate this issue for the attP end of the exchange; since MiMIC carries two attP sites flanking a yellow gene for use in conventional RMCE transgenesis, half of all inserts following step 1 of our method will be oriented with the yellow marker of MiMIC and the mini-white marker of P[acman] within the boundaries of the captured segment, which will then be lost from the recombinant chromosome following step 2 (Figure S3).
As a final consideration in adapting our strategy to other genomic regions, some potentially useful attP and/or FRT insertions may be located within exonic sequences and therefore may cause confounding phenotypes irrespective of alterations made within the captured segment. In our demonstration, the attP and FRT insertions were located in noncoding regions and caused no visible phenotypes as homozygotes prior to our manipulations. Following exchange, a small amount of transposon sequence remains in the genome at both ends of the captured segment, which could have an adverse effect in some positions in the genome. Thus, insertions in noncoding regions will likely be most useful in defining a captured segment for study.
We chose to establish our method for captured segment exchange based on a dual RMCE strategy employing both phiC31 and FLP due to the efficiency of the former and the larger number of potential targets available for the FLP/FRT system. However, other strategies are possible, as evidenced by our demonstration of segment exchange using two nearby attP insertions on homologous chromosomes. Given the expanding number of SSRs demonstrated to function in Drosophila and other eukaryotes (Nern et al. 2011) and the adaptability of RMCE strategies to diverse enzymes in flies and other organisms, future endeavors may generate new collections of SSR recognition-site insertions that could be adapted to our protocol. Indeed, large collections of SSR recognition-site insertions in other organisms may prove useful in developing similar strategies for genome manipulation in diverse model systems.
Supplementary Material
Acknowledgments
We thank the Bloomington Drosophila Stock Center, the Exelixis Collection at Harvard Medical School, the Drosophila Genomics Resource Center, the Biological Resources Branch of the National Cancer Institute Preclinical Repository, and the BACPAC Resource Center of the Children’s Hospital Oakland Research Institute for stocks and reagents; Bestgene Inc. for embryo injection services; and Chris Smith at the Mount Desert Island Biological Lab DNA Sequencing Core. Special thanks to Gary Struhl for the generous gift of anti-Tsh antibodies and to Michele Markstein for attP insertions. This work was supported by grants from the National Center for Research Resources (5P20RR016463-12) and the National Institute of General Medical Sciences (8 P20 GM103423-12), by funds from the Howard Hughes Medical Institute Undergraduate Science Program, and by Bowdoin College.
Footnotes
Communicating editor: J. Sekelsky
Literature Cited
- Anderson R. P., Voziyanova E., Voziyanov Y., 2012. Flp and Cre expressed from Flp-2A-Cre and Flp-IRES-Cre transcription units mediate the highest level of dual recombinase-mediated cassette exchange. Nucleic Acids Res. 40: e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer A., Bode J., 2001. Coping with kinetic and thermodynamic barriers: RMCE, an efficient strategy for the targeted integration of transgenes. Curr. Opin. Biotechnol. 12: 473–480 [DOI] [PubMed] [Google Scholar]
- Bateman J. R., Wu C. T., 2008. A simple polymerase chain reaction-based method for the construction of recombinase-mediated cassette exchange donor vectors. Genetics 180: 1763–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman J. R., Lee A. M., Wu C. T., 2006. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics 173: 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman J. R., Johnson J. E., Locke M. N., 2012. Comparing enhancer action in cis and in trans. Genetics 191: 1143–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., Liao G., He Y., Carlson J. W., et al. , 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda C. S., Dymecki S. M., 2004. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev. Cell 6: 7–28 [DOI] [PubMed] [Google Scholar]
- Choi C. M., Vilain S., Langen M., Van Kelst S., De Geest N., et al. , 2009. Conditional mutagenesis in Drosophila. Science 324: 54. [DOI] [PubMed] [Google Scholar]
- Court D. L., Swaminathan S., Yu D., Wilson H., Baker T., et al. , 2003. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene 315: 63–69 [DOI] [PubMed] [Google Scholar]
- Crosby M. A., Goodman J. L., Strelets V. B., Zhang P., Gelbart W. M., et al. , 2007. FlyBase: genomes by the dozen. Nucleic Acids Res. 35: D486–D491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., McMahon C., Chen J., Rong Y. S., 2008. A powerful method combining homologous recombination and site-specific recombination for targeted mutagenesis in Drosophila. Proc. Natl. Acad. Sci. USA 105: 13999–14004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor G. B., Nassif N. A., Johnson-Schlitz D. M., Preston C. R., Engels W. R., 1991. Targeted gene replacement in Drosophila via P element-induced gap repair. Science 253: 1110–1117 [DOI] [PubMed] [Google Scholar]
- Golic K. G., Golic M. M., 1996. Engineering the Drosophila genome: chromosome rearrangements by design. Genetics 144: 1693–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W. J., Golic K. G., 2003. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100: 2556–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R., Calos M. P., 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166: 1775–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R. M., Cai Z. L., Ho S. N., Pease L. R., 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8: 528–535 [PubMed] [Google Scholar]
- Huang J., Zhou W., Dong W., Watson A. M., Hong Y., 2009. From the cover: directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc. Natl. Acad. Sci. USA 106: 8284–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauth M., Spreafico F., Dethleffsen K., Meyer M., 2002. Stable and efficient cassette exchange under non-selectable conditions by combined use of two site-specific recombinases. Nucleic Acids Res. 30: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D. A., et al. , 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73: 56–65 [DOI] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S. E., Perrimon N., 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40: 476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. R., Chen J. L., Geyer P. K., Wu C. T., 1998. Two modes of transvection: enhancer action in trans and bypass of a chromatin insulator in cis. Proc. Natl. Acad. Sci. USA 95: 10740–10745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses K., Rubin G. M., 1991. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 5: 583–593 [DOI] [PubMed] [Google Scholar]
- Negre N., Brown C. D., Ma L., Bristow C. A., Miller S. W., et al. , 2011. A cis-regulatory map of the Drosophila genome. Nature 471: 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A., Pfeiffer B. D., Svoboda K., Rubin G. M., 2011. Multiple new site-specific recombinases for use in manipulating animal genomes. Proc. Natl. Acad. Sci. USA 108: 14198–14203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder M., Galli A., Rosen B., Skarnes W. C., Zeller R., et al. , 2010. Dual RMCE for efficient re-engineering of mouse mutant alleles. Nat. Methods 7: 893–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks A. L., Cook K. R., Belvin M., Dompe N. A., Fawcett R., et al. , 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292 [DOI] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018 [DOI] [PubMed] [Google Scholar]
- Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N., et al. , 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E., Blows F., Ashburner M., Bautista-Llacer R., Coulson D., et al. , 2004. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167: 797–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan S. K., Thomason L. C., Kuznetsov S. G., Court D. L., 2009. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 4: 206–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. C., Brown W. R., McEwan A. R., Rowley P. A., 2010. Site-specific recombination by phiC31 integrase and other large serine recombinases. Biochem. Soc. Trans. 38: 388–394 [DOI] [PubMed] [Google Scholar]
- Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A., et al. , 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287 [DOI] [PubMed] [Google Scholar]
- Venken K. J., Bellen H. J., 2005. Emerging technologies for gene manipulation in Drosophila melanogaster. Nat. Rev. Genet. 6: 167–178 [DOI] [PubMed] [Google Scholar]
- Venken K. J., Bellen H. J., 2007. Transgenesis upgrades for Drosophila melanogaster. Development 134: 3571–3584 [DOI] [PubMed] [Google Scholar]
- Venken K. J., He Y., Hoskins R. A., Bellen H. J., 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751 [DOI] [PubMed] [Google Scholar]
- Venken K. J. T., Schulze K. L., Haelterman N. A., Pan H., He Y., et al. , 2011. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8: 737–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S., Costantino N., Court D. L., Jenkins N. A., Copeland N. G., 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng R., Chen Y. W., Bushati N., Cliffe A., Cohen S. M., 2009. Recombinase-mediated cassette exchange provides a versatile platform for gene targeting: knockout of miR-31b. Genetics 183: 399–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.