Abstract
The Eyes Absent (EYA) proteins, first described in the context of fly eye development, are now implicated in processes as disparate as organ development, innate immunity, DNA damage repair, photoperiodism, angiogenesis, and cancer metastasis. These functions are associated with an unusual combination of biochemical activities: tyrosine phosphatase and threonine phosphatase activities in separate domains, and transactivation potential when associated with a DNA-binding partner. EYA mutations are linked to multiorgan developmental disorders, as well as to adult diseases ranging from dilated cardiomyopathy to late-onset sensorineural hearing loss. With the growing understanding of EYA biochemical and cellular activity, biological function, and association with disease, comes the possibility that the EYA proteins are amenable to the design of targeted therapeutics. The availability of structural information, direct links to disease states, available animal models, and the fact that they utilize unconventional reaction mechanisms that could allow specificity, suggest that EYAs are well-positioned for drug discovery efforts. This review provides a summary of EYA structure, activity, and function, as they relate to development and disease, with particular emphasis on recent findings.
Keywords: EYA, Eyes absent, Angiogenesis, Cancer, Organ development, Cell migration
Introduction
The Eyes Absent (EYA) proteins are components of a conserved regulatory network involved in cell-fate determination in organisms ranging from insects to humans. In Drosophila, where most of the initial characterization was conducted, this network is referred to as the retinal determination network and is composed of the genes twin of eyeless (toy), eyeless (ey), sine oculis (so), eyes absent (eya) and dachshund (dac). These genes are regulated by interconnected feedback loops and their protein products form complexes that play critical roles in fly eye development [1–5]. In higher animals, an analogous network variably composed of genes belonging to the Pax (corresponding to toy and ey), Six (corresponding to so), Eya (corresponding to eya), and Dach (corresponding to dac) families play key regulatory roles in the development of multiple organs including the eye, muscle, ears, heart, lungs, endocrine glands, placodes, pharyngeal pouches, craniofacial skeleton, and parathyroid. This network is often referred to as the Pax-Six-Eya-Dach network (PSEDN) to better reflect the vertebrate genes/proteins involved. Because of its deployment in multiple developmental contexts and its high degree of conservation through animal evolution, the PSEDN has been extensively studied.
A unique feature of the EYA proteins is that they combine several biochemical activities in a single polypeptide: independent protein tyrosine phosphatase (PTP) and threonine phosphatase domains, and a transcriptional activation domain. Each of these activities has been linked to distinct cellular functions, biological roles, and disease states. There is now an emerging understanding of the molecular mechanisms and regulation of EYA’s biochemical activities, the cellular pathways influenced by EYAs, and the biological context in which these processes occur. This is leading to a better understanding of the role of EYAs in both normal development and in disease, thereby raising the possibility that EYAs might be attractive molecular targets for therapeutic intervention. Here we focus on the EYA proteins, their structure, mechanism of action, and association with human development and disease.
EYA protein architecture and activities
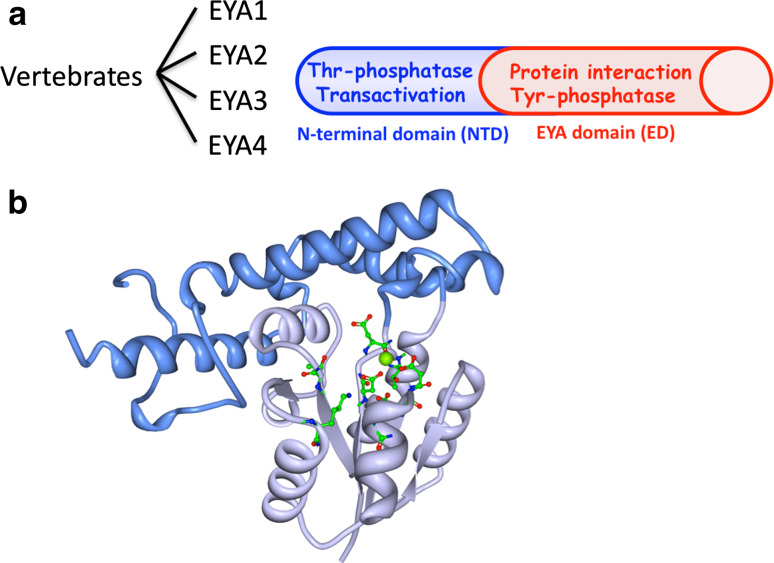
Vertebrates encode four EYA proteins (EYA1–4) that are characterized by a conserved C-terminal 271 amino-acid domain commonly referred to as the EYA domain (ED) (Fig. 1). Animal EYA proteins have a poorly conserved N-terminal domain (NTD) that ranges in size between 266 and 320 amino acids in vertebrates, but is 491 amino acids long in Drosophila. In contrast, plant EYA does not have an NTD.
Fig. 1.
Molecular architecture of the EYA proteins. a There are four vertebrate EYA proteins. They have a well-conserved C-terminal domain called the EYA domain (ED) and a poorly conserved N-terminal domain (NTD). The ED participates in protein interactions, notably with the SIX family of proteins that are part of the PSEDN. The ED also has tyrosine phosphatase activity. The NTD has transactivation and threonine phosphatase activities. b The three-dimensional structure of the EYA2 ED as determined by X-ray crystallography [14]. It has two subdomains referred to as the core (light blue) and cap (dark blue) domains. The active site residues are shown in green. A divalent metal ion (green sphere) in the active site participates in the catalytic reaction
The conserved EYA domain
The ED was originally described as a protein interaction domain involved in binding the SIX and DACH proteins [6–8]. It was later recognized that the ED also includes the sequence motifs characteristic of the large haloacid dehalogenase class of enzymes [7, 9, 10] and that it has tyrosine phosphatase activity [9, 10]. This observation linked the PSEDN to signal transduction and altered the conventional view that it was a purely transcriptional network. The EYAs represent a novel mechanistic class of PTPs as they do not have the signature Cys-containing motif that traditionally defines the large PTP family (reviewed in [11, 12]). Instead EYAs use an aspartate residue as a nucleophile and require a divalent metal ion in the active site to catalyze tyrosine phosphate hydrolysis. To date, the only other tyrosine phosphatase shown to share this reaction mechanism is the TFIIF associating component of CTD phosphatase/small CTD phosphatase (FCP1/SCP) [13].
Three-dimensional structural data are available for the EYA2 ED [14]. Like most HAD family enzymes, the EYA2 ED is composed of two subdomains: a “core” catalytic domain that includes all three conserved motifs that bear the catalytic residues, and an inserted helical “cap” domain (Fig. 1b). A divalent metal ion is found in the active site. The cap domain is positioned such that it forms part of the catalytic center, and is thus likely to participate in substrate binding and/or selection.
The N-terminal domain
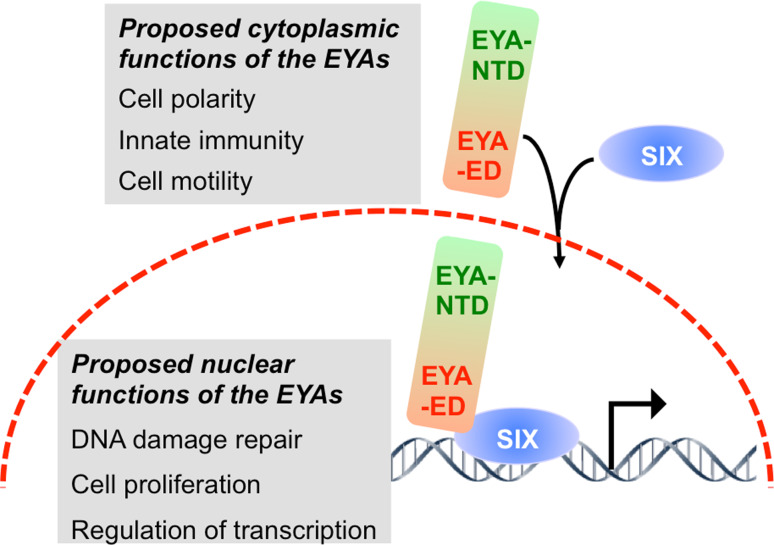
While the amino acid sequence in the NTD is very poorly conserved (Table 1), in all cases it is rich in Pro, Ser and Thr residues reminiscent of the proline/serine/threonine-rich transactivation domains [15]. Xu et al. [16] used a classical GAL4–DNA binding domain fusion of the NTD to demonstrate that it could modestly activate the expression of a CAT reporter. Subsequently Ohto et al. [6] showed that the EYA proteins can be localized on DNA via the homeodomain-containing SIX proteins and can activate transcription from SIX-responsive elements. Thus the prevailing model (Fig. 2) holds that the intrinsically cytosolic EYA proteins are translocated to the nucleus as complexes with SIX proteins, and that the SIX–EYA complex can activate transcription, whereas the SIX proteins by themselves are generally transcriptional repressors.
Table 1.
Amino acid sequence conservation among human EYA1, EYA2, EYA3, EYA4, and Drosophila (dm) Eya
| EYA1 | EYA2 | EYA3 | EYA4 | Dm Eya | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Percentage identity | Percentage similarity | Percentage identity | Percentage similarity | Percentage identity | Percentage similarity | Percentage identity | Percentage similarity | Percentage identity | Percentage similarity | |
| EYA1 | 82.7 | 92.3 | 72.1 | 84.9 | 88.6 | 95.2 | 69.1 | 83.6 | ||
| EYA2 | 39.4 | 55.6 | 68.4 | 82.7 | 79.8 | 91.2 | 64.4 | 82.5 | ||
| EYA3 | 26.8 | 44.8 | 28.8 | 36.5 | 73.2 | 85.7 | 61.8 | 77.5 | ||
| EYA4 | 54.3 | 68.9 | 29.7 | 43.9 | 25.3 | 48.7 | 78.2 | 84.4 | ||
| Dm Eya | 11.2 | 22.8 | 6.7 | 16.9 | 7.7 | 21.8 | 13.8 | 25.5 | ||
Italics EDs, bold NTDs.
Accession numbers for the protein sequences: EYA1 NP_000494.2, EYA2 NP_005235.3, EYA3 NP_001981.2, EYA4 AAH41063.1, dm Eya AAF52400.
Fig. 2.
EYA subcellular localization and functions. The EYAs are intrinsically cytoplasmic proteins. Upon interaction with the SIX proteins (with the exception of SIX3) they are translocated into the nucleus where they are localized on DNA via the homeodomain of the SIX proteins, and convert the SIX proteins into transcriptional activators. The proposed cellular functions for cytoplasmic and nuclear EYA are shown in the gray boxes and discussed in the text
Most recently the NTD has been shown to have threonine phosphatase activity [17, 18]. While the NTD bears no sequence resemblance to any known family of Thr phosphatases, Alanine replacement of a set of four relatively conserved tyrosine residues disrupts threonine phosphatase activity [17]. Whether this observation reflects involvement of the tyrosine residues in the catalytic function, or general disruption of the three-dimensional structure of the NTD remains to be determined.
Despite the apparent conservation of biochemical function across species, the NTDs of bilaterian EYAs bear little homology at the amino acid level to EYAs from protostomes. Furthermore, plant EYAs have no true NTD, but just a short stretch of 18–23 residues [19]. This raises the possibility that an ancestral plant EYA PTP gained an NTD and a transcriptional function in animals. Pertinent to this is the observation that plants do not encode orthologues of the other members of the PSEDN, and thus there is no conservation of this transcriptional regulatory cascade between plants and animals.
EYA: a protein phosphatase with dual specificity
Conventional dual specificity phosphatases (DUSPs) are characterized by their ability to dephosphorylate both phosphotyrosine and phosphoserine/threonine residues within the same substrate using the same catalytic domain [20, 21]. In contrast, EYA’s threonine phosphatase and tyrosine phosphatase activities exist in separate domains, and there is no evidence yet that EYAs can dephosphorylate both phosphothreonine and phosphotyrosine residues in the same substrate. Hence, EYAs do not fit the classical definition of DUSPs, but rather represent a unique and unusual class of phosphoprotein phosphatases with dual specificity. It remains to be established whether the NTD and EDs regulate each other’s catalytic activities.
To date the only truly validated substrate for the EYA tyrosine phosphatase activity is the minor histone protein H2AX (described in detail below) [22, 23]. No substrate has yet been identified for the threonine phosphatase activity. Given the growing list of cellular roles that are emerging for the EYA family of proteins, it is highly likely that multiple EYA substrates will be identified in the future.
EYA: a transcriptional activator-phosphatase
Early studies in Drosophila characterized the transcriptional role of EYA through genetic and/or biochemical interaction with the SO/SIX [24] and DAC/DACH [25] classes of transcription factors. SIX proteins physically interact with, and actively translocate EYAs from the cytoplasm to the nucleus [6, 24, 26]. Since EYA has no recognized DNA binding activity, but possesses a transactivation domain, it is widely accepted that EYAs act as transcriptional coactivators upon recruitment by the SIX protein [6, 16, 27]. A direct interaction between Eya and Dac is thought to underlie the synergistic induction of ectopic eyes in Drosophila [25], and a similar interaction has been reported between mouse EYA2 and DACH2 [26]; however, a later study revealed that the interaction between EYA and DACH may be mediated by the CREB binding protein (CBP) [8]. DACH is believed to be a transcriptional repressor that binds directly to specific DNA sites [28, 29] and recruits the coregulator transcriptional elongation regulator 1 (TCERG1) [30]. The DACH–CBP–EYA complex acts as a transactivator [8]. Similarly, multiple studies suggest that the SIX proteins are either repressors of transcription or weak activators. In either case, the presence of EYA converts them into strong activators of transcription [6, 7, 31, 32]. Moreover, no SIX- and DACH-independent transcriptional activities of EYA have yet been clearly described.
While GAL4–DNA-binding domain fusions of the EYA-NTD can activate transcription on their own [16], transactivation by a SIX–EYA complex is dependent on the EYA tyrosine phosphatase activity [7]. This observation is intriguing and implies that the N-terminal transactivation activity is somehow regulated by the C-terminal phosphatase activity, or that a substrate of the SIX–EYA complex is involved in transcriptional activation. The precise mechanism underlying this observation is yet to be determined. There has been some suggestion that the phosphatase activity of the EYAs may participate in a cytoplasmic cellular function while the transactivation activity of the SIX–EYA complex represents a nuclear function. This is borne out by evidence that cytoplasmic EYA4 plays a role in innate immunity via its threonine phosphatase activity [17] and that cytoplasmic EYA3 promotes cell motility via its tyrosine phosphatase activity [33]. However, the role of the EYA tyrosine phosphatase activity in DNA damage repair [22] is clearly a nuclear function.
In the Drosophila eye, EYA target genes include lozenge, hedgehog, eyeless, so and atonal (reviewed in [34]). In vertebrates, EYA target genes are implicated in the development of multiple organs and include Na + /K+ ATPase α1 subunit, myogenin, Igfbp5, aldolase A, c-myc, Gdnf, cyclin A1, cyclin D1, Slc12a2, p27Kip1, muscle creatine kinase, ezrin and Six2 [7, 35–43]. In the immune response to viral infection, EYA4 promotes the expression of interferon-beta (IFN-β) and CXCL10 [17, 44].
EYA binding proteins
In a study aimed at mapping the interactome of the worm Caenorhabditis elegans, EYA was found to have an unusually large number of interacting partners [45]. EYA binding proteins supported by experimental evidence are listed in Table 2. The SIX and DACH transcription factors remain the best-characterized and functionally validated EYA-binding proteins. Nuclear translocation of EYA can be prevented by EYA interaction with the alpha subunits of Gz and Gi proteins [46]. This might be relevant to eye development, as a mouse knockout of a G-coupled protein receptor recapitulates anterior ocular defects seen in patients with mutations in the EYA1 gene [47, 48]. On the other hand, EYA’s cotranscriptional activator function can be enhanced by its interaction with SIPL1 (shank-interacting protein-like 1) and RBCK1 (RBCC protein interacting with PKC1), and this interaction is important in craniofacial development in mouse and zebrafish [49]. In lung epithelial morphogenesis, EYA1 forms a complex with several polarity proteins including PAR3, PAR6, NUMB, LGN and MLNSC, through its direct binding to the zeta isoform of the atypical protein kinase C (aPKC-zeta) [50]. In response to DNA damage, EYA3 is phosphorylated by the ataxia telangiectasia mutated (ATM)/ATM Rad3-related (ATR) kinase, and upon activation, can bind to and dephosphorylate H2AX, a prerequisite for DNA damage repair [22, 51].
Table 2.
Experimentally validated EYA binding proteins [86]
| EYA binding partner | Validation method | Functional outcome | References |
|---|---|---|---|
| SIX/So and DACH/Dac | Yeast two-hybrid, genetics, immunoprecipitation | Nuclear translocation of EYA by SIX; increase in SIX transcriptional activity by EYA; alleviation of DACH repressor activity toward SIX; increase in SIX-DNA binding affinity | [8, 25, 86, 153] |
| Gαz and Gαi | Yeast two-hybrid, immunoprecipitation, immunofluorescence | Recruitment of EYA to plasma membrane and prevention of SIX-mediated nuclear translocation | [46, 154] |
| SIPL1 and RBCK1 | Yeast two-hybrid, immunoprecipitation | Enhancement of the transactivation potential of the SIX–EYA complex | [49] |
| aPKC-zeta | Immunoprecipitation | Dephosphorylation of aPKC-zeta and NUMB; regulation of polarity in the lung epithelium | [50] |
| ATM/ATR | Immunoprecipitation | Phosphorylation of EYA; interaction of EYA with γH2AX | [22, 51] |
| γH2AX | Immunoprecipitation, immunofluorescence | Dephosphorylation of pY142-γH2AX; initiation of DNA damage repair | [22, 23] |
| Abl kinase | Immunoprecipitation, genetics | Phosphorylation of Eya and retention in the cytoplasm | [52] |
| Nemo kinase | Immunoprecipitation, genetics | Phosphorylation of Eya; potentiation of transcriptional activity of the Eya–So complex | [53] |
| IPS-1, STING, NLRX1 | Immunoprecipitation | Dephosphorylation of a phospho-Ser/Thr substrate; enhancement of innate immune response | [17, 44] |
| SOX2 | Immunoprecipitation | Possibly acts with EYA1 in the generation of progenitor cells in the otocyst | [54] |
In Drosophila eye development, the interaction of Eya with the Abelson (Abl) tyrosine kinase (which phosphorylates Eya) is required for Eya to function as a cytoplasmic protein phosphatase [52]. The Drosophila MAP kinase family member Nemo regulates retinal determination genes by phosphorylating Eya, thereby potentiating the transcriptional activity of the Eya–So complex [53]. In its role as stimulator of the innate immune response against viruses, EYA4 forms a complex with the IFN-β promoter stimulator 1 (IPS-1), the stimulator of IFN genes (STING) and the nucleotide-binding oligomerization domain leucine-rich repeat containing X1 (NLRX1) in immune cells [17, 44, 52]. EYA1 also forms a complex with the transcription factor SOX2 in the sensory epithelium of the inner ear and in the future organ of Corti, implicating this interaction in the development of sensory progenitors as well as hair cell differentiation [54]. SOX2 is the only DNA-binding protein (other than the SIX/So proteins) thus far identified as an EYA interaction partner, raising the possibility that a SOX2–EYA1 complex could regulate expression of SOX2-dependent genes. While such SOX2–EYA1 targets are yet to be described, SOX2, SIX1 and EYA1 are known to interact and regulate Atoh1 expression through both SOX2 and SIX1 binding sites in the Atoh1 enhancer, and to specify hair cell fate in the developing ear [55].
Cellular functions of the EYAs
EYA in cell proliferation and survival
Embryonic growth and patterning is dependent upon the proper balance between proliferation and cell death. Drosophila germ-line eya mutations are embryonic lethal, and eye-specific mutations are characterized by massive cell death in the eye primordium [3, 4]. In C. elegans, loss of Eya1 by RNAi and deletion mutations result in early larval lethality with incomplete penetrance, and late embryos display pharyngeal malformation and excess cell death in the anterior region [56]. Zygotic depletion of Xeya3 induces local apoptosis in the anterior neural plate, and overexpression of exogenous Xeya3 is able to enlarge the neural plate by causing an over-proliferation of neural precursor cells [57]. Mice mutant for Eya display abnormal apoptosis and reduced cell proliferation during the development of multiple tissues including the kidney, muscle, and ear [7, 58, 59]. EYA has also been shown to regulate proliferation through negative modulation of Sonic hedgehog signaling in mouse lung epithelium [60]. Although the developmental role of the EYA proteins appears to be overwhelmingly antiapoptotic and proproliferative, there are instances in which it has a proapoptotic function. Overexpression of Eya triggers apoptotic cell death in 32D.3 murine myeloid cells [61], and the C. elegans SIX protein (CEH-34) and EYA1 cooperate to promote programmed cell death of a particular pharyngeal neuron by directly activating expression of the proapoptotic protein egl-1 [62].
EYA in cell migration
The process of cell migration is fundamental to embryogenesis, as the initial morula ball segregates into the three main germ layers and cells move relative to each other to shape and populate different tissues. The promigratory function of EYA was first reported in Drosophila, since mutant embryos displayed defects in germ cell migration and head morphogenesis [1, 63]. In C. elegans, Eya1 mutants are characterized by a loss of directionality in the migration of gonad cells [56]. In vertebrates, Eya transcripts are expressed in migrating cells, including muscle precursor cells, neural crest cells and their derivatives [57, 64–67], suggesting that EYA proteins may play a role in developmental cell migration. Overexpression of Eyas in breast epithelial cell lines increases single cell migration [33, 68], and siRNA-mediated depletion of EYA4 in malignant peripheral nerve sheath tumor (MPNST) cells [69], or EYA3 in endothelial cells [68], reduces their migration in Transwell assays. While all of these observations support a role for EYAs in promoting cell migration, the precise cellular signaling pathways involved have not yet been delineated. However, links between EYA phosphatase activity and processes fundamental to cell motility such as GTPase activation and cytoskeletal architecture have been observed in mammary epithelial cells [33].
EYA in DNA damage repair
The most validated substrate for the EYA tyrosine phosphatase activity thus far is the minor histone protein H2AX that links EYA to DNA damage repair [22, 23]. In both mouse embryonic kidney and human embryonic kidney cell lines, H2AX is dephosphorylated at the C-terminal tyrosine 142 (pY142) by EYA1, EYA2, and EYA3. This permits recruitment of the MDC1/MRN repair complex and tips the balance towards survival rather than cell death. While phosphorylation of H2AX at Ser139 following DNA damage has been well-established (reviewed in [70]), phosphorylation at tyrosine 142 has only recently been uncovered. Xiao et al. [71] have reported that the William-Beuren syndrome transcription factor (WSTF; also known as BAZ1B) phosphorylates H2AX tyrosine 142 utilizing a novel kinase domain. Furthermore, Xiao et al. [71] showed that H2AX is constitutively phosphorylated at tyrosine 142 in mouse embryonic fibroblasts and that Y142 phosphorylation levels fall, while Ser139 phosphorylation levels rise, upon DNA damage. However, Xie et al. [72] could not detect H2AX Y142 phosphorylation using mass spectrometric techniques, and they did not observe differences between the abilities of wild-type and Y142F H2AX to promote either MDC1 focus formation or homologous recombination in mouse embryonic stem cells. This discrepancy could reflect true cell-specific and/or condition-specific differences of biological interest, or merely technical differences in the referenced studies.
EYA and angiogenesis
Endothelial cell migration, a process promoted by EYA, is a prerequisite for angiogenesis—the formation of new blood vessels. EYA tyrosine phosphatase activity has recently been shown to contribute to sprouting angiogenesis in cell culture-based assays, ex vivo assays, and in a zebrafish model system [68]. Although angiogenesis is essential for vascular development throughout embryogenesis and in the formation of functionally distinct, tissue-specific vascular beds, there is only one report of a vascular defect upon Eya deletion; Eya1−/− embryos show hemorrhage around the large pulmonary vessels attributed to vascular smooth muscle defects that weaken the blood vessels [60]. It is possible that compensation by other isoforms of Eya mask more widespread vascular phenotypes, or that the phenotypes are subtle and thus escaped notice.
The mechanism by which EYA modulates angiogenesis has not yet been defined. However, recent evidence links the DNA damage response to hypoxia-induced angiogenesis [73]. Specifically, hypoxia induces replicative stress and stalled replication forks. This activates the ATR kinase, leading to phosphorylation of H2AX (to yield γ-H2AX; phosphorylated on Ser139), the recruitment of MDC1 and DNA repair. Thus the proliferative potential of endothelial cells is restored leading to neovascularization. Economopoulou et al. [73] have shown that inactivation of H2AX substantially impairs neovascularization in several hypoxic conditions including retinal disease, hindlimb ischemia and tumor growth. Given the established link between EYA and H2AX-mediated recruitment of MDC1 and other MRN components, it is possible that activation of EYA in hypoxic conditions, both during normal development and in disease, promotes DNA damage repair via H2AX tyrosine dephosphorylation. This in turn could promote angiogenesis.
Salient to this discussion, a recent report shows that SIX1 in breast cancer cells promotes lymphangiogenesis by upregulating VEGF-C expression [74]. A SIX1 binding site was reported in the VEGF-C promoter. As in other contexts, SIX1 activates VEGF-C transcription only in the presence of EYA2, thus implicating EYA2 in promoting lymphangiogenesis. This mechanism is likely to be distinct from the angiogenic role for EYA in endothelial cells described by Tadjuidje et al. [68].
EYA and developmental cell polarity
Oocyte polarity is a prerequisite to normal cleavage and the establishment of proper cell fate in animal embryos. Tissue polarity is important for proper differentiation of embryonic tissue and homeostasis of adult tissues [75, 76]. Drosophila Eya is a key repressor of polar cell fate during oogenesis [77], and EYA1 is involved in the control of cell polarity and mitotic spindle orientation in lung epithelium [50]. Therefore, EYA may play a role in polarity not only during the early cleavage stage, but also later in development when highly specialized epithelial tissues differentiate. aPKC-zeta is a proposed EYA1 tyrosine phosphatase substrate reported to play a role in specifying the apical–basal polarity of lung epithelial cells [60] since aPKC-zeta tyrosine phosphorylation levels are increased upon Eya1 knockdown. However, the literature provides little information on either tyrosine phosphorylation site(s) on aPKC-zeta or the role of aPKC-zeta tyrosine phosphorylation, hence the molecular mechanism by which EYA1 specifies cell polarity remains unclear.
EYA in innate immunity
Okabe et al. [17] identified EYA4 as a regulator of the innate immune response to challenge from either cytosolic nucleic acids or undigested DNA from apoptotic cells. In vitro, EYA4 leads to phosphorylation of IRF3 and upregulation of NF-κB, and enhanced induction of IFN-β and CXCL10. These authors propose that EYA4 initiates this response as a complex with RIG-1, TNF receptor-associated death domain, TNF receptor-associated factor 3 and NEMO (IKKγ). They further showed that the N-terminal threonine phosphatase activity of EYA4 is required for this function. While a role for EYA in nucleic acid-induced immunity is yet to be established in vivo, EYA as a mediator of such a response, possibly by dephosphorylating and inactivating an inhibitory kinase, is an attractive possibility.
Eyes Absent in development
In Drosophila, specific recessive eya mutations result in elimination of compound eyes in viable flies, while eya null mutations are embryonic lethal [4, 63, 78]. This could imply that Eya is required in other embryonic tissues and prior to eye development. In support of this, eya transcripts are expressed during embryogenesis of plants [19], squid and worms [56, 79], flies [3] and vertebrates [57, 64, 67, 80–83]. Expression data from flies and vertebrates are consistent with a requirement for EYA function during the entire process of animal embryogenesis, beginning from the oocyte in which the transcript is deposited during oogenesis [57, 77]. This correlates with the numerous defects observed upon genetic inactivation of Eya genes in mice (Table 3). Here we outline the role of EYA in the development of organs where there is a clearly established association with human developmental disorders.
Table 3.
Phenotypes of Eya mutant mice
| Mouse mutant | Phenotype | References |
|---|---|---|
| Eya1−/− | Die at birth; craniofacial and skeletal defects; ear malformation; dysmorphic/absent kidneys; thymus and parathyroid agenesis; thyroid hypoplasia; open eyelids; hypoplastic lungs | [59, 60, 155] |
| Eya2−/− | No external phenotype; viable and fertile | [156] |
|
Eya1−/− Eya2−/+ |
No diaphram; severe limb muscle hypoplasia | [156] |
| Eya3−/− | Viable and fertile with no external phenotype; reduced body length | [108] |
| Eya4−/− | Die shortly after birth; abnormal ear development and hearing deficiency; males sterile or reduced fertility | [105] |
EYA in eye development
Early studies showed that eya mutation leads to a lack of compound eyes in flies with otherwise normal head structures [84, 85]. Furthermore, forced ectopic expression of eya is sufficient to induce retinal development [5]. Despite this strong link between eya and retinal development in Drosophila, a no-eye phenotype has thus far not been reported in mouse Eya1, Eya2, Eya3 and Eya4 mutants. It is possible that functional redundancy in the vertebrate eye, where multiple Eyas are expressed in overlapping patterns, may underlie this discrepancy. In particular, Eya1 and Eya2 are expressed in a complementary fashion in the developing retina, with the lens placode being the only optic structure that only expresses Eya1 [16, 64]. EYA1 mutations have been reported in human patients with congenital cataracts [47], suggesting a role for EYA in human eye development. Moreover, transgenes of mouse Eya1, Eya2 and Eya3 can rescue the eyeless phenotype in the Drosophila eya mutant [86], suggesting that vertebrate EYAs are indeed endowed with eye specification potency. The recently uncovered role for EYAs in angiogenesis and cell migration may also point to roles in developmental retinal angiogenesis that will be uncovered upon more detailed and targeted analyses.
EYA in kidney development
EYA1 and EYA2 are expressed in fetal and adult human kidneys, respectively [79, 87]. Mouse Eya1 is expressed in the developing kidney, and Eya3 is expressed in the adult kidney [81, 87, 88], chick Eya1 is expressed in the nephrogenous mesenchyme [89], and Xenopus eya2 is expressed in the nephric mesoderm [82]. Moreover, Eya1-mutant homozygous mice lack kidneys, and molecular analysis suggests that Eya1 expression in the metanephric mesenchyme is required for the expression of the gene encoding glial-derived neurotrophic factor (Gdnf), which in turn is required to direct ureteric bud outgrowth [59, 90]. Six1-deficient mice lack kidneys, but have ureters. Detailed analysis has revealed that the initial expression of Gdnf is not lost; therefore, the ureteric bud emerges from the Wolffian duct, but fails to invade the mesenchyme, which subsequently leads to apoptosis in the mesenchyme [91, 92]. Six2-deficient mice have hypoplastic kidneys due to depletion of the progenitor cell population within the metanephric mesenchyme as a result of a premature and ectopic differentiation of mesenchymal cells into epithelia [93]. Since EYA1 is required very early in kidney development, a comparison of Eya1, Gdnf, Six1 and Pax2 mutant mice led to the suggestion that Eya1 probably functions at the top of the genetic hierarchy controlling kidney organogenesis [90]. Surprisingly, no Eya1 expression has been reported in the developing kidney of frog or fish [66, 67, 94]. Thus the function of EYA1 in nephrogenesis might be a characteristic of higher vertebrates, or another EYA may undertake this function in lower vertebrates as suggested by the expression of eya2 in Xenopus nephric mesoderm.
EYA in ear development
Disorders of the auditory system coexist with renal anomalies in humans with mutations of EYA genes [95–98], and EYA4 mutations cause late onset deafness [99]. Expression and loss of function data support a role for EYA in the development of the auditory system. The otic vesicle develops from the otic placode. In zebrafish, amphibians and chick, all placodes originate from a common precursor domain, the preplacodal region, marked by the expression of Six1 and Six4, and Eya1 and Eya2 [100]. In zebrafish, Eya1 is found in the developing otic primordium as early as the placodal stage [49, 66], and mutations in the eya1 gene or morpholino-mediated knockdown of the protein cause the dog-eared phenotype which is characterized by a defect in the formation of the inner ear [101]. In Xenopus, eya1 and eya2 transcripts are expressed in the otic placode [67, 82], and immunostaining reveals that the eya1 protein displays a distinctive expression pattern during multiple stages of ear development [102]. Moreover, overexpression of mutant forms of eya1 mRNA cause Xenopus embryos to develop with dysmorphogenesis of the otic vesicle, defects in the establishment of sensory tissue and defective otic innervations [103]. Mouse Eya1 is expressed in the developing otic vesicle [64] and Eya1-deficient mice lack ears [59]. It is suggested that EYA1 is required in a dose-dependent manner for proper ear development because hypomorphic mutations of the Eya1 gene result in inner ear malformations and hearing defects in mice and humans [54]. Furthermore, EYA1 and SIX1 can induce the putative neurosensory stem cells in the cochlea (GER cells) to differentiate into hair cells [55], while coexpressed EYA1, SIX1 and SOX2 can induce GER cells to differentiate into neurons [104]. Thus EYA1 (along with SIX1) initiates neuronal development in the inner ear. On the other hand, EYA4 is involved in middle ear development, with Eya4−/− mice having abnormal middle ear cavities and eustachian tube dysmorphology [105]. They are also more prone to otitis media, and could represent a valuable animal model for this common childhood disease.
EYA in heart development and function
Mutations in EYA4 cause dilated cardiomyopathy, a disorder characterized by ventricular dilation and impaired systolic function resulting in congestive heart failure and arrhythmia [106]. Cardiofacial syndrome, a combination of an asymmetric crying face and heart defects, associated with an EYA1 mutation has also been reported [107]. Consistent with these findings, transcripts of EYA1 and EYA2 are expressed in the adult human heart [79, 81, 87, 106]. In zebrafish, eya4 is expressed in both the embryonic and adult heart, and antisense morpholino-mediated depletion of eya4 causes a progressive ventral swelling due to pericardial edema, suggestive of cardiovascular dysfunction. Further analyses have revealed that eya4-depleted embryos have smaller ventricles than controls [106]. Although there is no report of cardiac expression of Eyes Absent during mouse embryogenesis, Eya2 and Eya3 are expressed in the adult heart [79, 81, 108]. Moreover, Eya3-deficient young adult mice show a defect in the electrophysiology of the heart, possibly suggesting a role for EYA3 in heart function [108]. EYA2 appears to be an important regulator of both pathological and physiological hypertrophy. Eya2 is upregulated during regression of cardiac hypertrophy and blocks phenylephrine-induced development of cardiomyocyte hypertrophy in vitro. Similarly, Eya2 is upregulated during recovery following transverse aortic constriction (a treatment that causes prominent cardiac hypertrophy) [109], suggesting that EYA2 may function during the regression of hypertrophy. Transgene-mediated overexpression of Eya2 inhibits development of cardiac hypertrophy, and prevents wall thinning, ventricular dilation and deterioration of cardiac function as well as fibrosis and inflammation in the heart under long-term pressure overload. In addition, this prevention of pathological hypertrophy and heart failure by EYA2 correlates with the elevation of genes involved in mitochondrial biogenesis and metabolism in transgenic mice [110]. Eya2 expression is upregulated in hearts with swimming exercise-induced physiological hypertrophy [111].
In summary, although there is no solid evidence for the involvement of EYAs in mammalian heart development, there is considerable evidence that EYA is critical for cardiac function. Pathological and physiological cardiac hypertrophy are known to have different molecular signatures [112], and the involvement of EYA in both suggests that EYA could play a cardioprotective role.
EYA in craniofacial development
The craniofacial complex (including the head, face and oral cavity) is mostly formed of tissue derived from the neural crest and mesoderm. Many congenital craniofacial malformations display major anomalies in neural crest cell patterning, but often they arise as a secondary consequence of anomalies in other tissues with which the neural crest cells interact during their development and migration (reviewed in [113]). Cranial placodes and neural crest cells also depend upon each other for proper development. Craniofacial anomalies are part of the phenotypic characteristics of EYA deficiency-related syndromes such as cardiofacial and otofaciocervical syndromes, indicative of a role for EYA in craniofacial development [97, 107, 114, 115]. Consistent with the human phenotypes, Eya transcripts are expressed in cranial placodes and/or their derivatives during the embryogenesis of lower vertebrates. In Xenopus, eya1 is expressed in all neurogenic placodes [67, 94], and reducing eya1 protein levels by injection of morpholino antisense oligonucleotides leads to reduced expression of neuronal marker genes such as neuroD in all neurogenic placodes (reviewed in [116]). Eya2 is expressed in multiple cranial placodes [82], and Eya3 is expressed in migrating cranial neural crest [57]. Zebrafish eya1 is expressed in cranial placodes [66], and chick EYA1 and EYA2 are expressed in cranial placodes and derivatives [65, 89]. In mice, Eya1 and Eya2 are extensively expressed in cranial placodes, [64], and Eya4 is expressed in the nasal placode [80]. Moreover, EYA1 and SIX1 have been directly implicated as promoters of the early steps of neurogenesis in mouse cranial placodes [117, 118]. A role for EYA in craniofacial development is further supported by the work of Landgraf et al. who showed that Eya1 interacts with Sipl1 and Rbck1, proteins important in craniofacial development whose knockdown causes zebrafish embryos to develop with a branchiootorenal (BOR) syndrome-like phenotype [49].
EYA and photoperiodism
An exciting new development is the observation that EYA3 is the first and strongest molecular response that is activated by a long photoperiod (light cycle) in birds [119], mice [120] and sheep [121]. It appears that Eya3 expression in the pars tuberalis always occurs about 12 h after the onset of a dark phase, and is directly suppressed by darkness [122]. Along with SIX1 and TEF, EYA3 synergizes to induce thyroid-stimulating hormone β (Tsh-β) expression in both sheep and mice. Tsh-β contributes to the conversion of the inactive thyroid hormone (T4) to its active form T3, which in turn influences the activity of neurons producing gonadotropin-releasing hormone that regulate levels of follicle-stimulating hormone and luteinizing hormone. Hence, photoperiod-regulated EYA3 levels can directly influence the reproductive cycles of seasonally breeding animals. This places the EYA and SIX proteins at the heart of a conserved transcriptional photoperiodic response in the pars tuberalis that mediates a rapid response to changes in day length. Rhythmic cycling of Eya expression has also been observed in corals where transcription appears to be under the control of an endogenous light-entrained clock [123]. Here the diurnal pattern of Eya expression continues in constant darkness and cycling of Eya expression has been observed in both larvae and adult corals. Since the reproductive cycles and other physiological functions of animals are linked to seasonal environmental changes, including light cycles, these observations open up a new biological context in which EYA proteins play a fundamentally important role.
Eyes Absent in human disease
Mutations in EYA genes have long been associated with human developmental disorders (Table 4). In addition there is now growing evidence that overexpression of EYAs is associated with several malignancies. Below we outline the documented links between EYA proteins and human diseases.
Table 4.
EYA and human disease
| Disease | Gene | Type of misregulation | References |
|---|---|---|---|
| Branchiootorenal syndrome | EYA1 | Loss of function mutations | [87, 95] |
| Otofaciocervical syndrome | EYA1 | Loss of function mutations | [97] |
| Cardiofacial syndrome | EYA1 | Loss of function mutations | [107] |
| Congenital cataract | EYA1 | Loss of function mutations | [47] |
| Wilms’ tumor | EYA1 | Overexpression | [137] |
| EBV-negative gastric cancer | EYA1 | Frequent methylation | [138] |
| Late-onset deafness | EYA4 | Loss of function mutations | [99] |
| Dilated cardiomyopathy | EYA4 | Deletion | [106] |
| Esophageal adenocarcinoma | EYA4 | Frequent methylation (overexpression) | [128] |
| Colon cancer | EYA4 | Overexpression | [130] |
| Colorectal cancer | EYA4 | Overexpression | [131] |
| Colorectal cancer | EYA2 | Silencing methylation | [139] |
| Epithelial ovarian cancer | EYA2 | Overexpression | [133] |
| Lung adenocarcinoma | EYA2 | Overexpression | [134] |
| Pancreatic ductal adenocarcinoma | EYA3 | Frequent deletion | [140] |
EYA in BOR and BOR-related syndromes
The first human Eya homologue (EYA1) was identified by positional cloning in the search for the gene responsible for BOR syndrome [87, 95, 98]. The autosomal dominant BOR syndrome with a prevalence of 1:40,000 is characterized by hearing loss, branchial fistulae, preauricular pits or tags, and renal abnormalities [124]. These multiorgan defects are recapitulated in Xenopus embryos in which the endogenous eya1 protein is depleted and replaced with exogenous protein bearing mutations similar to those seen in patients with BOR syndrome [103], suggesting that the observed human phenotypes are a direct consequence of the reported mutations. Some EYA1 mutations encountered in BOR patients have been shown to disrupt EYA1–SIX [125] and EYA1–SOX2 [54] interactions, but most of them affect the enzymatic activity of EYA without affecting protein translation or stability [103, 126]. In addition to BOR syndrome, EYA1 mutation has been reported in patients with congenital cataracts and ocular anterior segment anomalies [47]. A point mutation of the EYA1 gene leading to aberrant mRNA maturation has been reported in the otofaciocervical syndrome, which is characterized by trophic alterations of face and shoulder girdles in addition to the malformations seen in BOR [97]. A deletion within the EYA1 gene that leads to a premature truncation of the protein has been reported in cardiofacial syndrome, associated with an asymmetric crying face and congenital heart defects [107]. Frame-shift insertion and point mutations that create a truncation of EYA4 C-terminal domain are associated with late-onset deafness [99], and deletion of the whole ED as well as part of the transactivation domain of EYA4 has been identified in patients with a form of dilated cardiomyopathy and heart failure preceded by sensorineural hearing loss [106].
In light of the extensive involvement of the EYAs in organ development, it is rather surprising that the list of human diseases associated with their loss of function is so short, albeit characterized by defects in multiple organs. One explanation may be redundancy arising from overlapping expression patterns of EYA transcripts. Furthermore, a survey of the expression profile of human EYA transcripts using the UniGene database [127] reveals that EYAs are present throughout human development from the embryoid body stage through adulthood, which suggests that they may also function very early in development. Therefore, one cannot exclude the possibility that some human loss of function mutations of EYA may go unnoticed due to early embryonic lethality, a phenomenon that has been reported in flies and worms [1, 56].
EYA in cancer and related pathologies
Overexpression of EYAs has been reported in various human cancers (Table 4). EYA4 is overexpressed in MPNST [69], esophageal adenocarcinoma, and colon and colorectal cancers [128–131]. In esophageal squamous cell carcinoma, its expression level in the peripheral blood correlates with disease progression [132]. Moreover, depletion of EYA4 induces MPNST cells to undergo necrosis [69], suggesting that it promotes the survival of those tumor cells. EYA2 is overexpressed in epithelial ovarian cancers and lung adenocarcinoma [133, 134], and its overexpression has been correlated with poor prognosis. EYA2 is also overexpressed in breast cancers [133, 135] and correlates with a worse prognosis. EYA overexpression in mammary epithelial cells promotes transformation, migration and invasion [33], and EYA2 is required to mediate the prometastatic function of SIX1 in an established human breast epithelial cancer cell line [135]. Loss of a portion of chromosome 2 (2q37), encoding a microRNA (miR562) that regulates EYA1, has been reported in Wilms’ tumors and EYA1 mRNA has also been shown to be overexpressed in these tumors [136, 137]. EYA1 may also be involved in gastric cancer, as its gene is often methylated in Epstein-Barr virus-negative gastric cancers [138]. Despite this tendency of EYAs to be oncogenic, some reports suggest that the outcome of the deregulation of EYA expression may be context-dependent. For example, silencing methylations of the EYA2 gene have been reported in colorectal cancers as opposed to normal tissues [139], the EYA3 gene is frequently deleted in certain pancreatic ductal adenocarcinomas [140], and overexpression of EYAs (including human EYA2) can trigger the apoptotic program (an antitumorigenic process) in a murine myeloid cell line [61].
Since the EYA proteins have multiple biochemical activities, and these are apparently associated with different cellular functions, it is likely that EYAs contribute to tumor growth, metastasis, and angiogenesis by different mechanisms. Another intriguing possibility, in light of the proposed role of the EYA tyrosine phosphatase activity in DNA damage repair, is that elevated EYA levels might contribute to increased resistance to DNA-damaging therapeutic regimens commonly used in cancer treatment.
Interestingly, there appears to be a coordinated misregulation of EYA, SIX and DACH gene expression in many cancers. Specifically in breast cancer [134, 135] and MPNST [69], levels of SIX1 and either EYA2 or EYA4 are elevated while DACH levels are reduced [141–144]. This is consistent with the reported tumor-suppressor functions of the DACH proteins, and the tumor-promoting properties of the SIX and EYA proteins. The PSEDN may thus represent an instance in which a pathway involved in fetal organogenesis promotes tumorigenesis when reinstated in the adult.
EYA: a novel therapeutic target?
While most EYA-associated developmental disorders are linked to loss-of-function mutations, there is growing evidence that elevation of EYA levels (gain of function) is associated with cancers. In both in vivo and in vitro experiments EYAs promote proliferation and invasiveness of tumor cells [33, 68, 69, 133, 145]. Furthermore the tyrosine phosphatase activity is specifically associated with cell migration/invasion and angiogenesis [33, 68], and removal of endogenous EYA can prevent the metastasis of tumor cells overexpressing SIX1 [135], suggesting that even when they are not overexpressed EYAs are still required for metastasis. Together these observations would imply that inhibition of the EYA tyrosine phosphatase activity could be useful as a targeted mode of cancer therapy.
Cell proliferation and migration are prerequisites for angiogenesis, the generation of new blood vessels from preexisting ones. A role for angiogenesis in cancer progression has been long-established. Newly formed blood vessels not only supply the tumor cells with oxygen and nutrients that they need for growth, but also serve as routes for the dissemination of cancer cells during metastasis (for review, see [146]). The EYA tyrosine phosphatase activity has been shown to contribute to angiogenesis using in vitro, ex vivo and in vivo assays [68]. Hence EYA proteins could be positive contributors to tumor growth through their angiogenic function. Pathological neovascularization is not limited to tumor angiogenesis; it is also seen in proliferative retinopathies, arthritis and vascular tumors as a response to a local hypoxic environment [147]. In all these instances inhibition of the EYA PTP could be useful.
There is growing evidence that tumor cells with greater metastatic potential frequently show activation of DNA repair pathways. This has been reported for melanomas, bladder cancer and breast cancers [148, 149]. Furthermore, solid tumors are frequently treated with DNA-damaging therapies both in the form of chemotherapeutics and ionizing radiation. Since EYA promotes DNA damage repair, it is quite likely that elevated EYA levels would reduce the effectiveness of such treatment. Hence, EYA inhibitors could potentially be useful in combination regimens as sensitizing agents.
EYA as a druggable target
While the association between EYA tyrosine phosphatase activity and various disease states suggests that it is a good target for drug development, the identification of specific inhibitors of PTPs has been historically difficult. This is largely because there are over 100 PTPs encoded in the genome that share similar active site architecture, making the discovery of selective ligands/inhibitors challenging. EYAs do not suffer from this problem, being the only known members of their mechanistic class, and having an active site that is stereochemically distinct from that of all other PTPs. A general consensus (recent reviewed in [150]) in the field of drug development is that a “druggable target” must have two important features: (1) be linked to a disease and (2) have the potential to bind, with high affinity, a small molecule having the appropriate chemical properties. As described in this review, EYA has both of these properties. Being an enzyme it has a defined active site that can be targeted by small molecules. Structural information is available [14] permitting rational design of inhibitors, and specific EYA inhibitors have been identified. The most extensively validated EYA PTP inhibitors include the compounds benzbromarone and benzarone [68]. While their IC50 values are modest (<10 μM) in enzymatic assays, these compounds exhibit potency in cellular assays inhibiting cell motility and tubulogenesis in vitro, and angiogenic sprouting ex vivo. They have also been shown to be effective in an in vivo assay measuring vascular development in zebrafish. Hence, these compounds are proof-of-principle that inhibition of EYA can attenuate angiogenesis and are also valuable leads for the development of more potent and effective EYA inhibitors. This class of compounds also has the advantage that because of their history of usage for gout treatment, their long-term toxicity, pharmacokinetic and pharmacodynamics profiles are well-established. As a result they are excellent candidates for the old compound–new target–new use paradigm.
Other classes of EYA inhibitors identified through virtual screening have been reported [151]. These compounds bind in the active site and chelate the essential divalent metal ion of EYA2. While they have not yet been tested in cellular or in vivo assays, these studies point to the feasibility of both in silico and experimental screening for EYA-inhibitory small-molecule compounds.
Conclusions
EYAs are an unusual class of proteins that combine tyrosine phosphatase, threonine phosphatase and transactivation activities. Along with other components of the PSEDN, Eyes Absents have been conserved among widely disparate animal species ranging from amphioxus to humans [152]. EYAs are required for the development of multiple organs and have correspondingly been implicated in multiple developmental disorders. Increasing evidence points to an oncogenic, prometastatic and angiogenic function for the EYAs, and based on its proposed role in angiogenesis, it is likely that EYAs will be associated with vascular disorders.
Key questions regarding EYA regulation and function remain. The range of biological processes associated with EYAs (including, but not restricted to, cell proliferation and migration, angiogenesis, DNA damage repair, innate immunity, photoperiodism) would imply that the EYA proteins are an integral part of core signaling processes in the cell. Moreover, since many of these activities are described in the adult and not the developing embryo, EYAs clearly have a role in the maintenance of function. Other than H2AX, no substrate for the EYA’s two phosphatase activities has been thoroughly validated. Undoubtedly, yet-unidentified substrates exist and in light of the multiple cellular roles of EYAs they are likely to exist in both the nucleus and the cytoplasm. Pertinent to this, the mechanism by which EYA’s subcellular localization is regulated is of interest. It is generally believed that EYA translocation to the nucleus is predicated on the formation of a SIX-EYA complex. Furthermore, since EYA has no intrinsic DNA-binding ability, it needs to be localized on DNA by a binding partner such as the SIX proteins. Hence SIX–EYA complexes must act in concert both in a transcriptional regulation role and in any proposed nuclear phosphatase activity such as DNA damage repair. Thus it would be reasonable to assume that any nuclear function of the EYAs is SIX-dependent, or that unidentified EYA binding partners facilitate the movement of this acidic protein into the nucleus. The converse also appears to be true in some instances: the prometastatic and lymphangiogenic functions of SIX1 are EYA-dependent, and quite likely dependent on EYA tyrosine phosphatase activity. And finally, with the association between EYAs and an ever-growing number of disease states including developmental disorders, cardiac conditions, innate immune responses, pathological angiogenesis, and tumor growth and metastasis, inhibition of EYA tyrosine phosphatase activity is an attractive target for drug development.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (NEI014648) to R.S.H.
References
- 1.Bonini NM, Leiserson WM, Benzer S. Multiple roles of the eyes absent gene in Drosophila. Dev Biol. 1998;196(1):42–57. doi: 10.1006/dbio.1997.8845. [DOI] [PubMed] [Google Scholar]
- 2.Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring WJ. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;125(12):2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- 3.Leiserson WM, Benzer S, Bonini NM. Dual functions of the Drosophila eyes absent gene in the eye and embryo. Mech Dev. 1998;73(2):193–202. doi: 10.1016/s0925-4773(98)00052-5. [DOI] [PubMed] [Google Scholar]
- 4.Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72(3):379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- 5.Bonini NM, Bui QT, Gray-Board GL, Warrick JM. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development. 1997;124(23):4819–4826. doi: 10.1242/dev.124.23.4819. [DOI] [PubMed] [Google Scholar]
- 6.Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, Sato S, Kawakami K. Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol. 1999;19(10):6815–6824. doi: 10.1128/mcb.19.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, Rosenfeld MG. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426(6964):247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 8.Ikeda K, Watanabe Y, Ohto H, Kawakami K. Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol Cell Biol. 2002;22(19):6759–6766. doi: 10.1128/MCB.22.19.6759-6766.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rayapureddi JP, Kattamuri C, Steinmetz BD, Frankfort BJ, Ostrin EJ, Mardon G, Hegde RS. Eyes absent represents a class of protein tyrosine phosphatases. Nature. 2003;426(6964):295–298. doi: 10.1038/nature02093. [DOI] [PubMed] [Google Scholar]
- 10.Tootle TL, Silver SJ, Davies EL, Newman V, Latek RR, Mills IA, Selengut JD, Parlikar BE, Rebay I. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. 2003;426(6964):299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- 11.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7(11):833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 12.Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117(6):699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Moorhead GB, De Wever V, Templeton G, Kerk D. Evolution of protein phosphatases in plants and animals. Biochem J. 2009;417(2):401–409. doi: 10.1042/BJ20081986. [DOI] [PubMed] [Google Scholar]
- 14.Jung SK, Jeong DG, Chung SJ, Kim JH, Park BC, Tonks NK, Ryu SE, Kim SJ. Crystal structure of ED-Eya2: insight into dual roles as a protein tyrosine phosphatase and a transcription factor. FASEB J. 2010;24(2):560–569. doi: 10.1096/fj.09-143891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mermod N, O’Neill EA, Kelly TJ, Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989;58(4):741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- 16.Xu PX, Cheng J, Epstein JA, Maas RL. Mouse Eya genes are expressed during limb tendon development and encode a transcriptional activation function. Proc Natl Acad Sci USA. 1997;94(22):11974–11979. doi: 10.1073/pnas.94.22.11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okabe Y, Sano T, Nagata S. Regulation of the innate immune response by threonine-phosphatase of Eyes absent. Nature. 2009;460(7254):520–524. doi: 10.1038/nature08138. [DOI] [PubMed] [Google Scholar]
- 18.Sano T, Nagata S. Characterization of the threonine-phosphatase of mouse eyes absent 3. FEBS Lett. 2011;585(17):2714–2719. doi: 10.1016/j.febslet.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 19.Takeda Y, Hatano S, Sentoku N, Matsuoka M. Homologs of animal eyes absent (eya) genes are found in higher plants. Mol Gen Genet. 1999;262(1):131–138. doi: 10.1007/s004380051067. [DOI] [PubMed] [Google Scholar]
- 20.Patterson KI, Brummer T, O’Brien PM, Daly RJ. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J. 2009;418(3):475–489. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- 21.Pulido R, Hooft van Huijsduijnen R. Protein tyrosine phosphatases: dual-specificity phosphatases in health and disease. FEBS J. 2008;275(5):848–866. doi: 10.1111/j.1742-4658.2008.06250.x. [DOI] [PubMed] [Google Scholar]
- 22.Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458(7238):591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnan N, Jeong DG, Jung SK, Ryu SE, Xiao A, Allis CD, Kim SJ, Tonks NK. Dephosphorylation of the C-terminal tyrosyl residue of the DNA damage-related histone H2A.X is mediated by the protein phosphatase eyes absent. J Biol Chem. 2009;284(24):16066–16070. doi: 10.1074/jbc.C900032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development [published erratum appears in Cell 1998 Feb 20;92(4):following 585] Cell. 1997;91(7):881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- 25.Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila [see comments] Cell. 1997;91(7):893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- 26.Heanue TA, Reshef R, Davis RJ, Mardon G, Oliver G, Tomarev S, Lassar AB, Tabin CJ. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 1999;13(24):3231–3243. doi: 10.1101/gad.13.24.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silver SJ, Davies EL, Doyon L, Rebay I. Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol Cell Biol. 2003;23(17):5989–5999. doi: 10.1128/MCB.23.17.5989-5999.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J, Wang C, Wang Z, Dampier W, Wu K, Casimiro MC, Chepelev I, Popov VM, Quong A, Tozeren A, Zhao K, Lisanti MP, Pestell RG. Attenuation of Forkhead signaling by the retinal determination factor DACH1. Proc Natl Acad Sci USA. 2010;107(15):6864–6869. doi: 10.1073/pnas.1002746107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SS, Zhang R, Braunstein SE, Joachimiak A, Cvekl A, Hegde RS. Structure of the retinal determination protein dachshund reveals a DNA binding motif. Structure. 2002;10(6):787–795. doi: 10.1016/s0969-2126(02)00769-4. [DOI] [PubMed] [Google Scholar]
- 30.Zhou J, Liu Y, Zhang W, Popov VM, Wang M, Pattabiraman N, Sune C, Cvekl A, Wu K, Jiang J, Wang C, Pestell RG. Transcription elongation regulator 1 is a co-integrator of the cell fate determination factor Dachshund homolog 1. J Biol Chem. 2010;285(51):40342–40350. doi: 10.1074/jbc.M110.156141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patrick AN, Schiemann BJ, Yang K, Zhao R, Ford HL. Biochemical and functional characterization of six SIX1 branchio-oto-renal syndrome mutations. J Biol Chem. 2009;284(31):20781–20790. doi: 10.1074/jbc.M109.016832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu X, Rosenfeld MG. Transcriptional control of precursor proliferation in the early phases of pituitary development. Curr Opin Genet Dev. 2004;14(5):567–574. doi: 10.1016/j.gde.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 33.Pandey RN, Rani R, Yeo EJ, Spencer M, Hu S, Lang RA, Hegde RS. The Eyes Absent phosphatase-transactivator proteins promote proliferation, transformation, migration, and invasion of tumor cells. Oncogene. 2010;29(25):3715–3722. doi: 10.1038/onc.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jemc J, Rebay I. The eyes absent family of phosphotyrosine phosphatases: properties and roles in developmental regulation of transcription. Annu Rev Biochem. 2007;76:513–538. doi: 10.1146/annurev.biochem.76.052705.164916. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Perissi V, Liu F, Rose DW, Rosenfeld MG. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science. 2002;297(5584):1180–1183. doi: 10.1126/science.1073263. [DOI] [PubMed] [Google Scholar]
- 36.Spitz F, Demignon J, Porteu A, Kahn A, Concordet JP, Daegelen D, Maire P. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc Natl Acad Sci USA. 1998;95(24):14220–14225. doi: 10.1073/pnas.95.24.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coletta RD, Christensen K, Reichenberger KJ, Lamb J, Micomonaco D, Huang L, Wolf DM, Muller-Tidow C, Golub TR, Kawakami K, Ford HL. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc Natl Acad Sci USA. 2004;101(17):6478–6483. doi: 10.1073/pnas.0401139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawakami K, Ohto H, Ikeda K, Roeder RG. Structure, function and expression of a murine homeobox protein AREC3, a homologue of Drosophila sine oculis gene product, and implication in development. Nucleic Acids Res. 1996;24(2):303–310. doi: 10.1093/nar/24.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brodbeck S, Besenbeck B, Englert C. The transcription factor Six2 activates expression of the Gdnf gene as well as its own promoter. Mech Dev. 2004;121(10):1211–1222. doi: 10.1016/j.mod.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 40.Grifone R, Laclef C, Spitz F, Lopez S, Demignon J, Guidotti JE, Kawakami K, Xu PX, Kelly R, Petrof BJ, Daegelen D, Concordet JP, Maire P. Six1 and Eya1 expression can reprogram adult muscle from the slow-twitch phenotype into the fast-twitch phenotype. Mol Cell Biol. 2004;24(14):6253–6267. doi: 10.1128/MCB.24.14.6253-6267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Himeda CL, Ranish JA, Angello JC, Maire P, Aebersold R, Hauschka SD. Quantitative proteomic identification of six4 as the trex-binding factor in the muscle creatine kinase enhancer. Mol Cell Biol. 2004;24(5):2132–2143. doi: 10.1128/MCB.24.5.2132-2143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ando Z, Sato S, Ikeda K, Kawakami K. Slc12a2 is a direct target of two closely related homeobox proteins, Six1 and Six4. FEBS J. 2005;272(12):3026–3041. doi: 10.1111/j.1742-4658.2005.04716.x. [DOI] [PubMed] [Google Scholar]
- 43.Yu Y, Davicioni E, Triche TJ, Merlino G. The homeoprotein six1 transcriptionally activates multiple protumorigenic genes but requires ezrin to promote metastasis. Cancer Res. 2006;66(4):1982–1989. doi: 10.1158/0008-5472.CAN-05-2360. [DOI] [PubMed] [Google Scholar]
- 44.Sander LE, Blander JM. Innate immune cells cast an eye on DNA. J Mol Cell Biol. 2009;1(2):77–79. doi: 10.1093/jmcb/mjp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF, Jacotot L, Vaglio P, Reboul J, Hirozane-Kishikawa T, Li Q, Gabel HW, Elewa A, Baumgartner B, Rose DJ, Yu H, Bosak S, Sequerra R, Fraser A, Mango SE, Saxton WM, Strome S, Van Den Heuvel S, Piano F, Vandenhaute J, Sardet C, Gerstein M, Doucette-Stamm L, Gunsalus KC, Harper JW, Cusick ME, Roth FP, Hill DE, Vidal M. A map of the interactome network of the metazoan C. elegans. Science. 2004;303(5657):540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan X, Brass LF, Poncz M, Spitz F, Maire P, Manning DR. The alpha subunits of Gz and Gi interact with the eyes absent transcription cofactor Eya2, preventing its interaction with the six class of homeodomain-containing proteins. J Biol Chem. 2000;275(41):32129–32134. doi: 10.1074/jbc.M004577200. [DOI] [PubMed] [Google Scholar]
- 47.Azuma N, Hirakiyama A, Inoue T, Asaka A, Yamada M. Mutations of a human homologue of the Drosophila eyes absent gene (EYA1) detected in patients with congenital cataracts and ocular anterior segment anomalies. Hum Mol Genet. 2000;9(3):363–366. doi: 10.1093/hmg/9.3.363. [DOI] [PubMed] [Google Scholar]
- 48.Weng J, Luo J, Cheng X, Jin C, Zhou X, Qu J, Tu L, Ai D, Li D, Wang J, Martin JF, Amendt BA, Liu M. Deletion of G protein-coupled receptor 48 leads to ocular anterior segment dysgenesis (ASD) through down-regulation of Pitx2. Proc Natl Acad Sci USA. 2008;105(16):6081–6086. doi: 10.1073/pnas.0708257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Landgraf K, Bollig F, Trowe MO, Besenbeck B, Ebert C, Kruspe D, Kispert A, Hanel F, Englert C. Sipl1 and Rbck1 are novel Eya1-binding proteins with a role in craniofacial development. Mol Cell Biol. 2010;30(24):5764–5775. doi: 10.1128/MCB.01645-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El-Hashash AH, Turcatel G, Al Alam D, Buckley S, Tokumitsu H, Bellusci S, Warburton D. Eya1 controls cell polarity, spindle orientation, cell fate and Notch signaling in distal embryonic lung epithelium. Development. 2011;138(7):1395–1407. doi: 10.1242/dev.058479. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, Zhang H, Polakiewicz RD, Comb MJ. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci USA. 2007;104(50):19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiong W, Dabbouseh NM, Rebay I. Interactions with the abelson tyrosine kinase reveal compartmentalization of eyes absent function between nucleus and cytoplasm. Dev Cell. 2009;16(2):271–279. doi: 10.1016/j.devcel.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morillo SA, Braid LR, Verheyen EM, Rebay I. Nemo phosphorylates Eyes absent and enhances output from the Eya-sine oculis transcriptional complex during Drosophila retinal determination. Dev Biol. 2012;365(1):267–276. doi: 10.1016/j.ydbio.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zou D, Erickson C, Kim EH, Jin D, Fritzsch B, Xu PX. Eya1 gene dosage critically affects the development of sensory epithelia in the mammalian inner ear. Hum Mol Genet. 2008;17(21):3340–3356. doi: 10.1093/hmg/ddn229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahmed M, Wong EY, Sun J, Xu J, Wang F, Xu PX. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev Cell. 2012;22(2):377–390. doi: 10.1016/j.devcel.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Furuya M, Qadota H, Chisholm AD, Sugimoto A. The C. elegans eyes absent ortholog EYA-1 is required for tissue differentiation and plays partially redundant roles with PAX-6. Dev Biol. 2005;286(2):452–463. doi: 10.1016/j.ydbio.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 57.Kriebel M, Muller F, Hollemann T. Xeya3 regulates survival and proliferation of neural progenitor cells within the anterior neural plate of Xenopus embryos. Dev Dyn. 2007;236(6):1526–1534. doi: 10.1002/dvdy.21170. [DOI] [PubMed] [Google Scholar]
- 58.Zou D, Silvius D, Rodrigo-Blomqvist S, Enerback S, Xu PX. Eya1 regulates the growth of otic epithelium and interacts with Pax2 during the development of all sensory areas in the inner ear. Dev Biol. 2006;298(2):430–441. doi: 10.1016/j.ydbio.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23(1):113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- 60.El-Hashash AH, Al Alam D, Turcatel G, Bellusci S, Warburton D. Eyes absent 1 (Eya1) is a critical coordinator of epithelial, mesenchymal and vascular morphogenesis in the mammalian lung. Dev Biol. 2011;350(1):112–126. doi: 10.1016/j.ydbio.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clark SW, Fee BE, Cleveland JL. Misexpression of the eyes absent family triggers the apoptotic program. J Biol Chem. 2002;277(5):3560–3567. doi: 10.1074/jbc.M108410200. [DOI] [PubMed] [Google Scholar]
- 62.Hirose T, Galvin BD, Horvitz HR. Six and Eya promote apoptosis through direct transcriptional activation of the proapoptotic BH3-only gene egl-1 in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2010;107(35):15479–15484. doi: 10.1073/pnas.1010023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boyle M, Bonini N, DiNardo S. Expression and function of clift in the development of somatic gonadal precursors within the Drosophila mesoderm. Development. 1997;124(5):971–982. doi: 10.1242/dev.124.5.971. [DOI] [PubMed] [Google Scholar]
- 64.Xu PX, Woo I, Her H, Beier DR, Maas RL. Mouse Eya homologues of the Drosophila eyes absent gene require Pax6 for expression in lens and nasal placode. Development. 1997;124(1):219–231. doi: 10.1242/dev.124.1.219. [DOI] [PubMed] [Google Scholar]
- 65.Mishima N, Tomarev S. Chicken Eyes absent 2 gene: isolation and expression pattern during development. Int J Dev Biol. 1998;42(8):1109–1115. [PubMed] [Google Scholar]
- 66.Sahly I, Andermann P, Petit C. The zebrafish eya1 gene and its expression pattern during embryogenesis. Dev Genes Evol. 1999;209(7):399–410. doi: 10.1007/s004270050270. [DOI] [PubMed] [Google Scholar]
- 67.David R, Ahrens K, Wedlich D, Schlosser G. Xenopus Eya1 demarcates all neurogenic placodes as well as migrating hypaxial muscle precursors. Mech Dev. 2001;103(1–2):189–192. doi: 10.1016/s0925-4773(01)00355-0. [DOI] [PubMed] [Google Scholar]
- 68.Tadjuidje E, Wang TS, Pandey RN, Sumanas S, Lang RA, Hegde RS. The EYA tyrosine phosphatase activity is pro-angiogenic and is inhibited by benzbromarone. PLoS One. 2012;7(4):e34806. doi: 10.1371/journal.pone.0034806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miller SJ, Lan ZD, Hardiman A, Wu J, Kordich JJ, Patmore DM, Hegde RS, Cripe TP, Cancelas JA, Collins MH, Ratner N. Inhibition of Eyes absent homolog 4 expression induces malignant peripheral nerve sheath tumor necrosis. Oncogene. 2009;29(3):368–379. doi: 10.1038/onc.2009.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36(17):5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, Ishibe-Murakami S, Wang B, Tempst P, Hofmann K, Patel DJ, Elledge SJ, Allis CD. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457(7225):57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie A, Odate S, Chandramouly G, Scully R. H2AX post-translational modifications in the ionizing radiation response and homologous recombination. Cell Cycle. 2010;9(17):3602–3610. doi: 10.4161/cc.9.17.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Economopoulou M, Langer HF, Celeste A, Orlova VV, Choi EY, Ma M, Vassilopoulos A, Callen E, Deng C, Bassing CH, Boehm M, Nussenzweig A, Chavakis T. Histone H2AX is integral to hypoxia-driven neovascularization. Nat Med. 2009;15(5):553–558. doi: 10.1038/nm.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang CA, Jedlicka P, Patrick AN, Micalizzi DS, Lemmer KC, Deitsch E, Casas-Selves M, Harrell JC, Ford HL. SIX1 induces lymphangiogenesis and metastasis via upregulation of VEGF-C in mouse models of breast cancer. J Clin Invest. 2012;122(5):1895–1906. doi: 10.1172/JCI59858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edwards RG, Beard HK. Oocyte polarity and cell determination in early mammalian embryos. Mol Hum Reprod. 1997;3(10):863–905. doi: 10.1093/molehr/3.10.863. [DOI] [PubMed] [Google Scholar]
- 76.Plusa B, Hadjantonakis AK, Gray D, Piotrowska-Nitsche K, Jedrusik A, Papaioannou VE, Glover DM, Zernicka-Goetz M. The first cleavage of the mouse zygote predicts the blastocyst axis. Nature. 2005;434(7031):391–395. doi: 10.1038/nature03388. [DOI] [PubMed] [Google Scholar]
- 77.Bai J, Montell D. Eyes absent, a key repressor of polar cell fate during Drosophila oogenesis. Development. 2002;129(23):5377–5388. doi: 10.1242/dev.00115. [DOI] [PubMed] [Google Scholar]
- 78.Leiserson WM, Bonini NM, Benzer S. Transvection at the eyes absent gene of Drosophila. Genetics. 1994;138(4):1171–1179. doi: 10.1093/genetics/138.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duncan MK, Kos L, Jenkins NA, Gilbert DJ, Copeland NG, Tomarev SI. Eyes absent: a gene family found in several metazoan phyla. Mamm Genome. 1997;8(7):479–485. doi: 10.1007/s003359900480. [DOI] [PubMed] [Google Scholar]
- 80.Borsani G, DeGrandi A, Ballabio A, Bulfone A, Bernard L, Banfi S, Gattuso C, Mariani M, Dixon M, Donnai D, Metcalfe K, Winter R, Robertson M, Axton R, Brown A, van Heyningen V, Hanson I. EYA4, a novel vertebrate gene related to Drosophila eyes absent. Hum Mol Genet. 1999;8(1):11–23. doi: 10.1093/hmg/8.1.11. [DOI] [PubMed] [Google Scholar]
- 81.Zimmerman JE, Bui QT, Steingrimsson E, Nagle DL, Fu W, Genin A, Spinner NB, Copeland NG, Jenkins NA, Bucan M, Bonini NM. Cloning and characterization of two vertebrate homologs of the Drosophila eyes absent gene. Genome Res. 1997;7(2):128–141. doi: 10.1101/gr.7.2.128. [DOI] [PubMed] [Google Scholar]
- 82.Neilson KM, Pignoni F, Yan B, SA M. Developmental expression patterns of candidate cofactors for vertebrate six family transcription factors. Dev Dyn. 2010;239(12):3446–3466. doi: 10.1002/dvdy.22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fougerousse F, Durand M, Lopez S, Suel L, Demignon J, Thornton C, Ozaki H, Kawakami K, Barbet P, Beckmann JS, Maire P. Six and Eya expression during human somitogenesis and MyoD gene family activation. J Muscle Res Cell Motil. 2002;23(3):255–264. doi: 10.1023/a:1020990825644. [DOI] [PubMed] [Google Scholar]
- 84.Eissenberg JC, Ryerse JS. ey-2: A recessive eyeless mutation on the second chromosome of Drosophila melanogaster. Drosophila Inf Serv. 1991;70:266–268. [Google Scholar]
- 85.Sved J. Eyes Absent (eya) Drosophila Inf Serv. 1986;63:169. [Google Scholar]
- 86.Bui QT, Zimmerman JE, Liu H, Bonini NM. Molecular analysis of Drosophila eyes absent mutants reveals features of the conserved Eya domain. Genetics. 2000;155(2):709–720. doi: 10.1093/genetics/155.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, Bitner-Glindzicz M, Francis M, Lacombe D, Vigneron J, Charachon R, Boven K, Bedbeder P, Van Regemorter N, Weissenbach J, Petit C. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet. 1997;15(2):157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- 88.Kalatzis V, Sahly I, El-Amraoui A, Petit C. Eya1 expression in the developing ear and kidney: towards the understanding of the pathogenesis of branchio-oto-renal (BOR) syndrome. Dev Dyn. 1998;213(4):486–499. doi: 10.1002/(SICI)1097-0177(199812)213:4<486::AID-AJA13>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 89.Ishihara T, Ikeda K, Sato S, Yajima H, Kawakami K. Differential expression of Eya1 and Eya2 during chick early embryonic development. Gene Expr Patterns. 2008;8(5):357–367. doi: 10.1016/j.gep.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 90.Sajithlal G, Zou D, Silvius D, Xu PX. Eya 1 acts as a critical regulator for specifying the metanephric mesenchyme. Dev Biol. 2005;284(2):323–336. doi: 10.1016/j.ydbio.2005.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu PX, Zheng W, Huang L, Maire P, Laclef C, Silvius D. Six1 is required for the early organogenesis of mammalian kidney. Development. 2003;130(14):3085–3094. doi: 10.1242/dev.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nie X, Xu J, El-Hashash A, Xu PX. Six1 regulates Grem1 expression in the metanephric mesenchyme to initiate branching morphogenesis. Dev Biol. 2011;352(1):141–151. doi: 10.1016/j.ydbio.2011.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Self M, Lagutin OV, Bowling B, Hendrix J, Cai Y, Dressler GR, Oliver G. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 2006;25(21):5214–5228. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schlosser G, Ahrens K. Molecular anatomy of placode development in Xenopus laevis. Dev Biol. 2004;271(2):439–466. doi: 10.1016/j.ydbio.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 95.Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Levi-Acobas F, Cruaud C, Le Merrer M, Mathieu M, Konig R, Vigneron J, Weissenbach J, Petit C, Weil D. Clustering of mutations responsible for branchio-oto-renal (BOR) syndrome in the eyes absent homologous region (eyaHR) of EYA1. Hum Mol Genet. 1997;6(13):2247–2255. doi: 10.1093/hmg/6.13.2247. [DOI] [PubMed] [Google Scholar]
- 96.Kim SH, Shin JH, Yeo CK, Chang SH, Park SY, Cho EH, Ki CS, Kim JW. Identification of a novel mutation in the EYA1 gene in a Korean family with branchio-oto-renal (BOR) syndrome. Int J Pediatr Otorhinolaryngol. 2005;69(8):1123–1128. doi: 10.1016/j.ijporl.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 97.Rickard S, Parker M, van’t Hoff W, Barnicoat A, Russell-Eggitt I, Winter RM, Bitner-Glindzicz M. Oto-facio-cervical (OFC) syndrome is a contiguous gene deletion syndrome involving EYA1: molecular analysis confirms allelism with BOR syndrome and further narrows the Duane syndrome critical region to 1 cM. Hum Genet. 2001;108(5):398–403. doi: 10.1007/s004390100495. [DOI] [PubMed] [Google Scholar]
- 98.Vincent C, Kalatzis V, Abdelhak S, Chaib H, Compain S, Helias J, Vaneecloo FM, Petit C. BOR and BO syndromes are allelic defects of EYA1. Eur J Hum Genet. 1997;5(4):242–246. [PubMed] [Google Scholar]
- 99.Wayne S, Robertson NG, DeClau F, Chen N, Verhoeven K, Prasad S, Tranebjarg L, Morton CC, Ryan AF, Van Camp G, Smith RJ. Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Hum Mol Genet. 2001;10(3):195–200. doi: 10.1093/hmg/10.3.195. [DOI] [PubMed] [Google Scholar]
- 100.Sato S, Ikeda K, Shioi G, Ochi H, Ogino H, Yajima H, Kawakami K. Conserved expression of mouse Six1 in the pre-placodal region (PPR) and identification of an enhancer for the rostral PPR. Dev Biol. 2010;344(1):158–171. doi: 10.1016/j.ydbio.2010.04.029. [DOI] [PubMed] [Google Scholar]
- 101.Kozlowski DJ, Whitfield TT, Hukriede NA, Lam WK, Weinberg ES. The zebrafish dog-eared mutation disrupts eya1, a gene required for cell survival and differentiation in the inner ear and lateral line. Dev Biol. 2005;277(1):27–41. doi: 10.1016/j.ydbio.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 102.Bane BC, Van Rybroek JM, Kolker SJ, Weeks DL, Manaligod JM. EYA1 expression in the developing inner ear. Ann Otol Rhinol Laryngol. 2005;114(11):853–858. doi: 10.1177/000348940511401108. [DOI] [PubMed] [Google Scholar]
- 103.Li Y, Manaligod JM, Weeks DL. EYA1 mutations associated with the branchio-oto-renal syndrome result in defective otic development in Xenopus laevis. Biol Cell. 2010;102(5):277–292. doi: 10.1042/BC20090098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ahmed M, Xu J, Xu PX. EYA1 and SIX1 drive the neuronal developmental program in cooperation with the SWI/SNF chromatin-remodeling complex and SOX2 in the mammalian inner ear. Development. 2012;139(11):1965–1977. doi: 10.1242/dev.071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Depreux FF, Darrow K, Conner DA, Eavey RD, Liberman MC, Seidman CE, Seidman JG. Eya4-deficient mice are a model for heritable otitis media. J Clin Invest. 2008;118(2):651–658. doi: 10.1172/JCI32899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schonberger J, Wang L, Shin JT, Kim SD, Depreux FF, Zhu H, Zon L, Pizard A, Kim JB, Macrae CA, Mungall AJ, Seidman JG, Seidman CE. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat Genet. 2005;37(4):418–422. doi: 10.1038/ng1527. [DOI] [PubMed] [Google Scholar]
- 107.Shimasaki N, Watanabe K, Hara M, Kosaki K. EYA1 mutation in a newborn female presenting with cardiofacial syndrome. Pediatr Cardiol. 2004;25(4):411–413. doi: 10.1007/s00246-003-0271-3. [DOI] [PubMed] [Google Scholar]
- 108.Soker T, Dalke C, Puk O, Floss T, Becker L, Bolle I, Favor J, Hans W, Holter SM, Horsch M, Kallnik M, Kling E, Moerth C, Schrewe A, Stigloher C, Topp S, Gailus-Durner V, Naton B, Beckers J, Fuchs H, Ivandic B, Klopstock T, Schulz H, Wolf E, Wurst W, Bally-Cuif L, de Angelis MH, Graw J. Pleiotropic effects in Eya3 knockout mice. BMC Dev Biol. 2008;8:118. doi: 10.1186/1471-213X-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang DK, Choi BY, Lee YH, Kim YG, Cho MC, Hong SE, Kim do H, Hajjar RJ, Park WJ. Gene profiling during regression of pressure overload-induced cardiac hypertrophy. Physiol Genomics. 2007;30(1):1–7. doi: 10.1152/physiolgenomics.00246.2006. [DOI] [PubMed] [Google Scholar]
- 110.Lee SH, Yang DK, Choi BY, Lee Y-H, Kim S-Y, Jeong D, Hajjar RJ, Park WJ. The transcription factor Eya2 prevents pressure overload-induced adverse cardiac remodeling. J Mol Cell Cardiol. 2009;46(4):596–605. doi: 10.1016/j.yjmcc.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 111.Lee SH, Kim J, Ryu JY, Lee S, Yang DK, Jeong D, Kim JM, Hajjar RJ, Park WJ. Transcription coactivator Eya2 is a critical regulator of physiological hypertrophy. J Mol Cell Cardiol. 2012;52(3):718–726. doi: 10.1016/j.yjmcc.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 112.McMullen JR, Jennings GL. Differences between pathological and physiological cardiac hypertrophy: novel therapeutic strategies to treat heart failure. Clin Exp Pharmacol Physiol. 2007;34(4):255–262. doi: 10.1111/j.1440-1681.2007.04585.x. [DOI] [PubMed] [Google Scholar]
- 113.Walker MB, Trainor PA. Craniofacial malformations: intrinsic vs extrinsic neural crest cell defects in Treacher Collins and 22q11 deletion syndromes. Clin Genet. 2006;69(6):471–479. doi: 10.1111/j.0009-9163.2006.00615.x. [DOI] [PubMed] [Google Scholar]
- 114.Mercer C, Gilbert R, Loughlin S, Foulds N. Patient with an EYA1 mutation with features of branchio-oto-renal and oto-facio-cervical syndrome. Clin Dysmorphol. 2006;15(4):211–212. doi: 10.1097/01.mcd.0000204986.54366.7c. [DOI] [PubMed] [Google Scholar]
- 115.Estefania E, Ramirez-Camacho R, Gomar M, Trinidad A, Arellano B, Garcia-Berrocal JR, Verdaguer JM, Vilches C. Point mutation of an EYA1-gene splice site in a patient with oto-facio-cervical syndrome. Ann Hum Genet. 2006;70(Pt 1):140–144. doi: 10.1111/j.1529-8817.2005.00204.x. [DOI] [PubMed] [Google Scholar]
- 116.Schlosser G. Induction and specification of cranial placodes. Dev Biol. 2006;294(2):303–351. doi: 10.1016/j.ydbio.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 117.Zou D, Silvius D, Fritzsch B, Xu PX. Eya1 and Six1 are essential for early steps of sensory neurogenesis in mammalian cranial placodes. Development. 2004;131(22):5561–5572. doi: 10.1242/dev.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schlosser G, Awtry T, Brugmann SA, Jensen ED, Neilson K, Ruan G, Stammler A, Voelker D, Yan B, Zhang C, Klymkowsky MW, Moody SA. Eya1 and Six1 promote neurogenesis in the cranial placodes in a SoxB1-dependent fashion. Dev Biol. 2008;320(1):199–214. doi: 10.1016/j.ydbio.2008.05.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nakao N, Ono H, Yamamura T, Anraku T, Takagi T, Higashi K, Yasuo S, Katou Y, Kageyama S, Uno Y, Kasukawa T, Iigo M, Sharp PJ, Iwasawa A, Suzuki Y, Sugano S, Niimi T, Mizutani M, Namikawa T, Ebihara S, Ueda HR, Yoshimura T. Thyrotrophin in the pars tuberalis triggers photoperiodic response. Nature. 2008;452(7185):317–322. doi: 10.1038/nature06738. [DOI] [PubMed] [Google Scholar]
- 120.Masumoto KH, Ukai-Tadenuma M, Kasukawa T, Nagano M, Uno KD, Tsujino K, Horikawa K, Shigeyoshi Y, Ueda HR. Acute induction of Eya3 by late-night light stimulation triggers TSHbeta expression in photoperiodism. Curr Biol. 2010;20(24):2199–2206. doi: 10.1016/j.cub.2010.11.038. [DOI] [PubMed] [Google Scholar]
- 121.Dupre SM, Miedzinska K, Duval CV, Yu L, Goodman RL, Lincoln GA, Davis JR, McNeilly AS, Burt DD, Loudon AS. Identification of Eya3 and TAC1 as long-day signals in the sheep pituitary. Curr Biol. 2010;20(9):829–835. doi: 10.1016/j.cub.2010.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dardente H, Wyse CA, Birnie MJ, Dupre SM, Loudon AS, Lincoln GA, Hazlerigg DG. A molecular switch for photoperiod responsiveness in mammals. Curr Biol. 2010;20(24):2193–2198. doi: 10.1016/j.cub.2010.10.048. [DOI] [PubMed] [Google Scholar]
- 123.Brady AK, Snyder KA, Vize PD. Circadian cycles of gene expression in the coral, Acropora millepora. PLoS One. 2011;6(9):e25072. doi: 10.1371/journal.pone.0025072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Spruijt L, Hoefsloot LH, van Schaijk GH, van Waardenburg D, Kremer B, Brackel HJ, de Die-Smulders CE. Identification of a novel EYA1 mutation presenting in a newborn with laryngomalacia, glossoptosis, retrognathia, and pectus excavatum. Am J Med Genet A. 2006;140(12):1343–1345. doi: 10.1002/ajmg.a.31285. [DOI] [PubMed] [Google Scholar]
- 125.Buller C, Xu X, Marquis V, Schwanke R, Xu PX. Molecular effects of Eya1 domain mutations causing organ defects in BOR syndrome. Hum Mol Genet. 2001;10(24):2775–2781. doi: 10.1093/hmg/10.24.2775. [DOI] [PubMed] [Google Scholar]
- 126.Rayapureddi JP, Hegde RS. Branchio-oto-renal syndrome associated mutations in Eyes absent 1 result in loss of phosphatase activity. FEBS Lett. 2006;580(16):3853–3859. doi: 10.1016/j.febslet.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 127.Miller G, Fuchs R, Lai E. IMAGE cDNA clones, UniGene clustering, and ACeDB: an integrated resource for expressed sequence information. Genome Res. 1997;7(10):1027–1032. doi: 10.1101/gr.7.10.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zou H, Osborn NK, Harrington JJ, Klatt KK, Molina JR, Burgart LJ, Ahlquist DA. Frequent methylation of eyes absent 4 gene in Barrett’s esophagus and esophageal adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2005;14(4):830–834. doi: 10.1158/1055-9965.EPI-04-0506. [DOI] [PubMed] [Google Scholar]
- 129.Schatz P, Distler J, Berlin K, Schuster M. Novel method for high throughput DNA methylation marker evaluation using PNA-probe library hybridization and MALDI-TOF detection. Nucleic Acids Res. 2006;34(8):e59. doi: 10.1093/nar/gkl218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Osborn NK, Zou H, Molina JR, Lesche R, Lewin J, Lofton-Day C, Klatt KK, Harrington JJ, Burgart LJ, Ahlquist DA. Aberrant methylation of the eyes absent 4 gene in ulcerative colitis-associated dysplasia. Clin Gastroenterol Hepatol. 2006;4(2):212–218. doi: 10.1016/j.cgh.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 131.Kim YH, Lee HC, Kim SY, Yeom YI, Ryu KJ, Min BH, Kim DH, Son HJ, Rhee PL, Kim JJ, Rhee JC, Kim HC, Chun HK, Grady WM, Kim YS. Epigenomic analysis of aberrantly methylated genes in colorectal cancer identifies genes commonly affected by epigenetic alterations. Ann Surg Oncol. 2011;18(8):2338–2347. doi: 10.1245/s10434-011-1573-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li H, Diao TY, Zhou ZY, Yang FY, Ma Q, Li QH. Relationship between the expression of hTERT and EYA4 mRNA in peripheral blood mononuclear cells with the progressive stages of carcinogenesis of the esophagus. J Exp Clin Cancer Res. 2009;28:145. doi: 10.1186/1756-9966-28-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang L, Yang N, Huang J, Buckanovich RJ, Liang S, Barchetti A, Vezzani C, O’Brien-Jenkins A, Wang J, Ward MR, Courreges MC, Fracchioli S, Medina A, Katsaros D, Weber BL, Coukos G. Transcriptional coactivator Drosophila eyes absent homologue 2 is up-regulated in epithelial ovarian cancer and promotes tumor growth. Cancer Res. 2005;65(3):925–932. [PubMed] [Google Scholar]
- 134.Guo J, Liang C, Ding L, Zhou N, Ye Q. Drosophila Eyes absent homologue 2 is up-regulated in lung adenocarcinoma. Chin Ger J Clin Oncol. 2009;8(12):681–684. [Google Scholar]
- 135.Farabaugh SM, Micalizzi DS, Jedlicka P, Zhao R, Ford HL. Eya2 is required to mediate the pro-metastatic functions of Six1 via the induction of TGF-beta signaling, epithelial-mesenchymal transition, and cancer stem cell properties. Oncogene. 2012;31(5):552–562. doi: 10.1038/onc.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Drake KM, Ruteshouser EC, Natrajan R, Harbor P, Wegert J, Gessler M, Pritchard-Jones K, Grundy P, Dome J, Huff V, Jones C, Aldred MA. Loss of heterozygosity at 2q37 in sporadic Wilms’ tumor: putative role for miR-562. Clin Cancer Res. 2009;15(19):5985–5992. doi: 10.1158/1078-0432.CCR-09-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li CM, Guo M, Borczuk A, Powell CA, Wei M, Thaker HM, Friedman R, Klein U, Tycko B. Gene expression in Wilms’ tumor mimics the earliest committed stage in the metanephric mesenchymal-epithelial transition. Am J Pathol. 2002;160(6):2181–2190. doi: 10.1016/S0002-9440(10)61166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Matsusaka K, Kaneda A, Nagae G, Ushiku T, Kikuchi Y, Hino R, Uozaki H, Seto Y, Takada K, Aburatani H, Fukayama M. Classification of Epstein-Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res. 2011;71(23):7187–7197. doi: 10.1158/0008-5472.CAN-11-1349. [DOI] [PubMed] [Google Scholar]
- 139.Zou H, Harrington JJ, Shire AM, Rego RL, Wang L, Campbell ME, Oberg AL, Ahlquist DA. Highly methylated genes in colorectal neoplasia: implications for screening. Cancer Epidemiol Biomarkers Prev. 2007;16(12):2686–2696. doi: 10.1158/1055-9965.EPI-07-0518. [DOI] [PubMed] [Google Scholar]
- 140.Gutierrez ML, Munoz-Bellvis L, Abad Mdel M, Bengoechea O, Gonzalez–Gonzalez M, Orfao A, Sayagues JM. Association between genetic subgroups of pancreatic ductal adenocarcinoma defined by high density 500 K SNP-arrays and tumor histopathology. PLoS One. 2011;6(7):e22315. doi: 10.1371/journal.pone.0022315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nan F, Lu Q, Zhou J, Cheng L, Popov VM, Wei S, Kong B, Pestell RG, Lisanti MP, Jiang J, Wang C. Altered expression of DACH1 and cyclin D1 in endometrial cancer. Cancer Biol Ther. 2009;8(16):1534–1539. doi: 10.4161/cbt.8.16.8963. [DOI] [PubMed] [Google Scholar]
- 142.Popov VM, Wu K, Zhou J, Powell MJ, Mardon G, Wang C, Pestell RG. The Dachshund gene in development and hormone-responsive tumorigenesis. Trends Endocrinol Metab. 2010;21(1):41–49. doi: 10.1016/j.tem.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu K, Katiyar S, Li A, Liu M, Ju X, Popov VM, Jiao X, Lisanti MP, Casola A, Pestell RG. Dachshund inhibits oncogene-induced breast cancer cellular migration and invasion through suppression of interleukin-8. Proc Natl Acad Sci USA. 2008;105(19):6924–6929. doi: 10.1073/pnas.0802085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wu K, Li A, Rao M, Liu M, Dailey V, Yang Y, Di Vizio D, Wang C, Lisanti MP, Sauter G, Russell RG, Cvekl A, Pestell RG. DACH1 is a cell fate determination factor that inhibits cyclin D1 and breast tumor growth. Mol Cell Biol. 2006;26(19):7116–7129. doi: 10.1128/MCB.00268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Smith AN, Miller LA, Song N, Taketo MM, Lang RA. The duality of beta-catenin function: a requirement in lens morphogenesis and signaling suppression of lens fate in periocular ectoderm. Dev Biol. 2005;285(2):477–489. doi: 10.1016/j.ydbio.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 146.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 147.Chung AS, Ferrara N. Developmental and pathological angiogenesis. Annu Rev Cell Dev Biol. 2011;27:563–584. doi: 10.1146/annurev-cellbio-092910-154002. [DOI] [PubMed] [Google Scholar]
- 148.Sarasin A, Kauffmann A. Overexpression of DNA repair genes is associated with metastasis: a new hypothesis. Mutat Res. 2008;659(1–2):49–55. doi: 10.1016/j.mrrev.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 149.Kauffmann A, Rosselli F, Lazar V, Winnepenninckx V, Mansuet-Lupo A, Dessen P, van den Oord JJ, Spatz A, Sarasin A. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27(5):565–573. doi: 10.1038/sj.onc.1210700. [DOI] [PubMed] [Google Scholar]
- 150.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discovery. 2002;1(9):727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 151.Park H, Jung SK, Yu KR, Kim JH, Kim YS, Ko JH, Park BC, Kim SJ. Structure-based virtual screening approach to the discovery of novel inhibitors of eyes absent 2 phosphatase with various metal chelating moieties. Chem Biol Drug Des. 2011;78(4):642–650. doi: 10.1111/j.1747-0285.2011.01192.x. [DOI] [PubMed] [Google Scholar]
- 152.Kozmik Z, Holland ND, Kreslova J, Oliveri D, Schubert M, Jonasova K, Holland LZ, Pestarino M, Benes V, Candiani S. Pax-Six-Eya-Dach network during amphioxus development: conservation in vitro but context specificity in vivo. Dev Biol. 2007;306(1):143–159. doi: 10.1016/j.ydbio.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 153.Hu S, Mamedova A, Hegde RS. DNA-binding and regulation mechanisms of the SIX family of retinal determination proteins. Biochemistry. 2008;47(11):3586–3594. doi: 10.1021/bi702186s. [DOI] [PubMed] [Google Scholar]
- 154.Embry AC, Glick JL, Linder ME, Casey PJ. Reciprocal signaling between the transcriptional co-factor Eya2 and specific members of the G alpha i family. Mol Pharmacol. 2004;66(5):1325–1331. doi: 10.1124/mol.104.004093. [DOI] [PubMed] [Google Scholar]
- 155.Xu PX, Zheng W, Laclef C, Maire P, Maas RL, Peters H, Xu X. Eya1 is required for the morphogenesis of mammalian thymus, parathyroid and thyroid. Development. 2002;129(13):3033–3044. doi: 10.1242/dev.129.13.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Grifone R, Demignon J, Giordani J, Niro C, Souil E, Bertin F, Laclef C, Xu PX, Maire P. Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev Biol. 2007;302(2):602–616. doi: 10.1016/j.ydbio.2006.08.059. [DOI] [PubMed] [Google Scholar]