Abstract
EG1 is a modular glycoside hydrolase family 5 endoglucanase from Volvariella volvacea consisting of an N-terminal carbohydrate-binding module (CBM1) and a catalytic domain (CD). The ratios of soluble to insoluble reducing sugar produced from filter paper after 8 and 24 h of exposure to EG1 were 6.66 and 8.56, respectively, suggesting that it is a processive endoglucanase. Three derivatives of EG1 containing a core domain only or additional CBMs were constructed in order to evaluate the contribution of the CBM to the processivity and enzymatic mode of EG1 under stationary and agitated conditions. All four enzymatic forms exhibited the same mode of action on both soluble and insoluble cellulosic substrates with cellobiose as a main end product. An additional CBM fused at either the N or C terminus reduced specific activity toward soluble and insoluble celluloses under stationary reaction conditions. Deletion of the CBM significantly decreased enzyme processivity. Insertion of an additional CBM also resulted in a dramatic decrease in processivity in enzyme-substrate reaction mixtures incubated for 0.5 h, but this effect was reversed when reactions were allowed to proceed for longer periods (24 h). Further significant differences were observed in the substrate adsorption/desorption patterns of EG1 and enzyme derivatives equipped with an additional CBM under agitated reaction conditions. An additional family 1 CBM improved EG1 processivity on insoluble cellulose under highly agitated conditions. Our data indicate a strong link between high adsorption levels and low desorption levels in the processivity of EG1 and possibly other processive endoglucanses.
INTRODUCTION
Cellulose is the most abundant biopolymer on Earth and therefore represents both a vast carbon source for cellulolytic microorganisms in the biosphere and a renewable feedstock for industrial-scale conversion into fuels and other products. Efficient hydrolysis of cellulose depends on the synergistic action of three classes of enzymes, namely, cellobiohydrolases (CBHs; EC 3.2.1.91), endoglucanases (EGs; EC 3.2.1.4), and β-glucosidases (EC 3.2.1.21) (1). Processive CBHs are the major components of most cellulolytic systems and are responsible for the degradation of crystalline cellulose (2). Processivity is a feature common to many cellobiohydrolases and is thought to be a critical strategy for improving the catalytic efficiency for hydrolysis of crystalline substrates (3). Structure analyses have revealed that CBHs such as CBHI and CBHII from Trichoderma reesei have enclosed active-site tunnels for substrate binding and catalysis (4, 5). A single glucan chain enters the tunnel from one end, and disaccharides are cleaved off at the catalytic center during its passage.
EGs are typically nonprocessive enzymes that are expressed in smaller amounts and assist CBHs by randomly attacking internal sites in the cellulose chain, thereby generating new chain ends. Unlike CBHs, classical EGs, such as EGI of Fusarium oxysporum, have a relatively open active-site cleft (6), thereby permitting them to cleave bonds in the middle of polymer chains. However, some bacteria and fungi synthesize processive EGs, which cleave cellulose internally and also release soluble oligosaccharides before detaching from the polysaccharide (7–9). Processive EGs belong almost exclusively to the GH9 family of enzymes associated with bacterial systems (10). However, processive EGs belonging to the GH5 family, produced by the brown rot basidiomycete Gloeophyllum trabeum (11) and the marine bacterium Saccharophagus degradans 2-40 (12), have also been reported. Since both these cellulolytic systems lack significant CBH activity, processive EGs are thought to be functionally equivalent to the EGs and CBHs that together comprise other cellulolytic systems, thereby providing a CBH-independent cellulose-degrading mechanism.
In common with other cellulases, processive EGs have modular architectures generally comprising catalytic domains (CD) that are joined, via linker regions, to noncatalytic carbohydrate-binding modules (CBMs). CBMs have since been shown to be critical for the processivity of processive GH9 EGs (8, 13–15). For example, crystalline cellulose degradation and processivity were abolished by deletion of CBM3c of Thermomonospora fusca (16), the main function of which is to disrupt the cellulose chains in the crystalline cellulose substrate and to feed a single chain into the active site of the CD (8). Processive GH5 EGs from S. degradans 2-40 are also linked via a flexible linker to CBM6, which was expected to exhibit properties typical of a type B CBM such as CBM3c. However, processivity associated with GH5 EGs from S. degradans 2-40 appears to be independent of the CBM, since the processivity ratio of each enzyme was unaffected by the absence of CBM6 (12).
Volvariella volvacea is an atypical white rot basidiomycetous fungus, the cellulolytic system of which includes CBHs, EGs, and β-glucosidases (18, 19). One EG isoform, EG1, was previously characterized and shown to modify cellulose fibers (20). EG1 is a modular enzyme that contains a family 5 CD and a family 1 CBM. In this study, we investigated the processivity and mode of action of EG1 under stationary and agitated conditions and evaluated the contribution of the CBM to the processivity, catalytic activity, and enzymatic mode of the enzyme.
MATERIALS AND METHODS
Bacterial and yeast strains, growth media, and chemicals.
Pichia pastoris, strain KM71H, was used for the expression of EG1 and its derivatives. Escherichia coli DH5α was used for plasmid construction and propagation. Yeast growth media were prepared according to the Pichia expression system manual from Invitrogen. All chemicals were of reagent grade or higher and purchased from Sigma (St. Louis, MO) unless otherwise indicated. Regenerated amorphous cellulose (RAC) was prepared from Avicel as described by Zhang et al. (21).
Construction of plasmids.
The entire eg1 gene in plasmid pBluescript II KS-EG1 was used as the template for construction. The DNA fragment encoding the mature EG1 (amino acids 24 to 389; GenBank accession number AF329732) was amplified using plasmid pBluescript II KS-EG1 and the primer pair P1 (5′-AAAACTGCAGCCGTCCCAGTATGGGGACAAT-3′; PstI site underlined) and P2 (5′-GCTCTAGAGCCACGAATGGTTTCAAAGC-3′; XbaI site underlined). The truncated form that comprised the CD only was amplified by PCR with the primer pair P3 (5′-AAAACTGCAGGCGCCGGACCTACGACAA-3′; PstI site underlined) and P2 (5′-GCTCTAGAGCCACGAATGGTTTCAAAGC-3′; XbaI site underlined). These two amplified DNA fragments were digested with PstI and XbaI and ligated into similarly cut pPICZaB to generate the expression vectors pPICZaB-EG1 and pPICZaB-CD, respectively. pPICZaB-CBM-CBM-CD was constructed by inserting a fragment consisting of the CBM and a 25-amino-acid (GPTTTSSAPNPTSSGCPNATKFRFF) linker upstream of EG1. For this, the fragment encoding the CBM and linker was amplified by PCR using the primer pair P4 (5′-AAAACTGCAGCCGTCCCAGTATGGGGACAAT-3′; PstI site underlined) and P5 (5′-TTTCTGCAGCGAAGAATCTGAACTTGGTGGCA-3′; PstI site underlined). The fragment was then inserted into pPICZaB-EG1 after digestion with PstI. pPICZaB-CBM-CD-CBM was constructed by inserting a fragment consisting of the 25-amino-acid linker and CBM downstream of EG1. For this, the linker and CBM were amplified by PCR using the primer pair P6 (5′-AGCTTTCTAGAAGGCGCCGGACCTACGACAACCA-3′; XbaI site underlined) and P7 (5′-GAATCTGAACTTGGTGGCAT-3′) and the primer pair P8 (5′-ATGCCACCAAGTTCAGATTCGCCGTCCCAGTATGGGGACAAT-3′) and P9 (5′-TGTTCTAGAAAAGGCTGGCATTGGTGGTACCA-3′; XbaI site underlined), respectively. Then the fragment encoding linker and CBM was amplified by overlapping PCR using the primer pair P6 and P9 and the linker and CBM fragments as the template. The fragment was then inserted into pPICZaB-EG1 digested with XbaI. The scheme adopted for the construction of pPICZaB-CBM-CBM-CD and pPICZaB-CBM-CD-CBM is shown in Fig. S1 in the supplemental material. Fragments containing EG1, CD, CBM-CBM-CD, and CBM-CD-CBM in the expression vectors were all under the transcriptional control of the AOX1 (alcohol oxidase) promoter. All engineered EG1s were fitted with a 6-histidine tag to facilitate purification using affinity chromatography.
Enzyme expression and purification.
P. pastoris transformants were grown and harvested as previously described (9). Recombinant EG1, CD, CBM-CBM-CD, and CBM-CD-CBM, fitted with 6-histidine tags, were purified by affinity chromatography using nickel-nitrilotriacetic acid (Ni-NTA) agarose gel (Qiagen) according to the manufacturer's manual. Enzyme homogeneity and the molecular weights of purified EG1 and the three derivatives were estimated using 10% (wt/vol) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Circular dichroism spectra (CDS).
Spectra for EG1 and its derivatives (0.025 mg/ml) were recorded at 25°C over a range of 190 to 260 nm with a Chirascan circular dichroism spectrometer (Applied PhotoPhysics) using a 1-cm-path-length cuvette, a bandwidth of 1 nm, and a scanning speed of 120 nm min−1. Data were analyzed using CDNN CDS deconvolution software (Applied PhotoPhysics).
Protein concentration and assay of cellulase activity.
Protein was determined by measuring A280 values using extinction coefficients calculated from the predicted amino acid compositions (εEG1 = 101,670 M−1 cm−1, εCBM-CBM-CD = 120,620 M−1 cm−1, εCBM-CD-CBM = 120,620 M−1 cm−1). Cellulase activity on carboxymethylcellulose (CMC; Sigma) (low viscosity, 10 mg), regenerated amorphous cellulose (RAC) (5 mg), and filter paper (FP; Whatman no. 1 strip) (50 mg) was assayed in reaction mixtures (1.5 ml total volume) containing 100 mM potassium phosphate buffer (pH 7.5), enzyme, and substrate, incubated at 50°C for 30 and 60 min in an orbital shaking water bath operated at 0 to 400 rpm. Reducing sugar released was determined by the Somogyi-Nelson method. All assays were performed in triplicate, and one unit of activity was defined as the amount of enzyme that released 1 μmol of glucose equivalents from the substrate per min.
Assay of binding and desorption capacities.
Assays of the binding capacity of EG1 and its derivatives to cellulosic substrates were carried out in low-protein-binding Eppendorf tubes containing different concentrations of recombinant protein and 10 mg of cellulosic substrate in a final volume of 0.5 ml of 50 mM potassium phosphate buffer (pH 7.5). Reaction mixtures were incubated at 4°C with constant shaking in an inverted-action shaker (150 inversions per min). After 1 h, cellulosic substrates were removed by centrifugation (13,000 × g, 4°C), and the level of unbound protein remaining in the supernatant was determined by measuring the absorbance at 280 nm. Data were subjected to nonlinear regression analysis using a standard single-site binding model (GraphPad Prism, version 2.01).
Adsorption and desorption tests on EG1 and its derivatives involving FP were carried out under the same conditions as described above except that reaction mixtures containing a fixed amount of protein (0.685 mg/ml) were incubated at 4°C in an orbital shaker water bath operated at 100 to 400 rpm. After 1 h, reaction mixtures were diluted with one volume of 50 mM potassium phosphate buffer (pH 7.5) and incubated for a further 60 min as before. For greater accuracy, unbound protein remaining in the supernatant after the two incubation periods was measured using a bicinchoninic acid (BCA) protein assay kit (Pierce). Although the BCA assay also detects solubilized sugar, our experiments confirmed there were no hydrolysis products present after 60 min at 4°C. All measurements were performed at least in triplicate, and the bound and desorbed protein levels were calculated according to the method of Azevedo et al. (22).
Viscosity assay.
Reaction mixtures (15 ml total volume) containing purified enzyme (0.25 μg/ml)–2% low-viscosity CMC–100 mM potassium phosphate buffer (pH 7.5) were incubated at 50°C. Samples were taken periodically and boiled for 5 min, and viscosity values were determined based on the time of outflow at 23°C using an Ubbelohde viscometer tube (Fisher Scientific).
Analysis of hydrolysis products by TLC and HPAEC-PAD.
Hydrolysis of RAC and FP by EG1 and its derivatives was carried out for 24 h under the same conditions used for the activity assay. Ampicillin (25 μg/ml) and zeocin (25 μg/ml) were added to reaction mixtures to prevent microbial contamination. Samples (50 μl) were withdrawn at regular intervals, heated for 5 min in a boiling water bath to terminate the reaction, and centrifuged at 11,000 × g for 10 min. Supernatants were analyzed for monosaccharides, disaccharides, and oligosaccharides by thin-layer chromatography (TLC) according to a method described by Zhang et al. (23). Hydrolysis of cellooligosaccharides (cellotriose, cellotetraose, cellopentaose) by EG1 and its derivatives was carried according to a method described by Yoda et al. (24). Products were analyzed by TLC after 2 h of incubation at 50°C. FP hydrolysis products were quantitatively analyzed at 30°C using a Carbo-Pac PA200 column (3 by 250 mm) fitted to an ICS-3000 high-performance anion exchange chromatography system (Dionex) with pulsed amperometric detection (HPAEC-PAD). A dual mobile-phase system (A, 100 mM NaOH; B, 500 mM sodium acetate) was applied, and saccharides were eluted using a linear sodium acetate gradient (B: 0 to 24% in 40 min; 0.3 ml/min), followed by elution with 100 mM NaOH (15 min; 0.3 ml/min).
Processivity.
Processivity was determined from the distribution of reducing sugars between the soluble and insoluble products derived from FP as described by Irwin et al. (25). Enzyme reactions were performed under the standard conditions, and soluble and insoluble fractions were separated by centrifugation. Levels of reducing sugar in the supernatant fractions and in the remaining FP strips (after the latter had been washed three times with reaction buffer and resuspended in the initial volume of buffer) were determined by the Somogyi-Nelson method. The effect of different agitation speeds on processivity was determined by incubating reaction mixtures for 1 h on an orbital shaker water bath operated at 0 to 400 rpm.
RESULTS
Construction and production of EG1 and its derivatives.
Schematic representations of EG1 and its derivatives are shown in Fig. 1. All the engineered forms of EG1 were successfully expressed in P. pastoris and purified by affinity chromatography using Ni-trap columns. SDS-PAGE revealed that all purified enzymes migrated as single dominant bands with molecular masses of 48 kDa, 55 kDa, 60 kDa, and 45 kDa, corresponding to EG1, CBM-CBM-CD, CBM-CD-CBM, and CD, respectively (Fig. 2). These were higher than the predicted values (42.5 kDa, 49.2 kDa, 49.2 kDa, and 35.7 kDa for EG1, CBM-CBM-CD, CBM-CD-CBM, and CD, respectively), due perhaps to glycosylation.
Fig 1.
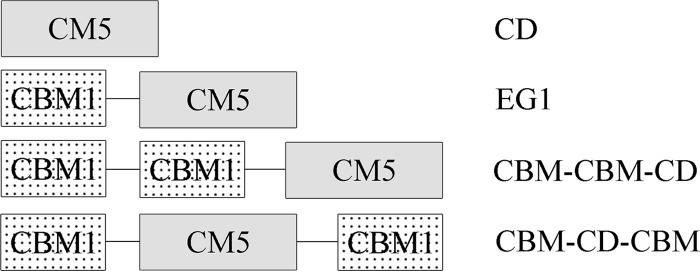
Schematic structures of EG1 and its derivatives.
Fig 2.
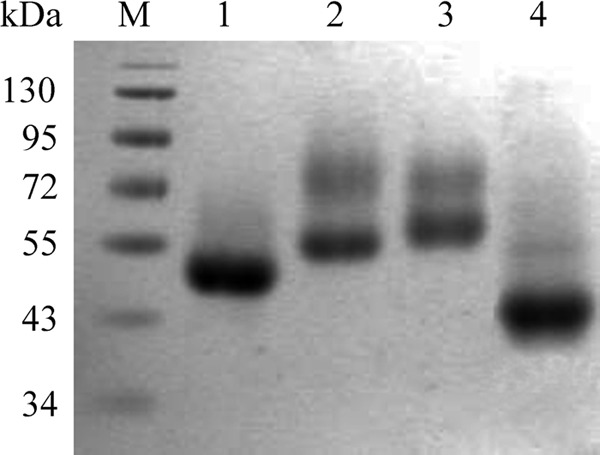
SDS-PAGE of EG1 and its derivatives. Lanes: M, protein markers; 1, EG1; 2, CBM-CBM-CD; 3, CBM-CD-CBM; 4, CD.
Circular dichroism spectra (CDS) were determined to eliminate the possibility that improper folding of CBM-CD-CBM and CD-CBM-CBM was responsible for the observed decreases in enzyme activity on CMC. Two clear minima at 210 and 222 nm were evident in all the spectra (see Fig. S2A and B in the supplemental material), in accordance with GH5 EG1 having both α-helix and β-sheet features. CBM-CBM-CD and CBM-CD-CBM spectra demonstrated relatively high and low contents of anti-parallel β-sheet and α-helix structures, respectively. Overall, CDS confirmed that the recombinant proteins exhibited marked secondary structure and were properly folded.
Activity and binding affinity of EG1 and its derivatives.
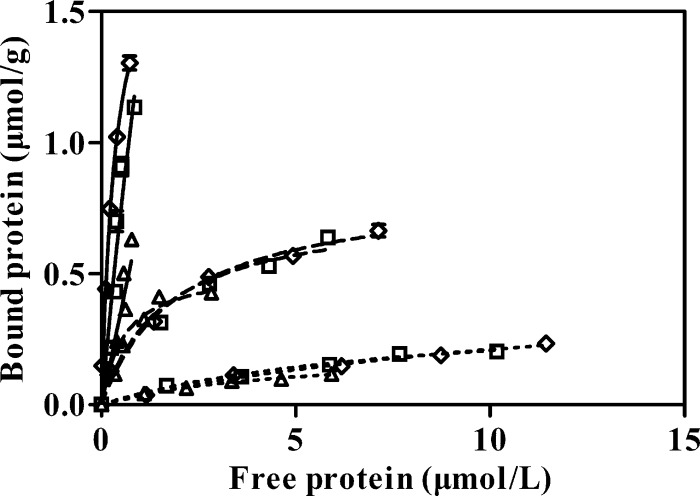
Both CBM-CBM-CD and CBM-CD-CBM displayed markedly lower specific activities than native EG1 tested using the insoluble substrates RAC (31.9% and 23.3%, respectively) and FP (50.7% and 38%, respectively) (Table 1). Specific activities recorded with the soluble substrate CMC were also lower (85.5% and 77.1% lower than the values for EG1) (Table 1). However, evaluation of the binding capacities of the three enzyme forms for Avicel, RAC, and FP revealed similar binding patterns in each case (Fig. 3).
Table 1.
Specific activity of EG1, CBM-CBM-CD, and CBM-CD-CBM on different substrates
| Enzyme | Specific activity (IU/μmol) |
||
|---|---|---|---|
| CMC | RAC | FP | |
| EG1 | 1470.0 | 531.3 | 7.1 |
| CBM-CBM-CD | 1127.8 | 169.6 | 3.6 |
| CBM-CD-CBM | 1016.4 | 124.0 | 2.7 |
Fig 3.
Binding isotherms of EG1 and its derivatives for different cellulosic substrates. Reaction mixtures containing different concentrations of recombinant protein (EG1, 1.5 to 13.8 μmol/liter; CBM-CBM-CD, 2.4 to 12.2 μmol/liter; CBM-CD-CBM, 1.5 to 7.1 μmol/liter) and 10 mg cellulosic substrate in a final volume of 0.5 ml of 50 mM potassium phosphate buffer (pH 7.5) were incubated at 4°C with constant agitation in an inversion-action shaker (150 inversions per min). Symbols: ◇, EG1; □, CBM-CBM-CD; Δ, CBM-CD-CBM. Bold lines, RAC; broken lines, FP; dotted lines, Avicel PH-101 (Fluka).
Enzymatic action mode of EG1 and its derivatives.
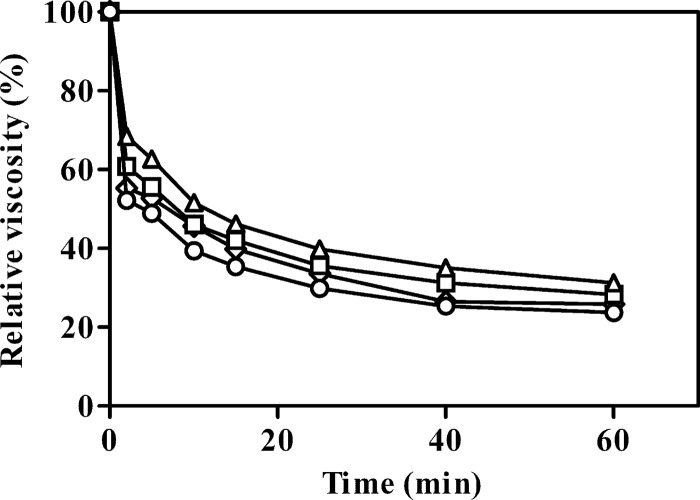
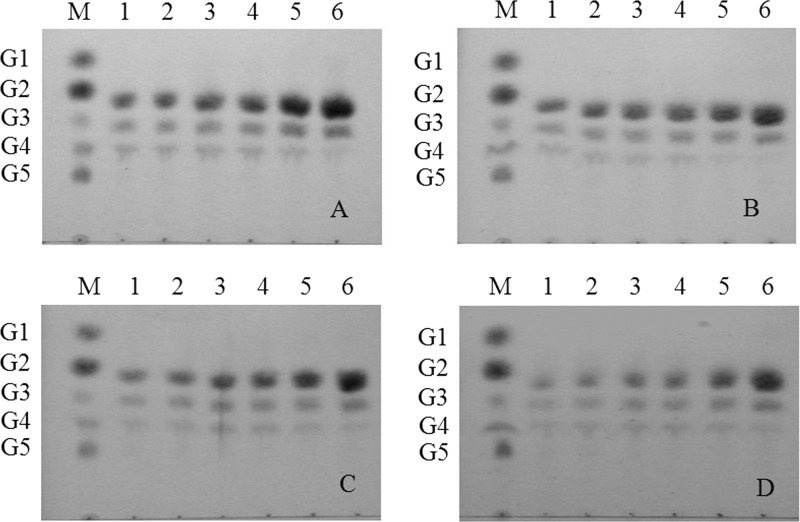
EG1 and its derivatives rapidly decreased the viscosity of CMC, indicating an endoaction hydrolytic mode (Fig. 4). TLC of the hydrolysis products released from insoluble celluloses and soluble cello-oligosaccharides revealed that all the enzyme forms displayed the same enzymatic mode of action on insoluble celluloses. After 24 h, cellobiose (G2) was the main product released from RAC, together with smaller quantities of cellotriose (G3) and cellotetraose (G4) (Fig. 5). None of the enzyme forms hydrolyzed cellotriose, whereas cellotetraose and cellopentaose were degraded into a mixture of cellobiose and cellotriose (Fig. 6). Quantitative analysis (using HPAEC-PAD) of the soluble hydrolysis products generated from FP following treatment with EG1 and its derivatives revealed a similar pattern (see Fig. S3 in the supplemental material). G2 was again the dominant product, and G3 and G4 were minor products (Table 2). G2, G3, and G4 mol percent values at 24 h ranged between 67.2 and 77.8%, 17.3 and 25.0%, and 4.3 and 7.8%, respectively. G2 levels generated from the hydrolysis of FP by EG1, CBM-CBM-CD, CBM-CD-CBM, and CD after 24 h were 6.61-, 10.41-, 5.30-, and 4.07-fold higher, respectively, than the levels recorded after 0.5 h.
Fig 4.
Viscosity of CMC solutions hydrolyzed by EG1 and its derivatives. Symbols: ◇, EG1; □, CBM-CBM-CD; Δ, CBM-CD-CBM; ○, CD.
Fig 5.
Thin-layer chromatography (TLC) of hydrolysis products from RAC hydrolyzed by EG1 (A), CD (B), CBM-CBM-CD (C), and CBM-CD-CBM (D). Lanes: M, glucose unit markers [glucose (G1), cellobiose (G2), cellotriose (G3), cellotetraose (G4), and cellopentaose (G5)]; 1 to 6, products after incubation for 0.5, 1, 1.5, 2, 6, and 24 h, respectively.
Fig 6.
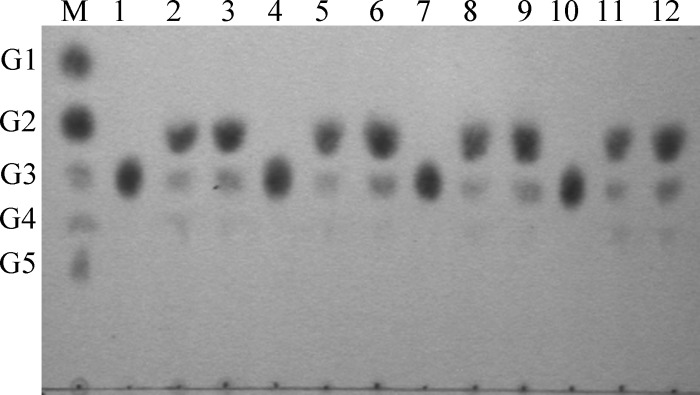
TLC of hydrolysis products from cello-oligosaccharides hydrolyzed by EG1 and its derivatives. Products released from oligoglucosides G3 to G5 by EG1 (lanes 1 to 3), CD (lanes 4 to 6), CBM-CBM-CD (lanes 7 to 9), and CBM-CD-CBM (lanes 10 to 12). Lane M: glucose unit markers [glucose (G1), cellobiose (G2), cellotriose (G3), cellotetraose (G4), and cellopentaose (G5)].
Table 2.
Oligosaccharide production following FP hydrolysis by EG1 and its derivativesa
| Protein | Oligosaccharide(s) | Production in μmol/liter (fold increase) or ratio at indicated time |
|||
|---|---|---|---|---|---|
| 0.5 h | 2 h | 8 h | 24 h | ||
| EG1 | G2 | 80.16 (1) | 252.82 (3.15) | 481.48 (6.01) | 529.57 (6.61) |
| G3 | 23.65 (1) | 60.71 (2.57) | 100.10 (4.23) | 117.29 (4.96) | |
| G4 | 6.62 (1) | 19.71 (2.98) | 30.75 (4.65) | 33.65 (5.09) | |
| G2/G3 | 3.39 | 4.16 | 4.81 | 4.52 | |
| CBM-CBM-CD | G2 | 32.25 (1) | 203.01 (6.29) | 280.69 (8.70) | 335.70 (10.41) |
| G3 | 11.18 (1) | 36.46 (3.26) | 60.63 (5.42) | 92.15 (8.24) | |
| G4 | 2.75 (1) | 10.58 (3.85) | 16.34 (5.94) | 22.28 (8.10) | |
| G2/G3 | 2.88 | 5.57 | 4.63 | 3.64 | |
| CBM-CD-CBM | G2 | 32.05 (1) | 71.40 (2.23) | 112.42 (3.51) | 169.76 (5.30) |
| G3 | 11.98 (1) | 27.74 (2.32) | 43.20 (3.61) | 63.28 (5.28) | |
| G4 | 3.29 (1) | 9.27 (2.82) | 13.82 (4.21) | 19.74 (6.01) | |
| G2/G3 | 2.68 | 2.57 | 2.60 | 2.68 | |
| CD | G2 | 59.19 (1) | 150.42 (2.54) | 194.22 (3.28) | 240.61 (4.07) |
| G3 | 19.03 (1) | 53.03 (2.79) | 66.71 (3.51) | 78.47 (4.12) | |
| G4 | 4.37 (1) | 12.59 (2.88) | 12.99 (2.97) | 14.21 (3.25) | |
| G2/G3 | 3.11 | 2.84 | 2.91 | 3.06 | |
G2, cellobiose; G3, cellotriose; G4, cellotetraose. Values in parentheses represent the fold increase values derived from the ratio of the molar amounts of oligosaccharide released at the indicated incubation time and at 0.5 h. G2/G3 values represent the fold increases derived from the ratio of the molar amounts of oligosaccharide released by G2 and G3.
Processivity of EG1 and its derivatives.
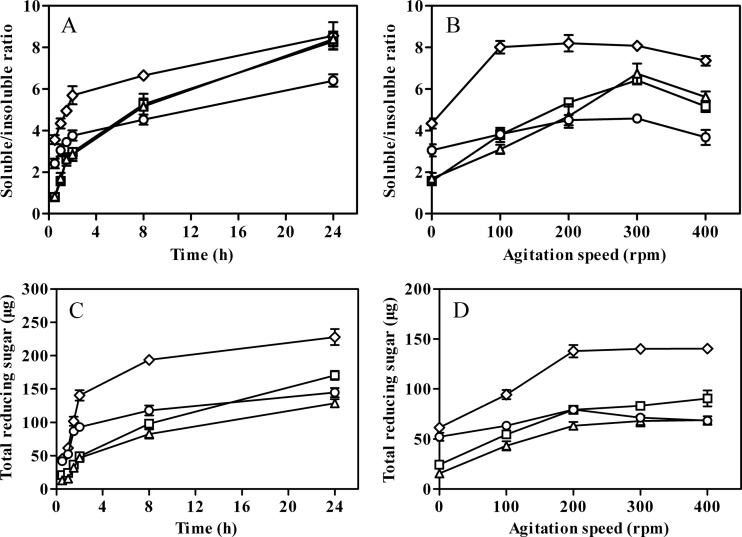
EG1 gave the highest soluble/insoluble product ratios under both stationary and agitated conditions. After 0.5 h of incubation, the ratio of soluble to insoluble products reached 3.56 in reaction mixtures containing EG1 and FP, increasing to 8.56 after 24 h (Fig. 7A). Corresponding values for CD, which lacked the family 1 CBM, were 2.42 and 6.40, respectively. Significantly lower processivity was observed during short-term (0.5 h) incubations in the case of enzyme derivatives with additional CBMs (0.79 and 0.83 for CBM-CBM-CD and CBM-CD-CBM, respectively). However, the ratios of soluble/insoluble products for both enzyme forms then increased markedly and reached values similar to EG1 after 24 h (Fig. 7A). The soluble/insoluble product ratios in reaction mixtures containing CD and FP ranged from 3.05 to 4.58 when agitation speeds were increased from 0 to 400 rpm, indicating that mechanical agitation had a moderate effect on CD processivity. Similarly, a 1.83-fold increase in processivity (4.34 versus 8.0) was observed for EG1 when the agitation level was increased from 0 to 100 rpm, but the soluble/insoluble product ratio decreased slightly at agitation speeds in excess of 200 rpm. In contrast, 4.09- and 3.99-fold increases in the soluble/insoluble product ratio were recorded for CBM-CBM-CD and CBM-CD-CBM, respectively, when the agitation level was increased from 0 to 300 rpm (Fig. 7B). Total levels of reducing sugar (soluble sugar plus reducing end groups generated in partially hydrolyzed, insoluble cellulose) in incubation mixtures containing filter paper under stationary and agitation conditions are shown in Fig. 7C and D.
Fig 7.
Processivity and reducing sugar released from FP by EG1 and its derivatives under stationary (A and C) and agitated (B and D) conditions. Symbols: ◇, EG1; □, CBM-CBM-CD; Δ, CBM-CD-CBM; ○, CD. Values shown are the means of the results of triplicate experiments ± standard errors of the means (SE).
Enzyme adsorption and desorption under agitated conditions.
The effects of different levels of agitation on enzyme adsorption to or desorption from FP are shown in Table 3. Adsorption of CD on FP was very low and completely reversible, whereas increased agitation resulted in slightly increased adsorption and desorption in the case of EG1. At low agitation speeds (100 rpm), enzyme derivatives with additional CBMs displayed lower adsorption but higher desorption capacities compared with EG1. When agitation speeds were increased, lower desorption capacities were observed for enzyme derivatives with additional CBMs whereas adsorption capacities differed only marginally.
Table 3.
Binding and desorption capacities of EG1 and its derivatives at different levels of mechanical agitation
| Protein | Protein or desorption category | Amt ± SE of bound or desorbed protein (mg) or % desorption at indicated speed (rpm)a |
|||
|---|---|---|---|---|---|
| 100 | 200 | 300 | 400 | ||
| EG1 | Protein bound after 1 h | 0.225 ± 0.010 | 0.238 ± 0.009 | 0.243 ± 0.004 | 0.255 ± 0.011 |
| Protein bound after 2 h | 0.202 ± 0.007 | 0.214 ± 0.008 | 0.216 ± 0.006 | 0.227 ± 0.008 | |
| Desorbed protein | 0.023 | 0.024 | 0.027 | 0.031 | |
| Desorption (%) | 10.2 | 10.1 | 11.1 | 12.2 | |
| CBM-CBM-CD | Protein bound after 1 h | 0.216 ± 0.004 | 0.203 ± 0.006 | 0.198 ± 0.008 | 0.192 ± 0.005 |
| Protein bound after 2 h | 0.153 ± 0.011 | 0.175 ± 0.009 | 0.185 ± 0.002 | 0.178 ± 0.010 | |
| Desorbed protein | 0.063 | 0.028 | 0.013 | 0.019 | |
| Desorption (%) | 29.2 | 13.8 | 6.6 | 9.9 | |
| CBM-CD-CBM | Protein bound after 1 h | 0.182 ± 0.003 | 0.184 ± 0.006 | 0.194 ± 0.007 | 0.181 ± 0.009 |
| Protein bound after 2 h | 0.146 ± 0.005 | 0.149 ± 0.004 | 0.165 ± 0.003 | 0.166 ± 0.005 | |
| Desorbed protein | 0.038 | 0.035 | 0.029 | 0.015 | |
| Desorption (%) | 20.9 | 19.2 | 14.9 | 18.5 | |
| CD | Protein bound after 1 h | 0.072 ± 0.003 | 0.079 ± 0.005 | 0.069 ± 0.002 | 0.061 ± 0.006 |
| Protein bound after 2 h | |||||
| Desorbed protein | 0.072 | 0.079 | 0.069 | 0.061 | |
| Desorption (%) | 100 | 100 | 100 | 100 | |
The binding and desorption values shown represent the means ± standard errors of the results of triplicate experiments.
DISCUSSION
In this paper, we describe the effects of additional CBMs and mechanical agitation on the processivity and enzymatic mode of a glycoside hydrolase family 5 endoglucanase (EG1) from V. volvacea. Of the cellulosic substrates tested, the native enzyme exhibited highest activity on CMC and was less active toward RAC and FP. EG1 and the derived forms rapidly decreased the viscosity of CMC, indicating an “endo” mode of action. However, unlike classical endoglucanases that randomly cleave cellulose polymers to form a variety of degradation products, the primary action of EG1 appeared to be the release of cellobiose as the major end product from insoluble cellulose substrates such as RAC and FP. Therefore, EG1 cleavage patterns on insoluble cellulose are the same as those of CelI from Clostridium thermocellum and Cel5H from S. degradans, both of which are also reported to release G2 as the major product from insoluble cellulose (7, 12). However, EG1 differs from a processive endoglucanase from Clostridium phytofermentans (10), Cel9R, a major component in the cellulosome of C. thermocellum (26), and CelZ from Clostridium stercorarium (27), all three of which released G4 as the major end product. Processivity is usually measured by comparing the ratio of reducing end groups in the soluble and insoluble fractions resulting from enzyme hydrolysis of filter paper (9, 25). The assay is based on the proviso that the percentage of soluble reducing sugars released by processive endoglucanases increases with longer incubation times whereas the number of reducing end groups in the insoluble fraction remains basically constant (28). Ratios of soluble to insoluble products for processive endoglucanases acting on FP for 16 h were estimated to be within the range of 3.1 to 10.3 (8, 28). In this study, the ratios of soluble to insoluble products generated by EG1 from FP ranged between 3.56 and 8.56 depending on the reaction time. Taken together, these data strongly suggest that EG1 acts as a processive endoglucanase on insoluble substrates, exhibiting both “endo” (on CMC) and “exo” types of activity. Further support for the idea of processive endoglucanases playing bifunctional roles is provided by the GH5 processive endoglucanases identified in the marine bacterium S. degradans and the brown-rot basidiomycete G. trabeum. Both degrade crystalline cellulose, and yet no cellobiohydrolase activity has been detected in either organism. However, V. volvacea has a complete cellulolytic system that includes CBHs, EGs, and β-glucosidases. Furthermore, EG1 has a different modular architecture whereby the CD is linked to a family 1 CBM instead of CBM6 as in Cel5H. The enzyme also exhibits very low (less than 32%) amino acid sequence identity to other GH5 processive endoglucanases, suggesting that EG1 represents a novel class of processive EGs.
CBMs described so far have been classified into 64 families based on similarities in their primary and tertiary structures (http://www.cazy.org/Carbohydrate-Binding-Modules.html) and differ widely in terms of binding kinetics and specificity (17, 29). Family 1 CBMs are found almost exclusively in fungi and are distinct in size and structure from CBMs assigned to other families. As type A CBMs, they share similar conformations, with a flat, hydrophobic surface containing three key aromatic residues that facilitate binding to crystalline cellulose. The surface containing these aromatic residues is generally regarded as hydrophobic and directs the enzyme to matching surfaces on crystalline cellulose 1β (30), which exposes the faces of β-d-glucopyranose rings in the chair conformation.
In order to clarify the role of the family 1 CBM in the processive hydrolysis of cellulose by EG1, we compared native EG1 with enzyme derivatives containing the CD only or additional CBMs in terms of catalytic activity, processivity, and mode of enzymatic action. Although the cleavage patterns on soluble cellodextrins and insoluble cellulose remained unchanged, removal of the CBM significantly reduced the ratio of soluble to insoluble products from FP, indicating a critical role for CBM in EG1 processivity. However, additional CBMs inserted on either side of the CD had an adverse effect on EG1 activity and processivity under stationary conditions, unlike bacterial glycoside hydrolases, where multiple CBMs act in synergy to bind the enzyme to the target ligand, thereby increasing the affinity and activity of the enzyme for the polysaccharide (29, 31). However, additional CBMs may not always enhance bacterial endoglucanase activity on solid substrates (32). It should be noted that, unlike the multiple CBMs associated with bacterial glycoside hydrolases, fungal cellulases usually contain only one family 1 CBM, indicating that additional CBMs are not required for efficient cellulose hydrolysis in natural environments. Recently, a gene encoding an atypical multimodular glycoside hydrolase family 45 endoglucanase (designated PpCel45A) bearing five different family 1 CBMs was identified in the P. pastoris GS115 genome (33). However, both binding of the enzyme to crystalline cellulose and hydrolysis of crystalline cellulose and cellohexaose were substantially enhanced when only a single CBM was present rather than five. This effect may be due to steric hindrance, whereby multiple family 1 CBMs prevent the exposure of the flat, hydrophobic surface required for interaction with cellulose chains.
Hydrolytic attack on insoluble cellulosic materials by cellobiohydrolases is thought to involve initial adsorption of the enzyme onto crystalline regions of the cellulose surface, formation of protein-cellulose complexes, catalysis of the initial hydrolytic event to generate glucose (G1), cellobiose (G2), cellotriose (G3), or cellotetraose (G4), subsequent processive attacks along the chain generating only G2, and, finally, dissociation of the enzyme from the cellulose chain (34). Accordingly, the degree of processivity exhibited by cellobiohydrolases has been determined by comparing the ratio of [G2]/([G1] plus [G3]) or [G2]/([G3] plus [G4]) (8, 23). In addition to this proposed sequence of events, processive endoglucanases also engage in a nonprocessive random attack on the less crystalline regions of exposed single cellulose chains, followed by processive hydrolysis resulting in G2 production (35). This increased generation of G2 (together with G3 and G4) would account for the significant increase in the soluble/insoluble reducing sugar ratio recorded after 24 h of hydrolysis of FP by EG1 and its derivatives (Table 2). We propose that ensuing changes in adsorption/desorption patterns caused by adding a second CBM affects the initial interaction between EG1 and the insoluble cellulose substrate and leads to enhanced random hydrolytic attack and lower processivity during the early stages of hydrolysis which, in turn, account for the observed lower soluble oligosaccharide production (Table 2, Fig. 7A). Although G2/G3 ratios were used to estimate substrate-binding modes and/or processivity, it should be emphasized that these ratios may simply reflect different preferences for two initial binding modes, which release dimers or trimers, respectively (35, 36). Interestingly, the four enzyme variants exhibited clear differences in their G2/G3 ratios during the 24-h hydrolysis period (Table 2), supporting the idea of a role for CBMs in enzyme activity and processivity.
Although multiple studies have shown mechanical agitation to have profound effects on cellulase activity and enzyme adsorption to and desorption from solid substrates, we have found no reports relating this parameter to cellulase processivity. Cellulase hydrolysis by classical endoglucanases (with or without a CBM) can be achieved via rapid and constant adsorption and desorption. Consequently, the more readily reversibly bound EGV and EGV core enzymes from Humicola insolens exhibit higher activity than the less mobile CenA and CenA core enzymes from Cellulomonas fimi (22). At first glance, EG1 and derivatives with additional CBMs appeared to exhibit similar binding properties, but experiments employing agitated conditions revealed significant differences in enzyme desorption properties. An additional family 1 CBM improved the processivity of EG1 on insoluble cellulose under highly agitated conditions. Although the limited data available preclude a detail description of the adsorption-desorption process, this suggests that the presence of additional CBMs may be advantageous in promoting extended association of the modular EG1 with the cellulosic substrate by decreasing the desorption level from insoluble celluloses under conditions of high-level agitation. Together, our data indicate a strong link between enzyme processivity and adsorption-desorption properties and that high levels of adsorption combined with low levels of desorption play an important role in the processivity of processive endoglucanases.
Supplementary Material
ACKNOWLEDGMENTS
We thank John Buswell, Institute of Edible Fungi, Shanghai Academy of Agricultural Sciences, for linguistic revision of the manuscript.
This work was supported by a research grant (no. 30671652) from the National Natural Science Foundation of China, a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, and the Doctorate Fellowship Foundation of Nanjing Forestry University.
Footnotes
Published ahead of print 30 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02725-12.
REFERENCES
- 1. Béguin P, Aubert JP. 1994. The biological degradation of cellulose. FEMS Microbiol. Rev. 13:25–58 [DOI] [PubMed] [Google Scholar]
- 2. Teeri TT. 1997. Crystalline cellulose degradation: new insight into the function of cellobiohydrolases. Trends Biotechnol. 15:160–167 [Google Scholar]
- 3. Kurasin M, Väljamäe P. 2011. Processivity of cellobiohydrolases is limited by the substrate. J. Biol. Chem. 286:169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Divne C, Sinning I, Ståhlberg J, Pettersson G, Bailey M, Siika-aho M, Margolles-Clark E, Teeri T, Jones TA. 1993. Crystallization and preliminary X-ray studies on the core proteins of cellobiohydrolase I and endoglucanase I from Trichoderma reesei. J. Mol. Biol. 234:905–907 [DOI] [PubMed] [Google Scholar]
- 5. Rouvinen J, Bergfors T, Teeri T, Knowles JK, Jones TA. 1990. Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science 249:380–386 [DOI] [PubMed] [Google Scholar]
- 6. Sulzenbacher G, Schulein M, Davies GJ. 1997. Structures of the endoglucanase I from Fusarium oxysporum: native, cellobiose and 3,4-epoxybutyl β-d-cellulobioside inhibited forms at 2.3 Å resolution. Biochemistry 36:5902–5911 [DOI] [PubMed] [Google Scholar]
- 7. Gilad R, Rabinovich L, Yaron S, Bayer EA, Lamed R, Gilbert HJ, Shoham Y. 2003. Ce1I, a noncellulosomal family 9 enzyme from Clostridium thermocellum, is a processive endoglucanase that degrades crystalline cellulose. J. Bacteriol. 185:391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li YC, Irwin DC, Wilson DB. 2007. Processivity, substrate binding, and mechanism of cellulose hydrolysis by Thermobifida fusca Ce19A. Appl. Environ. Microbiol. 73:3165–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reverbel-Leroy C, Pages S, Belaich A, Belaich JP, Tardif C. 1997. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J. Bacteriol. 179:46–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang XZ, Sathitsuksanoh N, Zhang YHP. 2010. Glycoside hydrolase family 9 processive endoglucanase from Clostridium phytofermentans: heterologous expression, characterization, and synergy with family 48 cellobiohydrolase. Bioresour. Technol. 101:5534–5538 [DOI] [PubMed] [Google Scholar]
- 11. Cohen R, Suzuki MR, Hammel KE. 2005. Processive endoglucanase active in crystalline cellulose hydrolysis by the brown rot basidiomycete Gloeophyllum trabeum. Appl. Environ. Microbiol. 71:2412–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Watson BJ, Zhang H, Longmire AG, Moon YH, Hutcheson SW. 2009. Processive endoglucanases mediate degradation of cellulose by Saccharophagus degradans. J. Bacteriol. 191:5697–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burstein T, Shulman M, Jindou S, Petkun S, Frolow F, Shoham Y, Bayer EA, Lamed R. 2009. Physical association of the catalytic and helper modules of a family-9 glycoside hydrolase is essential for activity. FEBS Lett. 583:879–884 [DOI] [PubMed] [Google Scholar]
- 14. Oliveira OV, Freitas LCG, Straatsma TP, Lins RD. 2009. Interaction between the CBM of Cel9A from Thermobifida fusca and cellulose fibers. J. Mol. Recognit. 22:38–45 [DOI] [PubMed] [Google Scholar]
- 15. Sakon J, Irwin DC, Wilson DB, Karplus PA. 1997. Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat. Struct. Biol. 4:810–818 [DOI] [PubMed] [Google Scholar]
- 16. Irwin DC, Shin DH, Zhang S, Barr BK, Sakon J, Karplus PA, Wilson DB. 1998. Roles of the catalytic domain and two cellulose binding domains of Thermomonospora fusca E4 in cellulose hydrolysis. J. Bacteriol. 180:1709–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cai YJ, Chapman SJ, Buswell JA, Chang ST. 1999. Production and distribution of endoglucanase, cellobiohydrolase, and beta-glucosidase components of the cellulolytic system of Volvariella volvacea, the edible straw mushroom. Appl. Environ. Microbiol. 65:553–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ding S, Ge W, Buswell JA. 2006. Cloning of multiple cellulase cDNAs from Volvariella volvacea and their differential expression during substrate colonization and fruiting. FEMS Microbiol. Lett. 263:207–213 [DOI] [PubMed] [Google Scholar]
- 20. Wu S, Ding S, Zhou R, Li Z. 2007. Comparative characterization of a recombinant Volvariella volvacea endoglucanase I (EG1) with its truncated catalytic core (EG1-CM), and their impact on the bio-treatment of cellulose-based fabrics. J. Biotechnol. 130:364–369 [DOI] [PubMed] [Google Scholar]
- 21. Zhang YHP, Cui JB, Lynd LR, Kuang LR. 2006. A transition from cellulose swelling to cellulose dissolution by o-phosphoric acid: evidences from enzymatic hydrolysis and supramolecular structure. Biomacromolecules 7:644–648 [DOI] [PubMed] [Google Scholar]
- 22. Azevedo H, Bishop D, Cavaco-Paulo A. 2000. Effects of agitation level on the adsorption, desorption, and activities on cotton fabrics of full length and core domains of EGV (Humicola insolens) and CenA (Cellulomonas fimi). Enzyme Microb. Technol. 27:325–329 [DOI] [PubMed] [Google Scholar]
- 23. Zhang XZ, Zhang Z, Zhu Z, Sathitsuksanoh N, Yang Y, Zhang YH. 2010. The noncellulosomal family 48 cellobiohydrolase from Clostridium phytofermentans ISDg: heterologous expression, characterization, and processivity. Appl. Microbiol. Biotechnol. 86:525–533 [DOI] [PubMed] [Google Scholar]
- 24. Yoda K, Toyoda A, Mukoyama Y, Nakamura Y, Minato H. 2005. Cloning, sequencing, and expression of a Eubacterium cellulosolvens 5 gene encoding an endoglucanase (Cel5A) with novel carbohydrate-binding modules, and properties of Cel5A. Appl. Environ. Microbiol. 71:5787–5793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Irwin DC, Spezio M, Walker LP, Wilson DB. 1993. Activity studies of eight purified cellulases: specificity, synergism, and binding domain effects. Biotechnol. Bioeng. 42:1002–1013 [DOI] [PubMed] [Google Scholar]
- 26. Zverlov VV, Schantz N, Schwarz WH. 2005. A major new component in the cellulosome of Clostridium thermocellum is a processive endo-beta-1,4-glucanase producing cellotetraose. FEMS Microbiol. Lett. 249:353–358 [DOI] [PubMed] [Google Scholar]
- 27. Bronnenmeier K, Staudenbauer WL. 1990. Cellulose hydrolysis by a highly thermostable endo-1,4-β-glucanase (Avicelase I) from Clostridium stercorarium. Enzyme Microb. Technol. 12:431–436 [Google Scholar]
- 28. Chiriac AI, Cadena EM, Vidal T, Torres AL, Diaz P, Pastor FI. 2010. Engineering a family 9 processive endoglucanase from Paenibacillus barcinonensis displaying a novel architecture. Appl. Microbiol. Biotechnol. 86:1125–1134 [DOI] [PubMed] [Google Scholar]
- 29. Carrard G, Koivula A, Soderlund H, Beguin P. 2000. Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc. Natl. Acad. Sci. U. S. A. 97:10342–10347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lehtiö J, Sugiyama J, Gustavsson M, Fransson L, Linder M, Teeri TT. 2003. The binding specificity and affinity determinants of family 1 and family 3 cellulose binding modules. Proc. Natl. Acad. Sci. U. S. A. 100:484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bolam DN, Xie H, White P, Simpson PJ, Hancock SM, Williamson MP, Gilbert HJ. 2001. Evidence for synergy between family 2b carbohydrate binding modules in Cellulomonas fimi xylanase 11A. Biochemistry 40:2468–2477 [DOI] [PubMed] [Google Scholar]
- 32. Liu W, Zhang XZ, Zhang Z, Zhang YH. 2010. Engineering of Clostridium phytofermentans endoglucanase Cel5A for improved thermostability. Appl. Environ. Microbiol. 76:4914–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Couturier M, Feliu J, Haon M, Navarro D, Lesage-Meessen L, Coutinho PM, Berrin JG. 2011. A thermostable GH45 endoglucanase from yeast: impact of its atypical multimodularity on activity. Microb. Cell Fact. 10:103 doi:10.1186/1475-2859-10-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fox JM, Levine SE, Clark DS, Blanch HW. 2012. Initial- and processive-cut products reveal cellobiohydrolase rate limitations and the role of companion enzymes. Biochemistry 51:442–452 [DOI] [PubMed] [Google Scholar]
- 35. Medve J, Karlsson J, Lee D, Tjerneld F. 1998. Hydrolysis of microcrystalline cellulose by cellobiohydrolase I and endoglucanase II from Trichoderma reesei: adsorption, sugar production pattern, and synergism of the enzymes. Biotechnol. Bioeng. 59:621–634 [PubMed] [Google Scholar]
- 36. Horn SJ, Sørbotten A, Synstad B, Sikorski P, Sørlie M, Vårum KM, Eijsink VG. 2006. Endo/exo mechanism and processivity of family 18 chitinases produced by Serratia marcescens. FEBS J. 273:491–503 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.