Abstract
Viral tumor models have significantly contributed to our understanding of oncogenic mechanisms. How transforming delta-retroviruses induce malignancy, however, remains poorly understood, especially as viral mRNA/protein are tightly silenced in tumors. Here, using deep sequencing of broad windows of small RNA sizes in the bovine leukemia virus ovine model of leukemia/lymphoma, we provide in vivo evidence of the production of noncanonical RNA polymerase III (Pol III)-transcribed viral microRNAs in leukemic B cells in the complete absence of Pol II 5′-LTR–driven transcriptional activity. Processed from a cluster of five independent self-sufficient transcriptional units located in a proviral region dispensable for in vivo infectivity, bovine leukemia virus microRNAs represent ∼40% of all microRNAs in both experimental and natural malignancy. They are subject to strong purifying selection and associate with Argonautes, consistent with a critical function in silencing of important cellular and/or viral targets. Bovine leukemia virus microRNAs are strongly expressed in preleukemic and malignant cells in which structural and regulatory gene expression is repressed, suggesting a key role in tumor onset and progression. Understanding how Pol III-dependent microRNAs subvert cellular and viral pathways will contribute to deciphering the intricate perturbations that underlie malignant transformation.
Keywords: noncoding RNA, oncogenesis, retrovirus silencing, HTLV-1
Bovine leukemia virus (BLV) is one of the most common infectious viruses of cattle with a worldwide distribution. Infection is life persistent, often subclinical, and results in the development of B-cell leukemia/lymphoma in ∼5% of BLV-infected animals after an average incubation period of 7 y (1, 2). BLV can be experimentally transmitted to sheep, which all develop B-cell tumors after a significantly shorter latency period (20 mo on average), making this an attractive experimental cancer model (3). BLV shares many common features in genomic organization and disease pathogenesis with human T-cell leukemia virus-1 (HTLV-1), a closely related deltaretrovirus that infects 20 million people worldwide and causes adult T-cell leukemia (ATL) (4).
Despite several decades of research, how BLV induces tumorigenesis remains poorly understood. After infection, BLV integrates into the genome of infected B cells and propagates into the host mainly through polyclonal expansion of BLV-carrying B cells (5). Eventually, a single clone emerges, leading to the accumulation of malignant B cells characterized by the monoclonal integration of BLV proviral DNA in the peripheral blood (B-cell leukemia) and/or diverse organs and tissues (B-cell lymphoma). However, what differentiates the emerging from the other clones remains largely unknown. Although Tax, the viral transactivator, is an essential contributor to the oncogenic potential of the virus, there is compelling evidence that expression of Tax is not sufficient for transformation (6–8). The identification of mutations in tumor-associated proviruses further proves that neither virus nor Tax expression is required for maintaining the transformed state (9–11). One of the most puzzling observations is that the BLV provirus appears transcriptionally silent in transformed B cells. Evidence from several studies indicates that both genetic and epigenetic changes account for the absence of 5′-LTR–driven RNA polymerase II (Pol II)-dependent transcription of the BLV genome in primary B-cell tumors, resulting in undetectable viral mRNA and protein (11–14).
Here, we demonstrate the abundant expression of a highly conserved BLV-encoded microRNA (miRNA) cluster in primary leukemic B cells. These viral miRNAs result from the transcription of five independent transcriptional units located in a particular BLV proviral region found to be dispensable for in vivo infectivity and considered functionally irrelevant thus far (2, 15, 16). While we were preparing this manuscript, Kincaid et al. (17) reported that BLV was capable of encoding miRNAs in vitro. We present evidence that, in primary B-cell tumors, BLV miRNA production is mediated by a noncanonical Pol III-dependent process in the complete absence of 5′-LTR Pol II-transcribed viral mRNAs/proteins and discuss the biological significance of this viral miRNA cluster in vivo.
Results
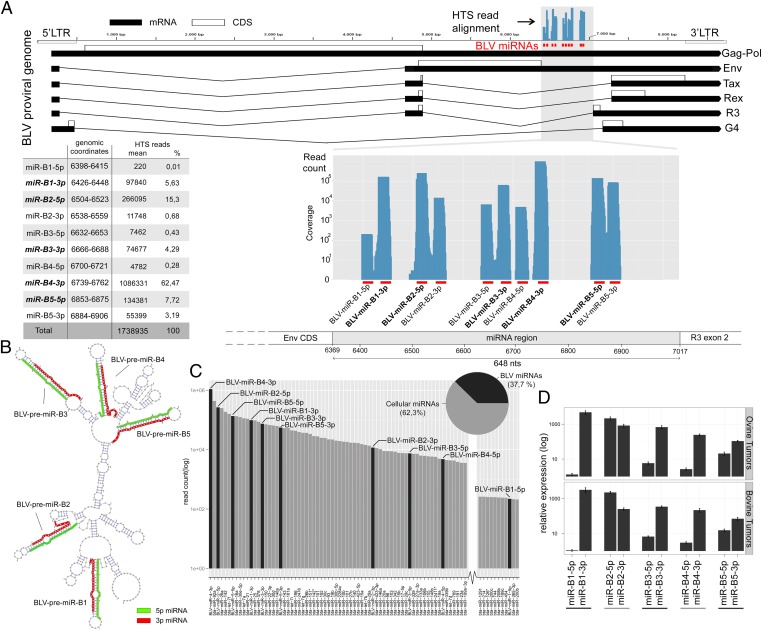
Deep Sequencing Reveals Abundant Expression of BLV miRNAs in Natural and Experimental B-Cell Tumors.
miRNAs regulate gene expression and play a key role in tumor development (18). To explore the in vivo miRnome of malignant B cells, we performed deep sequencing of small RNA libraries prepared from primary leukemic B cells and B-cell lymphoma isolated from BLV-infected sheep. Malignant tissues were composed of >93% pure B-cell populations characterized by monoclonal integration of the BLV provirus. We analyzed eight primary B-cell tumors and two malignant B-cell lines, YR2 and L267 (10, 12). Sequencing resulted in an average of 31,982,397 reads per sample (Table S1), of which at least 3,131,930 (9.79%) represent known cellular miRNAs. We also found 3,113,071 BLV-derived 20- to 23-nt-long reads (9.73%), which originated from the proviral genomic region located between the Env CDS and R3 exon 2 (Table S2, and Fig. 1A). We identified 10 unique BLV-mapping small RNAs derived from five predicted miRNA precursor hairpins BLV-premiR-B1 to BLV-premiR-B5 (Fig. 1B). Eight of these miRNAs correspond to the miRNAs recently shown to be encoded by BLV in vitro (17). The two additional BLV-mapping reads represent the less abundant star strands derived from BLV-premiR-B1 and BLV-premiR-B4, respectively. Surprisingly, BLV miRNAs represent ∼40% of the total miRNA pool in primary tumors (Fig. 1C and Fig. S1). The five predominant mature viral miRNAs belong to the top 15 most abundant miRNAs, supporting their biological relevance in vivo. Interestingly, BLV-miR-B4-3p expression was superior to miR-92a, a member of the cellular miR-17-92 cluster (oncomir-1), which we found up-regulated in BLV-infected tumor B cells, consistent with the observation in human B-cell lymphoma (19). Sequencing data were validated by stem–loop primer and QuantiMir-based quantitative RT-PCR (qRT-PCR). Abundant expression of all 10 BLV miRNAs was confirmed in 12 additional ovine experimental tumors and 8 B-cell lymphoma samples from naturally infected cattle (Fig. 1D). Viral miRNA copy numbers per cell were calculated with respect to standard curves generated with synthetic miRNAs, assuming 30 pg of total RNA per tumor cell. The level of expression ranged from 6,150 to >80,000 copies per cell (B1-5p and B4-3p, respectively). This confirms that all BLV miRNAs lie within the range of highly expressed cellular miRNAs with the most abundant members reaching outstanding levels in terms of copy numbers per cell (20).
Fig. 1.
Abundant BLV miRNA expression in tumor B cells and genomic location. (A) Small RNA HTS reads from 10 B-cell tumors mapped to the BLV proviral genome. (Upper) BLV provirus with viral miRNAs (red bars). Blue bars: read coverage. LTR, long terminal repeat; BLV mRNA (black boxes); protein coding regions (open boxes); Gag-Pol, genomic RNA/Gag-Pol precursor; Env, envelope; Tax, Rex, R3, and G4, regulatory proteins. (Lower) BLV miRNA region. Blue area: read counts and coverage; mature miRNAs are shown in bold. (B) BLV miRNA region predicted RNA secondary structure showing positions of 5p (green) and 3p (red) arm of each miRNA. Nt 1, position 6,369 (National Center for Biotechnology Information K02120). (C) Relative expression of viral and cellular miRNAs in tumor B cells: mean HTS read counts for the 120 most abundant miRNAs in eight primary B-cell tumors. Black bars: BLV miRNAs; gray bars: ovine miRNAs. (D) Abundant expression of BLV miRNAs in experimental and natural B-cell tumors: mean expression levels from triplicate qRT-PCR assays for 20 primary ovine B-cell tumors and 2 tumor B-cell lines (Upper), 8 bovine B-cell tumors (Lower). Shown is mean fold change ± SD.
If the BLV miRNAs fulfill an essential role during the life cycle of the virus, their sequences are expected to be highly conserved between viral isolates. To test this, we compared the nucleotide diversity (π: average difference per nucleotide site for all pairwise comparisons of available sequences) of the BLV miRNAs, with that of the tax gene (known to be essential) for (i) seven BLV isolates with both miRNA and tax sequences, and (ii) sets of 25 independent (nonpaired) BLV miRNA and tax sequences. For the BLV miRNA genomic region, we separately considered (i) the mature miRNAs, (ii) the star miRNAs, and when available (iii) the nonmiR sequences (5′ and 3′ flanking, intermiRNA and loops). For tax, we either considered (i) the entire tax coding region, or (ii) only the third codon positions for the part of tax that does not overlap with rex. The corresponding third codon positions are expected to evolve largely freely. This yielded the remarkable observation that the mature BLV miRNAs are characterized by a nucleotide diversity π = 0.0068, which is significantly lower than any of the other analyzed sequence segments including the full-length tax. The segments corresponding to the BLV miRNA star sequences evolve at the same rate as the full-length tax, whereas the nonmiR sequences evolve at a faster rate that does not significantly differ from the freely evolving tax third codon positions. Taken together, these data indicate that the BLV miRNAs are undergoing purifying selection that is stronger than that of a protein-coding sequence such as tax, which implies an essential function (Fig. S2 and Table S3).
Noncanonical BLV miRNA Biogenesis in Tumors.
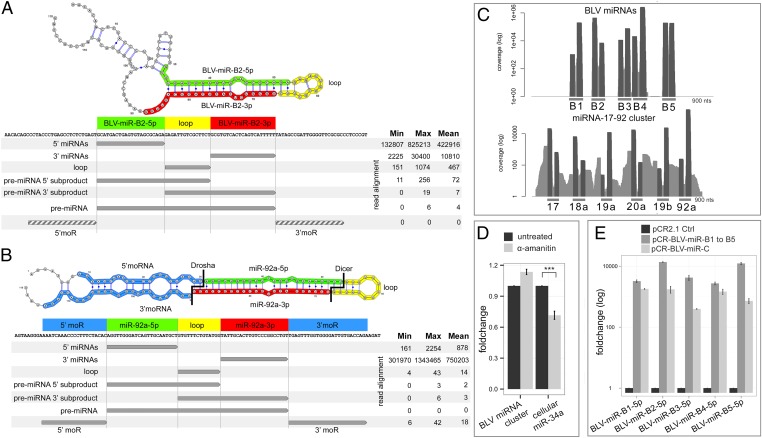
MiRNAs typically result from Pol II-dependent transcription of pri-miRNAs followed by Drosha- and Dicer-mediated processing (21). As shown in Fig. 1B, BLV miRNAs in vivo derive from five hairpins. The read positions over the predicted hairpins showed Dicer-like features. However, typical marks of Drosha processing were lacking. Both the 5′ ends of the 5p arms and the 3′ ends of the 3p arms either reached or overlapped the hairpin base. This is not compatible with Drosha excision, which typically takes place near the base but within the stem of premiRNAs and mainly generates a 2-nt 3′ overhang (22). Consistent with this common rule, alignments of ovine tumor HTS reads mapping the canonical miR cluster 17–92 on their precursor structures exhibit more central positions (Fig. S3).
To search for additional signatures of BLV miRNA biogenesis in transformed B cells, we performed deep sequencing of libraries prepared without prior size selection, making the capture of miRNA subproduct repertoires possible. We compared their distribution to those obtained for the cellular oncomiR cluster 17–92 (Table S4 and Fig. 2). As expected, we identified all five phased products miR-5p, miR-3p, loop, 5′ and 3′ miR offset reads (5′ moR and 3′ moR) as well as premiR 5′ and 3′ subproducts for the cellular miR cluster 17–92 members, indicative of both Drosha- and Dicer-dependent processing (23). In the case of BLV reads, besides the predominant mature and star BLV miRNAs, we found reads corresponding to byproducts of miRNA processing, such as cleaved terminal loops, premiRNA 5′ and 3′ products, consistent with Dicer-mediated processing (Table S4 and Fig. 2). However, although reads flanking the premiRNAs (moRs) were consistently detected in the case of the cellular miR 17–92 cluster, we did not identify these offset products in BLV miRNA-derived reads. Our high-throughput sequencing (HTS) data rule out a role for Drosha in processing the BLV miRNA transcripts in leukemic B cells and strongly argue that the Pol III-mediated transcription previously observed in vitro (17) is driving BLV miRNA expression in tumor B cells in vivo. Interestingly, as expected from Pol III-mediated transcription, the 5′ ends of the 5p reads showed a nearly uniform position. Furthermore, we detected the rare BLV miRNA precursors with a confident poly-T–rich 3′ terminator signature for Pol III, providing additional evidence that Pol III rather than Drosha defines the viral premiRNAs in B-cell tumors.
Fig. 2.
Noncanonical RNA Pol III-dependent BLV miRNA biogenesis. (A and B) Deep sequencing of broad windows of small RNA sizes reveals BLV miRNA and cellular miR 17–92 processing products. Read counts for mature miRNAs, miRNA processing subproducts, and premiRNAs for BLV-miR-B2 (A) and cellular miR 92a (B) precursors (10 tumors). 5p (green), 3p (red), loops (yellow), miRNA offset products (moRs) (blue). Gray arrows: tumor HTS reads. Dashed arrows: BLV sequences for which no mapping reads were found. (C) Coverage of BLV miRNA and cellular miR 17–92 reads aligned on proviral and sheep genome, respectively, showing absence of BLV moRs. (D) Treatment of transformed B cells with the RNA Pol II inhibitor α-Amanitin does not decrease BLV miRNA production: BLV miRNA and cellular miR-34a qRT-PCR of α-Amanitin–treated YR2 (1 µg/mL, 24 h). Mean fold change (n = 9) ± SD. ***P = 9.19 × 10−5. (E) Tumor-derived BLV miRNAs possess endogenous promoter elements: qRT-PCR of HelaT transfected with promoterless BLV miRNA cluster (pCR-BLV-miR-C) or individual BLV miRNA hairpin (pCR-BLV-miR-B1 to -B5) vectors. Expression relative to background amplification of control pCR2.1-transfected cells.
We next sought to experimentally confirm our deep sequencing data. First, treatment of malignant B cells with the Pol II inhibitor α-Amanitin did not decrease BLV miRNA production, supporting Pol III dependence of the virus-encoded cluster in tumors (Fig. 2D). This was further supported by transfection experiments using a promoterless vector (pCR2.1) in which the complete tumor-derived BLV miRNA cluster was cloned (pCR-BLV-miR-C). We found abundant expression of all BLV miRNAs, consistent with a Pol II-independent, Pol III-mediated transcriptional activity confined to the miRNA-encoding region (Fig. 2E). In addition, each individual BLV premiRNA (pCR-BLV miR-B1 to -B5) was sufficient for driving expression of the corresponding mature and star miRNA processing product, respectively. Last, Dicer dependence was investigated by lentiviral vector-mediated delivery of BLV miRNAs in cell lines deficient in the Dicer machinery and treatment of tumor B-cell lines with poly-L-lysine (PLL) (24) (Fig. S4). Both approaches resulted in a significant decrease in miRNA expression, confirming the HTS Dicer signature in B-cell tumors. In conclusion, we show that in tumors BLV miRNAs are processed from five independent transcriptionally self-sufficient Pol III units, which each possess intragenic control elements, in a Drosha-independent, Dicer-dependent manner.
BLV miRNAs Are Produced in the Absence of 5′-LTR–Dependent mRNA Transcripts, yet Transcriptional Reactivation Does Not Interfere with Their Expression.
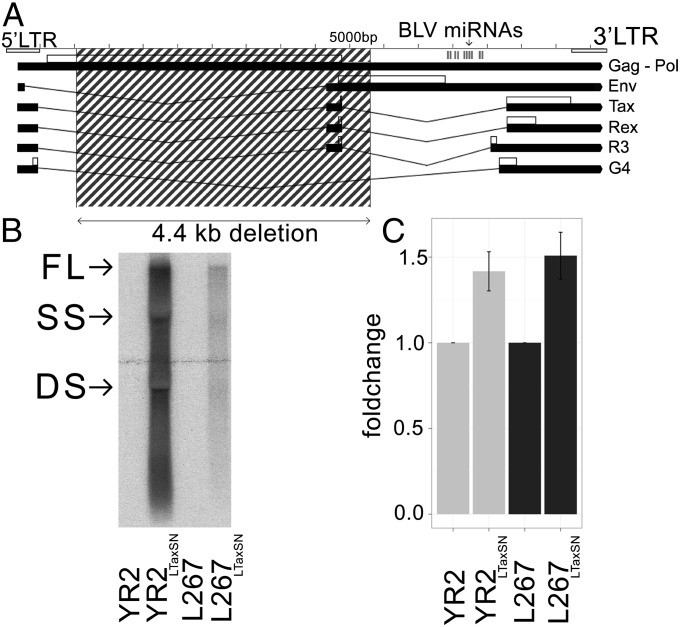
Viral mRNA expression is consistently suppressed in primary B-cell tumors and in in vitro-derived transformed B-cell lines. Experimental evidence of this transcriptional inactivity in the deep-sequenced B-cell tumors came from qRT-PCR assays, ex vivo culture/in vivo infection experiments, and the detection of repressive epigenetic marks on the viral 5′-LTR Pol II promoter (10–14). Thus, in B-cell tumors, abundant Pol III-driven viral miRNA expression is accompanied by undetectable 5′-LTR Pol II transcripts. The most convincing evidence of silencing came from a subset of primary tumors carrying proviruses with loss-of-function mutations in tax (11, 12) or with large deletions that compromise the ability to produce viral mRNA (Fig. 3A). Together, these observations provide conclusive evidence that BLV miRNAs can be released from a completely silent provirus in terms of 5′-LTR–driven mRNA transcription.
Fig. 3.
BLV miRNAs are produced in the absence of 5′-LTR–dependent viral mRNA in tumors, yet transcriptional reactivation does not interfere with their expression. (A) Structure of defective T1345 tumor provirus: the 4.4 kb deletion (dashed box) covers sequences required for virus/mRNA production but leaves an intact miRNA region. (B and C) Tax-induced 5′-LTR transcriptional reactivation in malignant B cells does not significantly alter BLV miRNA expression. (B) Northern blot analysis of activated (YR2LTaxSN, L267LTaxSN) or silent (YR2, L267) cell lines hybridized with a BLV 3′ probe. Arrows: full-length (FL), single-spliced (SS), and double-spliced (DS) mRNAs. (C) qRT-PCR analysis of BLV miRNA expression in silent and virus-producing tumor B-cell lines. Data are represented as mean fold change ± SD.
Treatment with epigenetic drugs or ectopic expression of Tax promotes rescue from latency (11, 13). We examined whether this 5′-LTR–dependent transcriptional reactivation would interfere with Pol III miRNA production in tumor cells. Tax-mediated reactivation of BLV gene expression did not noticeably alter BLV miRNA expression levels, as suggested by a nonsignificant 1.5-fold increase in qRT-PCR analysis (Fig. 3 B and C). Of note, we did not observe inverse trends in the relative abundance of BLV mRNA and miRNA transcripts, revealing that Pol III- and Pol II-mediated transcription are not mutually exclusive in tumor cells. This indicates that overlap of Pol III activity and 5′-LTR–driven Pol II transcription does not significantly alter the BLV miRNA abundance, supporting the conclusion that the five Pol III miRNA transcriptional units are controlled by a miRNA-specific 5′-LTR–independent regulation process.
Viral miRNAs Are Produced in BLV-Infected Asymptomatic Animals.
HTS analysis of B cells collected at the asymptomatic polyclonal stage in three sheep from which tumor-derived HTS data were available revealed abundant BLV miRNA reads, and this was validated by qRT-PCR (Tables S5 and S6, and Fig. S5 A and B). To examine whether the BLV miRNA level in these nonmalignant B cells would be within the range of BLV miRNAs in tumor B cells, we compared qRT-PCR data from animal-matched paired samples. From the B-cell percentage and BLV proviral load estimations at the time of peripheral blood mononuclear cell (PBMC) collection (Table S5), we extrapolated that the level of miRNA expression per B cell was not significantly different in the polyclonal B-cell pool that carries proviral DNA (viral load estimation of 16.1–22.5%) compared with tumor B cells. Furthermore, knowing the total proviral load, the B-cell percentage before purification and the tumor-specific proviral integration site (TIS), we estimated relative proviral copy numbers associated with the pretumoral clone compared with provirus integrated in B cells that would not give rise to tumor (TIS-specific qPCR). Considering that the tumor clone accounted for <0.02% of the total viral load in the polyclonal B-cell population isolated from animal M2531, we concluded that the contribution of the pretumoral clone to global miRNA expression observed at the asymptomatic stage was minimal (Fig. S5C). Interestingly, the relative abundance of BLV miRNAs was equivalent to that observed in transformed B cells (Fig. S6). We conclude that the BLV miRNA cluster is significantly expressed in vivo before leukemia onset and that this production is not restricted to the B-cell clone from which the tumor will arise.
BLV miRNAs Associate with Argonautes.
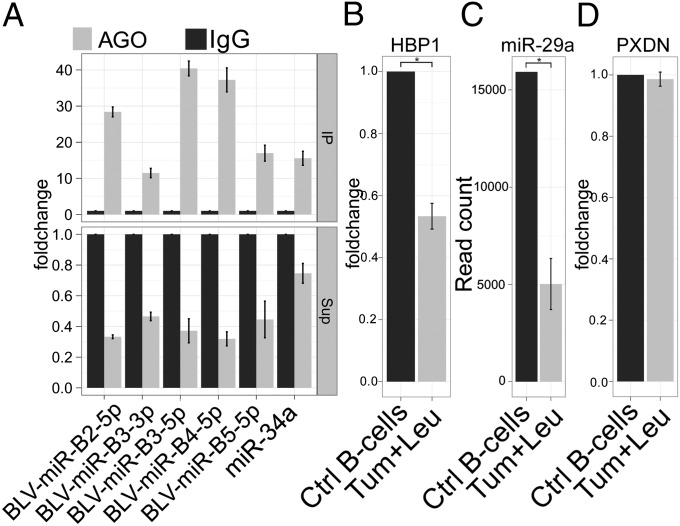
Because miRNAs must be associated with RNA-induced silencing complex (RISC) to have repressive function, we tested the association of the BLV miRNAs with Argonaute (Ago) proteins, the core component of the RISC (25). Ago immunoprecipitation (IP) experiments showed a significant enrichment (10- to 40-fold) of the complete set of BLV miRNAs in the Ago precipitate, whereas the miRNA levels in IP supernatants showed a significant decrease (Fig. 4A). These results show that, in B-cell tumors, BLV miRNAs associate with Argonautes, consistent with a critical function in posttranscriptional silencing.
Fig. 4.
BLV miRNA association with RISC and target identification. (A) BLV miRNAs are associated with Argonaute: BLV miRNA and cellular miRNA 34a qRT-PCR of Ago-IP fractions (upper graph) or supernatant after IP (lower graph) of YR2 tumor B-cell lysates. Relative expression: enrichment of target RNA by Ago IP over background IgG IP. (B) Microarray analysis showing decreased HBP1 in malignant B cells isolated from leukemic sheep (Leu + Tum) (7). Mean expression of twelve tumor samples to control purified B-cell pool (n = 3). (C) HTS showing decreased miR-29a–mapping read numbers in B cells from 10 leukemic relative to 3 uninfected sheep (DE-Seq). (D) Microarray analysis of PXDN as described in B shows no significant decrease compared with control B cells. *P < 0.001. Data are represented as mean fold change ± SD.
BLV miRNA Targeting of Host Cell mRNA: Investigation of HBP1 and PXDN in Primary Tumors.
HTS analysis of BLV-induced tumors confirmed that BLV-miR-B4-3p shares perfect nucleotide identity with the seed (positions 2–7) of the bovine miR-29 family (17). Interestingly, miR-29a down-regulates tumor suppressor HMG-box transcription factor 1 (HBP1) in human B-cell malignancies (26). We analyzed microarray transcriptomic data and found a twofold decrease in HBP1 expression (P value: 8 × 10−12) in primary B-cell tumors relative to B cells isolated from healthy sheep (Fig. 4B). Interestingly, both HTS and qRT-PCR analysis revealed a threefold decrease (P value: 0.0004) of miR-29a in ovine tumor B cells compared to controls, suggesting that BLV-miR-B4-3p and not cellular miR-29a may be responsible for HBP1 repression (Fig. 4C). In contrast, expression of peroxidasin homolog (PXDN), the second mRNA shown to be negatively regulated by BLV-miR-B4-3p in bovine 3′-UTR reporter assays and decreased by miR-29a overexpression in murine studies (17), was not altered in ovine tumors (Fig. 4D). These data point to HBP1 as a potential in vivo target but exclude PXDN. We conclude that miRNA target identification must take place in vivo, in the relevant host and cell type.
HTLV-1 and ATL.
We first observed that the predicted RNA secondary structure of the region located between the Env CDS and tax exon 2 in the proviral genome of the deltaretroviruses HTLV-1, HTLV-2, and simian T-cell leukemia virus (STLV) did not show typical miRNA-compatible hairpin structures (Fig. S7). Deep sequencing of leukemic CD4+ T cells from three ATL patients generated 20,120,958 reads per sample on average, of which 795,976 (4%) represent known cellular miRNAs, yet none mapped the HTLV-1 genome (NCBI NC_001436). Together, these computational and HTS data strongly suggest that, unlike BLV, HTLV-1 does not encode miRNA-like products from this particular proviral region in malignant ATL cells.
Discussion
Deep sequencing revealed the consistent and abundant in vivo expression of 10 miRNAs processed from a cluster of five hairpin structures. The BLV miRNA cluster is largely predominant in tumors, with ∼40% of the total cellular miRNA pool being of viral origin. At the individual level, the five mature BLV miRNAs range within the 15 most abundant miRNAs in malignant B cells. It is worth noting that some of them reach extremely elevated expression levels (>80,000 copies per cell), consistent with a robust regulatory capacity. BLV miRNAs are undergoing purifying selection that is stronger than that of a protein-coding sequence such as Tax, which implies an essential function. Our findings in primary tumors assign biological significance to the recent independent observation showing that BLV is capable of encoding miRNAs in vitro (17).
The identification of BLV miRNAs in tumors is remarkable for two reasons. First, the virus was assumed to be completely silent in leukemic B cells. BLV miRNAs produced in tumors do not fit the canonical Pol II-Drosha-dependent scheme of miRNA production. Their dependence on Pol III allows BLV to sustain miRNA expression while keeping Pol II protein-coding genes turned off. This strategy enlightens the hitherto puzzling BLV silencing dilemma. Second, BLV miRNAs are produced from a proviral region that is not required for in vivo infectivity. Retroviruses exploit their entire small (<10 kb) and genetically compact genome to achieve full functionality. So far, BLV was an exception to this rule. A significant portion of the BLV genome (650 bp, ∼7%) was found dispensable for viral infectivity in sheep and therefore assumed to lack a function. (2, 18, 19). The observation that abundantly expressed miRNAs lie within this segment and the remarkable finding that they undergo strong purifying selection assign functionality to this particular region.
Our data in primary tumors provide evidence that BLV miRNAs operate as independent self-sufficient transcriptional units in vivo. The characterization of viral miRNA biogenesis subproducts demonstrated the absence of moRs, which contrasts dramatically with the Drosha-confident signature observed for canonical miRNAs. Consistent with this, the depth of small RNA analysis permitted the identification of five BLV premiRNAs that possess the hallmarks of Pol III products: two typical type-II Pol III intragenic TFIII-c–interacting promoter elements located downstream of the transcription start site, and a poly-T–rich termination signal at the 3′ end of each precursor (17, 27). Transfection experiments with individual BLV premiRNA vectors and α-Amanitin treatment of tumor B cells reinforced this line of evidence. Of note, rescuing Pol II-driven viral mRNA transcription in malignant cells did not significantly alter the BLV miRNA levels, consistent with the conclusion that Pol III-mediated BLV miRNA production is independent of the transcriptional regulation of protein-coding viral information. Recent Chip-seq studies have revealed unexpected levels of complexity for Pol III regulation, including overlap with Pol II transcription (28). In B-cell tumors, the 5′-LTR–associated Pol II machinery upstream of the active Pol III-transcribed miRNA cluster appeared to have a minimal impact on miRNA transcription, consistent with epigenetic studies of Pol III target genes throughout the human genome (29).
Besides proviruses with intact coding potential, we found extensively deleted proviral genomes that were compromised in their ability to produce mRNAs but had kept an intact miRNA region. T1345 is a noticeable example. Apart from central proviral sequences, 5′-LTR control elements and 3′-located ORFs often carry genetic alterations that compromise transcription or critical protein function in tumors. In contrast, deletions that affect the BLV miRNA region were not identified, nor did we find a single example in which one or several BLV miRNAs were absent among 45 examined primary tumors. Among the BLV-encoded products, Tax attracted by far the most attention. Tax has been initially identified as the major viral oncoprotein. However, Tax expression is turned off in malignant B cells without affecting the tumor properties, suggesting that mechanisms underlying tumor maintenance do no longer require Tax but may depend on so far unidentified factors. We speculate that the tight silencing of Tax/BLV in malignant B cells reflects a viral strategy to escape immune surveillance. The transcriptionally self-sufficient and nonimmunogenic miRNAs in contrast can operate in the complete absence of Pol II-expressed viral immunogenic proteins. Of note, Tax is the major target of cytotoxic T lymphocytes (CTLs) in vivo, yet Tax-specific CTLs are not sufficient to prevent leukemia onset (30). Interestingly, the Pol III strategy used for miRNA processing grants BLV replication—essential for the virus life cycle—as mRNA transcripts will not be cleaved by this Drosha-independent process. Finally, significant levels of the BLV miRNA cluster were detected in asymptomatic animals, and this expression was not restricted to the pretumoral B-cell clone. It remains unclear whether transformed B cells are addicted to BLV miRNAs. However, (i) their abundant expression, (ii) association with Argonautes, and (iii) the strong purifying selection signature strongly suggest they have acquired important biological targets that they regulate to a substantial degree.
We consistently found the tumor suppressor HBP1 down-regulated in ovine tumors. Interestingly, HBP1 down-regulation in B-cell tumors is consistent with findings in murine and human leukemia in which miRNA-29a is the silencing trigger (26, 31, 32). Although we do not present conclusive evidence for a causal effect of miR-B4-3p, the reduced expression of miRNA-29a, the corresponding endogenous miRNA, suggests that its role is taken over by miR-B4-3p. The tumor suppressor PXDN, in contrast, was not confirmed as an in vivo target, although in vitro reporter assays and studies in mice were suggestive of this mechanism (17, 32). This underscores the importance of conducting miRNA target search in vivo, in the relevant host/cell type, keeping in mind that BLV miRNAs may act in combination with each other. Finally, BLV may use miRNAs to induce or maintain viral silencing. Viral miRNAs that contribute to latency have been mainly identified in DNA viruses and their production relies on Pol II (33–35) with the exception of murine herpes virus 68 (MHV68) miRNAs (36). Interestingly, several studies have revealed the existence of retroviral HIV-1–encoded miRNAs. Klase et al. (37) showed that processing of the HIV-1 TAR element creates a miRNA that contributes to viral latency. Further work is in progress to identify the in vivo target repertoire of the complete BLV miRNA cluster and elucidate its function. As a final note, deep sequencing of HTLV-1-induced ATLs did not uncover any comparable virus-encoded products, although we cannot rule out production of HTLV-1 miRNAs at other stages of the disease. Altogether BLV is thus far the only deltaretrovirus shown to encode tumor-associated miRNAs. Our findings in the BLV leukemia model lay the foundation for further functional studies of this particular class of miRNAs.
Materials and Methods
Small RNA libraries were prepared from 18- to 30-nt size-selected RNA (Illumina DGE Small RNA Sample Prep Kit). For capture of broader windows of RNA sizes, total RNA without prior size selection was processed using the Small RNA, version 1.5, Sample Prep Kit, and dsDNA libraries (120-200 bp) were PAGE-purified. Libraries were sequenced on GA II for 36 or 76 cycles (Illumina). miRNAs were quantified using custom TaqMan Stem–Loop MicroRNA assays (ABI) or QuantiMir qRT-PCR (SBI). Full detailed methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Groupe Interdisciplinaire Génoprotéomique Appliquée–Geno-Transcriptomics Platform, Benoit Hennuy for technical help, Raymond Cesaire (Centre Hospitalier Universitaire Fort de France) for ATL samples, and F. Kashanchi and K. Kehn-Hall (George Mason University) for Dicer and TRBP KO cell lines. This work was supported by Fonds National de la Recherche Scientifique, International Brachet Stiftung, Fondation Lambeau-Marteaux, Les Amis de l’Institut Bordet, Télévie Grants (to N.R. and M.M.). K.D. is supported by Fonds de la Recherche Scientifique Médicale, and F.M. by Institut National de la Santé et de la Recherche Médicale. A.V.d.B. is Collaborateur Scientifique Télévie.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE42316).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213842110/-/DCSupplemental.
References
- 1.Burny A, et al. Bovine Leukemia Virus: Biology and mode of transformation. In: Minson AC, Neil JC, McRae MA, editors. Viruses and Cancer. Cambridge, UK: Cambridge Univ Press; 1994. pp. 313–334. [Google Scholar]
- 2.Gillet N, et al. Mechanisms of leukemogenesis induced by bovine leukemia virus: Prospects for novel anti-retroviral therapies in human. Retrovirology. 2007;4:18. doi: 10.1186/1742-4690-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mammerickx M, Palm R, Portetelle D, Burny A. Experimental transmission of enzootic bovine leukosis to sheep: Latency period of the tumoral disease. Leukemia. 1988;2(2):103–107. [PubMed] [Google Scholar]
- 4.Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7(4):270–280. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- 5.Moulés V, et al. Fate of premalignant clones during the asymptomatic phase preceding lymphoid malignancy. Cancer Res. 2005;65(4):1234–1243. doi: 10.1158/0008-5472.CAN-04-1834. [DOI] [PubMed] [Google Scholar]
- 6.Willems L, et al. Cooperation between bovine leukaemia virus transactivator protein and Ha-ras oncogene product in cellular transformation. EMBO J. 1990;9(5):1577–1581. doi: 10.1002/j.1460-2075.1990.tb08277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klener P, et al. Insights into gene expression changes impacting B-cell transformation: Cross-species microarray analysis of bovine leukemia virus tax-responsive genes in ovine B cells. J Virol. 2006;80(4):1922–1938. doi: 10.1128/JVI.80.4.1922-1938.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szynal M, et al. Disruption of B-cell homeostatic control mediated by the BLV-Tax oncoprotein: Association with the upregulation of Bcl-2 and signaling through NF-kappaB. Oncogene. 2003;22(29):4531–4542. doi: 10.1038/sj.onc.1206546. [DOI] [PubMed] [Google Scholar]
- 9.Kettmann R, et al. Leukemogenesis by bovine leukemia virus: Proviral DNA integration and lack of RNA expression of viral long terminal repeat and 3′ proximate cellular sequences. Proc Natl Acad Sci USA. 1982;79(8):2465–2469. doi: 10.1073/pnas.79.8.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van den Broeke A, et al. Even transcriptionally competent proviruses are silent in bovine leukemia virus-induced sheep tumor cells. Proc Natl Acad Sci USA. 1988;85(23):9263–9267. doi: 10.1073/pnas.85.23.9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Den Broeke A, et al. In vivo rescue of a silent tax-deficient bovine leukemia virus from a tumor-derived ovine B-cell line by recombination with a retrovirally transduced wild-type tax gene. J Virol. 1999;73(2):1054–1065. doi: 10.1128/jvi.73.2.1054-1065.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merimi M, et al. Complete suppression of viral gene expression is associated with the onset and progression of lymphoid malignancy: Observations in bovine leukemia virus-infected sheep. Retrovirology. 2007;4:51. doi: 10.1186/1742-4690-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merimi M, et al. Suppression of viral gene expression in bovine leukemia virus-associated B-cell malignancy: Interplay of epigenetic modifications leading to chromatin with a repressive histone code. J Virol. 2007;81(11):5929–5939. doi: 10.1128/JVI.02606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colin L, et al. Chromatin disruption in the promoter of bovine leukemia virus during transcriptional activation. Nucleic Acids Res. 2011;39(22):9559–9573. doi: 10.1093/nar/gkr671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willems L, et al. In vivo infection of sheep by bovine leukemia virus mutants. J Virol. 1993;67(7):4078–4085. doi: 10.1128/jvi.67.7.4078-4085.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Florins A, et al. Even attenuated bovine leukemia virus proviruses can be pathogenic in sheep. J Virol. 2007;81(18):10195–10200. doi: 10.1128/JVI.01058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kincaid RP, Burke JM, Sullivan CS. RNA virus microRNA that mimics a B-cell oncomiR. Proc Natl Acad Sci USA. 2012;109(8):3077–3082. doi: 10.1073/pnas.1116107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ventura A, Jacks T. MicroRNAs and cancer: Short RNAs go a long way. Cell. 2009;136(4):586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42(8):1348–1354. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ragan C, Zuker M, Ragan MA. Quantitative prediction of miRNA-mRNA interaction based on equilibrium concentrations. PLoS Comput Biol. 2011;7(2):e1001090. doi: 10.1371/journal.pcbi.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 22.Han J, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125(5):887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 23.Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12(12):846–860. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- 24.Watashi K, Yeung ML, Starost MF, Hosmane RS, Jeang KT. Identification of small molecules that suppress microRNA function and reverse tumorigenesis. J Biol Chem. 2010;285(32):24707–24716. doi: 10.1074/jbc.M109.062976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutvagner G, Simard MJ. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9(1):22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 26.Han YC, et al. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med. 2010;207(3):475–489. doi: 10.1084/jem.20090831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santanam U, et al. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proc Natl Acad Sci USA. 2010;107(27):12210–12215. doi: 10.1073/pnas.1007186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orioli A, Pascali C, Pagano A, Teichmann M, Dieci G. RNA polymerase III transcription control elements: Themes and variations. Gene. 2012;493(2):185–194. doi: 10.1016/j.gene.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 29.White RJ. Transcription by RNA polymerase III: More complex than we thought. Nat Rev Genet. 2011;12(7):459–463. doi: 10.1038/nrg3001. [DOI] [PubMed] [Google Scholar]
- 30.Barski A, et al. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol. 2010;17(5):629–634. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van den Broeke A, et al. Cytotoxic responses to BLV tax oncoprotein do not prevent leukemogenesis in sheep. Leuk Res. 2010;34(12):1663–1669. doi: 10.1016/j.leukres.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Pekarsky Y, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66(24):11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 33.Cullen BR. Five questions about viruses and microRNAs. PLoS Pathog. 2010;6(2):e1000787. doi: 10.1371/journal.ppat.1000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umbach JL, et al. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454(7205):780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011;411(2):325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bogerd HP, et al. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral microRNAs. Mol Cell. 2010;37(1):135–142. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klase Z, et al. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.