Abstract
FST is frequently used as a summary of genetic differentiation among groups. It has been suggested that FST depends on the allele frequencies at a locus, as it exhibits a variety of peculiar properties related to genetic diversity: higher values for biallelic single-nucleotide polymorphisms (SNPs) than for multiallelic microsatellites, low values among high-diversity populations viewed as substantially distinct, and low values for populations that differ primarily in their profiles of rare alleles. A full mathematical understanding of the dependence of FST on allele frequencies, however, has been elusive. Here, we examine the relationship between FST and the frequency of the most frequent allele, demonstrating that the range of values that FST can take is restricted considerably by the allele-frequency distribution. For a two-population model, we derive strict bounds on FST as a function of the frequency M of the allele with highest mean frequency between the pair of populations. Using these bounds, we show that for a value of M chosen uniformly between 0 and 1 at a multiallelic locus whose number of alleles is left unspecified, the mean maximum FST is ∼0.3585. Further, FST is restricted to values much less than 1 when M is low or high, and the contribution to the maximum FST made by the most frequent allele is on average ∼0.4485. Using bounds on homozygosity that we have previously derived as functions of M, we describe strict bounds on FST in terms of the homozygosity of the total population, finding that the mean maximum FST given this homozygosity is 1 − ln 2 ≈ 0.3069. Our results provide a conceptual basis for understanding the dependence of FST on allele frequencies and genetic diversity and for interpreting the roles of these quantities in computations of FST from population-genetic data. Further, our analysis suggests that many unusual observations of FST, including the relatively low FST values in high-diversity human populations from Africa and the relatively low estimates of FST for microsatellites compared to SNPs, can be understood not as biological phenomena associated with different groups of populations or classes of markers but rather as consequences of the intrinsic mathematical dependence of FST on the properties of allele-frequency distributions.
DIFFERENTIATION among groups is one of the fundamental subjects of the field of population genetics. Comparisons of the level of variation among subpopulations with the level of variation in the total population have been employed frequently in population-genetic theory, in statistical methods for data analysis, and in empirical studies of distributions of genetic variation. Wright’s (Wright 1951) fixation indices, and FST in particular, have been central to this effort.
Wright’s FST was originally defined as the correlation between two randomly sampled gametes from the same subpopulation when the correlation of two randomly sampled gametes from the total population is set to zero. Several definitions of FST or FST-like quantities are now available, relying on a variety of different conceptual formulations but all measuring some aspect of population differentiation (e.g., Charlesworth 1998; Holsinger and Weir 2009). Many authors have claimed that one or another formulation of FST is affected by levels of genetic diversity or by allele frequencies, either because the range of FST is restricted by these quantities or because these quantities affect the degree to which FST reflects population differentiation (e.g., Charlesworth 1998; Nagylaki 1998; Hedrick 1999, 2005; Long and Kittles 2003; Jost 2008; Ryman and Leimar 2008; Long 2009; Meirmans and Hedrick 2011). For example, Nagylaki (1998) and Hedrick (1999) argued that measures of FST may be poor measures of genetic differentiation when the level of diversity is high. Charlesworth (1998) suggested that FST can be inflated when diversity is low, arguing that FST might not be appropriate for comparing loci with substantially different levels of variation. In a provocative recent article, Jost (2008) used the diversity dependence of forms of FST to question their utility as differentiation measures at all.
One definition that is convenient for mathematical assessment of the relationship of an FST-like quantity and allele frequencies is the quantity labeled GST by Nei (1973), which for a given locus measures the difference between the heterozygosity of the total (pooled) population, hT, and the mean heterozygosity across subpopulations, hS, divided by the heterozygosity of the total population:
| (1) |
In terms of the homozygosity of the total population, HT = 1 − hT, and the mean homozygosity across subpopulations, HS = 1 − hS, we can write
| (2) |
The Wahlund (1928) principle guarantees that HS ≥ HT and, therefore, because HS ≤ 1 and for a polymorphic locus with finitely many alleles, 0 < HT < 1, GST lies in the interval [0,1].
Using GST for their definition of FST, Hedrick (1999, 2005) and Long and Kittles (2003) pointed out that because hT < 1, FST cannot exceed the mean homozygosity across subpopulations, HS:
| (3) |
Hedrick (2005) obtained this result by considering a set of K equal-sized subpopulations, in which each allele is private to a single subpopulation. In the limit as K → ∞, a stronger upper bound on FST as a function of HS and K reduces to Equation 3 (see also Jin and Chakraborty 1995 and Long and Kittles 2003).
While Hedrick (1999, 2005) and Long and Kittles (2003) have clarified the relationship between FST and the mean homozygosity HS across subpopulations, their approaches do not easily illuminate the connection between FST and allele frequencies themselves. A formal understanding of the relationship between FST and allele frequencies would make it possible to more fully understand the behavior of FST in situations where markers of interest differ substantially in allele frequencies or levels of genetic diversity. Our recent work on the relationship between homozygosity and the frequency of the most frequent allele (Rosenberg and Jakobsson 2008; Reddy and Rosenberg 2012) provides a mathematical approach for formal investigation of bounds on population-genetic statistics in terms of allele frequencies. In this article, we therefore seek to thoroughly examine the dependence of FST on allele frequencies by investigating the upper bound on FST in terms of the frequency M of the most frequent allele across a pair of populations. We derive bounds on FST given the frequency of the most frequent allele and bounds on the frequency of the most frequent allele given FST. We consider loci with arbitrarily many alleles in a pair of subpopulations. Using theory for the bounds on homozygosity given the frequency of the most frequent allele, we obtain strict bounds on FST given the homozygosity of the total population. Our analysis clarifies the relationships among FST, allele frequencies, and homozygosity, providing explanations for peculiar observations of FST that can be attributed to allele-frequency dependence.
Model
We examine a polymorphic locus with at least two alleles in a setting with K subpopulations that contribute equally to a total population. Denote the number of distinct alleles by I, the frequency of allele i in population k by pki, and the mean frequency of allele i across populations by We primarily report our results in terms of homozygosities, which can be easily transformed into heterozygosities.
We consider FST formulated as a property of nonnegative numbers between 0 and 1 such that within populations, the allele frequencies sum to 1 ( for each k). This formulation is the same as the formulation of Nei’s GST, which we hereafter denote by F. We have (Nei 1973)
where
and
The assumption that the locus is polymorphic guarantees that HT < 1. The assumption that I, the number of distinct alleles at the locus, is finite guarantees that HT > 0 (and hence, HS > 0 because HS ≥ HT). Thus, 0 < HT < 1 and 0 < HS ≤ 1.
We assume that all allele frequencies are the parametric allele frequencies of the population under consideration. Thus, the frequency of an allele is the probability of drawing the allele from the parametric frequency distribution; homozygosity is then the probability that two independent random draws carry the same allelic type, and heterozygosity is the probability that two independent random draws carry different allelic types. We emphasize that in our formulation, F, HT, and HS are functions of the parametric allele frequencies, and our interest is in the properties of these functions and their relationships with the allele frequencies; we do not investigate their estimation from data, nor do we consider how evolutionary models affect the underlying allele frequencies involved in their computation.
We focus on the case of two subpopulations (K = 2). In this case, the allele frequencies are denoted p1i for population 1 and p2i for population 2. For each i from 1 to I, let σi = p1i + p2i be the sum across populations of the frequency of allele i. Each σi lies in (0, 2), and the number of alleles I counts only those alleles with σi > 0. We denote . Without loss of generality, we place the alleles in decreasing order, such that σ1 ≥ σ2 ≥ … ≥ σI. We denote the frequency of the most frequent allele in the total pooled population by M = σ1/2, and we find it convenient to express some results in terms of σ1 and others in terms of M. Because and each σi is positive, we have 1/I ≤ M < 1.
Let δi = |p1i − p2i| be the absolute difference between p1i and p2i. We can write the homozygosity of the total population as
and the mean homozygosity across subpopulations as
We then have (Boca and Rosenberg 2011)
| (4) |
In other words, F can be computed solely using the allele-frequency sums and differences between the two populations.
Bounds on F
Our goal is to study the relationship between F and M in the general case of I alleles in two populations. For convenience, we write F as a function of σ1, keeping in mind that σ1/2 = M, and we begin by considering the special case in which I = 2.
Bounds on F for two alleles
This case has two alleles, with frequencies p11 and p12 in population 1, and p21 and p22 in population 2 (Table 1). The frequency of the second allele is p12 = 1 − p11 in population 1 and p22 = 1 − p21 in population 2. Using Equation 4, we have a simple expression for F (Weir 1996; Rosenberg et al. 2003):
| (5) |
We determine the upper and lower bounds of F in terms of the frequency of the most frequent allele M = σ1/2. Because the alleles are arranged to satisfy σ1 ≥ σ2 and because σ1 + σ2 = 2, σ1 must lie in [1, 2). For the lower bound on F as a function of σ1, we note that if allele 1 has the same frequency in both populations, then p11 = p21 = σ1/2. The frequency of allele 2 will also be the same in the two populations, p12 = p22 = 1 − σ1/2, and δ1 and δ2 will both equal zero. For these allele frequencies, we see that HS = HT, and it is clear from Equation 5 that F(σ1) ≥ 0 for all values of σ1 in [1, 2), with equality if and only if p11 = p21 = σ1/2.
Table 1 . Notation for two alleles in two populations.
| Allele |
|||
|---|---|---|---|
| Population | 1 | 2 | Sum |
| 1 | p11 | p12 | 1 |
| 2 | p21 | p22 | 1 |
| Sum | σ1 | σ2 | 2 |
| Absolute difference | δ1 | δ2 | — |
For the upper bound, we first note that because δ1 = 2p11 − σ1 when p11 ≥ p21 and δ1 = 2p21 − σ1 when p21 ≥ p11,
| (6) |
with equality if and only if p11 = 1 or p21 = 1. Using Equations 5 and 6, we have
Thus, the upper bound on F as a function of σ1 is achieved when the allele frequencies of the two populations differ as much as possible, that is, when (p11, p21) = (1, σ1 − 1) or (p11, p21) = (σ1 − 1, 1). The bounds on F are
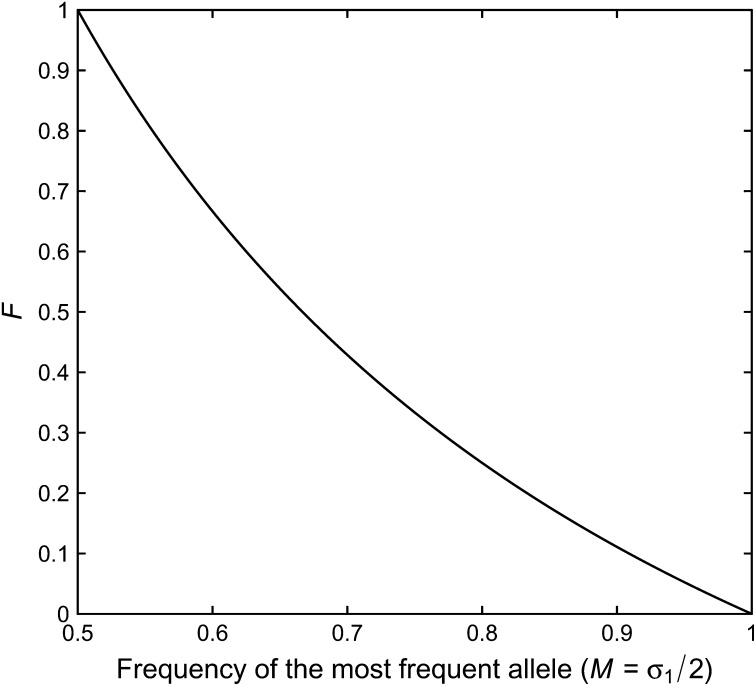
| (7) |
Figure 1 shows the upper bound as a function of the most frequent allele, illustrating a monotonic decline from q(1/2) = 1 to q(1) = 0.
Figure 1 .
The upper bound on F as a function of the frequency M of the most frequent allele, for the two-allele case. The upper bound is computed from Equation 7. The lower bound on F is 0 for all values of M.
Lower bound on F for an unspecified number of alleles
For any number of alleles I and any set of σi, by noting that the denominator of F in Equation 4 is positive and that the numerator is , we see that Equation 4 takes the value of zero if and only if for each i, p1i = p2i = σi/2. Thus, the lower bound on F as a function of σ1 is achieved when the allele frequencies are the same in both populations for all I alleles. Thus, F = 0 is attainable for any value of σ1 in (0, 2).
Upper bound on F for an unspecified number of alleles
The upper bound on F as a function of σ1 has different properties for σ1 ε (0, 1) and for σ1 ε [1, 2). We begin with σ1 ε (0, 1).
Using Equation 4, we can rearrange F(σ1) to obtain
| (8) |
As we assume that the locus of interest is polymorphic, both the numerator and denominator in the fraction in Equation 8 are positive. Fix and . Because the same quantity is subtracted in the numerator and denominator from quantities that must exceed it (2 in the numerator, in the denominator), the fraction is maximized when is minimized, that is, when . In other words, given , for fixed and , F(σ1) is maximal when each allele is found only in one of the two subpopulations.
To complete the maximization of F(σ1) as a function of σ1, it remains to maximize and . These two maximizations can be performed separately, as no allele appears in both subpopulations. Further, by symmetry, and must have the same maximum.
Define . The number of alleles I is unspecified; we search for an upper bound over all possible values I ≥ 2 and discover that the maximum occurs when each subpopulation has I = J distinct alleles. Because p1i + p2i ≤ σ1 and because for each i, at the maximum of F(σ1), each allele has either p1i = 0 or p2i = 0, it suffices to maximize subject to and p1i ≤ σ1 for all i. This maximization is the same problem considered in Rosenberg and Jakobsson (2008, Lemma 3), which demonstrates that the maximum occurs if and only if the locus has J − 1 alleles of frequency σ1 and one remaining allele of frequency 1 − (J − 1)σ1.
Lemma 3 of Rosenberg and Jakobsson (2008) yields 1 − σ1(J − 1)(2 − Jσ1) for each of the two maxima, on and on . We then conclude
| (9) |
with equality if and only if the locus has 2J alleles, J of which occur only in the first subpopulation and the other J of which occur only in the second population, and each subpopulation has J − 1 alleles of frequency σ1 and one allele of frequency 1 − (J − 1)σ1. Because , we have
| (10) |
For the case of σ1 ε [1, 2), we separate terms in Equation 4 for the first and subsequent alleles:
| (11) |
The upper bound on F, given σ1, occurs when , , and are maximized. To maximize , note that as in the two-allele case (Equation 6), for σ1 ε [1, 2), , with equality if and only if p11 = 1 or p21 = 1.
Next, for any i, δi ≤ σi, with equality if and only if p1i = 0 or p2i = 0. Then
| (12) |
where the last step follows from the fact that . Equality in the second step requires that among the σi with i ≥ 2, only one can be positive, namely σ2, by the assumption that the alleles are labeled in decreasing order of frequency. Thus, equality occurs in both inequalities if and only if σ2 = 2 − σ1 and either p12 or p22 is 0.
We have therefore found that given σ1 ε [1, 2), , , and are all maximized under exactly the same conditions—when (p11, p12, p21, p22) = (1, 0, σ1 − 1, 2 − σ1) or (σ1 − 1, 2 − σ1, 1, 0). Replacing the terms , and in Equation 11 using inequalities 6 and 12, we have
| (13) |
with equality if and only if p11 = 1 or p21 = 1 and σ2 = 2 − σ1. This result matches the two-allele case: when σ1 ε [1, 2), the case of an unspecified number of alleles reduces to the case of two alleles.
Summarizing our results, the bounds of F are
| (14) |
where
| (15) |
| (16) |
Note that the upper bound on F is continuous at σ1 = 1, as .
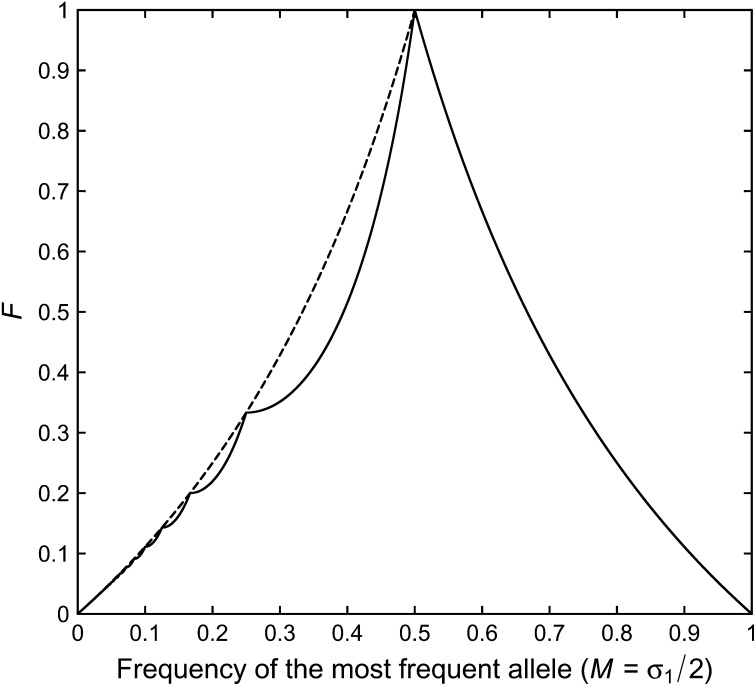
The upper bound on F is shown as the solid line in Figure 2. The plot illustrates that the upper bound on F(σ1) has a piecewise structure on (0, 1), with changes in shape occurring when σ1 is equal to the reciprocal of an integer. Similarly to the bounds examined by Rosenberg and Jakobsson (2008), for each J ≥ 2, Q(σ1) is monotonically increasing on the interval [1/J, 1/(J − 1)), where has the constant value J. Further, Q(σ1) is continuous at the boundaries 1/J between intervals, with Q(1/J) = 1/(2J − 1). On [1, 2), the upper bound has a simple monotonic decline according to q(σ1).
Figure 2 .
The upper bound on F (solid line) as a function of the frequency M of the most frequent allele, for the general case of any number of alleles. The upper bound is computed from Equations 15 and 16. The dashed line shows Equation 21, which the upper bound touches when M = 1/(2J) for integers J ≥ 2. The lower bound on F is 0 for all values of M.
Properties of the Upper Bound on F
The region between 0 and the upper bound on F exactly circumscribes the set of possible values of F as a function of σ1, as the upper bound is strict. We now explore a series of features of the upper bound on F as a function of σ1.
The space between the upper and lower bounds on F
The mean maximum F across the range of possible frequencies for the most frequent allele gives a sense of the maximal F attainable on average, when M is uniformly distributed. This mean can be obtained by evaluating the area of the region between the lower and upper bounds on F.
Because the lower bound on F is zero over the entire interval σ1 ε (0, 2), we need to determine only the area A under the upper bound on F. We integrate Q(σ1) for σ1 ε (0, 1) and q(σ1) for σ1 ε [1, 2),
| (17) |
The first integral can be computed as a sum over intervals [1/J, 1/(J − 1)) for J ≥ 2. On each such interval, has a fixed value of J. We then have
In the Appendix, we show that
| (18) |
By numerically evaluating the sum in Equation 18, we obtain an approximation .
The second term in Equation 17 is
| (19) |
and the area under q(σ1) for σ1 ε [1, 2) is ∼0.3862944.
Summing the values for the two integrals, the area A under the upper bound on F is ∼0.7170751. Considering F as a function of M = σ1/2 rather than σ1, F is confined to a region with area ∼0.3585376. This area under the curve is the mean maximal value of F across the space of values of M, and it is substantially less than 1. Thus, on average, F is constrained within a narrow range, and across most of the space of possible values for the frequency of the most frequent allele, F cannot achieve large values. For example, only over half the range—for M between 1/4 and 3/4—is it possible for F to exceed 1/3.
Jagged points touch a simple curve
For σ1 ε [1, 2), the upper bound on F is a smooth function q(σ1). For σ1 ε (0, 1), however, the upper bound is a jagged curve. At σ1 = 1/J for any integer J ≥ 2, that is, at the “jagged points” where the upper bound is not differentiable, Q(σ1) coincides with the reflection of q(σ1) across the line σ1 = 1. We have
| (20) |
because when σ1 = 1/J. Thus, for σ1 = 1/J, Q(σ1) touches the curve
| (21) |
The dashed line in Figure 2 plots q*(σ1) on (0, 1).
Because q*(σ1) on (0, 1) is the reflection of q(σ1) on [1, 2) across the line σ1 = 1, the area under q*(σ1) on (0, 1) is the same as the area of q(σ1) on [1, 2), or 2 ln 2 − 1. Thus, on the interval (0, 1), the space between q*(σ1) and Q(σ1) is
| (22) |
The contribution made by M to the upper bound on F
We denote by F1(σ1) the contribution of the most frequent allele to F(σ1). By this quantity, we mean the term in F(σ1) contributed by the difference between populations in the frequency of the most frequent allele. From Equation 4, F(σ1) can be written
| (23) |
If the ith term in the summation is denoted Fi(σ1), our interest is in the value of F1(σ1) obtained at the set of allele frequencies that maximizes F(σ1).
For σ1 in the interval (0, 1), defining , the maximum has 2J − 2 alleles with frequency σ1 and two alleles with frequency 1 − (J − 1)σ1: J − 1 alleles with frequency σ1 and one allele with frequency 1 − (J − 1)σ1 in each subpopulation. The value of at the maximum is . Denoting the contribution F1(σ1) to F(σ1) at the maximum by Q1(σ1), we have
| (24) |
In the Appendix, we evaluate . The expression is unwieldy, but it provides a numerical approximation .
For σ1 ε [1, 2), at the maximum of F(σ1), , and we have
| (25) |
The area under q1(σ1) is
Summing the areas under Q1(σ1) and q1(σ1), the total area B under F1 as σ1 ranges from 0 to 2 is
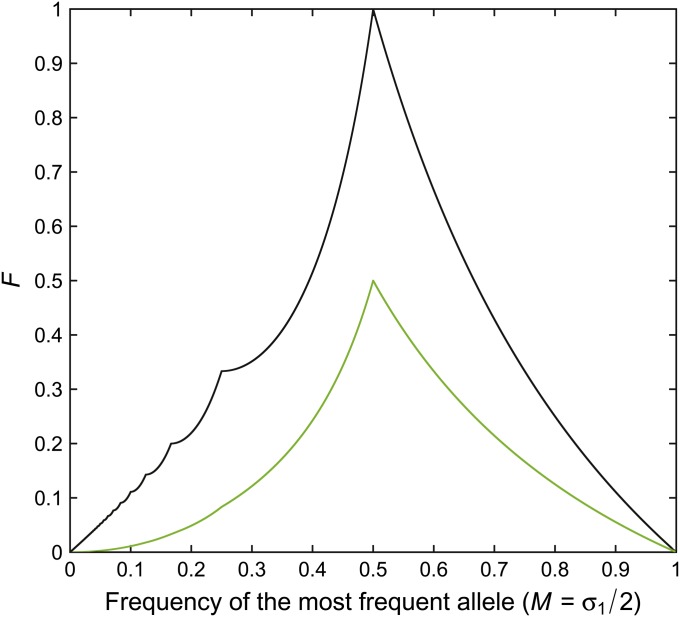
If we instead consider M = σ1/2, we find that F1 is confined to ∼0.1607997 of the space of possible pairs of values (M, F). The fraction of the area A under the upper bound on F contributed by the most frequent allele over the entire interval σ1 ε (0, 2) is B/A ≈ 0.4484877. This quantity can be interpreted as the mean contribution of the most frequent allele to the maximum value of F, and it indicates a substantial role for the most frequent allele. Indeed, for σ1 ε [1, 2), q1(σ1)/q(σ1) = 1/2. The contribution made by the most frequent allele to the upper bound on F appears in Figure 3.
Figure 3 .
The contribution to F, at the upper bound, that is made by the most frequent allele (green line). The contribution by the most frequent allele is computed from Equations 24 and 25. For comparison, the upper bound on F is shown as a black line.
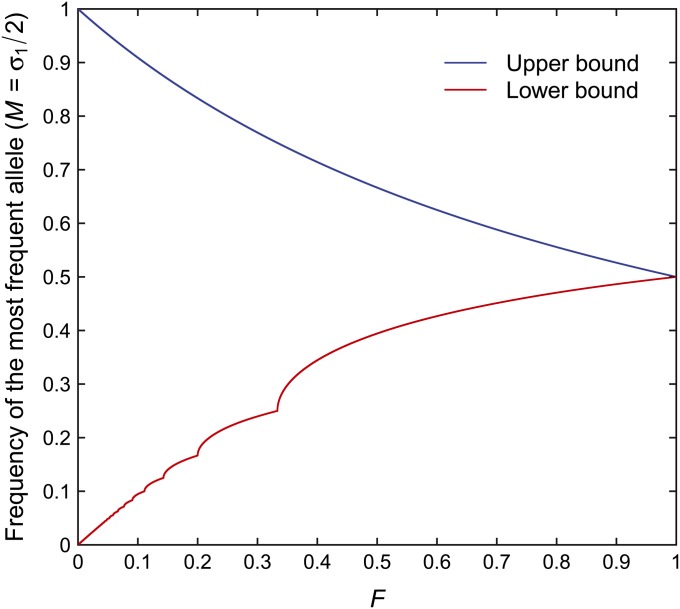
Bounds on M
Our derivation of the bounds on F as functions of the frequency M of the most frequent allele enables us to provide bounds on M as functions of F by taking the inverse of the functions q(σ1) and Q(σ1). For 0 < F < 1, we show that the bounds on the frequency of the most frequent allele in terms of F are
| (26) |
At the trivial case of F = 1, σ1 must equal 1, and for F = 0, σ1 lies in the open interval (0, 2).
Bounds on σ1 for two alleles
We first consider the two-allele case. By definition of σ1, regardless of the value of F, σ1 can be no smaller than 1, and when σ1 = 1, . For any F ε [0, 1], it is possible to choose allele frequencies p11 and p21 so that and σ1 = p11 + p21 = 1. We simply set and . Thus, the lower bound of σ1(F) = 1 can be achieved across the full domain F ε [0, 1].
For the upper bound on σ1, recall that the upper bound on F in terms of σ1 (Equation 7) is a continuous monotonically decreasing function on the interval σ1 ε [1, 2). We can therefore obtain the upper bound on σ1 as the inverse of this function. Thus, for F ε [0, 1], the bounds on σ1 are:
| (27) |
The corresponding bounds on M = σ1/2 appear in Figure 4.
Figure 4 .
The upper and lower bounds on the frequency M of the most frequent allele as functions of F, for the two-allele case. The bounds are computed from Equation 27.
Lower bound on σ1 for an unspecified number of alleles
For the general case, we obtain lower and upper bounds on F, considering all possible choices for the number of distinct alleles. It is useful to first recall that the function Q(σ1) for the upper bound on F for σ1 ε (0, 1) is monotonically increasing, while the function q(σ1) for the upper bound on F for σ1 ε [1, 2) is monotonically decreasing. We can therefore invert Q(σ1) and q(σ1), so that the lower bound on σ1 as a function of F is obtained by solving Q(σ1) = F for σ1 and the upper bound by solving q(σ1) = F for σ1. For the lower bound, we perform the inversion piecewise. For integers J ≥ 2, if σ1 ε [1/J, 1/(J − 1)), then Q(σ1) ε [1/(2J − 1), 1/(2J − 3)). Therefore, for J ≥ 2, if Q ε [1/(2J − 1), 1/(2J − 3)), then the lower bound on σ1 lies in [1/J, 1/(J − 1)). For this interval on Q, ⌈(1 + Q)/(2Q)⌉ = J, and in this region, the lower bound on σ1, which we term r(F), also satisfies ⌈r(F)⌉ = J. We solve Equation 10 for σ1 for Q ε[1/(2J − 1), 1/(2J − 3)), where both and ⌈(1 + Q)/(2Q)⌉ are equal to J:
| (28) |
A negative root is discarded because it yields values that are incompatible with the definition that σ1 ≥ σi for all i > 1. The upper and lower bounds appear in Figure 5.
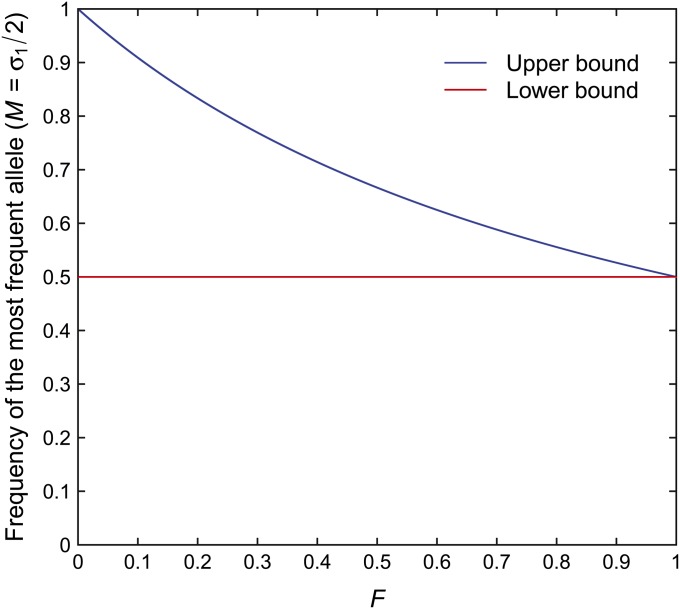
Figure 5 .
The upper and lower bounds on the frequency M of the most frequent allele as functions of F, for the general case of any number of alleles. The bounds are computed from Equations 29 and 28.
Upper bound on σ1 for an unspecified number of alleles
From Equation 13 and Figure 2, we see that for any F ε [0, 1], the upper bound on σ1 is ≥⩾1. Because Equation 13 is continuous and monotonically decreasing, we can take the inverse of this function to compute the upper bound on σ1 as a function of F. The upper bound R(F) on σ1 is
| (29) |
the same upper bound as for the two-allele case (Equation 27).
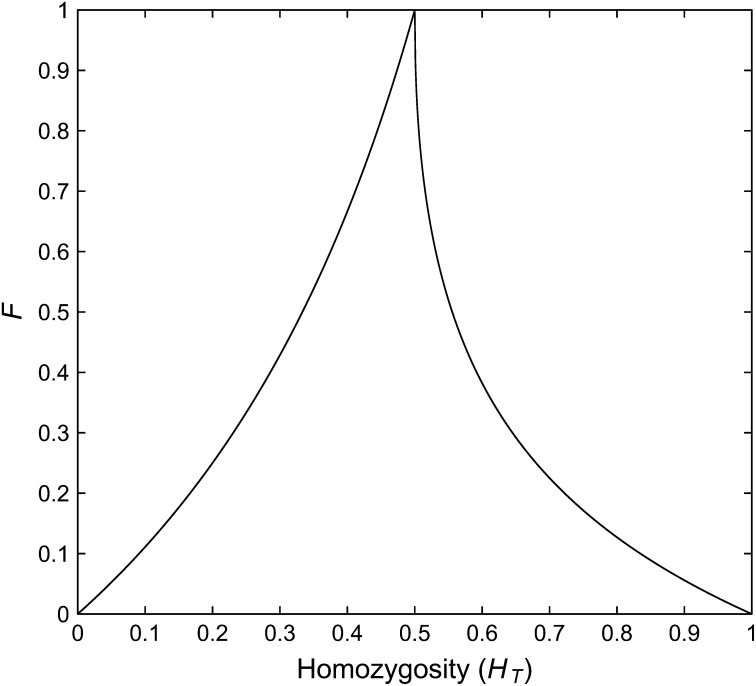
F and Homozygosity of the Total Population
The relationship between F and the frequency of the most frequent allele can be used together with the relationship between homozygosity and the frequency of the most frequent allele (Rosenberg and Jakobsson 2008; Reddy and Rosenberg 2012), to find a relationship between F and homozygosity, again in the setting of two populations. The homozygosity that we consider, H in Rosenberg and Jakobsson (2008), corresponds to the homozygosity of the total pooled population HT. We first note that given any HT ε (0, 1), the lower bound on F is zero. For example, for any HT, F = 0 is obtained by using the equality condition in Theorem 1ii of Rosenberg and Jakobsson (2008) to specify a list of allele frequencies with sum of squares HT and then assigning that same list of frequencies to both of the component subpopulations.
Upper bound on F given HT for an unspecified number of alleles
Rosenberg and Jakobsson (2008) showed that the value of HT constrains the frequency M of the most frequent allele to a narrow range. We have already determined the upper bound on F as a function of M. Thus, we can obtain an upper bound on F as a function of HT by taking the maximum value of the upper bound over the range of possible values of M allowed under the results of Rosenberg and Jakobsson (2008) for a given value of HT. This approach does not guarantee that the upper bound on F that we obtain in terms of HT is strict; nevertheless, the approach happens to produce a strict bound for HT ε [1/2, 1). For HT ε (0, 1/2), it is possible to produce a strict bound by writing F in terms of HT.
To obtain the bound for HT ε (0, 1/2), we substitute for in Equation 4 to write
| (30) |
Because , we obtain the bound
| (31) |
Given HT, equality is obtained in Equation 31 when . In other words, for HT ε (0, 1/2), F is maximized when each allele occurs in only one of the two populations. To see that the upper bound is strict, note that when , labeling the homozygosities of the two populations by H1 and H2, HT = (H1 + H2)/4. As HT < 1/2, 2HT < 1, and we can choose H1 = H2 = 2HT. Using the equality condition in Theorem 1ii of Rosenberg and Jakobsson (2008), we can specify a set L of exactly ⌈(2HT)−1⌉ allele frequencies whose sum of squares is HT. We then construct a set of 2⌈(2HT)−1⌉ alleles. In population 1, the first ⌈(2HT)−1⌉ alleles in the set have exactly the allele frequencies in L and the next ⌈(2HT)−1⌉ alleles have frequency 0. In population 2, the first ⌈(2HT)−1⌉ alleles have frequency 0, and the next ⌈(2HT)−1⌉ alleles have the frequencies in L.
For HT ε [1/2, 1), HT/(1 − HT) ≥ 1, so Equation 31 provides only the trivial bound of F ≤ 1, and another approach is needed. For any HT ε [1/2, 1), using Theorem 1ii of Rosenberg and Jakobsson (2008), M ≥ 1/2. For M ≥ 1/2, the upper bound on F as a function of σ1 is monotonically decreasing in σ1, and consequently, the upper bound on F as a function of HT is obtained by evaluating q(σ1) at the smallest value of σ1 permitted by HT. Theorem 1ii of Rosenberg and Jakobsson (2008) indicates that this smallest allowed σ1 satisfies
By replacing σ1/2 in Equation 16 with this expression, we have
| (32) |
where the last step follows from the fact that when HT ε [1/2, 1).
For HT ε [1/2, 1), the set of allele frequencies that achieves the minimum M as a function of HT and the set that achieves the maximum F as a function of M coincide. Given HT, M is minimized by setting , , and for all i ≥ 2. If these mean frequencies are distributed between the two populations such that or , then the upper bound on F is achieved.
Figure 6 shows our upper bound on F as a function of the total homozygosity HT. If HT is low, and particularly if HT is high, then F is restricted to small values. High values of F are possible only when HT is near 1/2. In fact, using Equations 31 and 32, F can exceed 1/2 only if HT lies in (1/3, 5/9).
Figure 6 .
The upper bound on F as a function of HT. The upper bound is computed from Equations 31 and 32. The lower bound on F is 0 for all values of HT.
The space between the upper and lower bounds on F given HT
In the same manner as in our investigation of the bounds on F as a function of M, we evaluate the area of the region between the upper and lower bounds on F to find the mean maximum F across the range of possible values of HT.
Because the lower bound on F is zero over the entire interval HT ε (0, 1), it suffices to evaluate the area A under the upper bound on F. This area is
| (33) |
The first term has indefinite integral −HT − ln(1 − HT) and evaluates to ln 2 − 1/2. The second term has indefinite integral and evaluates to 3/2 − 2 ln 2, so that A = 1 − ln 2 ≈ 0.3068528.
Note that F is substantially more constrained when HT ε [1/2, 1) than when HT ε [0, 1/2). The difference between the areas under the upper bound for HT ε [0, 1/2) and for HT ε [1/2, 1) is 3 ln 2 − 2 ≈ 0.0794415, a sizeable fraction of the sum of the two areas. Twice the difference in areas, or 6 ln 2 − 4 ≈ 0.1588831, is the expectation of the difference between the maximum value of F for a value of HT chosen uniformly at random from (0, 1/2) and the maximum value of F for a value of HT chosen uniformly at random from [1/2, 1).
Application to Data
We illustrate the bounds on F, M, and HT for a series of examples using human polymorphism data from Rosenberg et al. (2005) and Li et al. (2008). For each example, for each locus, we assume that the allele frequencies in the data sets are parametric allele frequencies. The parametric allele frequencies are obtained in each of a pair of populations, and they are then averaged to obtain parametric allele frequencies for the total population. F, M, and HT are then computed. The data set of Rosenberg et al. (2005) considers 1048 individuals genotyped for 783 microsatellites, and the data set of Li et al. (2008) considers 938 unrelated individuals genotyped for single-nucleotide polymorphisms (SNPs); for all analyses, we restrict our attention to the 935 individuals found in both data sets. For the Li et al. (2008) data, we examine only 640,034 SNPs studied by Pemberton et al. (2012).
Example 1: Africans and Native Americans
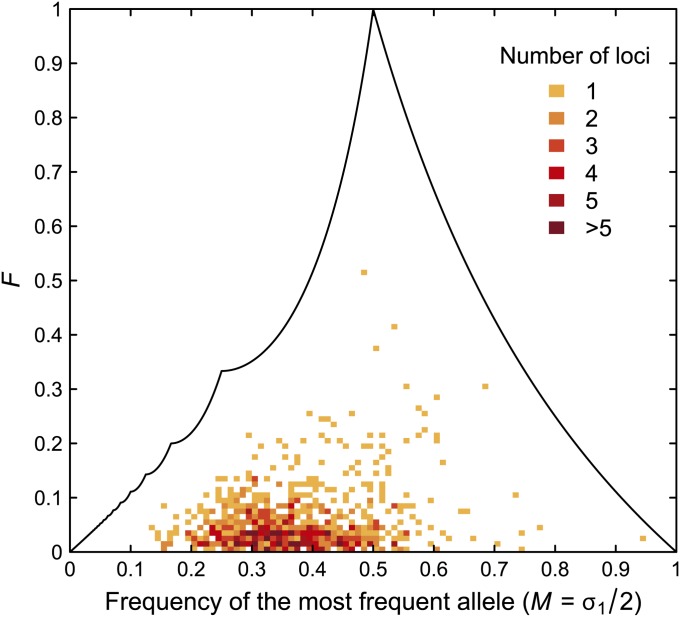
Our first example considers microsatellites in 101 Africans and 63 Native Americans, and it is chosen to illustrate a relatively wide range of values of F, M, and HT. Figure 7 shows F and M, demonstrating that for the comparison of Africans and Native Americans, F < 0.1 for most of the 783 loci. The mean value of F is 0.05 with standard deviation 0.06, and the mean value of M is 0.37 with standard deviation 0.11.
Figure 7 .
F and the frequency of the most frequent allele (M) for 101 Africans and 63 Native Americans. At each of 783 microsatellite loci, allele frequencies are computed separately for the two population groups, and the total allele frequency is the average of the two group frequencies. Each bin has size 0.01 × 0.01, and the upper bound on F as a function of M is shown for comparison.
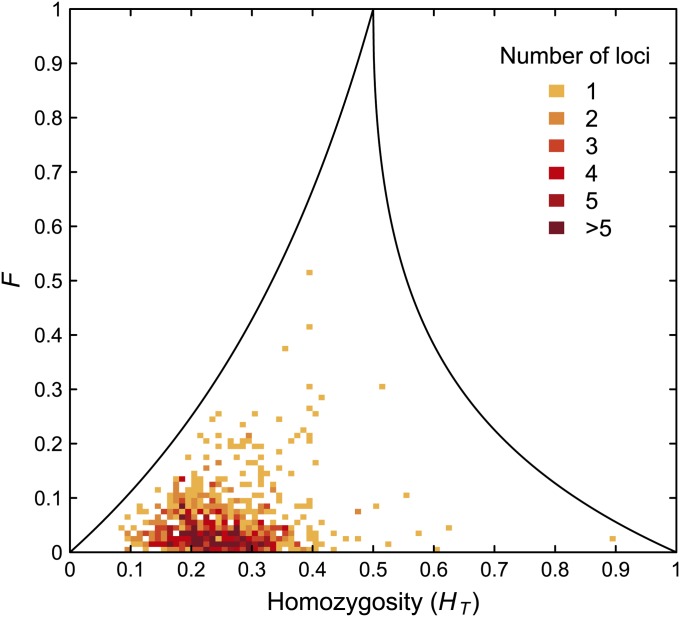
Similarly, Figure 8 plots F and HT for the 783 loci. The mean HT is 0.25 with standard deviation 0.08. In both Figures 7 and 8, relatively few loci approach the upper bound on F.
Figure 8 .
F and homozygosity (HT) for 101 Africans and 63 Native Americans. At each of 783 microsatellite loci, allele frequencies are computed separately for the two population groups, and the total allele frequency is the average of the two group frequencies. Each bin has size 0.01 × 0.01, and the upper bound on F as a function of HT is shown for comparison.
Example 2: High-diversity and low-diversity populations
The bounds on F as a function of M and HT indicate that genetic diversity in a pair of populations has a strong effect on the value of F between them. To illustrate this point, we compare the values of F obtained from two populations each with high within-population diversity to those obtained from two populations with lower within-population diversity.
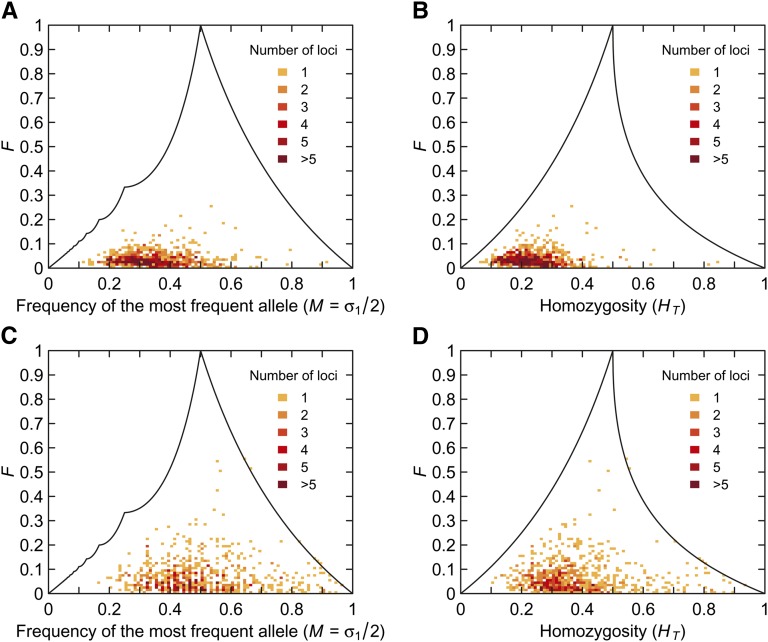
The Yoruba and Mbuti Pygmy populations are two African populations with high genetic diversity; the Colombian and Pima populations are Native American populations with lower diversity. Figure 9A shows F and M computed from the Yoruba and Mbuti Pygmy populations, and Figure 9B shows F and HT. The mean value of F is 0.04 with standard deviation 0.03, the mean value of M is 0.35 with standard deviation 0.11, and the mean value of HT is 0.24 with standard deviation 0.08.
Figure 9 .
Relationships among F, M, and HT, for pairs of African and Native American populations. (A) F and M for 21 Yoruba and 15 Mbuti Pygmy individuals. (B) F and HT for 21 Yoruba and 15 Mbuti Pygmy individuals. (C) F and M for 7 Colombian and 14 Pima individuals. (D) F and HT for 7 Colombian and 14 Pima individuals. In each plot, at each of 783 microsatellite loci, allele frequencies are computed separately for the two populations, and the total allele frequency is the average of the two population frequencies. Each bin has size 0.01 × 0.01, and the upper bound on F is shown for comparison.
By contrast, in corresponding plots for the less diverse Colombian and Pima populations, higher values of F, M, and HT are apparent (Figure 9, C and D). In particular, because M and HT tend to be nearer to 1/2, larger values of F are possible. The mean values of M and HT are much closer to 1/2 than in the African groups; the mean M is 0.50 with standard deviation 0.15, and the mean HT is 0.38 with standard deviation 0.15. As is suggested by the fact that F can attain its largest values when M and HT lie near 1/2, the mean value of F for the Native American groups is nearly twice as high as in the African groups (mean 0.07, standard deviation 0.07).
Example 3: Single-nucleotide polymorphisms
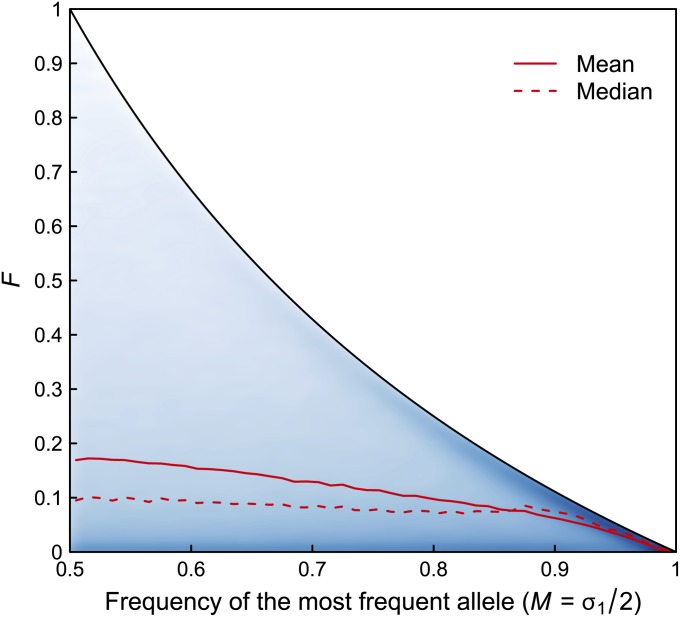
Our third example considers SNPs in the same set of Africans and Native Americans for which microsatellites were examined in Figures 7 and 8. Figure 10 shows the joint distribution of F and M as well as the mean and median of F for intervals of M ranging from 1/2 to 1 with width 0.01. Mean values of F decrease with M for M ε (1/2, 1), and this decrease is correlated with the decreasing value of the bound on F as a function of M (r = 0.94). Compared with the mean, the median value of F is less correlated with the value of the bound, although it also declines with increasing M (r = 0.77).
Figure 10 .
Smoothed scatterplot of F as a function of M for 101 Africans and 63 Native Americans, using SNP data. The shading reflects a two-dimensional kernel density estimate using a Gaussian kernel with bandwidth set to 0.007; the density was set to 0 outside the bounds on F as a function of M. For each of 640,034 SNP loci, allele frequencies are computed separately for the two population groups, and the total allele frequency is the average of the two group frequencies. The mean and median of F are computed for 50 bins of width 0.01 ranging from M = 1/2 to M = 1. The upper bound on F as a function of HT is shown for comparison.
For biallelic markers, for M > 1/2, at least one of the two alleles must appear in both populations, and the upper bound on F occurs when one of the populations has only one allele. In Figure 10, for high values of M, more SNPs approach the upper bound on F than for low values of M. This result indicates that SNPs with high values of M are more likely to have an allele found in one but not the other of the two populations.
Discussion
The range of F depends on the level of diversity in the markers considered. In this article, we have further shown that not only does diversity constrain the range of F, the frequency of the most frequent allele has a strong influence on the values that F can take. When the frequency of the most frequent allele is small or large, F is restricted to small values far from one (Figure 2). In fact, considering all possible values of M, F is restricted on average to only ∼35.85% of the space of possibilities. This extreme reduction in range for F can be viewed as a consequence of our result that about half of the contribution to the maximal F arises from the most frequent allele (exactly half for σ1 ε [1,2)). Using results from Rosenberg and Jakobsson (2008) on the relationship between homozygosity and the frequency of the most frequent allele, we have described a link between F and homozygosity of the total population (HT) via separate relationships of F and homozygosity to the frequency of the most frequent allele. F is restricted by HT even further than by M, to only ∼30.69% of the space of possibilities.
Our work extends knowledge of the connection between F and genetic diversity, providing a framework for interpreting a variety of features of values of F measured in population-genetic data. We have presented empirical computations that illuminate recently observed phenomena in human population genetics. In particular, even without a formal understanding of the ways in which evolutionary processes and the population-genetic models that encode them give rise to values of M, HT, and F, the mathematical constraints linking these quantities can aid in interpreting the patterns found in the data.
Low FST values in human populations from Africa
Estimates of FST in human populations have been low in Africa compared with other geographic regions, such as among Native Americans (Rosenberg et al. 2002; Tishkoff et al. 2009). This pattern appears to belie the extensive genetic differentiation known to exist among African populations. For example, using microsatellite loci, Tishkoff et al. (2009) identified a number of genetically distinctive subgroups of African populations despite confirming that FST in Africa has an unexpectedly small value. The apparent discrepancy between the extensive genetic differentiation among populations in Africa and counterintuitively low values of FST can be explained using our results. Because Africa has high within-population genetic diversity—including microsatellite homozygosities well below 1/2 in many populations (Tishkoff et al. 2009, Figure S2B)—the maximum FST for comparisons of African populations at microsatellite loci is relatively constrained compared with the maximum FST for comparisons of groups that have less within-population diversity and mean homozygosities nearer 1/2. Figure 9 shows that FST values comparing African populations are more constrained by M and HT than are those comparing Native American populations. Thus, the observation for microsatellites of low FST in African populations can be attributed to high within-population genetic diversities.
That FST is more tightly constrained for high-diversity populations than for populations where HT ≈ 1/2 has an additional consequence. When considering two pairs of populations with the same FST value and HT < 1/2, it is likely that a pair of populations with higher within-group diversity is more differentiated than is a pair of populations with relatively low within-group diversity. In other words, the higher the level of genetic diversity within a population, the greater the extent to which raw values of FST underpredict the intuitive level of differentiation among subpopulations; the result of Tishkoff et al. (2009) exactly follows this pattern.
Lower FST values for microsatellites than for SNPs
Computations of FST in human populations have generally found that FST estimates based on multiallelic loci such as microsatellites are lower than those obtained from biallelic loci such as SNPs (e.g., Rosenberg et al. 2002; Li et al. 2008). This observation is apparent in the difference between FST-like computations from nearly the same sets of individuals for microsatellites and for SNPs. When separating human populations into seven geographic regions and computing the within-population component of genetic variation, a quantity analogous to 1 − FST, Rosenberg et al. (2002) obtained an estimate of 0.941 with microsatellites, whereas Li et al. (2008) obtained 0.889 with SNPs. Our results provide a simple explanation for this difference. The SNPs of Li et al. (2008) each have only two alleles, so for each locus, the frequency of the most frequent allele is at least 1/2; further, the minor alleles tend to be common, such that many of the loci have M near 1/2. By contrast, the microsatellites in the study of Rosenberg et al. (2002) have ∼12 alleles on average, so M is typically smaller than 1/2 and often much smaller (Rosenberg and Jakobsson 2008). Thus, for microsatellites, because of lower frequencies of the most frequent allele and higher levels of genetic diversity, the maximum value of F is substantially more constrained than the corresponding maximum of F for SNPs (Figure 2). We can explain the difference in the magnitudes of the Rosenberg et al. (2002) and Li et al. (2008) FST values via this phenomenon.
Recently, attention has increasingly focused on biallelic sites for which the rarer allele has low frequency (Keinan and Clark 2012; Nelson et al. 2012; Tennessen et al. 2012). In our terms, these are sites for which the frequency of the most frequent allele, M, is high. Because F is tightly constrained for high values of M, we might expect that when FST is calculated using sites with rare minor alleles, small FST values will be produced. Indeed, Figure 10 shows that when F is used to compare Africans with Native Americans at SNP loci, mean values of F decrease as M increases from 1/2 to 1.
Conclusions
Measures of FST have often been used for making inferences about such phenomena as population structure, migration patterns, and range expansions. However, we have found that without a proper understanding of the dependence of FST on diversity and allele frequencies, FST can potentially produce puzzling or misleading results. We have described mathematical relationships between FST, the frequency of the most frequent allele, and homozygosity that are useful for interpreting the properties of differentiation measures when features of allele frequencies and diversity statistics vary across loci or populations—as they inevitably do in typical scenarios.
Beginning with Charlesworth (1998), Nagylaki (1998), and Hedrick (1999), recent studies have noted that FST is constrained by diversity, and the issue was described as early as in the work of Sewall Wright (Wright 1978, p. 82). Jost (2008) generated new interest in the dependence of FST on diversity, illustrating that the dependence can produce substantial discord between intuitions about and measurements of differentiation levels. Jost (2008) also used a multiplicative definition of diversity to propose a pair of new differentiation indices that have the feature of reaching their maximum value if and only if each allele is private to a single subpopulation. In our view, the key to choosing and applying measures of differentiation lies not in “fixation on an index” (Long 2009), be it FST, the measures of Jost (2008), or other indices that have recently been proposed (Meirmans and Hedrick 2011), but in developing an understanding of the ways in which possible statistics relate both to intuitive aspects of differentiation and to mathematical features of allele frequencies and genetic diversity. In this context, FST remains of particular interest on the basis of its long history of use in population genetics and its connection to features of biological models (Whitlock 2011). Our examples provide only a few among many ways in which the mathematical properties we have obtained for FST can be used to interpret its behavior in the analysis of empirical data.
Acknowledgments
We thank S. Boca and J. VanLiere for numerous discussions of this work. Financial support was provided by the Swedish Research Council, the Erik Philip Sörensen Foundation, the Burroughs Wellcome Fund, a Stanford Graduate Fellowship, and U.S. National Institutes of Health grants GM081441 and HG005855.
Appendix
The appendix provides the derivations of two integrals described in the main text.
Integral (Equation 18)
To obtain , we first note that for any integer k ≥ 1, if 1/(k + 1) ≤ σ1 < 1/k. We have
Defining , we then have
| (A1) |
Integral (with Q1 as in Equation 24)
To obtain , we first note that for any integer k ≥ 1, when . We have
The second term can be decomposed, defining
We have
| (A2) |
Footnotes
Communicating editor: M. A. Beaumont
Literature Cited
- Boca S. M., Rosenberg N. A., 2011. Mathematical properties of Fst between admixed populations and their parental source populations. Theor. Popul. Biol. 80: 208–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 1998. Measures of divergence between populations and the effect of forces that reduce variability. Mol. Biol. Evol. 15: 538–543 [DOI] [PubMed] [Google Scholar]
- Hedrick P. W., 1999. Perspective: highly variable loci and their interpretation in evolution and conservation. Evolution 53: 313–318 [DOI] [PubMed] [Google Scholar]
- Hedrick P. W., 2005. A standardized genetic differentiation measure. Evolution 59: 1633–1638 [PubMed] [Google Scholar]
- Holsinger K. E., Weir B. S., 2009. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat. Rev. Genet. 10: 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Chakraborty R., 1995. Population structure, stepwise mutations, heterozygote deficiency and their implications in DNA forensics. Heredity 74: 274–285 [DOI] [PubMed] [Google Scholar]
- Jost L., 2008. GST and its relatives do not measure differentiation. Mol. Ecol. 17: 4015–4026 [DOI] [PubMed] [Google Scholar]
- Keinan A., Clark A. G., 2012. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science 336: 740–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Z., Absher D. M., Tang H., Southwick A. M., Casto A. M., et al. , 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319: 1100–1104 [DOI] [PubMed] [Google Scholar]
- Long J. C., 2009. Update to Long and Kittles’s “Human genetic diversity and the nonexistence of biological races (2003): fixation on an index. Hum. Biol. 81: 799–803 [DOI] [PubMed] [Google Scholar]
- Long J. C., Kittles R. A., 2003. Human genetic diversity and the nonexistence of biological races. Hum. Biol. 75: 449–471 [DOI] [PubMed] [Google Scholar]
- Meirmans P. G., Hedrick P. W., 2011. Assessing population structure: FST and related measures. Mol. Ecol. Resources 11: 5–18 [DOI] [PubMed] [Google Scholar]
- Nagylaki T., 1998. Fixation indices in subdivided populations. Genetics 148: 1325–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., 1973. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 70: 3321–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. R., Wegmann D., Ehm M. G., Kessner D., Jean P. S., et al. , 2012. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science 337: 100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton T. J., Absher D., Feldman M. W., Myers R. M., Rosenberg N. A., et al. , 2012. Genomic patterns of homozygosity in worldwide human populations. Am. J. Hum. Genet. 91: 275–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S. B., Rosenberg N. A., 2012. Refining the relationship between homozygosity and the frequency of the most frequent allele. J. Math. Biol. 64: 87–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N. A., Jakobsson M., 2008. The relationship between homozygosity and the frequency of the most frequent allele. Genetics 179: 2027–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N. A., Pritchard J. K., Weber J. L., Cann H. M., Kidd K. K., et al. , 2002. Genetic structure of human populations. Science 298: 2381–2385 [DOI] [PubMed] [Google Scholar]
- Rosenberg N. A., Li L. M., Ward R., Pritchard J. K., 2003. Informativeness of genetic markers for inference of ancestry. Am. J. Hum. Genet. 73: 1402–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N. A., Mahajan S., Ramachandran S., Zhao C., Pritchard J. K., et al. , 2005. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 1: 660–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryman N., Leimar O., 2008. Effect of mutation on genetic differentiation among nonequilibrium populations. Evolution 62: 2250–2259 [DOI] [PubMed] [Google Scholar]
- Tennessen J. A., Bigham A. W., O’Connor T. D., Fu W., Kenny E. E., et al. , 2012. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337: 64–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff S. A., Reed F. A., Friedlaender F. R., Ehret C., Ranciaro A., et al. , 2009. The genetic structure and history of Africans and African Americans. Science 324: 1035–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlund S., 1928. Zusammensetzung von Populationen und Korrelationerscheinungen vom Standpunkt der Vererbungslehre aus Betrachtet. Hereditas 11: 65–106 [Google Scholar]
- Weir B. S., 1996. Genetic Data Analysis II. Sinauer, Sunderland, MA [Google Scholar]
- Whitlock M. C., 2011. /article/back/ref-list/ref/citation/inline-formula and D do not replace FST Mol. Ecol. 20: 1083–1091 [DOI] [PubMed] [Google Scholar]
- Wright S., 1951. The genetical structure of populations. Ann. Eugen. 15: 323–354 [DOI] [PubMed] [Google Scholar]
- Wright S., 1978. Evolution and the Genetics of Populations, Volume 4: Variability Within and Among Natural Populations University of Chicago Press, Chicago [Google Scholar]