Abstract
ETS family transcription factors are evolutionarily conserved downstream effectors of Ras/MAPK signaling with critical roles in development and cancer. In Drosophila, the ETS repressor Yan regulates cell proliferation and differentiation in a variety of tissues; however, the mechanisms of Yan-mediated repression are not well understood and only a few direct target genes have been identified. Yan, like its human ortholog TEL1, self-associates through an N-terminal sterile α-motif (SAM), leading to speculation that Yan/TEL1 polymers may spread along chromatin to form large repressive domains. To test this hypothesis, we created a monomeric form of Yan by recombineering a point mutation that blocks SAM-mediated self-association into the yan genomic locus and compared its genome-wide chromatin occupancy profile to that of endogenous wild-type Yan. Consistent with the spreading model predictions, wild-type Yan-bound regions span multiple kilobases. Extended occupancy patterns appear most prominent at genes encoding crucial developmental regulators and signaling molecules and are highly conserved between Drosophila melanogaster and D. virilis, suggesting functional relevance. Surprisingly, although occupancy is reduced, the Yan monomer still makes extensive multikilobase contacts with chromatin, with an overall pattern similar to that of wild-type Yan. Despite its near-normal chromatin recruitment, the repressive function of the Yan monomer is significantly impaired, as evidenced by elevated target gene expression and failure to rescue a yan null mutation. Together our data argue that SAM-mediated polymerization contributes to the functional output of the active Yan repressive complexes that assemble across extended stretches of chromatin, but does not directly mediate recruitment to DNA or chromatin spreading.
Keywords: enhancer, Drosophila development, signaling pathways
DYNAMIC regulation of gene expression during development requires the combined and coordinated action of transcriptional activators and repressors across multiple cis-regulatory modules (CRMs). Research over the last decade has led to a growing appreciation of the existence and importance of both short-range linear and long-range three-dimensional chromatin interactions to overall regulation of gene expression (Hong et al. 2008; Frankel et al. 2010; Perry et al. 2010, 2011; Bulger and Groudine 2011; Dunipace et al. 2011; He et al. 2011). However, the molecular determinants underlying long-range transcriptional regulation remain poorly understood.
One long-standing hypothesis of transcriptional repression is that the biochemical ability of a factor to polymerize might drive spreading of repressive complexes along the chromatin, thereby providing a mechanism of long-range repression (Courey and Jia 2001; Roseman et al. 2001). Well-studied examples include multiple Polycomb Group (PcG) corepressors and the ETS family transcriptional repressors TEL1 (ETV6) and Yan, all of which carry a strong oligomerization domain termed the sterile α-motif (SAM) (Kim et al. 2001, 2002, 2005; Tran et al. 2002; Qiao et al. 2004; Qiao and Bowie 2005). In vitro, the isolated SAM domains from these proteins form helical, head-to-tail polymers whose overall structural homology suggests a common mode of function. In vivo, mutations that disrupt SAM-mediated self-association have been shown to reduce or ablate repression activity of both the PcG and the ETS proteins in a variety of cultured cell and transgenic overexpression assays (Roseman et al. 2001; Song et al. 2005; Zhang et al. 2010; Robinson et al. 2012). Genome-wide occupancy analyses of two polymerization-competent PcG proteins in Drosophila, Polycomb (Pc) and Polyhomeotic (Ph), have shown that chromatin occupancy “spreads” over regions ranging from several to hundreds of kilobases (Negre et al. 2006; Schwartz et al. 2006; Tolhuis et al. 2006). Comparable studies have not been performed yet for either human TEL1 or Drosophila Yan, and although it is widely inferred, it has not been demonstrated that SAM-mediated oligomerization drives the long-range PcG chromatin occupancy patterns.
Here we have focused on the ETS family repressor Yan that acts downstream of receptor tyrosine kinase signaling in Drosophila to orchestrate a proper balance between proliferation and differentiation in a variety of tissues. Thus depending on context, loss of yan leads to overproliferation or inappropriate cell fate specification, while overexpression of a constitutively active form can block the induction of a variety of neural, epithelial, and mesodermal cell fates (Rebay and Rubin 1995; Rogge et al. 1995; Halfon et al. 2000; Hsu and Schulz 2000). In-depth investigation of a small number of direct transcriptional targets identified from genetic studies has led to the suggestion that Yan functions as a short-range passive repressor that competes with the ETS family activator Pointed (Pnt) for access to GGA(A/T) ETS consensus-binding motifs within specific cis-regulatory enhancers (Klambt 1993; Scholz et al. 1993; O’Neill et al. 1994). Competition between Yan and Pnt is regulated by MAPK activation, which attenuates Yan-mediated repression while stimulating Pnt-mediated activation (Gabay et al. 1996). These regulatory interactions have been proposed to provide a bistable switch that must be flipped for a cell to commit to a fate (Graham et al. 2010).
To test the model that Yan self-association through its SAM domain can induce spreading of repression complexes over extended stretches of chromatin and to gain further insight into Yan-mediated regulation of gene expression during development, we compared the global chromatin occupancy profile of endogenous wild-type Yan to that of a recombineered genomic transgene carrying a missense mutation in the SAM domain that restricts the Yan protein to a monomeric form. Consistent with the starting chromatin spreading model, we find that wild-type Yan binds at developmentally important genes as clusters of densely packed peaks that span multiple kilobases, a pattern that is conserved between Drosophila melanogaster and D. virilis. However, although binding is reduced, Yan monomers still exhibit prominent multikilobase chromatin occupancy patterns, an unexpected result in light of the limited ability of Yan monomers to rescue the null allele or repress gene expression. Based on these findings, we propose a revised model in which SAM-mediated polymerization of Yan does not provide the primary chromatin recruitment mechanism, but is instead required for the function, stability, and/or maintenance of long-range repressive complexes. Given that SAM-mediated chromatin spreading has also been proposed to underlie the extensive chromatin interactions and long-range repression of PcG proteins, our findings may have broader implications with respect to the relationship between polymerization, long-range chromatin occupancy, and transcriptional repression.
Materials and Methods
Fly strains, genotypic selection of yan mutant embryos, and rescue experiments
For chromatin immunoprecipitation (ChIP)–quantitative (q)PCR (ChIP-qPCR) verification of the ChIP-chip results, ∼400 stage-11 GFP-negative yan null embryos were hand selected from the cross yanER433/CyO, twist-Gal4, UAS-GFP (CTG)× yanE833/CTG. For YanV105R ChIP (∼800 embryos per replicate) and rescue experiments, GFP-negative stage-11 embryos were selected from the cross yanER433/CTG; YanV105R × yanE833/CTG; YanV105R. Controls were similarly selected from the cross yanER433/CTG; YanWT × yanE833/CTG; YanWT. For the genetic rescue experiments, embryos that hatched were counted as rescued to larval stage. Embryos that did not hatch were hand removed from their chorion and vitelline membrane, incubated in a 1:1 solution of glycerol:acetic acid overnight, mounted in Hoyers:lactic acid, and dried at 65° for ∼18 hr before imaging on a wide-field microscope.
Chromatin immunoprecipitation
w1118 embryos were collected; aged to stage 5–7 (2h10–3h10) or stage 11 (5h20–7h20, D. melanogaster; 10–12 hr at room temperature, D. virilis); dechorionated in 50% bleach; cross-linked in 10 ml of cross-linking solution (50 mM HEPES, pH 7.6, 1 mM EDTA, 0.5 mM EGTA, 100 mM NaCl, 1.8% formaldehyde) and 30 ml of n-Heptane for 15 min; washed in Stop Solution (PBS, 0.1% Triton X-100, 125 mM Glysine), PBS/T (PBS, 0.1% Triton X-100), and Wash Solution (10 mM HEPES, 10 mM EDTA, 0.5 mM EGTA, 0.25% Triton X-100); homogenized in ChIP lysis buffer [50 mM HEPES, 140 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, protease inhibitor tablet (Roche)]; and chromatin sonicated to a final size of 500 bp, using a Fisher Scientific (Pittsburgh) Sonic Dismembrator sonicator (Model 500) with nine cycles at 15% amplitude for 15 sec (0.9 sec on/0.1 sec off). Clarified lysates were incubated either with guinea pig anti-Yan (1:500) or with anti-GFP (1:200; Invitrogen, Carlsbad, CA) or were mock-treated with no antibody overnight at 4°. Gamma-bind sepharose beads (GE Healthcare) were added to the lysates and incubated for 4 hr at 4° to precipitate the DNA–protein complexes. Beads were washed in ChIP lysis buffer, high-salt ChIP lysis buffer (500 mM NaCl in ChIP lysis buffer), and TE (10 mM Tris, pH 8, 1 mM EDTA). Chromatin was washed off beads with TE/SDS (10 mM Tris, pH 8, 1 mM EDTA, 1% SDS) and reverse cross-linked at 65° overnight. ChIP DNA was purified using the QIAquick PCR purification kit (QIAGEN, Valencia, CA).
Following reverse cross-linking, native ChIP DNA was amplified via the linker-mediated PCR (LM-PCR) method (Lee et al. 2006), using Klenow DNA Polymerase (New England Biolabs, Beverly, MA) and ligation with a linker produced by annealing two oligos: 5′-/5Phos/AGAAGCTTGAATTCGAGCAGTCAG-3′ and 5′-CTGCTCGAATTCAAGCTTCT-3′. After adding the linker, DNA was amplified using the 20-mer primer and QIAquick purified. The dNTP mixture used in the amplification reaction contained a 3:7 ratio of dUTP:dTTP so that the products could be fragmented by Uracil DNA Glycosylase and APE1 (Affymetrix). Fragmentation, labeling, and hybridization were performed as described in the Affymetrix ChIP Assay Protocol. For ChIP-seq, after purification of native DNA, an adenine residue was added with Klenow [3′-5′ exo-] enzyme. Adaptors from Illumina for LM-PCR were ligated to the end of DNA molecules and the 200-bp product of the reaction was extracted and purified from a 2% agarose gel. Eighteen cycles of PCR were performed using phusion polymerase (Finnzyme F-530S) and the Illumina oligos. The product was purified by gel electrophoresis.
Genome-wide binding profiles and data analysis
Raw data are available as a GEO dataset (Series: GSE34038 and GSE34040) and were mapped to the April 2006 D. melanogaster genome. Three biological replicates of immunoprecipitated vs. no antibody control mock-treated ChIP samples hybridized on Drosophila Genomic Tiling Array 2.0R (Affymetrix) microarrays were analyzed with quantile normalization plus scaling and bandwidth of 200 bp, using Affymetrix tiling analysis software (TAS). Analysis using model-based analysis of tiling arrays (MAT) (Johnson et al. 2006) was performed using the following parameters: bandwidth, 200; MaxGap, 100; Minprobe, 10 (Roy et al. 2010). Peak calling and visual data inspection from both TAS and MAT genome-wide binding profiles were performed in the Integrated Genome Browser (IGB) (Affymetrix), using a top 3% or top 5% threshold with a minrun of 300 bp and maxgap of 100 bp (Supporting Information, Table S1). Statistical co-occurrence of binding peaks was analyzed using the Cooccur R package (Huen and Russell 2010). ChIP-chip data are shown as a −10 log10 P-value of the three biological replicates. The most significant peaks are expected to have a high consistency between replicates and thus a high P-value and, conversely, peaks with low significance represent those with low consistency between replicates (Landt et al. 2012).
Two biological replicates of ChIP and an input sample were sequenced on one lane of Illumina each and high-throughput sequencing was performed on an Illumina Genome Analyzer with the standard Illumina 36 cycles. The quality-filtered 36-bp reads were aligned using efficient local alignment of nucleotide data (ELAND). Following standard practice in the field (Robertson et al. 2007; Rozowsky et al. 2009; Zhong et al. 2010), the two deep-sequencing IP reads were combined to increase the number of reads and analyzed using model-based analysis of ChIP-seq (MACS) (Zhang et al. 2008) vs. input for peak calling, using a genome size of 1.2 × 108 and P-values 1 × 10−5 or 1 × 10−10. MACS-predicted Yan-bound regions and summits were used for transcription factor occupancy comparisons and motif analysis in Figures 4 and 6, respectively. Smoothed kernel density generated by the ChIP-seq analysis R package spp (Kharchenko et al. 2008) with bandwidth = 200 and step = 100 was used for visualization of peaks in Figures 1, 3, and 5.
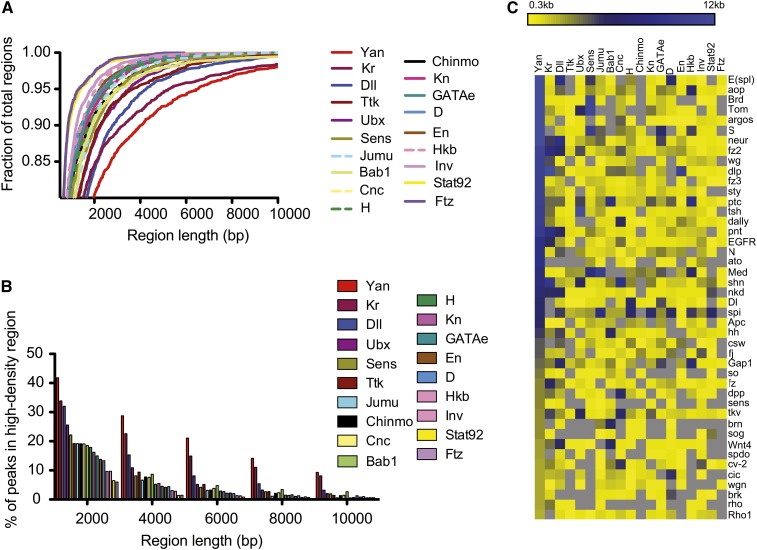
Figure 4 .
Meta-analysis of modENCODE transcription factors reveals high-density type binding is most prominent with Yan. (A) Cumulative frequency plot of transcription factor (TF)-bound region lengths. Using a threshold of the top 3% (MAT score), bound regions were defined for each transcription factor. Using our algorithm for defining HDRs, the lengths of bound regions occupied at 40% are plotted as a cumulative frequency distribution. Across the genome, Yan (red line) exceeds other transcription factors in the length of its bound regions. Kr, Jumu, H, GATAe, and Hkb data sets are from 0- to 8-hr embryo ChIP-chip; Sens data are ChIP-chip data from 4- to 8-hr embryos; and all other TF data sets, with the exception of Yan, are from 0- to 12-hr embryo ChIP-chip. (B) Percentage of TF-bound peaks that fall within HDRs, defined as regions of increasing length (bp) where TF occupancy ≥40%. (C) Meta-analysis of TF chromatin occupancy across the EGFR–Notch–Wingless pathway genes reveals extensive occupancy by Yan, but not by other transcription factors. The maximum length of bound regions at each gene was determined and plotted as a heat map for each transcription factor. Gray indicates no binding.
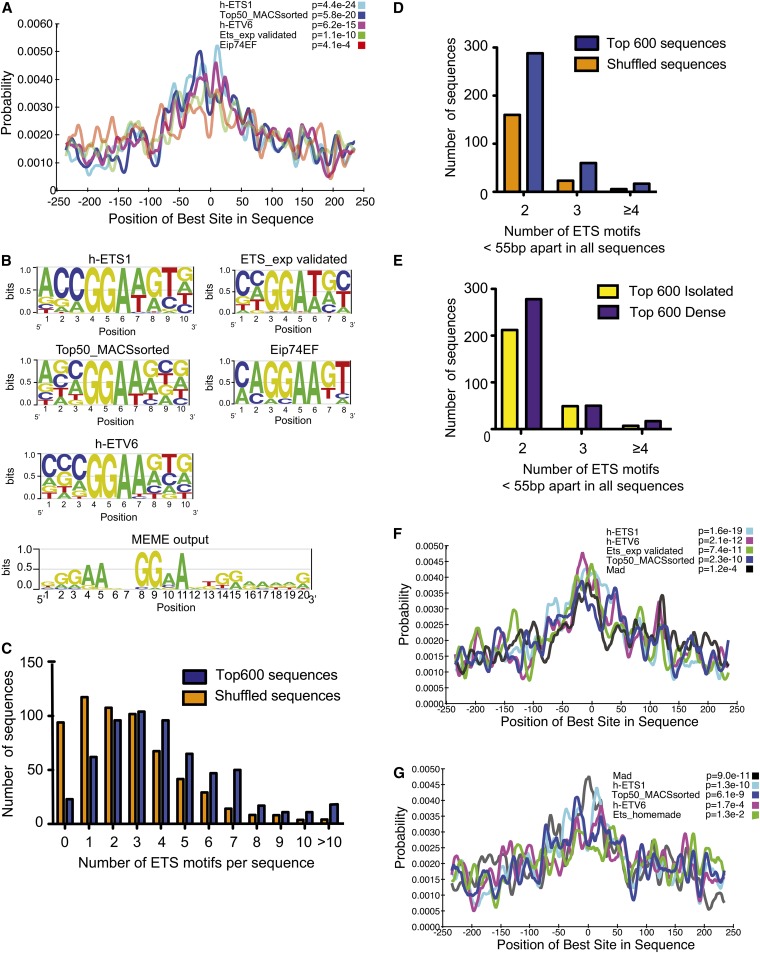
Figure 6 .
Motif analysis reveals clustering of ETS motifs in Yan-bound regions. (A) CentriMo analysis of the top 600 Yan-bound regions reveals central enrichment of the h-ETS1 motif and a motif derived from regions flanking GGAA/T sequences located within 72 bp around the top 50 ChIP-seq summits. (B) The PWMs of ETS motifs used to perform CentriMo analysis. MEME analysis of ChIP-seq peaks reveals clustering of GGAA/T motifs. (C) Scanning sequences bound by Yan at stage 11 (blue) with the PWM derived from the top 50 ChIP-seq sequences revealed enrichment of ETS motifs compared to a control set of sequences (orange). (D) Analysis of the gap between ETS motifs in either Yan-bound sequences (blue) or control sequences (orange) revealed an enrichment in clustering of ETS motifs. A cluster is defined as the number of ETS motifs found within 55 bp of each other (x-axis). (E) Clustering of ETS motifs in peaks associated with HDRs (purple) over isolated peaks (yellow). y-axis shows percentage of sequences with the number of motifs within 55 bp of each other (x-axis). (F and G) CentriMo analysis of known transcription factor PWMs, using the FlyReg (based on FlyReg Drosophila DNAse I Footprint Database v2.0), idmmpmm2009, and dmmpmm2009 (Kulakovskii and Makeev 2009; Kulakovskiy et al. 2009) databases, found within the top 600 isolated peaks (F) or top 600 peaks associated with HDRs (G) revealed that with the exception of Mad central enrichment, only ETS motifs were centrally enriched in either data set.
Figure 1 .
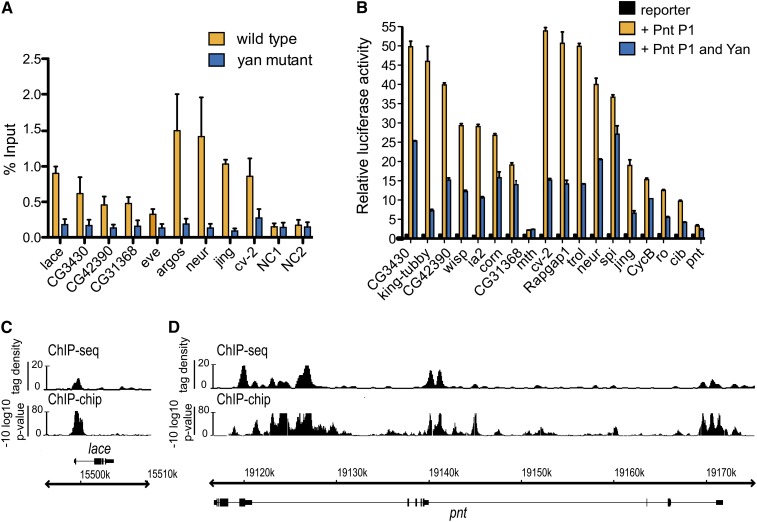
Yan associates with chromatin both at isolated peaks and within clusters of densely packed peaks. (A) ChIP enrichment is abolished in yanE833/yanER433 null mutant embryos relative to wild-type embryos. NC: negative control sites. Error bars are standard deviations of three independent ChIP experiments. lace, CG3430, CG42390, and CG31368 are classified as single-peak genes; eve, argos, neur, jing, and cv-2 are multiple-peak genes. (B) Transcription assays using luciferase reporters carrying Yan-binding regions identified in the ChIP-chip experiment. Reporters can be activated by Pnt and repressed by Yan. CG3430, king-tubby, CG42390, wisp, ia2, CG31368, and mth are single-peak genes; corn, cv-2, Rapgap1, trol, neur, spi, jing, cycB, ro, cib, and pnt are multiple-peak genes. Error bars are standard deviations of three independent experiments. (C and D) Occupancy patterns of wild-type Yan at the lace and pnt loci provide examples of isolated peak and clustered peak binding, respectively. ChIP-seq data are shown as smoothed tag density with a scale of number of reads per million. Gene structures are shown below ChIP patterns with gene names above or below the genome coordinates to depict + or – strands, respectively.
Figure 3 .
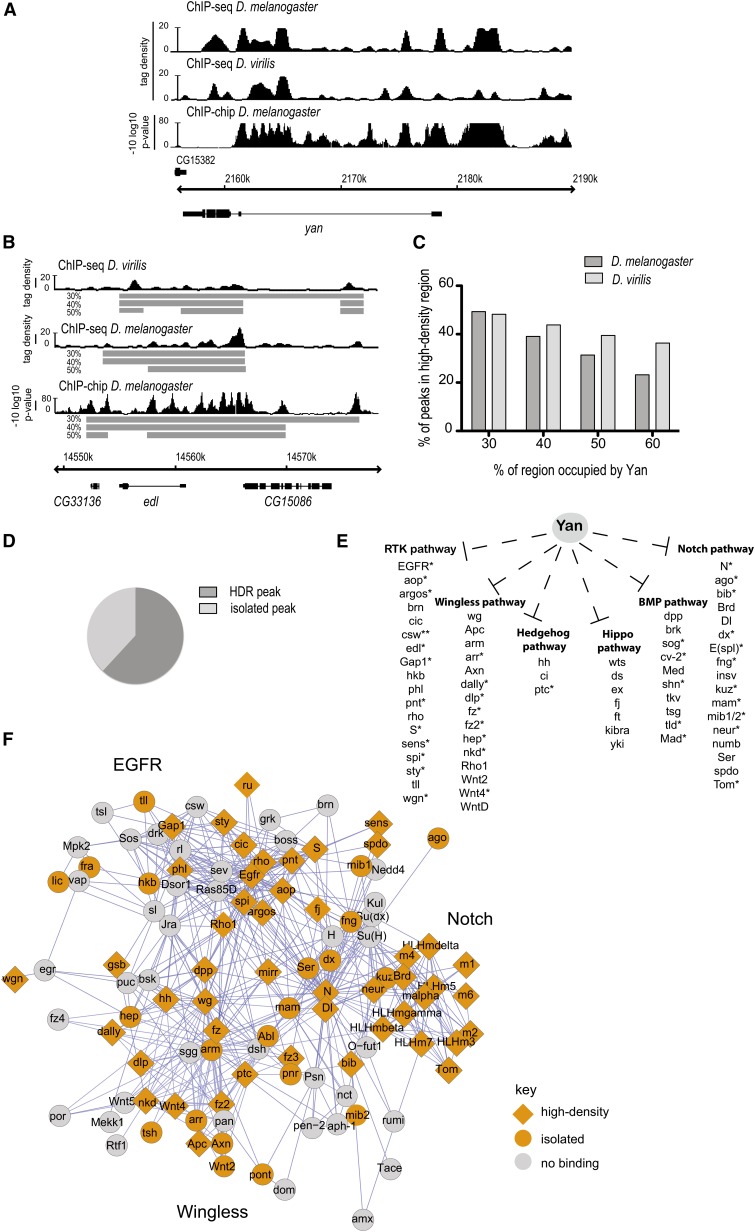
Conservation of Yan high-density regions in D. virilis. (A) ChIP-chip and ChIP-seq patterns at the yan locus highlight the conservation between D. melanogaster and D. virilis. (B) Defining high-density regions (HDRs) at the edl locus. Boxes indicate HDRs at the different percentages of occupancies. (C) Quantification of the prevalence of HDR occupancy across the genome in D. melanogaster and D. virilis. (D) D. melanogaster classification of peaks that are conserved in D. virilis. (E) Yan binds core components of multiple signaling pathways. *, genes assigned to Yan-bound peaks in both D. melanogaster and D. virilis. **, peaks close to csw just missed our statistical cutoff. (F) Cytoscape analysis of interactions within and between the EGFR, Notch, and Wg signaling pathways using the D. melanogaster stage-11 data set. Putative targets are colored orange, with orange diamonds representing genes associated with high-density Yan occupancy. In gray are genes not assigned to a Yan-bound peak. Interactions depicted are STRING 9.0 data, derived from experimentally determined protein–protein and genetic interactions as well as computationally predicted interactions.
Figure 5 .
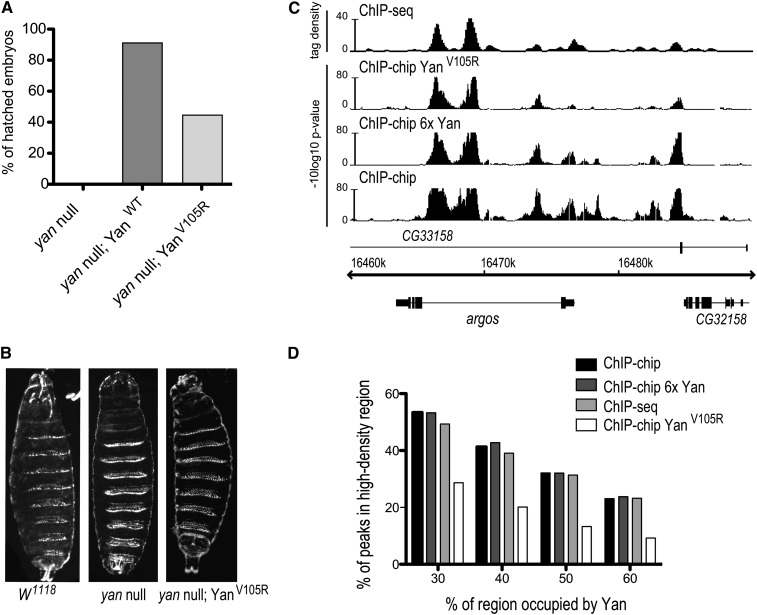
Yan self-association is not required for high-density occupancy patterns. (A) YanV105R has reduced genetic rescue ability relative to YanWT. The numbers of embryos that survived to hatching were counted for each genotype: (1) yanE833/yanER433, (2) yanE833/yanER433; YanWT, and (3) yanE833/yanER433; YanV105R. N > 240. (B) Cuticle preparations of (1) W1118, (2) yanE833/yanER433, and (3) yanE833/yanER433; YanV105R embryos reveal that YanV105R embryos die with an anterior open phenotype. (C) ChIP patterns of YanV105R, 6×Yan and wild-type Yan at the argos locus. ChIP-seq data are shown as smoothed tag density with a scale of number of reads per million. YanV105R and 6×Yan have broadly similar patterns to those of wild-type Yan, with high-density type binding to chromatin within clusters of multiple peaks. (D) Quantification of the prevalence of HDR occupancy at defined thresholds within each data set.
Identification of high-density regions and genomic assignments
Using a cutoff of the top 3% of Yan-bound peaks called by TAS and a homemade clustering algorithm (available upon request), the genome was separated into Yan-bound or unbound regions for each chromosome. Yan chromatin occupancy (bound length/total length) was calculated for each Yan-bound region. High-density regions (HDRs) were defined as regions >2 kb in which the density of Yan occupancy was >40%. The limits for each HDR were defined when the next Yan-bound peak was at a far enough distance that including it would decrease Yan occupancy levels below the 40% threshold (Figure S8). All peaks that did not fall into an HDR were classified as isolated.
For comparison between data sets, this analysis was carried out using thresholds of 30, 40, 50, and 60% Yan density. To calculate the percentage of total peak length within HDRs in Table S1, the total base-pair length of Yan-bound peaks within HDRs was divided by the total base-pair length of all peaks in the genome; this number thus represents the fraction of total Yan chromatin occupancy that falls into HDRs. Yan-bound peaks that did not overlap with a 5′-UTR, a 3′-UTR, an exon, or an intron (FlyBase r5.40) were annotated as intergenic. Gene assignments within 3 kb of Yan peaks were made using BDGP5.25 RefSeq genes (15,842 genes including noncoding RNAs).
Analysis of D. virilis data
We utilized the UCSC genome browser LiftOver software to translate each uniquely mapped D. virilis solexa sequencing read into the D. melanogaster genome, using default parameters with the exception of match = 0.5 to account for the large evolutionary distance between D. melanogaster and D. virillis (Kuhn et al. 2009).
Quantitative PCR
ChIP signals were quantified using the QuantiTech SYBR Green PCR Kit (QIAGEN). A standard curve for each primer pair (Table S10) was generated using serial dilutions of genomic DNA. The relative amounts of input, no antibody mock-treated, and immunoprecipitated DNA were determined based on the standard curves, and the ChIP signals were calculated as IP/input ratios.
Transcription assays
Reporter constructs were generated by inserting putative Yan-binding sequences (Table S9) upstream of the luciferase gene coding sequence. The inserts were chosen based on the ChIP-chip signals and predicted ETS binding sites, using CISTER (Frith et al. 2001). Primer sequences used for PCR amplification are available upon request. Transfection of Drosophila S2 cells grown in Gibco Sf-900 serum-free medium (Invitrogen) and transcription assays were as previously described (Zhang et al. 2010).
GO analysis
For stage 11 and stage 5–7, the top 2000 genes based on maximum peak P-values were functionally classified with Gene Ontology terms, using DAVID (Huang et al. 2009a,b). For the D. virilis data set, all genes assigned to Yan-bound regions were classified. Only functional clusters with a Bonferroni-corrected P-value <0.001 were regarded as significantly enriched. Panther (Thomas et al. 2003) was used to assess enrichment of signaling pathways (P < 0.05). Cytoscape (Cline et al. 2007) was used to map the interactions from String 9.0 (Szklarczyk et al. 2011) between members of the Notch, Wnt, and RTK signaling pathways. Comparisons between single-peak, multiple-peak, or high-density genes were limited to the stage 11 D. melanogaster data set.
Motif identification
Positional weight matrices were derived from either (a) all occurrences of GGAA/T and the flanking six residues found within the top 50 D. melanogaster ChIP-seq sequences (36 bp either side of the MACS predicted summit) or (b) all experimentally validated GGAA/T motifs within the Yan target genes eve, prospero, lozenge, and D-pax2. CentriMo analysis (Bailey and Machanick 2012) was performed with the top-scoring 600 ChIP-seq peaks, the top 600 isolated and the top 600 HDR sequences using positional weight matrix (PWM) motif databases from FlyReg (based on FlyReg Drosophila DNAse I Footprint Database v2.0), and idmmpmm2009 and dmmpmm2009 (Kulakovskii and Makeev 2009; Kulakovskiy et al. 2009). MAST (Bailey and Gribskov 1998) was used to predict binding sites (P < 0.0001) in a set of Yan-bound sequences derived from the top 600 ChIP-seq peaks vs. a set of control sequences derived from shuffling the regions within the genome, using BedTools (Quinlan and Hall 2010). Motif searches were also performed on the top 600 isolated peaks and the top 600 peaks associated with HDRs. MEME analysis was carried out on 600 sequences consisting of 100 bp around the MACS-identified ChIP-seq summit (P < 1 × 10−10). DREME analysis (Bailey 2011) was carried out on the top 600 isolated or HDR ChIP-seq peak sequences. Motif analysis of the genomic sequences used in transcription reporter assays was performed using MAST with a PWM derived from the top 50 Yan-bound sequences and a PWM for Mad from dmmpmm2009 (Kulakovskiy et al. 2009).
Recombineering
The V105R missense mutation was introduced into a BAC carrying the wild-type (WT) yan locus (2L: 215,618–2,188,108), using the Counter-Selection BAC Modification Kit (Gene Bridges, Heidelberg, Germany) following the manufacturer’s protocol. The YanWT and YanV105R BAC constructs were YFP or GFP tagged at the C terminus, respectively. Subcloning via the BAC Subcloning Kit (Gene Bridges) into the transformation vector and site-specific integration into the attP2 site on the third chromosome (Groth et al. 2004) were performed as described in Ludwig et al. (2011). YanWT was also integrated into the attP40 site on the second chromosome to enable construction of the 6×Yan stock that carries both copies of the endogenous yan gene plus four copies of YanWT, two on the second and two on the third chromosome.
Embryo staining, quantification of eve expression, and protein immunoblots
Embryos were collected for 2 hr, aged to stage 11 at 25°, genotyped as described above using GFP fluorescence of the CTG balancer, and stained as previously described (Zhang et al. 2010) with guinea-pig anti-Yan (1:2000) or mouse anti-Eve mAb 3C10 (1:10; Hybridoma Bank, Iowa City, IA). Using a Zeiss LSM 510 confocal microscope, 20 or 36 serial 0.8-μm z-sections were maximum projected using LSM software for analysis of Eve and Yan expression, respectively. For calculating the relative mean intensity of Eve expression in the mesodermal clusters of yanE833/ER443;YanWT and yan E833/ER443;YanV105R embryos, the mean background pixel intensity was subtracted from the mean pixel intensity for the cluster (measured using ImageJ with a box of defined size) and normalized by dividing by the average mean pixel intensity for Eve expression in wild-type embryos. For protein immunoblots, ∼100 stage-11 embryos per genotype (6×Yan or hand-selected yanE833;YanWT and yan E833;YanV105R) were homogenized in 50 ml SDS sample buffer (250 mM Tris-Cl, pH 8, 10% SDS, 50% glycerol, 5% b-mercaptoethanol, 0.04% bromophenol blue), passed through a 27-gauge needle 10 times, and boiled for 10 min prior to running on an 8% SDS–PAGE gel and transfer to PVDF as previously described (Zhang et al. 2010). The blots were probed with anti-GFP (1:1000; Invitrogen) and anti-tubulin (1:1000; Sigma, St. Louis).
Results
Yan chromatin occupancy spreads across multiple kilobases
To characterize the genome-wide binding profile of Yan and identify a more comprehensive list of targets, we performed a ChIP-on-chip analysis of endogenous Yan in stage-11 embryos, a developmental window in which both Yan is highly expressed and its signaling mediates critical developmental events (O’Keefe et al. 1997; Riesgo-Escovar and Hafen 1997; Scholz et al. 1997; Price and Lai 1999; Halfon et al. 2000). High-confidence–bound regions (Table S1 and Table S2) were mapped to genes within 3 kb, yielding 2901 potential Yan target genes. Approximately 10% of Yan-bound regions fall within known promoter regions, while 44% and 39% map to intronic or intergenic regions, respectively, consistent with the expected locations of cis-regulatory elements (Figure S1). Although the criterion for assigning Yan-bound regions to target genes will misassign CRMs that act over large distances and will generate false positives in gene-dense regions, it is broadly accurate since the assignments included known Yan target genes such as eve, argos, mae, prospero, and mir7 (Gabay et al. 1996; Golembo et al. 1996; Flores et al. 2000; Halfon et al. 2000; Qiao et al. 2004; Vivekanand et al. 2004; Li and Carthew 2005). Further, the putative targets include 104 genes previously shown by microarray analysis to be responsive to perturbation of the EGFR pathway in either follicle cells or third-instar wing discs (Table S3) (Jordan et al. 2005; Butchar et al. 2012).
To validate the ChIP-chip results, we performed ChIP followed by qPCR at selected Yan-bound regions from stage-11 wild-type or hand-selected yan null mutants. Although there is no evidence of maternally provided Yan (Rogge et al. 1995; Gabay et al. 1996), yan null embryos do not die until after stage 16, permitting analysis at stage 11. All nine regions tested showed enriched Yan occupancy in wild-type embryos, but not in yan null mutants (Figure 1A). Confirming antibody specificity, immunostaining of wild-type and yan null embryos revealed only background-level fluorescence in the mutants (Figure S2). Yan occupancy patterns were also independently verified at selected targets by ChIP-qPCR analysis of embryos carrying GFP-tagged wild-type Yan, using an anti-GFP antibody (Figure S3). Finally we showed that Yan-bound regions contain elements capable of responding to both Yan and Pnt by cloning candidate CRMs with predicted ETS-binding clusters [identified by Cister (Frith et al. 2001); see also Table S9] upstream of the firefly luciferase coding sequence and measuring activity in transfected cultured S2 cells. Sixteen of the 18 reporters tested were significantly activated by Pnt and repressed by Yan (Figure 1B).
In agreement with the starting prediction that Yan may spread linearly across the DNA, a prominent feature of the genome-wide occupancy profile was that Yan binding occurs not only as isolated peaks (Figure 1C) but also as clusters of densely packed peaks that span multiple kilobases (Figure 1D). Several controls were performed to ensure that these spreading patterns were not the result of experimental artifacts. First, we used PCR analysis to confirm efficient sonication of the genomic DNA into fragments of an average size of ≤500 bp (Figure S4). Second, a follow-up experiment using ChIP-seq revealed similar binding patterns (Figure 1, C and D), ruling out hybridization artifacts with the ChIP-chip platform. Thus, as our protocol is standard for the field (see Materials and Methods) and the results are reproducible between independent experiments and platforms, the observed patterns are unlikely to be the result of excessive cross-linking (Table S1).
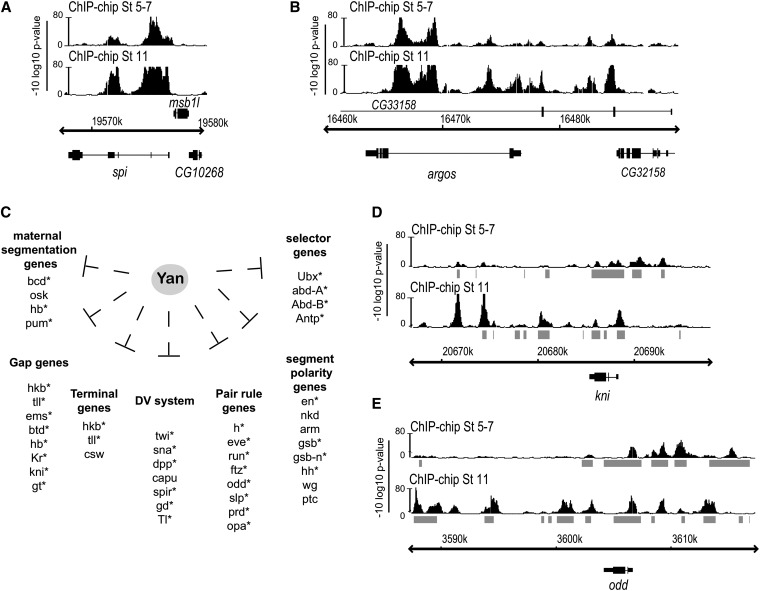
To determine whether the phenomenon of multikilobase Yan chromatin occupancy is stage specific, we profiled stage 5–7 embryos, a developmental time point in which the function of Yan remains largely uncharacterized despite its prominent expression (Price and Lai 1999). We separated putative target genes into two bins based on whether single or multiple Yan-bound peaks were detected and then compared the assignments at stage 5–7 vs. stage 11. Overall, Yan-binding patterns were broadly similar across the two time points with ∼60% of genes bound by Yan at both stages retaining their classification as either a single- or a multiple-peak gene. Thus multikilobase spreading appears to be a general characteristic of Yan occupancy, rather than a stage-specific anomaly (Figure 2, A and B).
Figure 2 .
Yan binding patterns are dynamic during development. (A and B) ChIP-chip patterns of spi and argos provide representative examples of loci that retain their classification as multiple-peak genes and have identical binding profiles at stage 5–7 and stage 11. (C) Yan putative targets include genes regulating axial patterning of the early embryo. Asterisks depict loci with temporally dynamic Yan chromatin occupancy patterns. (D and E) ChIP-chip patterns of representative loci with changing patterns of Yan chromatin occupancy. Shaded bars indicate DNAse-accessible chromatin at stage 5 or 11 (Li et al. 2011).
However, ∼11% of genes that retained their classification of having a single isolated Yan-bound peak, and almost 50% of genes classified as having a clustered multiple-peak Yan-binding profile, displayed stage-specific differences in chromatin occupancy. A striking example of such potential differential regulation was observed at genes involved in axial patterning of the early embryo, a developmental context in which Yan function has not been previously implicated. Thus, the majority of the anterior–posterior segmentation network, including maternal-effect, terminal, gap, pair-rule, segment polarity, and homeotic selector genes, was identified as putative Yan targets at both stage 5–7 and stage 11 (Figure 2C). Analysis of gene expression profiles available from the Berkley Drosophila Genome Project (Tomancak et al. 2002, 2007) revealed that with the exception of bcd and osk, these genes have dynamic expression profiles throughout the embryonic stages we have profiled and are expressed in regions of the embryo that overlap with yan’s expression profile (Table S4). At ∼80% of these genes, temporally dynamic binding patterns of Yan chromatin association were detected (Figure 2C, asterisks). For example, at the knirps (kni) locus, Yan binding shifted from multiple peaks upstream of the transcriptional unit in early embryos to multiple peaks across and downstream of the gene at stage 11 (Figure 2D). At the odd-skipped (odd) locus, we observed both loss and gain of individual peaks between stages 5–7 and 11 (Figure 2E). At each of these examples, stage-specific Yan binding appeared closely correlated with DNAse-accessible chromatin (Li et al. 2011). Across the genome, Yan-bound peaks overlapped with stage 5 and stage 11 DNAse-accessible chromatin by ∼70 and 87%, respectively, suggesting that the dynamic patterns of Yan recruitment to DNA may result from changes in chromatin conformation at different stages of development.
Yan chromatin association patterns are conserved in D. virilis
If chromatin binding across multikilobase regions is important for Yan-mediated regulation of its target genes, then both this tendency and the specific pattern at a given target gene should be evolutionarily conserved. To address this, we performed ChIP-seq from stage-11 D. virilis embryos, a species that despite diverging from D. melanogaster >60 million years ago exhibits a conserved pattern of embryonic Yan expression (Price and Lai 1999). Analysis of Yan chromatin occupancy patterns using LiftOver data mapped to the D. melanogaster genome (see Materials and Methods) revealed similar patterns across both species, with conservation of both the putative target genes bound by Yan and the actual Yan-binding footprint within multipeak regions (Figure 3A).
To facilitate comparisons of these data sets, we developed an algorithm to quantify the prevalence of clusters of Yan-bound peaks spanning multiple kilobases, defining a region of chromatin as an HDR if the length of Yan-bound peaks divided by the total length of the bound region was greater than a density threshold (see Materials and Methods). An example of an HDR at the edl locus, defined at increasing density thresholds, is given in Figure 3B. Across the genome, a similar fraction of Yan-bound peaks are assigned to HDRs at each density threshold within both data sets. For example, at 40% Yan occupancy, 39% of Yan-bound peaks are within HDRs in the melanogaster data set compared to 44% of Yan-bound peaks within the virilis data set (Figure 3C). Further, 40% of Yan-bound peaks present in the D. virilis data set overlap with peaks called within the D. melanogaster data set. Of these shared peaks, >60% are assigned to HDRs in the D. melanogaster data set (Figure 3D). This relatively high degree of conservation of HDR-type Yan-bound regions suggests that these CRMs could be involved in regulation of conserved expression patterns that are subject to strong selection.
In agreement with this hypothesis, analysis of gene ontology (GO) terms associated with putative target genes assigned to Yan-bound regions from D. melanogaster and D. virilis revealed that while single-peak genes are not significantly enriched for GO terms, genes associated with multiple peaks or HDRs are enriched for terms reflecting involvement in many aspects of development, with significant overrepresentation of several signal transduction pathways (Figure 3E and Table S5, Table S6, and Table S7). Both single-peak and high-density genes were among those validated by qPCR and transcription assays (Figure 1, A and B, and Figure S3), suggesting both categories include biologically relevant target genes. Thus the differences in complexity of Yan occupancy patterns likely reflect the requirement for more complicated regulation of key signaling and patterning genes.
Particularly striking pathway signatures were observed within the EGFR, Wingless, and Notch signaling networks in both species (Figure 3E). To highlight this, and using just the melanogaster data, we mapped the interactions between the canonical members of these three pathways, using a cytoscape STRINGWSClient plug-in (Cline et al. 2007; Szklarczyk et al. 2011); genes with Yan-bound peaks are highlighted in orange (Figure 3F). Analysis of the network suggests that Yan influences the expression of many core components of each signaling pathway predominantly via high-density type binding (Figure 3F, orange diamonds). Further, genes associated with HDRs appear more highly connected within the EGFR–Wingless–Notch network, showing an average of 13.3 interactions relative to 7.4 for the remaining Yan-bound genes (Figure 3F, orange circles).
Yan exceeds other transcription factors in high-density type chromatin binding
Previous studies of other sequence-specific transcription factors have noted multipeak regions analogous to the Yan-bound HDRs we have described (Li et al. 2008; Zinzen et al. 2009; Negre et al. 2011; Slattery et al. 2011). To compare the extent and prevalence of HDR patterns of chromatin association, we carried out a meta-analysis of modENCODE ChIP-chip data for 19 other sequence-specific transcriptional regulators (modENCODE Consortium et al. 2010).
First, using our HDR algorithm, we determined for each transcription factor the frequency with which bound peaks cluster into HDRs and the length distribution of bound regions. To compare between data sets we assessed the top 3% of bound regions for each transcription factor. Across the genome, Yan exceeds all other transcription factors in both HDR frequency and length, with only Kruppel (Kr) approaching comparable levels (Figure 4, A and B). Distalless (Dll) and Ultrabithorax (Ubx) exhibit prominent, but less prevalent and extensive HDR binding (Figure 4, A and B).
Second, focusing on the EGFR–Notch–Wingless signaling network genes (Figure 3F), we asked whether multikilobase Yan occupancy might correlate with clustered binding of other transcription factors. However, unlike Yan, none of the modENCODE transcription factors analyzed, including those with prominent genome-wide HDR signatures such as Kr, Dll, and Ubx (Figure 4, A and B), exhibit extensive occupancy at the canonical EGFR, Notch, and Wingless signaling network genes (Figure 4C).
SAM-mediated oligomerization is not the primary determinant of high-density Yan chromatin association patterns
The high-density chromatin association patterns we observe with Yan appear consistent with the hypothesis that SAM-mediated oligomerization could induce long-range spreading of repression complexes (Qiao et al. 2004; Qiao and Bowie 2005). Two additional predictions of this chromatin-spreading model are that increasing Yan levels should increase polymer formation and lead to even more extensive chromatin occupancy, while blocking SAM-mediated self-association should reduce the extent of occupancy. To test these predictions, we generated transgenic animals carrying either six copies of the wild-type yan gene (two endogenous copies plus four genomic transgenes; referred to as 6×Yan) or a version of yan into which the V105R missense mutation in the EH surface of the SAM domain was introduced. The V105R mutation blocks polymer formation in vitro and increases Yan diffusion rates in fluorescent recovery after photobleaching assays in Drosophila cells, consistent with loss of polymer formation (Qiao et al. 2004; Zhang et al. 2010). To ensure correct spatio-temporal patterns and levels of expression, a recombineered BAC clone spanning the entire yan genomic locus was used to generate both the wild-type and the V015R transgenes (see Materials and Methods). Immunostaining revealed normal patterns of Yan protein expression, with levels elevated in 6×Yan embryos and reduced in YanV105R mutants; the latter showed diffuse staining in both the cytoplasm and the nucleus (Figure S5), consistent with our previous finding that loss of SAM-mediated polymerization increases Yan nuclear export (Zhang et al. 2010). Whereas the control YanWT transgene fully rescued the yan null mutant, >50% of yan−/−;YanV105R embryos died just before hatching with an anterior open phenotype (Figure 5, A and B). The few escaper YanV105R animals that survived to adulthood were infertile and died after a few days (data not shown). Elevated expression of eve and an increased number of Eve-positive mesodermal cells in yan−/−;YanV105R embryos relative to control yan−/−;YanWT embryos (Figure S6) confirmed impaired repression of Yan target gene expression. These results agree with previous experiments with overexpressed cDNA transgenes that showed the V105R mutation, while not compromising DNA binding, abrogates the ability of Yan to repress gene expression and to supply full in vivo function (Qiao et al. 2004; Zhang et al. 2010).
Surprisingly, ChIP-chip analysis of both 6×Yan and hand-selected yan−/−;YanV105R stage-11 embryos revealed broadly similar occupancy profiles to those of wild-type embryos, suggesting that SAM-mediated polymerization is not the primary determinant of Yan occupancy patterns (Figure 5C). We quantified the prevalence of HDR-type binding and found that while the 6×Yan condition did not increase the frequency or extent of HDR occupancy genome-wide (Figure 5, C and D, and Table S1), quantitative changes in YanV105R binding patterns relative to wild type were apparent. Thus 20% of YanV105R peaks fall in dense occupancy regions at a 40% threshold, compared to 41% of wild-type Yan peaks, with a reduced median HDR length of 3.3 kb compared to 4.1 kb for wild-type Yan (Figure 5D). While the amplitude of bound peaks was generally reduced in the V105R sample, presumably reflecting the lower Yan protein levels (Figure S5), and may account for some of the reduction in HDR occupancy, the loss of Yan-bound peaks in the V105R data set was significant across a range of bound thresholds (Figure S7). We therefore propose that SAM-mediated polymerization is not, as previously suggested, the primary determinant for Yan spreading over extended chromatin domains, but rather contributes to the formation, stabilization, and function of the molecular complexes that occupy HDRs.
ETS and MAD motifs are enriched in Yan-bound sequences
Given the unexpected discovery that SAM-mediated self-association appears unlikely to be the primary determinant of Yan chromatin occupancy, an alternate hypothesis is that Yan could bind and spread along DNA via direct recognition of consensus ETS-binding sites located throughout the Yan-bound regions. To explore how differences in the number and/or spacing of ETS motifs contribute to Yan chromatin-binding patterns, we performed central motif enrichment analysis using CentriMo, a visualization and statistical analysis tool that identifies the region of maximum central enrichment in a set of ChIP-seq peak regions and displays the positional distributions of predicted sites (Bailey and Machanick 2012). Analysis of the top 600 ChIP-seq sequences revealed that while all ETS motifs tested are centrally enriched over flanking regions, motifs derived from the top 50 MACS-sorted ChIP-seq peaks (see Materials and Methods for details) and the human (h)-ETS1 motif (Wei et al. 2010) both show a stronger central enrichment and occur more frequently than the Drosophila ETS family Eip74EF motif (Kulakovskii and Makeev 2009), the experimentally derived TEL1 motif (h-ETV6 (Wei et al. 2010), or a motif derived from 16 experimentally validated ETS motifs within the Yan target genes prospero, eve, D-pax2, and lozenge (Flores et al. 2000; Halfon et al. 2000; Xu et al. 2000; Behan et al. 2002) (Figure 6, A and B).
Analysis of the top 600 ChIP-seq peaks using a positional weight matrix derived from the top 50 sequences revealed a 1.5-fold enrichment of ETS-binding sites over that of a control genomic shuffled sequence (Quinlan and Hall 2010) (Figure 6C). Using a window of 55 bp, the maximum distance between ETS sites that supports cooperative in vitro binding of the mammalian homolog of Yan, TEL1 (Green et al. 2010), we observed a 1.8-fold enrichment in the total number of sequences containing two adjacent motifs and a 2.7-fold enrichment in the number of sequences containing three or more adjacent motifs in the experimental vs. control data sets (Figure 6D). In agreement with this finding, MEME, a motif discovery tool (Bailey and Elkan 1994), identified a tandem GGAA/T repeat motif from a randomized set of 600 sequences composed of 100 bp around the MACs-predicted ChIP-seq summits (47 sites, P = 1.5 × 10−6; Figure 6B).
To identify DNA sequence characteristics specific to HDRs, we separated the top MACS-sorted sequences from HDRs and isolated Yan-bound peaks. Although the analysis reveals an equivalent density of ETS motifs within high-density and isolated peak regions, there is increased clustering of ETS motifs in HDRs, with a 1.3-fold enrichment over the isolated-peak regions (Figure 6E). We speculate that the extent of clustering of ETS-binding motifs may help stabilize Yan binding to chromatin in high-density clusters of peaks. However, the complexity of the chromatin occupancy patterns we have observed together with the temporal dynamics across development suggests additional modes of Yan recruitment to DNA are likely required.
Although our meta-analysis of modENCODE ChIP-chip data sets did not reveal likely candidates, recruitment of Yan to multikilobase regions may occur in conjunction with one or more DNA-binding transcription factors. If correct, then motifs for other transcription factors should be enriched in our data sets. Returning to the central motif enrichment analysis on our top 600 MACS-sorted sequences, we noted that while ETS motifs have high central enrichment in both the HDR and the isolated peak data sets (Figure 6, F and G), the site-probability curve is broader in the HDR data set, a feature that has been interpreted as indicative of cooperative binding to another transcription factor (Bailey and Machanick 2012). Of the transcription factors analyzed in Figure 4, A and B, that have available motif matrices, namely, Kr, Ttk, Ubx, Bab1, H, En, and Hkb, we did not observe significant enrichment in our data sets either by CentriMo or by DREME analysis (Figure 6, F and G, and Table S8). Instead, the binding motif for Mad, which along with Medea, is the primary transcriptional effector of the Dpp signaling pathway (Sekelsky et al. 1995; Hudson et al. 1998), was not only enriched in both the Yan HDR and isolated peak data sets, but also the most centrally enriched motif in the HDR data set (Figure 6G). This correlation raises the possibility of a link between Mad binding and HDR-type Yan recruitment. Supporting this, analysis of the genomic regions tested by transcription assay (Figure 1B) revealed putative MAD-binding sites in 13/18 of the reporters tested. Further, clustered MAD and ETS sites were identified in 12/13 of these regions with overlapping sites found in neur, cib, and Rapgap1 reporters (Table S9). Unfortunately, although Mad ChIP assays have been carried out (MacArthur et al. 2009), the data set quality is low with few peaks called, preventing us from comparing actual Mad- and Yan-bound regions. However, analysis of Medea ChIP data (MacArthur et al. 2009) reveals a 63% overlap of stage-11 Yan-bound peaks with Medea-bound regions, suggesting further investigation of Yan–Mad/Medea interactions might provide insight into mechanisms of Yan chromatin association.
Discussion
In this study, we have used genome-wide chromatin occupancy analysis to test the hypothesis that Yan’s ability to self-associate through its SAM domain enables it to spread across long distances on chromatin. Mechanistically, the chromatin-spreading model predicts that the initial recruitment of Yan monomers, either individually or cooperatively, to clusters of high-affinity ETS binding sites would nucleate polymer formation. The resulting increase in local Yan concentration would enable interactions with lower-affinity sites in flanking DNA that would not normally be bound by Yan monomers, thereby allowing stable repression complexes to assemble across multiple kilobases of DNA (Courey and Jia 2001; Kim et al. 2001, 2002; Roseman et al. 2001; Tran et al. 2002; Qiao et al. 2004; Qiao and Bowie 2005; Song et al. 2005; Zhang et al. 2010). Such large polymeric complexes could repress transcription by passively blocking access to large stretches of DNA and/or by actively recruiting associated factors. Consistent with the spreading model, a significant fraction of Yan occupancy occurs as multikilobase clusters of densely packed peaks.
Before considering the implications of this finding further, it is important to note that some of the complexity of Yan occupancy patterns undoubtedly results from distinct enhancers being bound in different cell types. However, both the general conservation of occupancy patterns across development, despite the presence of very different cell types, and the strength of the ChIP signals suggest that extreme cell-to-cell heterogeneity in Yan binding is unlikely to explain the full profile. While formal confirmation will require cell type-specific ChIP analyses, our interpretation that complex transcription factor chromatin occupancy can occur within a single tissue or cell type and still be detected in a whole-embryo ChIP assay is supported by recent work from the Furlong and Mann laboratories (Agelopoulos et al. 2012; Bonn et al. 2012). Thus for the purpose of the ensuing discussion, we assume this interpretation is correct.
Our finding that introduction of the V105R missense mutation into the Yan SAM domain only modestly reduced chromatin occupancy, despite drastically compromising function, leads us to propose that SAM-mediated polymerization is not the primary determinant of multikilobase chromatin spreading but instead contributes to active repression. Although it is formally possible that the modest chromatin occupancy differences detected between wild-type and monomeric YanV105R could fully explain the loss of functionality, the essentially identical multikilobase occupancy profiles observed across critical direct target genes such as aos (Figure 4C) argue against this. We also cannot rule out the possibility that YanV105R can polymerize via an unknown, SAM-independent mechanism sufficiently to establish long-range chromatin contacts. However, our prior FRAP studies in Drosophila cells demonstrated that the Yan monomer diffuses significantly faster than wild-type Yan (Zhang et al. 2010), consistent with a loss of polymer formation following mutation of the SAM interface.
A number of nonmutually exclusive mechanistic explanations can be envisioned for why Yan monomers appear significantly less functional, despite relatively wild-type patterns of chromatin recruitment. One possibility is that although the DNA-binding affinity of YanV105R is comparable to that of wild-type Yan in vitro (Tootle et al. 2003; Qiao et al. 2004; Song et al. 2005), in vivo, the absence of self-association might destabilize cooperative DNA binding sufficiently to abrogate function. Indeed, in vitro studies of Yan’s mammalian homolog Tel1 have demonstrated that cooperative DNA binding allows dimers to bind to linear templates carrying tandem ETS sites more stably than monomers (Green et al. 2010). We also observe a preference for clustered ETS-binding sites within Yan-bound regions, suggesting that a proportion of Yan chromatin association could reflect cooperative binding to multiple ETS sites. Indeed, previous work has shown the importance of homotypic clustering of binding sites for the cooperative recruitment of transcription factors (Lebrecht et al. 2005; Segal et al. 2008; Borok et al. 2010). Thus although the YanV105R monomers would still be recruited to high-affinity sites, loss of self-association–mediated cooperativity might destabilize DNA binding. The resulting reduced probability of occupancy could explain the lower but broadly similar ChIP signal obtained with YanV105R relative to wild-type Yan and the apparent failure to assemble a functional or stable repression complex.
Alternatively, oligomerization might promote or stabilize three-dimensional chromatin conformations that are critical for repression. In this scenario, we predict that abolishing SAM-mediated self-association would only minimally reduce linear chromatin spreading but significantly impair Yan’s repressive function. Investigation into the correlation between Yan-bound HDRs and three-dimensional chromatin conformation will be required to explore this idea. Very speculatively, if such contacts exist, they might not only influence transcription of a single locus, but also could provide a mechanism for coordinate regulation across multiple genes. The particularly striking high-density Yan occupancy observed across components of multiple-signaling pathways, including the Notch, Wingless, and EGFR networks, make these appealing contexts for further exploration of such ideas.
Finally, heterotypic interactions with other corepressor proteins, chromatin remodeling factors, or transcription factors could explain both the recruitment and the function of chromatin-bound Yan. For example, a protein–protein interaction that was critical to transcriptional repression but either specifically blocked by the V105R mutation or dependent on the presence of Yan oligomers could explain why monomeric YanV105R is less able to repress transcription despite its relatively normal chromatin recruitment. Regardless of the mechanism of Yan’s recruitment to DNA, the similar occupancy profiles and distinct functions of YanWT and YanV105R suggest a predominantly active rather than passive mode of Yan-mediated repression.
What might be the functional significance of extensive Yan occupancy across a locus with respect to regulation of gene expression? While formally there could be none, the fact that such patterns occur predominantly at essential signaling factors or developmental regulators and are highly conserved at these loci in distantly related Drosophila species argues otherwise. The prevailing model of Yan function is that competition with Pnt for binding to clustered ETS motifs provides a binary switch that dictates whether a target gene is repressed or activated (Gabay et al. 1996; Rohrbaugh et al. 2002; Vivekanand et al. 2004; Graham et al. 2010). Consistent with this, we showed that 16/18 Yan-bound regions selected based on high probability of clustered ETS-binding sites can be both activated by Pnt and repressed by Yan in cultured cell transcription assays. However, genome-wide, the complexity of the Yan occupancy pattern together with the relatively modest enrichment of ETS sites in HDRs vs. isolated peaks and the absence of ETS motifs in many high-confidence Yan-bound sequences suggests that there are other modes of Yan recruitment and function. Arguing for a more complex mode of regulation, the yan loss-of-function phenotype (Nusslein-Volhard and Wieschaus 1980; Rogge et al. 1995; Olson et al. 2011) is not consistent with broad “all or nothing” regulation of the developmentally important genes that we identify as putative Yan targets in our ChIP data sets. However, since to date there are no available ChIP data for Pnt, we cannot rule out the possibility that Yan and Pnt act in combination at all targets.
One speculative idea is that broad binding of Yan across loci that encode critical signaling pathway components could provide a tuning mechanism that both prevents premature pathway activation in response to stochastic transcription factor binding or subthreshold signaling and primes the system for rapid response once signaling exceeds a critical threshold. Alternatively, HDRs might not confer direct regulation, but instead could provide staging grounds for preassembly, storage, or sequestration of transcriptional complexes that are quickly mobilized to specific CRMs as needed. The fact that other transcriptional repressors, most strikingly Kr, also exhibit prominent HDR-type occupancy suggests that future investigation into the functional significance of such patterns could provide new insight into conserved mechanisms of gene regulation.
Supplementary Material
Acknowledgments
We thank the University of Chicago Microarray Facility and Argonne National Laboratory for ChIP-chip hybridization and deep sequencing, respectively. We thank Alec Victorsen and Ralf Kittler for recombineering the Yan constructs; Aaron Mitchell-Dick for assistance in screening for transgenics; and Misha Ludwig, Manu, and Rick Fehon for assistance with Eve-YFP quantification. We thank members of the Rebay, Fehon, and White laboratories for helpful discussions and Jean-Francois Boisclair-Lachance, Trevor Davis, Thomas Graham, Charlene Hoi, and Matthew Slattery for comments on the manuscript. K.P.W., R.W.C., and I.R. are supported by National Institutes of Health (NIH) grant P50 GM081892. Additional funding for I.R. is provided by NIH grant R01 GM80372. J.Z. is a recipient of a Women’s Board Fellowship of the University of Chicago. J.L.W. is supported by an award from the American Heart Association.
Footnotes
Communicating editor: P. K. Geyer
Literature Cited
- Agelopoulos M., McKay D. J., Mann R. S., 2012. Developmental regulation of chromatin conformation by Hox proteins in Drosophila. Cell Rep. 1: 350–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., 2011. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Elkan C., 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36 [PubMed] [Google Scholar]
- Bailey T. L., Gribskov M., 1998. Methods and statistics for combining motif match scores. J. Comput. Biol. 5: 211–221 [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Machanick P., 2012. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 40: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behan K. J., Nichols C. D., Cheung T. L., Farlow A., Hogan B. M., et al. , 2002. Yan regulates Lozenge during Drosophila eye development. Dev. Genes Evol. 212: 267–276 [DOI] [PubMed] [Google Scholar]
- Bonn S., Zinzen R. P., Girardot C., Gustafson E. H., Perez-Gonzalez A., et al. , 2012. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 44: 148–156 [DOI] [PubMed] [Google Scholar]
- Borok M. J., Tran D. A., Ho M. C., Drewell R. A., 2010. Dissecting the regulatory switches of development: lessons from enhancer evolution in Drosophila. Development 137: 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M., Groudine M., 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchar J. P., Cain D., Manivannan S. N., McCue A. D., Bonanno L., et al. , 2012. New negative feedback regulators of EGFR signaling in Drosophila. Genetics 191: 1213–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline M. S., Smoot M., Cerami E., Kuchinsky A., Landys N., et al. , 2007. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2: 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Jia S., 2001. Transcriptional repression: the long and the short of it. Genes Dev. 15: 2786–2796 [DOI] [PubMed] [Google Scholar]
- Dunipace L., Ozdemir A., Stathopoulos A., 2011. Complex interactions between cis-regulatory modules in native conformation are critical for Drosophila snail expression. Development 138: 4075–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores G. V., Duan H., Yan H., Nagaraj R., Fu W., et al. , 2000. Combinatorial signaling in the specification of unique cell fates. Cell 103: 75–85 [DOI] [PubMed] [Google Scholar]
- Frankel N., Davis G. K., Vargas D., Wang S., Payre F., et al. , 2010. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466: 490–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith M. C., Hansen U., Weng Z., 2001. Detection of cis-element clusters in higher eukaryotic DNA. Bioinformatics 17: 878–889 [DOI] [PubMed] [Google Scholar]
- Gabay L., Scholz H., Golembo M., Klaes A., Shilo B. Z., et al. , 1996. EGF receptor signaling induces pointed P1 transcription and inactivates Yan protein in the Drosophila embryonic ventral ectoderm. Development 122: 3355–3362 [DOI] [PubMed] [Google Scholar]
- Golembo M., Schweitzer R., Freeman M., Shilo B. Z., 1996. Argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development 122: 223–230 [DOI] [PubMed] [Google Scholar]
- Graham T. G., Tabei S. M., Dinner A. R., Rebay I., 2010. Modeling bistable cell-fate choices in the Drosophila eye: qualitative and quantitative perspectives. Development 137: 2265–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green S. M., Coyne H. J., 3rd, McIntosh L. P., Graves B. J., 2010. DNA binding by the ETS protein TEL (ETV6) is regulated by autoinhibition and self-association. J. Biol. Chem. 285: 18496–18504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R., Calos M. P., 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166: 1775–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon M. S., Carmena A., Gisselbrecht S., Sackerson C. M., Jimenez F., et al. , 2000. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell 103: 63–74 [DOI] [PubMed] [Google Scholar]
- He Q., Bardet A. F., Patton B., Purvis J., Johnston J., et al. , 2011. High conservation of transcription factor binding and evidence for combinatorial regulation across six Drosophila species. Nat. Genet. 43: 414–420 [DOI] [PubMed] [Google Scholar]
- Hong J. W., Hendrix D. A., Levine M. S., 2008. Shadow enhancers as a source of evolutionary novelty. Science 321: 1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T., Schulz R. A., 2000. Sequence and functional properties of Ets genes in the model organism Drosophila. Oncogene 19: 6409–6416 [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A., 2009a Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A., 2009b Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hudson J. B., Podos S. D., Keith K., Simpson S. L., Ferguson E. L., 1998. The Drosophila Medea gene is required downstream of dpp and encodes a functional homolog of human Smad4. Development 125: 1407–1420 [DOI] [PubMed] [Google Scholar]
- Huen D. S., Russell S., 2010. On the use of resampling tests for evaluating statistical significance of binding-site co-occurrence. BMC Bioinformatics 11: 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W. E., Li W., Meyer C. A., Gottardo R., Carroll J. S., et al. , 2006. Model-based analysis of tiling-arrays for ChIP-chip. Proc. Natl. Acad. Sci. USA 103: 12457–12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan K. C., Hatfield S. D., Tworoger M., Ward E. J., Fischer K. A., et al. , 2005. Genome wide analysis of transcript levels after perturbation of the EGFR pathway in the Drosophila ovary. Dev. Dyn. 232: 709–724 [DOI] [PubMed] [Google Scholar]
- Kharchenko P. V., Tolstorukov M. Y., Park P. J., 2008. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 26: 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. A., Phillips M. L., Kim W., Gingery M., Tran H. H., et al. , 2001. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 20: 4173–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. A., Gingery M., Pilpa R. M., Bowie J. U., 2002. The SAM domain of polyhomeotic forms a helical polymer. Nat. Struct. Biol. 9: 453–457 [DOI] [PubMed] [Google Scholar]
- Kim C. A., Sawaya M. R., Cascio D., Kim W., Bowie J. U., 2005. Structural organization of a Sex-comb-on-midleg/polyhomeotic copolymer. J. Biol. Chem. 280: 27769–27775 [DOI] [PubMed] [Google Scholar]
- Klambt C., 1993. The Drosophila gene pointed encodes two ETS-like proteins which are involved in the development of the midline glial cells. Development 117: 163–176 [DOI] [PubMed] [Google Scholar]
- Kuhn R. M., Karolchik D., Zweig A. S., Wang T., Smith K. E., et al. , 2009. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 37: D755–D761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakovskii I. V., Makeev V., 2009. Integration of data obtained by different experimental methods to determine the motifs in DNA sequences recognized by transcription-regulating factors. Biofizika 54: 965–974 [PubMed] [Google Scholar]
- Kulakovskiy I. V., Favorov A. V., Makeev V. J., 2009. Motif discovery and motif finding from genome-mapped DNase footprint data. Bioinformatics 25: 2318–2325 [DOI] [PubMed] [Google Scholar]
- Landt S. G., Marinov G. K., Kundaje A., Kheradpour P., Pauli F., et al. , 2012. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22: 1813–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrecht D., Foehr M., Smith E., Lopes F. J., Vanario-Alonso C. E., et al. , 2005. Bicoid cooperative DNA binding is critical for embryonic patterning in Drosophila. Proc. Natl. Acad. Sci. USA 102: 13176–13181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. I., Johnstone S. E., Young R. A., 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1: 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Carthew R. W., 2005. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell 123: 1267–1277 [DOI] [PubMed] [Google Scholar]
- Li X. Y., MacArthur S., Bourgon R., Nix D., Pollard D. A., et al. , 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 6: e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Y., Thomas S., Sabo P. J., Eisen M. B., Stamatoyannopoulos J. A., et al. , 2011. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome Biol. 12: R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M. Z., Manu R., Kittler K., White P., Kreitman M., 2011. Consequences of eukaryotic enhancer architecture for gene expression dynamics, development, and fitness. PLoS Genet. 7: e1002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur S., Li X. Y., Li J., Brown J. B., Chu H. C., et al. , 2009. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 10: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- modENCODE Consortium, S. Roy, J. Ernst, P. V. Kharchenko, P. Kheradpour et al, 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N., Hennetin J., Sun L. V., Lavrov S., Bellis M., et al. , 2006. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 4: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N., Brown C. D., Ma L., Bristow C. A., Miller S. W., et al. , 2011. A cis-regulatory map of the Drosophila genome. Nature 471: 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C., Wieschaus E., 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801 [DOI] [PubMed] [Google Scholar]
- O’Keefe L., Dougan S. T., Gabay L., Raz E., Shilo B. Z., et al. , 1997. Spitz and Wingless, emanating from distinct borders, cooperate to establish cell fate across the Engrailed domain in the Drosophila epidermis. Development 124: 4837–4845 [DOI] [PubMed] [Google Scholar]
- Olson E. R., Pancratov R., Chatterjee S. S., Changkakoty B., Pervaiz Z., et al. , 2011. Yan, an ETS-domain transcription factor, negatively modulates the Wingless pathway in the Drosophila eye. EMBO Rep. 12: 1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill E. M., Rebay I., Tjian R., Rubin G. M., 1994. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78: 137–147 [DOI] [PubMed] [Google Scholar]
- Perry M. W., Boettiger A. N., Bothma J. P., Levine M., 2010. Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 20: 1562–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M. W., Boettiger A. N., Levine M., 2011. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 108: 13570–13575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. D., Lai Z., 1999. The yan gene is highly conserved in Drosophila and its expression suggests a complex role throughout development. Dev. Genes Evol. 209: 207–217 [DOI] [PubMed] [Google Scholar]
- Qiao F., Bowie J. U., 2005. The many faces of SAM. Sci. STKE 2005: re7. [DOI] [PubMed] [Google Scholar]
- Qiao F., Song H., Kim C. A., Sawaya M. R., Hunter J. B., et al. , 2004. Derepression by depolymerization; structural insights into the regulation of Yan by Mae. Cell 118: 163–173 [DOI] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay I., Rubin G. M., 1995. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell 81: 857–866 [DOI] [PubMed] [Google Scholar]
- Riesgo-Escovar J. R., Hafen E., 1997. Drosophila Jun kinase regulates expression of decapentaplegic via the ETS-domain protein Aop and the AP-1 transcription factor DJun during dorsal closure. Genes Dev. 11: 1717–1727 [DOI] [PubMed] [Google Scholar]
- Robertson G., Hirst M., Bainbridge M., Bilenky M., Zhao Y., et al. , 2007. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods 4: 651–657 [DOI] [PubMed] [Google Scholar]
- Robinson A. K., Leal B. Z., Chadwell L. V., Wang R., Ilangovan U., et al. , 2012. The growth-suppressive function of the polycomb group protein polyhomeotic is mediated by polymerization of its sterile alpha motif (SAM) domain. J. Biol. Chem. 287: 8702–8713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge R., Green P. J., Urano J., Horn-Saban S., Mlodzik M., et al. , 1995. The role of yan in mediating the choice between cell division and differentiation. Development 121: 3947–3958 [DOI] [PubMed] [Google Scholar]
- Rohrbaugh M., Ramos E., Nguyen D., Price M., Wen Y., et al. , 2002. Notch activation of yan expression is antagonized by RTK/pointed signaling in the Drosophila eye. Curr. Biol. 12: 576–581 [DOI] [PubMed] [Google Scholar]
- Roseman R. R., Morgan K., Mallin D. R., Roberson R., Parnell T. J., et al. , 2001. Long-range repression by multiple polycomb group (PcG) proteins targeted by fusion to a defined DNA-binding domain in Drosophila. Genetics 158: 291–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N., et al. , 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky J., Euskirchen G., Auerbach R. K., Zhang Z. D., Gibson T., et al. , 2009. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotechnol. 27: 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz H., Deatrick J., Klaes A., Klambt C., 1993. Genetic dissection of pointed, a Drosophila gene encoding two ETS-related proteins. Genetics 135: 455–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz H., Sadlowski E., Klaes A., Klambt C., 1997. Control of midline glia development in the embryonic Drosophila CNS. Mech. Dev. 64: 137–151 [DOI] [PubMed] [Google Scholar]
- Schwartz Y. B., Kahn T. G., Nix D. A., Li X. Y., Bourgon R., et al. , 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Segal E., Raveh-Sadka T., Schroeder M., Unnerstall U., Gaul U., 2008. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature 451: 535–540 [DOI] [PubMed] [Google Scholar]
- Sekelsky J. J., Newfeld S. J., Raftery L. A., Chartoff E. H., Gelbart W. M., 1995. Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics 139: 1347–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M., Ma L., Negre N., White K. P., Mann R. S., 2011. Genome-wide tissue-specific occupancy of the Hox protein Ultrabithorax and Hox cofactor Homothorax in Drosophila. PLoS ONE 6: e14686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Nie M., Qiao F., Bowie J. U., Courey A. J., 2005. Antagonistic regulation of Yan nuclear export by Mae and Crm1 may increase the stringency of the Ras response. Genes Dev. 19: 1767–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., et al. , 2011. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39: D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. D., Kejariwal A., Campbell M. J., Mi H., Diemer K., et al. , 2003. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 31: 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B., de Wit E., Muijrers I., Teunissen H., Talhout W., et al. , 2006. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 38: 694–699 [DOI] [PubMed] [Google Scholar]
- Tomancak P., Beaton A., Weiszmann R., Kwan E., Shu S., et al. , 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3: RESEARCH0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P., Berman B. P., Beaton A., Weiszmann R., Kwan E., et al. , 2007. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 8: R145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootle T. L., Lee P. S., Rebay I., 2003. CRM1-mediated nuclear export and regulated activity of the Receptor Tyrosine Kinase antagonist YAN require specific interactions with MAE. Development 130: 845–857 [DOI] [PubMed] [Google Scholar]
- Tran H. H., Kim C. A., Faham S., Siddall M. C., Bowie J. U., 2002. Native interface of the SAM domain polymer of TEL. BMC Struct. Biol. 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivekanand P., Tootle T. L., Rebay I., 2004. MAE, a dual regulator of the EGFR signaling pathway, is a target of the Ets transcription factors PNT and YAN. Mech. Dev. 121: 1469–1479 [DOI] [PubMed] [Google Scholar]
- Wei G. H., Badis G., Berger M. F., Kivioja T., Palin K., et al. , 2010. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 29: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Kauffmann R. C., Zhang J., Kladny S., Carthew R. W., 2000. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell 103: 87–97 [DOI] [PubMed] [Google Scholar]
- Zhang J., Graham T. G., Vivekanand P., Cote L., Cetera M., et al. , 2010. Sterile alpha motif domain-mediated self-association plays an essential role in modulating the activity of the Drosophila ETS family transcriptional repressor Yan. Mol. Cell. Biol. 30: 1158–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., et al. , 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M., Niu W., Lu Z. J., Sarov M., Murray J. I., et al. , 2010. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 6: e1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen R. P., Girardot C., Gagneur J., Braun M., Furlong E. E., 2009. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462: 65–70 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.