Abstract
The estimation of dominance effects requires the availability of direct phenotypes, i.e., genotypes and phenotypes in the same individuals. In dairy cattle, classical QTL mapping approaches are, however, relying on genotyped sires and daughter-based phenotypes like breeding values. Thus, dominance effects cannot be estimated. The number of dairy bulls genotyped for dense genome-wide marker panels is steadily increasing in the context of genomic selection schemes. The availability of genotyped cows is, however, limited. Within the current study, the genotypes of male ancestors were applied to the calculation of genotype probabilities in cows. Together with the cows’ phenotypes, these probabilities were used to estimate dominance effects on a genome-wide scale. The impact of sample size, the depth of pedigree used in deriving genotype probabilities, the linkage disequilibrium between QTL and marker, the fraction of variance explained by the QTL, and the degree of dominance on the power to detect dominance were analyzed in simulation studies. The effect of relatedness among animals on the specificity of detection was addressed. Furthermore, the approach was applied to a real data set comprising 470,000 Holstein cows. To account for relatedness between animals a mixed-model two-step approach was used to adjust phenotypes based on an additive genetic relationship matrix. Thereby, considerable dominance effects were identified for important milk production traits. The approach might serve as a powerful tool to dissect the genetic architecture of performance and functional traits in dairy cattle.
Keywords: dominance, genotype probabilities, genomic selection, dairy cattle
IN the context of genomic selection in dairy cattle, an abundance of bulls has been genotyped by applying genome-wide dense marker panels. In 2010, the European reference population comprised >17,000 bulls representing >20 million daughters (Lund et al. 2010; Liu et al. 2011). In addition to their utilization in genomic prediction, these data are extensively used in genome-wide association studies to unravel the genetic factors affecting performance and functional traits. The expression of these traits is naturally limited to female individuals and thus, the phenotypes used in association studies are usually breeding values of sires based on performance data of many daughters. Such a structure of data allows only the estimation of allele substitution effects. There is no direct possibility to distinguish between additive and dominance effects. For the detection of these allelic interactions, genotypes and phenotypes had to be known in the same individuals. Compared to the bulls, the availability of genotype data for cows is limited. With the increasing number of genotyped bulls, genotypes of male ancestors become available for many cows, enabling the derivation of genotype probabilities. Within the current study, these probabilities were converted to additive and dominance coefficients suitable for regression analysis analogous to the procedures commonly applied to QTL mapping in resource populations (Haley and Knott 1992). The derivation of coefficients is not applied to intermarker intervals based on recombination fractions, but to unknown genotypes at given SNP marker positions. At a specific marker locus with two alleles A and B, the probabilities of the possible genotypes AA, AB, and BB can be deduced in cows based on the male ancestor’s genotypes and the allele frequencies in the population. The approach implies a loss of statistical power compared to the utilization of real genotype data. A large number of genotyped bulls and an extensive number of daughters per sire, however, should compensate for these limitations.
Alternative methods to deduce genotype probabilities include “peeling” algorithms based on the ideas of Elston and Stewart (1971), Monte Carlo methods (e.g., Guo and Thompson 1992; Henshall and Tier 2003), or Bayesian approaches. These methods are computationally infeasible for the very large data sets as used within the current study. Approaches to impute genotypes based on phase information are inapplicable because this would require at least partly genotyped females. We conducted a series of simulation studies to discover both the capabilities and limitations of the method and to evaluate the importance of factors like allele frequencies, sample size, and size of daughter groups. Subsequently, the approach was validated in a large German Holstein data set. The method presented herein enhances the utilization of existing large data sets to dissect the genetic architecture of important performance and functional traits in dairy cattle.
Materials and Methods
Derivation of genotype probabilities
To calculate the three genotype probabilities , , and for a biallelic SNP marker with two alleles A and B and the respective allele frequencies p and q in a cow, we considered pedigrees including at least a genotyped sire and a maternal grandsire (Figure 1). For any genotyped bull, the probability to transmit allele A is in the homozygous , for the heterozygous , and for the alternative homozygous state . The probability to transmit allele B is . This information is unavailable for the female individuals. Therefore, the respective population frequency of the allele was used as an approximation for the transmission probability from the maternal granddam to the dam of the cow under consideration. The genotype probabilities for the dam, are the conditional probabilities emerging from this approximation and the known genotypes of the maternal grandsires as summarized in Table 1. The allele frequencies were estimated from the bull data set, assuming that the allele frequencies in the cows are not considerably different from those observed in the bulls. For a given cow, nine possible scenarios exist, conditional on the genotypes of the sire and the maternal grandsire (Table 2). The respective genotype probabilities can be calculated once and assigned to the cows by looking up the precalculated values. However, based on the genotype probabilities presented in Table 1, the transmission probability for allele A of the dam conditional on the genotype of the maternal grandsire, can be calculated as the sum of the probability of being homozygous AA and half the probability of being heterozygous:
The second summand within the numerator in this case is equivalent to the transmission probability of the maternal grandsire, . Thus, the three equations can be jointly expressed as
Equivalently, the transmission probability for allele B, can be calculated as
The probabilities for the homozygous genotypes in the cow are finally calculated from the sire’s and dam’s transmission probabilities as follows:
The probability of being heterozygous is simply calculated as . It is also possible to directly calculate the cow’s probability of being heterozygous as
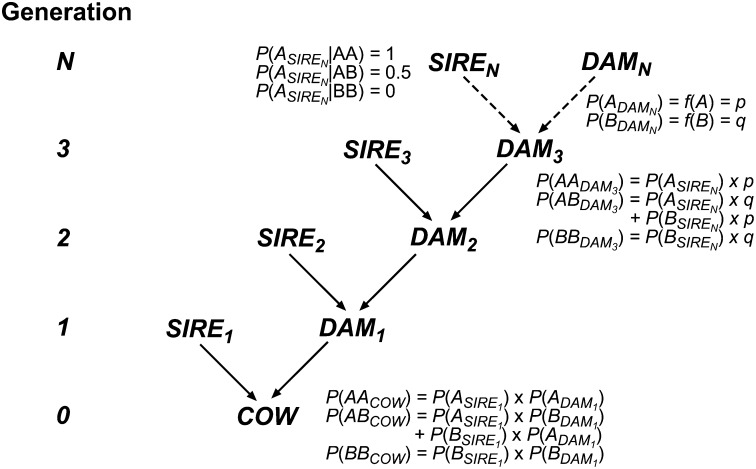
Figure 1 .
Graphical representation of the pedigree structure used in the calculation of genotype probabilities. N determines the generation in relation to the cow under consideration as used in the formulas to derive probabilities. P(Genotype) with genotypes AA, AB, and BB is the genotype probability and P(Allele) with alleles A and B is the transmission probability for the respective allele.
Table 1 . Genotype probabilities for the dam of the cow under consideration.
| Genotype GS | ||||
|---|---|---|---|---|
| AA | 1 | p | q | 0 |
| AB | ||||
| BB | 0 | 0 | p | q |
The probabilities were calculated from the allele frequencies and and the probability of the grandsire (GS) to transmit allele A, .
Table 2 . Conditional genotype probabilities of a cow.
| Genotype sire | |||
|---|---|---|---|
| Genotype maternal grandsire | AA | AB | BB |
| AA | |||
| AB | |||
| BB | |||
The nine possible scenarios for the genotype probabilities of a cow conditional on the genotypes of the animal’s sire and maternal grandsire as well as the frequency of the rare allele A, are shown. The probabilities were calculated by multiplying the respective transmission probabilities of the sire and dam of the cow under consideration.
Referring to Table 1 and considering the example of a sire with genotype AA and a maternal grandsire with genotype BB, the probability can be calculated as
(see Table 2).
This calculation would allow distinguishing between different parental origins of the alleles, resulting in the two probabilities and , which could be used to analyze imprinting effects. This was, however, out of the scope of the current study.
For practical reasons, all calculations were based only on the paternal transmission probability and the frequency of the rare allele representing the minor allele frequency (MAF). and were thus replaced by and , respectively. The approach can readily be adapted for deeper pedigrees. With the inclusion of a genotyped maternal great-grandsire (GGS), i.e., three ancestral generations, the probabilities for the homozygous genotypes of the cow under consideration can be calculated similarly to the procedure described above. The transmission probabilities in the granddam are and . Multiplication by the transmission probabilities of the grandsire gives the genotype probabilities of the dam, which can then be used to calculate the transmission probabilities. Analogous to the initial approach with two ancestral generations, the probabilities are
Thus, the probabilities for the homozygous genotypes in the cow under consideration can be calculated as
The inclusion of a further generation (great-great-grandsire, GGGS) consequently leads to the following transmission probabilities in the dam:
To generalize the approach for any number N of ancestral generations, the generations were numbered such that the cow under consideration represents generation “0” (Figure 1). The parents of the cow are generation 1 (SIRE1, sire; and DAM1, dam), the maternal grandparents are generation 2 [SIRE2, grandsire (GS); and DAM2], and the parents of DAM2 are generation 3. Defining N as the number of ancestral generations, the genotype probabilities can be calculated as follows:
The probabilities were transformed to regressors for additive and dominance effects with and , which can be used in a simple linear regression model of the form
where μ is the mean, yi represents the phenotype of the ith animal, addi and domi are the additive and dominance coefficients calculated from the genotype probabilities, β1 and β2 are the respective regression coefficients, and ei is a random residual term with .
Simulation studies
To verify the functionality of our approach and to determine the necessary sample size to sufficiently detect dominance effects, populations of completely unrelated cows were simulated. This was achieved by assigning an individual sire and grandsire per cow exclusively. A single biallelic marker in complete linkage disequilibrium (LD) with a nearby QTL was simulated. The sire genotypes for that marker were sampled from a population in Hardy–Weinberg equilibrium, given a MAF of 0.3. By utilizing the presented method for deriving genotype probabilities, the two coefficients add and dom were calculated for each cow based on the genotypes of sire, damsire, and the allele frequency. Furthermore, we assigned a true genotype for every cow by sampling genotypes given the probabilities presented in Table 2. This “true” genotype was used as a reference for the assignment of genetic effects to the simulated QTL. Phenotypes were calculated by assuming a defined phenotypic variance multiplied by a given heritability of h2 = 0.25, leading to the additive genetic variance. The sizes of the simulated QTL effects were determined as a multiple of the additive genetic standard deviations (), with the additive effect being a half of the genetic standard deviation and the dominance effect making up half the size of the additive effect. Finally, a random residual effect was assigned to every phenotype.
Subsequently, the influence of various properties of the created population and of the features of the simulated QTL on the power of detection was tested by separately varying the sample size, the number of considered generations, the minor allele frequency, the LD between QTL and marker, the fraction of variance explained by the QTL, and the degree of dominance. The effect of sample size was analyzed by a stepwise increase of up to 106 cows. To achieve a better resolution across the entire range of N, a reduced heritability of 0.2 and a smaller QTL effect of 0.2 additive genetic standard deviations were applied. The latter scenario was carried out with true genotypes as well as with genotype probabilities. The different scenarios are described in Table 3. For each scenario, 1000 iterations were performed and the power was defined as the rate of positives in the total number of iterations. The significance criterion was fixed to 5% probability of error after a Bonferroni correction to account for multiple testing. For this correction, 40,000 informative markers were assumed, which is a realistic scenario for the application of the Illumina 50k SNP Chip in Holstein cattle (see, e.g., Habier et al. 2010).
Table 3 . Detailed description of the single-marker simulation scenarios.
| Scenario | Tested parameter | h2 | QTL effect | Sample size | MAF | LD | d/|a| | Range of modified parameter | Explanatory notes |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Sample size | 0.2 | 0.2σA | — | 0.3 | r2 = 1 | 0.5 | N = 50,000–1,000,000 (14 steps) | Heritability and effect size were reduced to achieve a higher resolution across the entire range of N. True genotypes were compared with genotype probabilities. |
| 2 | No. considered generations | 0.25 | 0.5σA | N = 5,000–100,000 (19 steps) | 0.3 | r2 = 1 | 0.5 | 2 sires vs. 3 sires | Genotype probabilities were calculated using a genotyped sire and grandsire only or including an additional great-grandsire in an increasing sample size. |
| 3 | Minor allele frequency | 0.25 | 0.5σA | 10,000 vs. 100,000 | — | r2 = 1 | 0.5 | MAF = 0.025–0.5 (19 steps) | MAF was increased stepwise in two different sample sizes. |
| 4 | LD between QTL and marker | 0.25 | 0.5σA | N = 5,000–100,000 (19 steps) | 0.3/0.5 | — | 0.5 | r2 = 0.4/0.6/0.8 | An increasing sample size was analyzed for three different r2 values and two different MAFs. |
| 5 | Variance explained by the QTL | 0.25 | — | 10,000 vs. 100,000 | 0.3 | r2 = 1 | 1 | = 0.01–0.2 (14 steps) | was increased stepwise in two different sample sizes. |
| 6 | Degree of dominance | 0.25 | 0.5σA | 10,000 vs. 100,000 | 0.3 | r2 = 1 | — | d/a = 0.05–2 (39 steps) | d/|a| was increased stepwise in two different sample sizes. True genotypes were compared with genotype probabilities within these samples. |
For each scenario, the range of the modified parameter is given together with the fixed parameters. The QTL effects are expressed as additive genetic standard deviations (σA). MAF and LD stand for minor allele frequency and linkage disequilibrium, respectively. The degree of dominance was calculated as the ratio of dominance effect and absolute additive effect (d/|a|).
A second set of simulations was conducted to determine the influence of relatedness among animals and resulting stratification, which might lead to a high rate of false-positive signals. To this end, we repeated the simulations in a multimarker scenario including related ancestors of the cows. A sample of 100,000 cows was created, descending from initially 100 sires and 100 maternal grandsires. The phenotypes of the cows were assumed to consist of the combination of marker effects and additional polygenic effects received from the ancestors. A trait of midrange heritability (h2 = 0.3) was simulated with the genetic variance in the cows equally explained by the bull’s breeding values and the marker effects. The sires contributed half and the maternal grandsires one-quarter of their respective breeding values. A total of 150 markers with minor allele frequencies ranging from 0.05 to 0.5 were defined. One-tenth of the variance explained by markers was equally assigned to 10 of the markers, therefore each simulating a strong QTL. The dominance ratio was randomly assigned within these 10 markers. The remainder of markers was left without any effect. Subsequently, the effect of a varying family size was tested. For this purpose, the number of descendants per sire was varied from 50 to 5000 daughters, leading to different proportions of cows with identical sires and maternal grandsires, thus representing groups of animals with uniform genotype probabilities. For each family size, the specificity was calculated as the number of true negatives divided by the sum of true negatives and false positives.
To correct for relationship among animals as a source of stratification, especially due to sires with a very large number of daughters, a single-marker linear mixed-model regression using residual maximum likelihood (REML) was applied. Therefore, a relationship matrix based on dam, sire, and damsire was used with the model
where y is a vector of phenotypes, b is a vector of fixed effects consisting of the intercept and the coefficients add and dom, and X is the incidence matrix for the fixed effects. The vector a represents the random effects with , where A is the additive genetic relationship matrix. Z is the incidence matrix for the random effects and e is a vector of residuals with , with I representing an identity matrix. To test for significance, a Wald t-test was applied, where the denumerator degrees of freedom were numerically estimated (Kenward and Roger 1997).
Application to a real data set
To validate the procedures with real data, phenotypic and pedigree data of 847,000 German Holstein cows were obtained from the Vereinigte Informationssysteme Tierhaltung w.V. (VIT). The phenotypic data consisted of yield deviations (YDs) for the traits milk, protein, and fat yield as well as somatic cell score (SCS) in the first lactation representing the deviations of the cow’s yields from the population mean adjusted for nongenetic fixed and random effects. The phenotypic data are summarized in Table 4. Genotype information was available for 3200 Holstein bulls, genotyped with the Illumina BovineSNP50 BeadChip (Illumina, San Diego), featuring a total of 54,001 SNPs. The genotyping was done in the context of the GenoTrack research program funded by the German Federal Ministry of Education and Research. SNPs were filtered for their minor allele frequency and missing genotypes per locus. The thresholds were set to 0.05 and 0.1, respectively. Based on available pedigree information, a sample of ∼470,000 cows per trait with complete phenotypic data and genotyped sires and maternal grandsires was assembled (Table 4). The cows descend from 2081 bulls of which 1916 occur as sire and 1981 as grandsire. A total of 86,254 unique sire–damsire combinations were observed. The data set consisting of the bull’s genotypes, the pedigree, and the yield deviation of the cows is available in Supporting Information, File S2. For this sample, the add and dom coefficients were calculated according to the procedure developed within this study and phenotypes were regressed onto the coefficients by applying a simple linear regression model as outlined above.
Table 4 . Description of the real data set.
| μ | Min | Max | σP | N cows | |
|---|---|---|---|---|---|
| Phenotype summary | |||||
| Milk yield | 7.61 × 10−9 | −1,426.85 | 1,284.05 | 213.54 | 469,454 |
| Fat yield | 7.26 × 10−10 | −125.86 | 114.04 | 16.75 | 470,260 |
| Protein yield | −6.75 × 10−13 | −63.22 | 54.86 | 8.71 | 469,171 |
| Somatic cell score | 5.57 × 10−12 | −3.60 | 3.61 | 0.42 | 471,093 |
| Summary of family structure | |||||
| Daughters per sire | 283.0 | 1 | 18,030 | ||
| Granddaughters per sire | 274.7 | 1 | 33,139 | ||
| Cows per combination of sire and grandsire | 6.3 | 1 | 2,411 |
The top section summarizes the mean (μ), minimum (min), and maximum (max) values of the YD as well as their respective standard deviation (σP) and the number of cows with available phenotypes. The bottom section summarizes the family structure.
Furthermore, the mixed-model approach as described above was applied to correct for family-based stratification. This approach is computationally challenging on a genome-wide scale. Thus, a two-step approach was applied, which is equivalent to the previously described genome-wide rapid association using mixed model and regression (GRAMMAR) approach (Aulchenko et al. 2007). It offers a way to efficiently reduce the time-consuming computations required for extensive pedigrees. In the first step, phenotypes were adjusted for a polygenic effect by applying the mixed model, but omitting the effects for add and dom. The residuals coming from this step were used as phenotypes in a subsequent linear regression. With this two-step approach, the traits SCS, milk yield (MY), fat yield (FY), and protein yield (PY) were analyzed on a genome-wide scale.
To validate the two-step approach, chromosome 14 (BTA14) was analyzed by applying the full linear mixed model including a polygenic term as well as the coefficients for add and dom. BTA14 was chosen for this purpose, because the centromeric region of this chromosome contains a major QTL for milk production traits caused by polymorphisms in the acylCoA-diacylglycerol-acyltransferase (DGAT1) gene (Riquet et al. 1999; Grisart et al. 2002; Spelman et al. 2002; Winter et al. 2002; Thaller et al. 2003; Weller et al. 2003). This QTL segregates within the analyzed population and was thus used as a reference to evaluate the two-step approach as compared to a full model. Furthermore, those SNPs considered as genome-wide significant at a threshold of P ≤ 0.01 after Bonferroni correction were reanalyzed by applying a full model including a polygenic effect and the coefficients for add and dom at the same time.
Implementation
All data-handling procedures including genotype integrity checks, recoding, and allele frequency calculation were performed using unix shell scripts together with the genome analysis toolset plink (Purcell et al. 2007). The derivation of genotype probabilities and simulation routines as well as regression models and test statistics were realized using the R statistical environment (R Development Core Team 2008). The R-script with the simulation routines as well as the R- and Shell-scripts used for data analysis can be found in File S1. The linear mixed models were fitted using REML as implemented in ASReml 3.0 (VSN International) (Gilmour et al. 1995).
Results and Discussion
Simulations
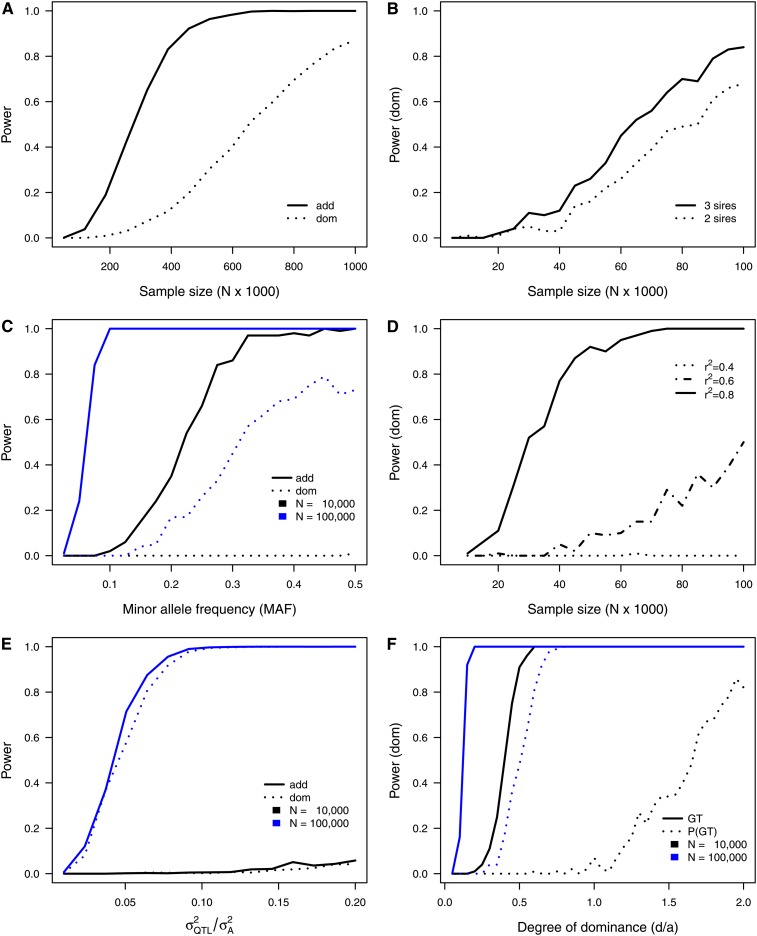
The results of the single-marker simulation scenarios are summarized in Figure 2, illustrating the major impacts on the power to detect additive and dominance effects, using genotype probabilities. All parameters under consideration substantially influenced the power of detection. Under a scenario with a heritability of 0.2 and an effect size of 0.2 with d/|a| = 0.5 (scenario 1, Table 3), a sample size of 800,000 cows was needed to achieve a power of 0.7 for the detection of dominance effects (Figure 2A). With an effect size of 0.5 and a heritability of 0.25 the same power can be achieved with 100,000 animals (Figure 2B). Approximately half of the sample size is needed to detect the corresponding additive effects (Figure 2A). When using the true genotypes, powers of 0.94 and 0.69 for additive and dominance effects, respectively, are achieved with 50,000 cows. A sample size of ∼100,000 animals results in a power of 1 for both effects under these assumptions (not shown in Figure 2). Using genotype probabilities, the necessary sample size is 10 times as large as needed with real genotype data. This clearly reflects the decrease in power arising from the impreciseness of genotype probabilities and the need to compensate this with a larger sample size. Under realistic conditions the power is even lower, because the simulated marker was assumed to be in complete LD with the QTL and the MAF was fixed to 0.3. From Figure 2D it arises that with the reduction of r2 from 0.8 to 0.6 the power to detect dominance effects drops from 1 to ∼0.5 (scenario 4, Table 3) when assuming a MAF of 0.5. This scenario was also carried out using the standard MAF of 0.3, but the power was considerably lower (not shown in Figure 2D). The substantial dependency between power and LD is reflecting the fact that only parts of the QTL variance can be explained by markers in incomplete LD. A marker showing an LD of r2 = 0.5 can capture approximately half of the QTL effect, meaning that the sample size must be doubled to capture an effect of a given size. Regarding the MAF, it can be seen from Figure 2C that a frequency of 0.5 results in the highest power to detect dominance effects (scenario 3, Table 3). This ideal situation cannot be expected under practical conditions. Thus, we set the MAF to 0.3 in the other simulations. For the additive effect, the MAF is not a limiting factor as in a sample of 100,000 cows the power equals 1 even for a MAF of 0.1. However, there might be a limitation to the strong interdependency between MAF and LD shown above. When there is incomplete LD between QTL and marker, the results are very sensitive to low minor allele frequencies. This has to be elucidated in further studies.
Figure 2 .
Graphical representation of the results obtained from single-marker simulation scenarios according to Table 3. (A) Power to detect additive (add) and dominance (dom) effects conditional on sample size (scenario 1). (B) Power to detect dominance effects conditional on sample size and the number of sires used in calculating genotype probabilities (scenario 2). (C) Power to detect additive (add) and dominance (dom) effects conditional on the minor allele frequency (MAF) in two different sample sizes (scenario 3). (D) Power to detect dominance effects conditional on sample size assuming different LD (r2) between marker and QTL (scenario 4). (E) Power to detect additive (add) and dominance (dom) effects conditional on the proportion of additive genetic variance explained by the QTL () in two different sample sizes (scenario 5). (F) Power to detect dominance effects conditional on the degree of dominance (d/|a|) and compared between true genotypes (GT) and genotype probabilities [P(GT), scenario 6].
Another important parameter is the proportion of variance explained by the QTL (scenario 5, Table 3). For a given degree of dominance of d/|a|= 1, a QTL explaining ∼5% of the additive genetic variance would be detectable with a power of ∼0.6 in a sample of 100,000 cows (Figure 2E). Assuming a heritability of 0.25, this would correspond to 1.25% of the phenotypic variance. That is a realistic value in dairy cattle (Hayes and Goddard 2001; Khatkar et al. 2004), leading to the conclusion that the approach should work well even for QTL with smaller effects. The power curves for additive and dominance effects as shown in Figure 2E are congruent, because the degree of dominance was set to 1. Figure 2F shows the dependency of the power to detect dominance under different degrees of dominance for a given total QTL effect (scenario 6, Table 3). From Figure 2 it can be seen that genotype probabilities calculated for a sample of 100,000 cows are sufficient to detect a degree of dominance of ∼0.33 at the given total QTL effect with a power of ∼0.8. Regarding the sample sizes discussed above, the detection of dominance effects even at a low degree of dominance is possible with the approach. Furthermore, Figure 2F impressively illustrates the inferiority of genotype probabilities in small sample sizes. In a sample of 10,000 cows, a power of 1 can be achieved at d/|a| = 1 with true genotypes, while the application of genotype probabilities results in a power close to zero for this d/|a|.
A part of the decrease in power arising from the use of genotype probabilities can be compensated by the utilization of deeper pedigrees in the estimation of these probabilities. When using N ancestral generations, a proportion of of the available information arises from the allele frequency and is thus equal for all animals. With the inclusion of two ancestral generations, i.e., a genotyped sire and damsire, one-quarter of the information comes from the allele frequency. When including a genotyped great-grandsire, only one-eighth arises from the frequency. Figure 2B illustrates the impact of two vs. three genotyped male ancestors on the power to detect dominance effects, reflecting this dependency (scenario 2, Table 3). The inclusion of the third sire increases the power by 0.1 and 0.16 for 50,000 and 100,000 cows, respectively.
From the results of these single-marker simulation scenarios it can be concluded that for traits of midrange heritability, at least strong QTL effects can be detected with markers being in LD with the QTL in the range of r2 > 0.4. However, the necessary sample size for detecting dominance effects is substantially >100,000 cows. A reliable power to detect smaller dominance effects might under practical conditions even require a sample size of >1,000,000 cows. This is, however, not limiting. Phenotypic data sets of this size are available on the basis of national evaluation and increasing reference samples in genomic selection schemes provide the necessary genotypic information. Furthermore, the simple regression approach presented herein is computationally not challenging. In reality, however, the finite number of sire–damsire combinations, representing the source of information for the genotype probabilities, will be the limiting factor in the sense of information content.
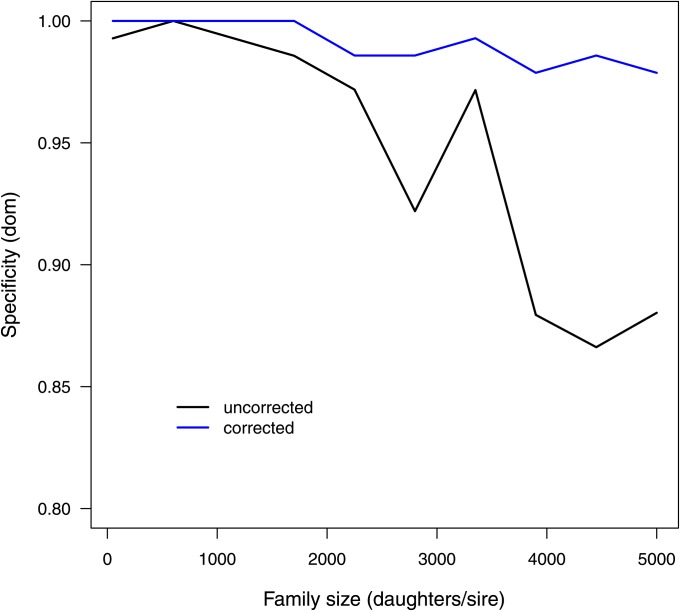
The second simulation was conducted as a multimarker scenario and focused on a more realistic population with family-based stratification. Expectedly, this led to the occurrence of false positives in addition to those arising at random as type I error. This is reflected by the specificity to detect dominance as depicted in Figure 3. When increasing the number of daughters per sire from 50 to 5000, the specificity drops from ∼1 to ∼0.85. To obtain reliable results, a correction for this stratification is necessary. A rather simple approach would be the inclusion of the fixed effect of the sire and possibly also of the grandsire. This is hampered by the fact that deduced genotypes are a direct function of ancestral genotypes. When using two ancestral generations, all cows descending from the same combination of sire and grandsire have equal genotype probabilities. Correcting for the fixed effects of sire and grandsire is thus inadequate. A valid correction is inevitably linked to the availability of a more independent source of information. A mixed model using a pedigree-based relationship matrix to include a polygenic effect meets this criterion. Figure 3 outlines the impact of family sizes and correction on the specificity to detect dominance effects. Even though there is no substantial decrease in specificity with increasing family size in simulated data, the correction using the linear mixed-effects model including the relationship matrix noticeably improves specificity. This is much more pronounced in real data as is shown below, emphasizing the importance of properly accounting for stratification.
Figure 3 .
Graphical representation of the specificity to detect dominance effects as obtained from the multimarker simulation scenario. Compared are the specificities conditional on family size (daughters per sire) without any correction for relatedness (black) and after correction applying a linear mixed model (blue).
Real data analyses
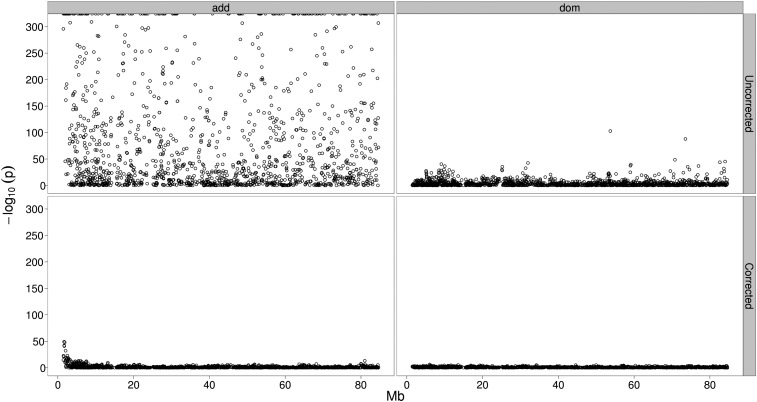
In a first attempt to validate our approach we analyzed cattle chromosome 14 (BTA14) for the trait milk yield without accounting for stratification. This chromosome was chosen, because DGAT1 located in the centromeric region is the underlying gene of a major QTL, which we attempted to detect based on genotype probabilities. The additive effect estimators for the markers in close proximity to DGAT1 showed results correctly reflecting the strong additive effect known to originate from this locus. However, the attempt was hampered by a striking lack of specificity due to stratification. The results are characterized by extremely low P-values along the entire chromosome, especially for the additive coefficients, while the dominance part is less influenced. These results are depicted in the top part of Figure 4. To analyze a possible effect of sample size, we started with 10,000 cows and sequentially increased the number up to 470,000, representing the entire data set (data not shown). Larger samples did not lead to an improvement; the specificity substantially decreased with an increasing number of cows. When reaching the maximum sample size, false positives were completely covering the association signal. The subsequent correction applying a mixed model resulted in an effective adjustment. The bottom part of Figure 4 shows the corrected results for additive and dominance signals, using the complete data set of 470,000 cows with genotype probabilities. The direct comparison of the results without and with correction impressively illustrates the effectiveness of the approach. After adjustment, the aforementioned DGAT-QTL can easily be detected at a position of ∼0.5 Mb.
Figure 4 .
Analysis of BTA14 for the trait milk yield in a real data set comprising 469,454 cows. The y-axis represents the negative decadic logarithm of the P-values for the additive (left) and dominance (right) effects after Bonferroni correction. The x-axis gives the position on BTA14 in megabase pairs according to genome build UMD 3.1. The top panel represents the results without correction. Especially for the additive effects, there is a striking lack of specificity. The bottom panel depicts the results obtained by applying mixed-model correction.
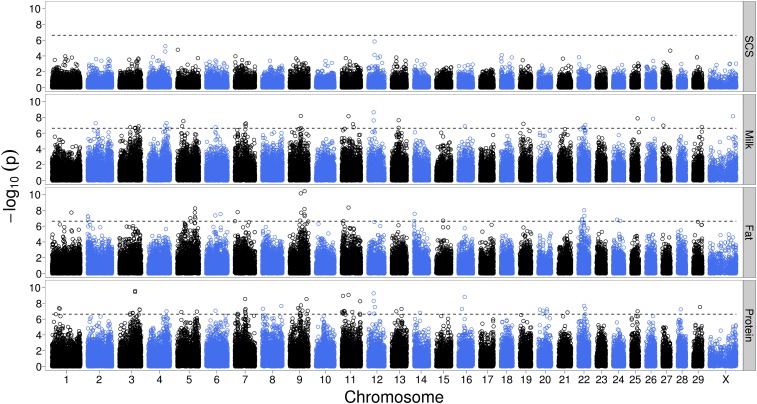
In a next step, we conducted analyses on a genome-wide scale for the traits SCS, MY, FY, and PY, applying the substantially faster two-step approach. The direct comparison of the results for BTA14 revealed almost identical results for dominance effects. The preadjustment of phenotypes, however, dramatically increased the sensitivity for additive signals (data not shown). Therefore, it works as a quick scan for dominance. It might be worth investigating whether this can be improved by the utilization of deeper pedigrees in the calculation of the relationship matrix. Figure 5 is an illustration of the result of a genome scan for dominance effects. Applying a significance threshold of P ≤ 0.01 after Bonferroni correction, there were no significant results for SCS. For MY, FY, and PY, 29 SNPs on 15 chromosomes, 30 SNPs on 11 chromosomes, and 59 SNPs on 17 chromosomes with significant dominance effects were detected, respectively (Table S1). Some chromosomal regions, especially on BTA9 and BTA22, exhibit significant dominance effects for all analyzed yield traits. To further characterize these findings, additional analyses for FY and PY were performed including MY as a covariable in the preadjustment, aiming to reflect the corresponding content traits. Notably, those regions on BTA9 and -22 affecting FY also significantly affected fat content, while those affecting PY had no significant dominance effect for protein content (data not shown). Thus, it can be concluded that the effect in PY is due to an effect on MY, while fat content is directly affected. These two chromosomes seem to harbor QTL for milk yield and fat content displaying considerable dominance effects. QTL for yield traits have been previously described on BTA9 (Wiener et al. 2000) as well as on BTA22 (Ashwell et al. 2004; Harder et al. 2006). However, these studies used genotyped sires and daughter-based phenotypes and did thus not identify any dominance effects. Furthermore, significant dominance effects were detected on BTA14 within the DGAT1 region. Applying the full model as described below, a dominance effect of 0.063 phenotypic standard deviations (σp) along with a significant additive effect of 0.202 σp was found for marker ARS-BFGL-NGS-100480 at 4.36 Mb. This is in general accordance with the divergent effect of the heterozygous condition compared to the mean of the homozygous states found in a study using 1035 Holstein cows genotyped for two DGAT1 polymorphisms (Kuehn et al. 2007).
Figure 5 .
Manhattan plots representing the results of a genome scan for dominance, applying correction for relatedness among animals in a two-step mixed-model approach. Analyzed were the traits somatic cell score (SCS), milk yield, fat yield, and protein yield. Shown are the negative decadic logarithms of the raw P-values with a Bonferroni-corrected 1% genome-wide significance level (dashed line).
As the dependent variables in this approach are residuals from the previous step, the effect estimators cannot be interpreted readily. Thus, an analysis applying the full model is necessary for the markers considered significant to obtain reliable effect estimators. This was conducted for the markers showing significant dominance effects with P ≤ 0.01 for MY, FY, and PY. The effect estimators obtained from this model are exemplarily shown for BTA9, -14, and -22 in Table 5 (see also Table S1). The markers on BTA9 are distributed across a large chromosomal region ranging from 46.5 to 100.3 Mb. They are characterized by notably high degrees of dominance. For milk yield, there is a clear peak around 62 Mb. The highest dominance effect of 0.065σp for this trait was found at marker Hapmap35559-SCAFFOLD35632_12026 at 62.96 Mb. The degree of dominance (d/|a|) = 1.754 and no significant additive effect was detected applying the full model. This is even more pronounced for PY. For this trait, the same marker shows a dominance effect of 0.072σp and a d/|a| value of 12.466. Such signals might be missed in studies using breeding values as phenotypes. In this case, however, there are adjacent markers, for which considerable additive effects were detected (Table 5). The same applies for BTA22, where the d/|a| values are generally smaller compared to those for BTA9. In total, there are 10, 12, and 26 SNPs with d/|a| values >2 for MY, FY, and PY, respectively (Table S1). For none of these markers was a significant additive effect observed when applying the full model. For each analyzed yield trait, there is at least one marker showing an additive effect of >0.1σp with P ≤ 0.01. These effects would probably have been detectable in studies using daughter-based breeding values, but the considerable dominance effects would be missed. Large-scale studies using breeding values or daughter yield deviations exhibit high statistical power and have successfully been applied to the identification of numerous QTL in dairy cattle. However, our results emphasize the need to use direct phenotypes to better understand the genetic architecture of traits.
Table 5 . Results from applying the full mixed model to the real data set for selected markers and chromosomes.
| Milk yield | Fat yield | Protein yield | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | bp | Add | Dom | d/|a| | Add | Dom | d/|a| | Add | Dom | d/|a| |
| Hapmap33674-BTA-156367 | 9 | 46,531,368 | — | — | — | — | — | — | 0.032 | 0.064 | 1.961 |
| BTB-01878346 | 9 | 54,937,110 | — | — | — | — | — | — | 0.072** | 0.066 | 0.918 |
| BTB-01576221 | 9 | 56,786,292 | — | — | — | — | — | — | 0.098* | 0.072 | 0.734 |
| BTB-01573160 | 9 | 58,011,347 | — | — | — | 0.032 | 0.055 | 1.703 | — | — | — |
| BTB-01352997 | 9 | 61,883,731 | 0.158* | 0.064 | 0.409 | 0.070** | 0.075 | 1.081 | — | — | — |
| Hapmap35559-SCAFFOLD35632_12026 | 9 | 62,961,279 | 0.037 | 0.065 | 1.754 | — | — | — | 0.006 | 0.072 | 12.466 |
| ARS-BFGL-NGS-113585 | 9 | 64,207,421 | — | — | — | 0.018 | 0.064 | 3.563 | — | — | — |
| ARS-BFGL-NGS-119476 | 9 | 70,904,362 | — | — | — | — | — | — | 0.005 | 0.060 | 11.968 |
| ARS-BFGL-NGS-118161 | 9 | 82,306,396 | — | — | — | 0.079** | 0.106 | 1.337 | — | — | — |
| BTB-00402956 | 9 | 82,586,467 | — | — | — | 0.012 | 0.069 | 5.754 | — | — | — |
| BTB-00400339 | 9 | 82,901,727 | — | — | — | 0.036 | 0.087 | 2.408 | — | — | — |
| BTA-84397-no-rs | 9 | 82,927,247 | — | — | — | 0.016 | 0.063 | 4.035 | — | — | — |
| ARS-BFGL-NGS-112933 | 9 | 93,552,330 | — | — | — | — | — | — | 0.010 | 0.080 | 8.269 |
| Hapmap49345-BTA-85101 | 9 | 100,304,045 | — | — | — | — | — | — | 0.236* | 0.297 | 1.256 |
| ARS-BFGL-NGS-107379 | 14 | 2,054,457 | — | — | — | 0.094* | 0.023 | 0.245 | — | — | — |
| ARS-BFGL-NGS-100480 | 14 | 4,364,952 | — | — | — | 0.202* | 0.063 | 0.311 | — | — | — |
| ARS-BFGL-NGS-113395 | 14 | 34,189,618 | — | — | — | — | — | — | 0.030 | 0.044 | 1.448 |
| ARS-BFGL-NGS-100124 | 22 | 21,431,682 | — | — | — | 0.051 | 0.077 | 1.530 | — | — | — |
| ARS-BFGL-NGS-65462 | 22 | 21,455,286 | — | — | — | 0.051 | 0.078 | 1.530 | — | — | — |
| Hapmap48103-BTA-86065 | 22 | 26,073,419 | 0.027 | 0.054 | 1.974 | — | — | — | — | — | — |
| Hapmap60793-rs29018735 | 22 | 31,702,243 | 0.152* | 0.086 | 0.565 | — | — | — | — | — | — |
| ARS-BFGL-NGS-60299 | 22 | 32,234,944 | 0.150* | 0.094 | 0.626 | 0.117* | 0.109 | 0.938 | 0.143* | 0.104 | 0.725 |
| Hapmap47345-BTA-60915 | 22 | 33,766,195 | — | — | — | 0.121* | 0.089 | 0.737 | — | — | — |
| ARS-BFGL-NGS-98103 | 22 | 38,727,002 | — | — | — | — | — | — | 0.073** | 0.060 | 0.825 |
| ARS-BFGL-NGS-82909 | 22 | 38,815,613 | 0.056 | 0.057 | 1.009 | — | — | — | — | — | — |
Given are the additive (Add) and dominance (Dom) effects obtained from the full model as well as the degrees of dominance (d/|a|) for SNP markers showing significant (P ≤ 0.01) dominance effects in the two-step genome scan. The markers on BTA9, -14, and -22 are exemplarily summarized (for a full list see Table S1). Effects are given in phenotypic standard deviations (σP). Chr and bp are the cattle chromosome and the position in base pairs according to genome build UMD 3.1. Significance of additive effects: *P ≤ 0.01; **P ≤ 0.05.
Conclusion
The detection of dominance effects relies on the utilization of direct phenotypes, while classical QTL mapping approaches in dairy cattle employed breeding values of sires based on daughter performance. The implementation of genomic selection schemes in dairy cattle breeding provides a large number of bulls genotyped for dense genome-wide marker panels. Within the current study, these genotypes were used to derive genotype probabilities in female descendants of these bulls. Using these probabilities it is possible to detect significant dominance effects on a genome-wide scale, relying on a large sample of phenotyped cows. Together with a two-step mixed model approach to account for relationship among animals, the approach is computationally not challenging and applicable to data sets of >106 cows. The application of this approach could substantially enhance our knowledge about the genetic architecture of performance and functional traits in dairy cattle. The introduced method could also be extended for use in genomic prediction, including the enlargement of reference populations or the prediction of individual performance accounting for nonadditive genetic effects.
Supplementary Material
Acknowledgments
This study was financially supported by the German Federal Ministry for Education and Research in the context of the Funktionelle Genomanalyse im Tierischen Organismus (FUGATO+, or Functional Genome Analysis in Animal Organisms) project GenoTrack (grant 0315134A) and the competence networks Synbreed (grant 0315528F) and Phänomics (grant 0315536D).
Footnotes
Communicating editor: I. Hoeschele
Literature Cited
- Ashwell M. S., Heyen D. W., Sonstegard T. S., Van Tassell C. P., Da Y., et al. , 2004. Detection of quantitative trait loci affecting milk production, health, and reproductive traits in Holstein cattle. J. Dairy Sci. 87: 468–475 [DOI] [PubMed] [Google Scholar]
- Aulchenko Y. S., de Koning D. J., Haley C., 2007. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics 177: 577–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston R. C., Stewart J., 1971. A general model for the genetic analysis of pedigree data. Hum. Hered. 21: 523–542 [DOI] [PubMed] [Google Scholar]
- Gilmour A. R., Thompson R., Cullis B. R., 1995. Average information REML: an efficient algorithm for variance parameter estimation in linear mixed models. Biometrics 51: 1440–1450 [Google Scholar]
- Guo S. W., Thompson E. A., 1992. A Monte Carlo method for combined segregation and linkage analysis. Am. J. Hum. Genet. 51: 1111–1126 [PMC free article] [PubMed] [Google Scholar]
- Grisart B., Coppieters W., Farnir F., Karim L., Ford C., et al. , 2002. Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res. 12: 222–231 [DOI] [PubMed] [Google Scholar]
- Habier D., Tetens J., Seefried F. R., Lichtner P., Thaller G., 2010. The impact of genetic relationship information on genomic breeding values in German Holstein cattle. Genet. Sel. Evol. 42: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley C. S., Knott S. A., 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324 [DOI] [PubMed] [Google Scholar]
- Harder B., Bennewitz J., Reinsch N., Thaller G., Thomsen H., et al. , 2006. Mapping of quantitative trait loci for lactation persistency traits in German Holstein dairy cattle. J. Anim. Breed. Genet. 123: 89–96 [DOI] [PubMed] [Google Scholar]
- Hayes B., Goddard M. E., 2001. The distribution of the effects of genes affecting quantitative traits in livestock. Genet. Sel. Evol. 33: 209–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henshall J. M., Tier B., 2003. An algorithm for sampling descent graphs in large complex pedigrees efficiently. Genet. Res. 81: 205–212 [DOI] [PubMed] [Google Scholar]
- Kenward M. G., Roger J. H., 1997. The precision of fixed effects estimates from restricted maximum likelihood. Biometrics 53: 983–997 [PubMed] [Google Scholar]
- Khatkar M. S., Thomson P. C., Tammen I., Raadsma H. W., 2004. Quantitative trait loci mapping in dairy cattle: review and meta-analysis. Genet. Sel. Evol. 36: 163–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn C., Edel C., Weikard R., Thaller G., 2007. Dominance and parent-of-origin effects of coding and non-coding alleles at the acylCoA-diacylglycerol-acyltransferase (DGAT1) gene on milk production traits in German Holstein cows. BMC Genet. 8: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Seefried F. R., Reinhardt F., Rensing S., Thaller G., et al. , 2011. Impacts of both reference population size and inclusion of a residual polygenic effect on the accuracy of genomic prediction. Genet. Sel. Evol. 43: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, M. S., A. P. W. de Roos, A. G. de Vries, T. Druet, V. Ducroqc et al., 2010 Improving genomic prediction by EuroGenomics collaboration, p. 150 in Proceedings of the 9th World Congress on Genetics Applied Livestock Production, German Society for Animal Science, Leipzig, Germany. [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2008 R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Riquet J., Coppieters W., Cambisano N., Arranz J. J., Berzi P., et al. , 1999. Fine-mapping of quantitative trait loci by identity by descent in outbred populations: application to milk production in dairy cattle. Proc. Natl. Acad. Sci. USA 96: 9252–9257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelman R. J., Ford C. A., McElhinney P., Gregory G. C., Snell R. G., 2002. Characterization of the DGAT1 gene in the New Zealand dairy population. J. Dairy Sci. 85: 3514–3517 [DOI] [PubMed] [Google Scholar]
- Thaller G., Kramer W., Winter A., Kaupe B., Erhardt G., et al. , 2003. Effects of DGAT1 variants on milk production traits in German cattle breeds. J. Anim. Sci. 81: 1911–1918 [DOI] [PubMed] [Google Scholar]
- Weller J. I., Golik M., Seroussi E., Ezra E., Ron M., 2003. Population-wide analysis of a QTL affecting milk-fat production in the Israeli Holstein population. J. Dairy Sci. 86: 2219–2227 [DOI] [PubMed] [Google Scholar]
- Wiener P., Maclean I., Williams J. L., Woolliams J. A., 2000. Testing for the presence of previously identified QTL for milk production traits in new populations. Anim. Genet. 31: 385–395 [DOI] [PubMed] [Google Scholar]
- Winter A., Kramer W., Werner F. A., Kollers S., Kata S., et al. , 2002. Association of a lysine-232/alanine polymorphism in a bovine gene encoding acyl-CoA:diacylglycerol acyltransferase (DGAT1) with variation at a quantitative trait locus for milk fat content. Proc. Natl. Acad. Sci. USA 99: 9300–9305 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.