Abstract
Yarrowia lipolytica, located at the frontier of hemiascomycetous yeasts and fungi, is an excellent candidate for studies of metabolism evolution. This yeast, widely recognized for its technological applications, in particular produces volatile sulfur compounds (VSCs) that fully contribute to the flavor of smear cheese. We report here a relevant global vision of sulfur metabolism in Y. lipolytica based on a comparison between high- and low-sulfur source supplies (sulfate, methionine, or cystine) by combined approaches (transcriptomics, metabolite profiling, and VSC analysis). The strongest repression of the sulfate assimilation pathway was observed in the case of high methionine supply, together with a large accumulation of sulfur intermediates. A high sulfate supply seems to provoke considerable cellular stress via sulfite production, resulting in a decrease of the availability of the glutathione pathway's sulfur intermediates. The most limited effect was observed for the cystine supply, suggesting that the intracellular cysteine level is more controlled than that of methionine and sulfate. Using a combination of metabolomic profiling and genetic experiments, we revealed taurine and hypotaurine metabolism in yeast for the first time. On the basis of a phylogenetic study, we then demonstrated that this pathway was lost by some of the hemiascomycetous yeasts during evolution.
INTRODUCTION
Yarrowia lipolytica is one of the most extensively studied nonconventional yeasts (1). This nonpathogenic yeast is frequently found in foods such as cheese and sausages (2). The strong proteolytic, lipolytic, and esterasic (split of esters into acid and alcohol) activities (3–5) of Y. lipolytica are particularly interesting for cheese aromatization, because this yeast produces large quantities of aroma precursors from caseins and milk fat hydrolysis, leading to various aromatic compounds. As a consequence, it has already been reported that Y. lipolytica produces a wider variety and quantity of volatile sulfur compounds (VSCs) than other commonly found cheese-ripening yeasts, such as Debaryomyces hansenii and Kluyveromyces lactis (6, 7). Its recurrent presence in soft cheeses due to inoculation from the environment (e.g., brine, ripening shelves, and personnel) is therefore indicative of its noteworthy adaptation to the cheese biotope and its positive effect on the aromatic quality of various soft cheeses (8).
The organoleptic qualities of ripened cheeses particularly depend on volatile sulfur compound production. The low odor thresholds of these compounds make them important contributors to the cheese odor and aroma. In cheese, VSCs arise essentially from the catabolism of methionine and cysteine contained in caseins (9). Since methionine is the main sulfur amino acid found in cheese curd, its catabolism has been investigated extensively in several cheese-ripening yeasts and bacteria with respect to VSC production (10, 11). It is well established that in the cheese ecosystem, VSCs arise primarily from the degradation of methionine to methanethiol (MTL), with the latter being converted subsequently to other sulfur-bearing compounds, including MTL oxidation to other products, such as dimethyl disulfide (DMDS) and dimethyl trisulfide (DMTS), and other VSCs, such as thioesters and thioethers (9).
In yeasts, methionine-to-MTL conversion proceeds via a two-step degradation pathway, initiated by a nonspecific aminotransferase, leading to the formation of the transamination product α-keto-methylthiobutyric acid (KMBA), which is subsequently converted to MTL (12). In Y. lipolytica, a branched-chain aminotransferase gene (YlBCA1) was amplified, resulting in an increased aminotransferase activity together with higher VSC production in the genetically modified strain than in the parental one (6). Lopez del Castillo Lozano et al. (13) demonstrated that H2S production is closely linked to cysteine consumption in several cheese-ripening microorganisms. In yeasts, cysteine and methionine catabolism may take place via the same degradation sequence. However, until recently, the enzymes and corresponding genes involved in VSC production have not been studied extensively.
Despite an increasing scientific literature concerning Y. lipolytica, a general study of its sulfur metabolism is not available. Major differences in global metabolism between Y. lipolytica and other ripening yeasts, such as D. hansenii and K. lactis, have already been highlighted. Whereas these yeasts first assimilate lactate as a carbon and energy source, followed by amino acids, Y. lipolytica preferentially degrades amino acids (14). Since amino acid degradation products have a major impact on cheese organoleptic properties, the study of sulfur metabolism in Y. lipolytica is of major interest for understanding VSC production.
Since the hemiascomycetous yeasts are separated by large evolutionary distances (15), we previously carried out an in silico study of 11 organisms of this phylum in order to shed light on variations in sulfur metabolism pathways (16). This previous work gave us strong bases to perform a complete inventory of sulfur metabolism in Y. lipolytica. We present here, for the first time, an overview of sulfur metabolism in Y. lipolytica, combining transcriptomics, metabolite profiling, and volatile compound measurement.
MATERIALS AND METHODS
Strain and culture conditions.
The Yarrowia lipolytica strain 1E07 (isolated from a Livarot cheese) was chosen for its interesting biotechnological properties during cheese ripening. This strain was grown in a defined sulfur-free medium (SM) (17) supplemented with sulfur sources as follows: 10 mM l-methionine, 1 mM l-cystine, or 10 mM (NH4)2SO4 for high concentrations and 10 μM l-methionine, 1 μM l-cystine, or 10 μM (NH4)2SO4 for low concentrations. Cysteine, which is very reactive, can spontaneously dimerize and form cystine. We consequently used cystine instead of cysteine to improve the control of the sulfur supply.
One hundred milliliters of SM supplemented with a sulfur substrate was inoculated from a preculture carried out in the same medium (inoculation size = 1 × 106 CFU ml−1). To avoid differences in the growth stage and stress inductions or limitations, we maintained the cells in exponential phase for 10 generations in a defined medium by seeding the cells into fresh medium after 2 generations, according to the method of Godard et al. (18). Since the cells were harvested during exponential-phase growth and at a low cell density (∼5 × 106 CFU ml−1), we could thus consider that changes in medium composition and oxygen availability were minimal during cell culture and that cells were harvested in a steady state of exponential growth. The specific growth rate was appreciably the same under all of the conditions studied (0.28 ± 0.01 h−1 [value obtained from three repetitions for each condition]). These precautions ensured highly reproducible growth conditions (17).
All cultures were carried out in 500-ml flasks at 25°C with orbital agitation (150 rpm). Three independent cultures were made for each sulfur condition. Samples for transcriptomic, metabolomic, and gas chromatography-mass spectrometry (GC-MS) experiments were taken from these cultures and then stored at −80°C.
Transcriptome analysis.
RNA extraction followed by cDNA synthesis and labeling was performed as described by Hébert et al. (17). The expression profiles of Y. lipolytica grown in the presence of various sulfur sources were analyzed using DNA microarrays (Eurogentec, Seraing, Belgium). Hybridizations were performed for 17 h at 65°C in dedicated microchambers with 100 pmol of the different labeled samples. Dye swaps were included in the experimental design. Array scanning was carried out with a DNA microarray scanner from Agilent in extended dynamic range (XDR), with a resolution of 5 nm per pixel. For further analysis, intensity-dependent normalization was performed with a global loess (19) procedure, followed by subtraction of the log ratio median calculated over the values for an entire block from each individual log ratio, using the anapuce package (20) of R software (www.R-project.org). Differential analysis was performed using a mixture model that identifies clusters of genes with equal variance (21). The raw P values were adjusted by the Bonferroni method, which controls the familywise error rate. Genes were considered to be regulated significantly if they had a P value of <0.05. Finally, we selected genes with at least a 2-fold change in expression level.
These data were validated by quantitative real-time PCR as described by Hébert et al. (17). The primers used are listed in Table S1 in the supplemental material.
Volatile sulfur compound extraction and analysis.
The volatile compounds were analyzed by GC-MS as described by Forquin et al. (22). The production of H2S in each culture was measured according to the method developed by Lopez del Castillo Lozano et al. (13).
Extraction of intracellular metabolites and identification by LC-MS.
Intracellular metabolites of Y. lipolytica cells were extracted as follows. Exponentially growing cells (1 × 108 yeast cells) were centrifuged. The cells were washed with 20 ml of cold ultrapure water to prevent contamination with SM and then centrifuged again. The cells were then stored at −80°C. The cells were resuspended with 1 ml of 1% formic acid. The solution was then incubated for 10 min at 95°C and centrifuged for 30 min at 4°C. The supernatant was lyophilized and stored at −80°C. Samples were resuspended in water containing 0.1% formic acid prior to injection. Chromatographic separation was performed on a Discovery HS-F5 column (2.1 mm × 250 mm × 5 μm) from Supelco Analytical (Interchim, Montluçon, France) by using a Surveyor liquid chromatography (LC) system (ThermoFisher Scientifics, Courtaboeuf, France). Before injection, samples were stored at 4°C in the autosampler tray. Separations were carried out using the following gradient at 200 μl/min: 0 to 3 min, 0% B; 3 to 20 min, 0 to 100% B; 20 to 25 min, 100% B; and 25 to 45 min, 0% B. Solvent A was water and solvent B was acetonitrile, both containing 0.1% formic acid. The column temperature was set to 30°C.
Mass spectrometric detection was performed using an LTQ/Orbitrap hybrid mass spectrometer (ThermoFisher Scientifics, Courtaboeuf, France) fitted with an electrospray source operated in the positive ionization mode. The detection was achieved from 75 to 1,000 u (u = unified atomic mass unit) at the maximum resolving power of 30,000 (expressed as the full width at half the maximum for an ion at 400 u). The mass spectrometer was operated with a capillary voltage of 4 kV and a capillary temperature of 275°C. Nitrogen was used as the sheath and auxiliary gas, with pressures set at 45 and 10 (arbitrary units), respectively.
Data processing and statistical analysis for LC-MS.
All data were processed using the Qualbrowser module of Xcalibur, version 2.0.7 (Thermo Fisher Scientific, Les Ulis, France), and its chemical formula generator was used to provide elemental compositions. Automatic peak detection from LC-MS chromatograms was performed using XCMS software (23), version 1.14.1, running under R, version 2.8.1. The R language was installed on a Dell eight-core Intel Xeon 3.00-GHz processor with 16 Gb RAM running under Linux (Centos 5.2 ×86_64). The matchedFilter algorithm was used, and default values were set for all parameters except for fwhm, step, steps, mzdiff, mzwid, bw, and snthresh, which were set at 25, 0.01, 3, 0.1, 0.1, 5, and 3, respectively. A list of the areas of the peaks detected was generated for each sample by using a combination of the retention time (RT) and the m/z ratio as an identifier. An arbitrary number was assigned to each of these RT-m/z pairs, in order of elution. The data were then combined into a single matrix by aligning peaks with the same mass-RT pair together from each data file in the data set. The XCMS data sets were annotated using tools developed in-house: the spectral database of the laboratory and an informatics tool for automatic query of metabolic and metabolomic public databases with the measured accurate masses.
Commercial compounds were also analyzed in order to confirm metabolite identification on the bases of chromatographic retention time and MS/MS spectrum similarities (concentration of 5 μg/ml). Table 1 summarizes the commercial compounds analyzed and eventually detected, as well as compounds detected in our biological samples.
Table 1.
Metabolites related to sulfur metabolism detected by high-performance LC coupled to LTQ-Orbitrap MSa
| Compound | Commercial product | Sample |
|---|---|---|
| Methionine | + | + |
| Cystathionine | + | + |
| Cysteine | + | + |
| γ-Glutamylcysteine | + | + |
| Reduced glutathione | + | + |
| Cysteinylglycine | + | + |
| Spermidine | + | + |
| Serine | + | + |
| Taurine | + | + |
| O-Acetylserine | + | − |
| KMBA | + | − |
| Homocysteine | + | − |
| S-Adenosylhomocysteine | + | − |
| Cystine | + | − |
| Oxidized glutathione | + | − |
| Spermine | + | − |
| S-Adenosylmethionine | − | − |
| Hypotaurine | / | + |
| O-Acetylhomoserine | / | + |
| 5-Methylthioadenosine | / | + |
| S-Adenosylmethioninamine | / | − |
| 1,2-Dihydroxy-3-keto-5-methyl-thioptene | / | − |
+, detected; −, not detected; /, product not commercially available.
Microarray data accession number.
The complete experimental data set from microarray analysis was deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE39834.
RESULTS
Experimental design.
In order to study sulfur metabolism in the technological yeast Y. lipolytica, we hypothesized that in an excess of sulfur, yeast metabolism is oriented towards sulfur amino acid catabolism, whereas with a limited sulfur supply, metabolism is oriented towards biosynthesis of sulfur amino acids. Consequently, the studied yeast was grown with different sulfur sources—methionine (M), cystine (C), and sulfate (S)—at high (H) and low (L) concentrations, at a ratio of 1,000/1. The cells were collected in the exponential phase, and their growth parameters appeared very similar under all tested conditions. This experimental design was used throughout this study for transcriptomic, metabolomic, and VSC analyses. To make the reading easier, we described Y. lipolytica orthologs by using the gene names of Saccharomyces cerevisiae. The “low-level” sample was taken as the control to compare gene expression between sulfur catabolism and anabolism by calculating the HM/LM, HC/LC, and HS/LS ratios.
Modulation of gene expression according to sulfur supply.
Among the 6,448 genes of Y. lipolytica, we observed 516 genes that were differentially expressed (∼8%). Excluding the genes of unknown function, we can assume that almost 25% of less-expressed genes under high-sulfur conditions are related to sulfur transport and metabolism (see Table S2 in the supplemental material). This result indicates that yeast restricts sulfur assimilation and anabolism under conditions of sulfur excess. Moreover, genes of unknown function represented approximately one-third of differentially expressed genes, creating opportunities to find new genes involved in sulfur metabolism. The majority of sulfur-related genes were regulated in the three sulfur sources (Table 2). Consequently, we described the three conditions at the same time, using the following notation: H/L(M and/or C and/or S). Table 3 and Table 4 provide a summary of all the genes related to sulfur metabolism that were differentially expressed under at least one of our experimental conditions. The validation of the transcriptomic experiment by qPCR is available in Table S1.
Table 2.
Breakdown of sulfur-related genes expressed differentially depending on sulfur supply
| Sulfur source | Notation in texta | No. (%) of differentially expressed genes under high- vs low-sulfur conditions |
|---|---|---|
| All sulfur sources | H/L(M,C,S) | 23 (47) |
| Methionine and cystine | H/L(M,C) | 7 (14) |
| Methionine and sulfate | H/L(M,S) | 1 (2) |
| Cystine and sulfate | H/L(C,S) | 0 (0) |
| Methionine only | H/L(M) | 17 (35) |
| Cystine only | H/L(C) | 0 (0) |
| Sulfate only | H/L(S) | 1 (2) |
H, high; L, low; M, methionine; C, cystine; S, sulfate.
Table 3.
Transcriptomic response of Y. lipolytica transporters to sulfur supplya
| S. cerevisiae gene name | Y. lipolytica gene name | Ratio Met | P value for Met | Ratio Cys | P value for Cys | Ratio Sul | P value for sulfate | Function |
|---|---|---|---|---|---|---|---|---|
| SUL1-SUL2 | YALI0B17930g | 0.01 | 1.39E−12 | 0.09 | 0 | 0.16 | 0 | High-affinity sulfate permease |
| SSU1 | YALI0E24167g | 5.23 | 2.83E−12 | Sulfite pump, required for efficient sulfite efflux | ||||
| MUP1 | YALI0F25795g | 0.05 | 1.39E−12 | 0.20 | 1.13E−11 | 0.27 | 8.71E−08 | High-affinity methionine permease |
| YALI0D16137g | 0.02 | 0 | 0.03 | 0 | 0.04 | 0 | ||
| YALI0F03498g | 0.46 | 4.74E−02 | 0.37 | 1.54E−03 | ||||
| YALI0D19646g | 0.32 | 2.22E−07 | ||||||
| YALI0F07018g | ||||||||
| TPO1 | YALI0F07062g | 0.43 | 1.61E−03 | Polyamine transporter | ||||
| YALI0A15576g | ||||||||
| YALI0F08063g | ||||||||
| YALI0E21241g | ||||||||
| YALI0A18902g | ||||||||
| DUR3 | YALI0E28622g | 0.03 | 1.39E−12 | 0.03 | 0 | 0.06 | 0 | Transporter for both urea and polyamines |
| YALI0E33583g | 0.02 | 0 | 0.04 | 1.41E−12 | 0.06 | 0 | ||
| YALI0B04202g | ||||||||
| YALI0C15807g | ||||||||
| GAP1 | YALI0F19866g | 6.55 | 2.78E−12 | 3.93 | 1.02E−08 | 2.32 | 4.08E−03 | General amino acid permease |
| YALI0B19800g | 0.44 | 8.69E−04 | ||||||
| YALI0B16522g | ||||||||
| YALI0C17237g | ||||||||
| YALI0E10219g | ||||||||
| YALI0C09889g | ||||||||
| YALI0B19492g | ||||||||
| OPT2 | YALI0F09691g | 0.11 | 0 | 0.20 | 7.07E−12 | 0.35 | 3.96E−04 | Oligopeptide transporter |
| YALI0A00110g | 0.29 | 1.35E−09 | ||||||
| YALI0C18491g | ||||||||
| YALI0B07898g | ||||||||
| YALI0C20823g | ||||||||
| YALI0F30041g | ||||||||
| YALI0F18964g | ||||||||
| YALI0B04642g | ||||||||
| YALI0F13959g | ||||||||
| YALI0A03949g | ||||||||
| YALI0E19294g | ||||||||
| YALI0B02090g | ||||||||
| YALI0E17589g | ||||||||
| YALI0F10241g | ||||||||
| YALI0B02398g | ||||||||
| YALI0C22616g | ||||||||
| JEN1 | YALI0C15488g | 26.58 | 1.39E−12 | 5.43 | 0 | 3.61 | 2.35E−07 | Lactate transporter, required for uptake of lactate and pyruvate |
| YALI0D20108g | ||||||||
| YALI0D24607g | ||||||||
| YALI0E32901g | ||||||||
| YALI0C21406g | ||||||||
| YALI0B19470g | ||||||||
| SEO1 | YALI0C17281g | 0.14 | 0 | 0.10 | 0 | 0.14 | 0 | Putative allantoate permease |
| YALI0C17303g | 0.06 | 0 | 0.03 | 0 | 0.04 | 0 | ||
| YALI0F13607g | 0.05 | 0 | 0.22 | 1.55E−11 | 0.17 | 0 | ||
| YALI0E35200g | 0.33 | 2.28E−07 | ||||||
| YALI0C00627g | ||||||||
| YALI0B01904g | ||||||||
| YIL166c | YALI0D00407g | 0.01 | 0 | 0.03 | 1.41E−12 | 0.03 | 0 | Similarity to allantoate permease |
| YALI0B16412g | 0.36 | 1.71E−04 | 0.44 | 1.49E−02 | 0.34 | 1.81E−04 |
Values described in the text (P values of ≤0.05 and ratios of ≤0.5 or ≥2) are listed in this table. Ratio Met, ratio of high to low methionine results; Ratio Cys, ratio of high to low cystine results; and Ratio Sul, ratio of high to low sulfate results.
Table 4.
Transcriptomic response of Y. lipolytica sulfur metabolism and catabolism to sulfur supplya
| S. cerevisiae gene name | Y. lipolytica gene name | Ratio Met | P value for Met | Ratio Cys | P value for Cys | Ratio Sul | P value for Sul | Function |
|---|---|---|---|---|---|---|---|---|
| Sulfur metabolism genes | ||||||||
| MET 3 | YALI0B08184g | 0.03 | 0 | 0.15 | 9.89E−12 | 0.29 | 7.35E−04 | ATP sulfurylase |
| MET 16 | YALI0B08140g | 0.02 | 4.17E−12 | 0.12 | 1.41E−12 | 0.16 | 1.42E−12 | 3′-Phosphoadenylsulfate reductase |
| GSH2 | YALI0C17831g | 0.34 | 5.23E−07 | 0.32 | 1.62E−05 | 0.37 | 3.38E−05 | Glutathione synthetase |
| SPE3 | YALI0E33143g | 2.63 | 2.03E−05 | 2.66 | 2.60E−04 | 2.32 | 1.59E−02 | Spermidine synthase |
| ADI1 | YALI0A14498g | 0.26 | 9.32E−11 | 0.34 | 2.12E−05 | 0.41 | 5.09E−03 | Acireductone dioxygenease |
| ARO9 | YALI0C05258g | 0.21 | 0 | 0.16 | 0 | 0.28 | 2.31E−07 | Aromatic aminotransferase II |
| MET10 | YALI0E16368g | 0.18 | 0 | 0.33 | 2.49E−05 | Subunit alpha of assimilatory sulfite reductase | ||
| MET17 | YALI0D25168g | 0.11 | 0 | 0.36 | 6.74E−04 | O-Acetylhomoserine-O-acetylserine sulfhydrylase | ||
| MET30 | YALI0B09977g | 0.38 | 3.66E−05 | 0.48 | 2.85E−02 | F-box protein, controls sulfur metabolism; regulates Met4p | ||
| MET4 | YALI0D04466g | 0.19 | 2.78E−12 | 0.32 | 8.72E−06 | Transcriptional activator, regulation of the sulfur amino acid pathway | ||
| MET 2 | YALI0E00836g | 0.27 | 1.26E−09 | l-Homoserine-O-acetyltransferase | ||||
| MET 14 | YALI0E00418g | 0.40 | 1.14E−04 | Adenylylsulfate kinase | ||||
| MET 1 | YALI0A01133g | 0.32 | 3.00E−07 | Uroporphyrin-III C-methyltransferase | ||||
| MET13 | YALI0B00572g | 0.40 | 6.99E−05 | Major isozyme of methylenetetrahydrofolate reductase | ||||
| SAM1 SAM2 | YALI0B14509g | 0.30 | 1.84E−08 | S-Adenosylmethionine synthetase | ||||
| YGR012W | YALI0E08536g | 0.34 | 1.31E−06 | Putative cysteine synthetase | ||||
| YALI0F14047g | 0.38 | 3.08E−04 | ||||||
| Genes potentially involved in methionine catabolism | ||||||||
| BAT1 | YALI0D01265g | 2.43 | 1.85E−04 | 2.13 | 4.29E−02 | Mitochondrial branched-chain amino acid aminotransferase | ||
| JEN1 | YALI0C15488g | 26.58 | 1.39E−12 | 5.43 | 0 | 3.61 | 2.35E−07 | Lactate transporter |
| Sulfur-related genes | ||||||||
| YALI0A06743g | 0.16 | 0 | 0.19 | 1.41E−12 | 0.24 | 1.56E−11 | Glutathione transferase | |
| YALI0F25575g | 0.08 | 2.78E−12 | 0.15 | 0 | 0.33 | 5.13E−05 | ||
| JLP1 | YALI0F22825g | 0.01 | 1.39E−12 | 0.05 | 0 | 0.06 | 0 | Fe(II)-dependent sulfonate/alpha-ketoglutarate dioxygenase |
| YALI0A12177g | 0.17 | 0 | 0.09 | 1.41E−12 | 0.14 | 0 | ||
| YALI0D17622g | 0.03 | 0 | 0.03 | 1.41E−12 | 0.05 | 0 | ||
| YALI0B21472g | 0.02 | 0 | 0.07 | 2.83E−12 | 0.10 | 0 | ||
| YALI0A21439g | 0.21 | 1.39E−12 | 0.29 | 3.33E−06 | 0.42 | 4.23E−02 | ||
| YALI0D21098g | 0.10 | 0 | 0.06 | 1.41E−12 | 0.19 | 0 | ||
| FMO1 | YALI0D21076g | 0.12 | 1.39E−12 | 0.11 | 2.83E−12 | 0.12 | 0 | Flavin-containing monooxygenase, catalyzes oxidation of biological thiols to maintain the endoplasmic reticulum redox ratio for correct folding of disulfide-bonded proteins |
| YHR112c | YALI0C22088g | 0.25 | 4.17E−12 | 0.44 | 2.87E−02 | Protein of unknown function, similarity to CYS3, STR3, MET17, and STR2 | ||
| PRX1 | YALI0A19426g | 0.13 | 0 | 0.43 | 2.45E−02 | Mitochondrial peroxiredoxin with thioredoxin peroxidase activity | ||
| YALI0F08195g | 0.29 | 2.60E−09 | ||||||
| MXR1 | YALI0B03916g | 0.12 | 0 | Methionine-S-sulfoxide reductase | ||||
| YALI0E20119g | 0.49 | 1.75E−02 | ||||||
| TSA1 | YALI0B15125g | 0.40 | 4.47E−04 | Thioredoxin peroxidase | ||||
| TRX1 | YALI0E23540g | 0.46 | 9.56E−03 | Cytoplasmic thioredoxin |
Values described in the text (P values of ≤0.05 and ratios of ≤0.5 or ≥2) are listed in this table. Ratio Met, ratio of high to low methionine results; Ratio Cys, ratio of high to low cystine results; Ratio Sul, ratio of high to low sulfate results.
(i) Transporters.
Table 3 deals with the genes involved in transport. Because of its considerable proteolytic activities, Y. lipolytica possesses a wide variety of transporters. This yeast frequently presents two or more genes that have sequence similarities with a given transporter gene of the model yeast S. cerevisiae. Thus, it is difficult to identify the orthologs of S. cerevisiae genes encoding transporters with the same functional specificity in Y. lipolytica. We can hypothesize that among each Y. lipolytica transporter family, each gene may encode a protein with a specific substrate specificity and/or affinity.
We observed that when Y. lipolytica has some orthologs for one transporter, they are not regulated in the same way. Of the five Y. lipolytica orthologs of the S. cerevisiae high-affinity methionine permease (encoded by MUP1), four were regulated under our conditions. Two of these genes were repressed under H/L(M,C,S) conditions, while the other two were repressed only under H/L(M,S) and H/L(S) conditions. The high-affinity sulfate transporter (encoded by SUL1-SUL2) of Y. lipolytica was highly repressed in H/L(M,C,S).
Polyamine metabolism is directly linked to sulfur metabolism. It was therefore not surprising to observe the repression of several genes (e.g., DUR3 and TPO1) involved in polyamine transport. However, the orthologs of these two genes are not similarly regulated. Of the four orthologs of DUR3, two were repressed in H/L(M,C,S), whereas of the four orthologs of TPO1, one was repressed in H/L(M).
Only 2 of the 16 orthologs of the S. cerevisiae oligopeptide transporter (encoded by OPT2) were repressed, in H/L(M,C,S) and in H/L(M). In Y. lipolytica, there are eight orthologs of the GAP1 gene, which encodes general amino acid transporters in S. cerevisiae. These genes could encode transporters with different specificities, as reported for Candida albicans by Kraidlova et al. (24). Two of these genes were regulated under our conditions. One was repressed in H/L(M), while the other was overexpressed in H/L(M,C,S). We also observed that the sulfite exporter (encoded by SSU1) was specifically overexpressed in H/L(S).
We demonstrated that one Y. lipolytica gene was overexpressed in H/L(M,C,S), particularly under high-methionine conditions. This gene, annotated as encoding a lactate transporter, presents very limited sequence similarity with lactate transporters encoded by JEN1 in S. cerevisiae (only in a quarter of the sequence). Among this family of six genes, YALI0C15488g was the only one that was differentially expressed. Taking the particular state of Y. lipolytica transporters into account, we hypothesized that this gene could play a role in sulfur metabolism.
We observed that transporters probably involved in sulfur assimilation were repressed under one or more of our conditions (H/liter), suggesting that the sulfur supply is in excess under conditions of high supply. The differences in regulation in the same group of orthologs suggest that the regulation and specificity of Y. lipolytica transporters are very complex.
(ii) Sulfate assimilation.
We observed that numerous genes involved in sulfate assimilation are regulated (Table 4). Concerning the first step of sulfate activation, both the MET3 (encoding an ATP sulfurylase) and MET14 (encoding an APS kinase) genes were repressed, in H/L(M,C,S) and in H/L(M), respectively. Regarding the following sulfate reduction stage, the MET16 gene, encoding a PAPS reductase, was repressed in H/L(M,C,S). The next enzymatic step is carried out by a sulfite reductase, which is a heterodimer encoded by MET5 and MET10. Only MET10 was repressed in H/L(M,C). We also observed that sulfite reductase needs siroheme to be functional and that MET1, whose product is involved in siroheme biosynthesis, is repressed in H/L(M). Sulfur metabolism interacts with aspartate metabolism through production of homoserine, which is the last common intermediate of sulfur and threonine metabolism. MET2, which encodes a homoserine-O-acetyltransferase, is repressed in H/L(M). The latter enzyme synthesizes O-acetylhomoserine (OAH). The O-acetylhomoserine sulfhydrylase, encoded by MET17, leads to homocysteine production (precursor of methionine and cysteine). This gene is repressed in H/L(M,C).
(iii) Cysteine and glutathione metabolism.
Surprisingly, the direct and reverse transsulfuration pathways were not regulated under all conditions tested. However, two genes (YALI0E08536g and YALI0F14047g) which are orthologs of the S. cerevisiae gene YGR012w, encoding cysteine synthases involved in the second pathway of cysteine metabolism (O-acetylserine [OAS] pathway), were repressed in H/L(M). The GSH2 gene, encoding glutathione synthase, was repressed in H/L(M,C,S).
(iv) Methionine metabolism.
Methionine leads to the production of S-adenosylmethionine (SAM) via the methyl cycle. The gene leading to SAM synthesis was repressed in H/L(M). While MET6 was not regulated, the repression in H/L(M) of MET13, involved in the synthesis of the MET6 cofactor, led to an indirect repression of methionine synthesis. The methionine salvage pathway leads to polyamine biosynthesis and production of 5-methylthioadenosine (5MTA), which can be recycled into methionine. The SPE3 gene, leading to spermidine synthesis, was overexpressed in H/L(M,C,S). Two genes of the methionine salvage pathway were repressed in H/L(M,C,S): ADI1 and ARO9.
(v) Sulfur regulation in H/L(M,C,S).
Among the eight genes involved in the sulfur regulation network (16), two genes (MET4 and MET30) were repressed in H/L(M,C). MET4 encodes the transcriptional activator of sulfur metabolism, and MET30 encodes an F-box protein involved in the regulation of Met4p.
(vi) Catabolism in H/L(M,C,S).
We investigated the overexpression of aminotransferases potentially involved in methionine catabolism, i.e., the branched-chain aminotransferases encoded by BAT1 and BAT2 and the aromatic aminotransferases encoded by ARO8 and ARO9. In light of our results, only one of all the candidate genes seems to be particularly interesting. Indeed, this gene (YALI0D01265g), which is the ortholog of BAT1, was overexpressed in H/L(M,C) (Table 4).
Volatile sulfur compounds and intracellular sulfur metabolites.
The major VSC produced was dimethyl disulfide (DMDS), which was produced under all tested conditions. However, its production was 47.3- and 2.4-fold higher, on average, in HM and HS, respectively (Table 5). The concentration of DMDS was nearly unchanged in cystine and even lower in HC. Concerning other aromatic compounds, dimethyl trisulfide (DMTS) and hydrogen sulfide (H2S) were produced only in HM and HS (Table 5). Furthermore, the wide variety of aromatic compounds obtained in HM underlines the remarkable aromatic properties of Y. lipolytica.
Table 5.
Production of volatile sulfur compounds depending on sulfur supplya
| Compound | Ratio Met | Ratio Cys | Ratio Sul | Concn (μg/kg) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| HM | LM | HC | LC | HS | LS | ||||
| DMDS | 47.3 | 0.8 | 2.4 | 74.78 ± 3.77 | 1.58 ± 0.43 | 1.3 ± 0.21 | 1.7 ± 0.23 | 3.85 ± 0.29 | 1.61 ± 0.16 |
| DMTS | NS | NS | NS | 2.38 ± 0.87 | 0.14 ± 0.05 | ||||
| Methanethiol | NS | NS | NS | 0.40 ± 0.02 | |||||
| DMS | NS | NS | NS | 0.36 ± 0.03 | |||||
| Methional | NS | NS | NS | 0.31 ± 0.07 | |||||
| H2Sb | NS | NS | NS | 23.60 ± 3.60 | 23.53 ± 0.39 | ||||
Results are the mean values ± standard errors of the means for three independent cultures. H, high; L, low; M, methionine; C, cystine; S, sulfate; Ratio Met, HM/LM ratio; Ratio Cys, HC/LC ratio; Ratio Sul, HS/LS ratio; NS, not significant.
In milligrams per kilogram.
A list of metabolites of interest is given in Table 1, which summarizes the detection by LC coupled to LTQ-Orbitrap MS of sulfur intermediates in samples and their availability as commercial references. Some commercially available metabolites that were detectable were never observed experimentally, i.e., these metabolites did not accumulate in our samples. This was the case for homocysteine, oxidized glutathione, cystine, spermine, S-adenosylhomocysteine, O-acetylserine, and KMBA. It is important that homocysteine was detected in Brevibacterium aurantiacum samples by use of a similar extraction protocol (22). With the experimental protocol utilized, the SAM pool was not detected in either the commercial compound solution or our samples. The presence of some intermediates (not commercially available) has been investigated. We identified hypotaurine, O-acetylhomoserine, and 5-methylthioadenosine in our samples. Their identities were confirmed by MS/MS. Data concerning intermediates of sulfur metabolism are given in Table 6. These data are reported further and discussed throughout this paper.
Table 6.
Metabolomic response of Y. lipolytica to sulfur supplya
| Compound | Area |
Ratio |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| HM | LM | HC | LC | HS | LS | Met | Cys | Sul | |
| 5-Methylthioadenosine | 1.30E+09 [1.19, 1.37]E+09 | 3.92E+08 [3.38, 4.72]E+08 | 4.04E+07 [3.32, 4.94]E+07 | 2.54E+08 [1.85, 2.92]E+08 | 9.54E+07 [8.24, 10.70]E+07 | 3.40E+08 [2.60, 4.13]E+08 | 3.32 | 0.16 | 0.28 |
| Cystathionine | 1.74E+08 [1.48, 2.24]E+08 | 1.64E+07 [1.21, 1.88]E+07 | 1.93E+06 [1.45, 2.80]E+06 | 8.87E+06 [4.72, 12.70]E+06 | 5.75E+06 [5.07, 6.71]E+06 | 1.01E+07 [0.92, 1.13]E+07 | 10.67 | 0.22 | 0.57 |
| Hypotaurine | 2.89E+07 [2.53, 3.55]E+07 | 1.45E+06 [1.39, 1.55]E+07 | 2.68E+06 [2.22, 3.21]E+06 | 9.03E+05 [6.29, 11.70]E+05 | 1.76E+06 [0.97, 2.22]E+06 | 1.16E+06 [0.05, 2.12]E+06 | 19.91 | 2.97 | |
| Spermidine | 4.37E+08 [4.30, 4.49]E+08 | 3.56E+08 [3.13, 4.06]E+08 | 2.34E+08 [2.29, 2.39]E+08 | 2.70E+08 [2.43, 3.04]E+08 | 3.68E+08 [3.19, 4.03]E+08 | 3.75E+08 [3.36, 4.14]E+08 | 1.23 | 0.87 | |
| Taurine | 8.72E+06 [7.66, 9.70]E+06 | 1.98E+06 [1.80, 2.07]E+06 | 2.65E+06 [2.47, 2.81]E+06 | 1.11E+06 [0.83, 1.28]E+06 | 3.15E+06 [2.29, 4.04]E+06 | 1.50E+06 [0.46, 3.47]E+06 | 4.41 | 2.39 | |
| Cysteine | 1.23E+07 [1.11, 1.33]E+07 | 6.36E+06 [6.03, 6.70]E+06 | 5.82E+06 [5.26, 6.28]E+06 | 4.89E+06 [3.87, 5.81]E+06 | 1.58E+05 [0.76, 2.49]E+05 | 4.35E+06 [3.00, 5.24]E+06 | 1.94 | 0.04 | |
| γ-Glutamylcysteine | 4.24E+06 [3.49, 5.08]E+06 | 6.96E+06 [6.80, 7.23]E+06 | 5.17E+06 [4.63, 6.26]E+06 | 5.01E+06 [3.40, 5.87]E+06 | 8.36E+05 [5.50, 12.50]E+05 | 7.44E+06 [5.05, 11.80]E+06 | 0.61 | 0.11 | |
| Methionine | 1.21E+08 [0.94, 1.94]E+08 | 1.79E+07 [1.76, 1.82]E+07 | 9.13E+06 [8.43, 9.76]E+06 | 1.22E+07 [0.83, 1.48]E+07 | 1.78E+07 [1.37, 2.13]E+07 | 1.80E+07 [1.38, 2.54]E+07 | 6.75 | ||
| Serine | 2.03E+07 [1.86, 2.31]E+07 | 5.53E+07 [5.39, 5.79]E+07 | 4.08E+07 [3.57, 4.97]E+07 | 3.63E+07 [2.64, 4.45]E+07 | 4.86E+07 [3.62, 5.72]E+07 | 5.29E+07 [4.77, 5.74]E+07 | 0.37 | ||
| Glutathione | 4.67E+08 [4.18, 5.43]E+08 | 5.03E+08 [4.73, 5.53]E+08 | 4.55E+08 [4.01, 5.50]E+08 | 3.81E+08 [2.96, 4.25]E+08 | 2.22E+08 [2.00, 2.52]E+08 | 5.05E+08 [4.31, 6.50]E+08 | 0.44 | ||
| Cysteinylglycine | 5.82E+07 [5.16, 6.73]E+07 | 6.45E+07 [5.79, 7.43]E+07 | 5.96E+07 [5.31, 7.10]E+07 | 5.01E+07 [4.23, 5.46]E+07 | 3.32E+06 [2.31, 3.96]E+06 | 6.01E+07 [4.87, 7.60]E+07 | 0.06 | ||
| O-Acetylhomoserine | 9.43E+04 [3.27, 12.90]E+04 | 9.20E+04 [4.82, 11.73]E+04 | 4.67E+04 [1.77, 8.77]E+04 | 4.70E+04 [3.40, 6.44]E+04 | 2.99 + 04 [0.76, 4.48]E+04 | 1.68E+04 [4.29, 14.59]E+04 | |||
H, high; L, low; M, methionine; C, cystine; S, sulfate; Ratio Met, HM/LM ratio; Ratio Cys, HC/LC ratio; Ratio Sul, HS/LS ratio. The areas indicated are means for three independent experiments. Extreme values are indicated in brackets.
(i) Methionine.
Under HM supplementation conditions, some sulfur intermediates were highly accumulated. We observed that the intracellular pools of cystathionine and methionine were 10.67-fold and 6.75-fold higher, respectively, in HM than in LM. The pools of 5-methylthioadenosine, cysteine, and spermidine were also higher in HM than in LM, but to a lesser extent (ratios of 3.32, 1.94, and 1.23, respectively). In contrast, the pools of γ-glutamylcysteine and serine were lower in HM than in LM (ratios of 0.61 and 0.37, respectively). Moreover, we observed pools of two specific sulfur compounds: hypotaurine and taurine. Their pools were higher in HM than in LM (ratios of 19.91 and 4.41, respectively).
(ii) Cystine.
In HC versus LC, we observed a slight increase in hypotaurine and taurine (ratios of 2.97 and 2.39, respectively). In contrast, pools of 5-methylthioadenosine, cystathionine, and spermidine were lower in HC than in LC (ratios of 0.16, 0.22, and 0.87, respectively).
(iii) Sulfate.
In HS versus LS, all pools of glutathione pathway intermediates were decreased (ratios of 0.04 for cysteine, 0.57 for cystathionine, 0.11 for γ-glutamylcysteine, 0.44 for glutathione, and 0.06 for cysteinylglycine). The pool of 5-methylthioadenosine was also lower in HS than in LS (ratio of 0.28).
DISCUSSION
Considering the three sulfur sources tested, we observed that Yarrowia lipolytica developed a specific response to each substrate. Consequently, we first discuss the effects of each sulfur source, using three summary diagrams (genes with no modified expression under any of our six conditions are listed in Table S3 in the supplemental material). We then discuss VSC production. Finally, we conclude with the specificities of Y. lipolytica sulfur metabolism.
Specific response to each sulfur supply. (i) Accumulation of sulfur intermediates with a methionine supply.
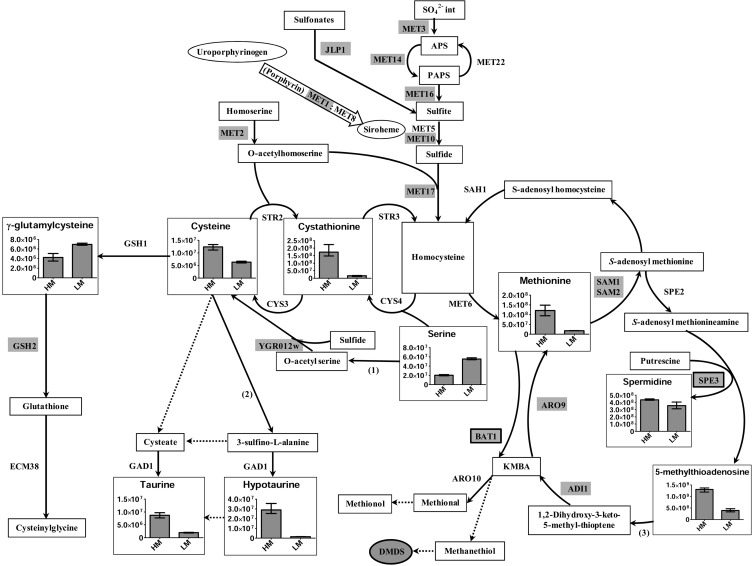
With a high methionine supply, we observed an expected repression of the sulfate assimilation pathway (Fig. 1). The intracellular excess of sulfur is distributed between numerous sulfur intermediates, such as methionine, 5-methylthioadenosine, cystathionine, and cysteine pools. Under high-methionine conditions, the serine and γ-glutamylcysteine pools are decreased. The decrease in the serine pool could be a consequence of the increase in the intracellular methionine pool. In fact, since the reverse transsulfuration pathway leading to cysteine biosynthesis is not regulated, the increase in the methionine pool could reorientate the sulfur flux toward cysteine synthesis. This could provoke a decrease in the serine pool, since it is a precursor of cysteine. Considering the two cysteine biosynthesis pathways (reverse transsulfuration and the OAS pathway), only the OAS pathway is regulated under high-methionine conditions. Interestingly, this pathway is absent in S. cerevisiae. K. lactis, a hemiascomycetous yeast that is more closely related to S. cerevisiae, also possesses these two pathways (16). Meanwhile, we observed an absence of regulation of the OAS pathway and a regulation of the transsulfuration pathway in K. lactis (17) that was identical to that observed in S. cerevisiae (25). This suggests that cysteine biosynthesis could be regulated differently in Y. lipolytica. Considering these differences in regulation, we hypothesize that the OAS pathway, being regulated in Y. lipolytica, plays the main role compared to the transsulfuration pathway.
Fig 1.
Overview of sulfur metabolism in Y. lipolytica under high- versus low-methionine conditions. (1) Serine-O-acetyltransferase. (2) Cysteine dioxygenase. The genes encoding these two enzymes are absent in S. cerevisiae. (3) Methionine salvage pathway. Genes highlighted in gray are repressed under high-methionine conditions. Genes in black boxes and highlighted in gray are overexpressed under high-methionine conditions. The reactions carried out by unidentified or assortments of enzymes are indicated with dotted arrows. VSCs produced at a higher level are surrounded in black and highlighted in gray. Histograms are utilized only for compounds with significant differences. Values and error bars in histograms represent means with extreme values.
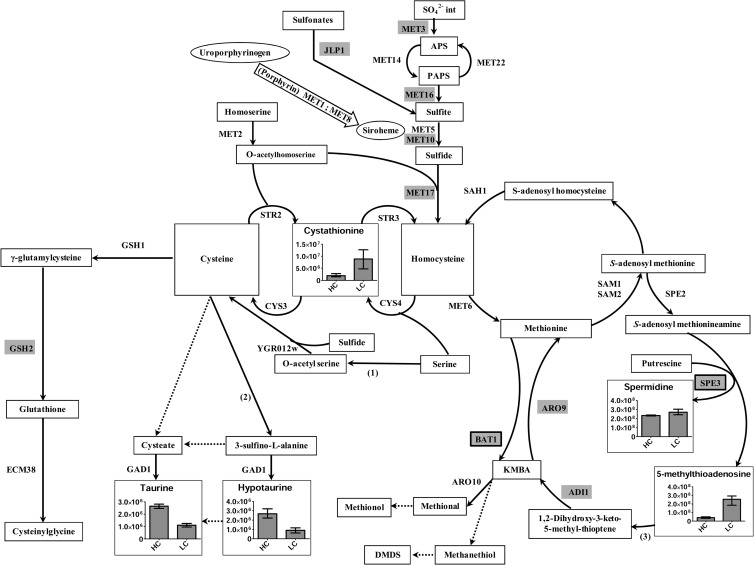
(ii) Maintaining sulfur metabolism in a steady state with a cystine supply.
With a high cystine supply, we observed a repression of the sulfate assimilation pathway (Fig. 2). The intracellular pools are rather stable, suggesting that cystine metabolism is highly regulated. In the well-studied yeast S. cerevisiae, it was determined that the genes involved in sulfur metabolism are regulated mainly by the cysteine pool (26). Sulfur excess seems to be directed to hypotaurine and taurine synthesis, thus maintaining intermediates of glutathione synthesis in a steady state.
Fig 2.
Overview of sulfur metabolism in Y. lipolytica under high- versus low-cystine conditions. (1) Serine-O-acetyltransferase. (2) Cysteine dioxygenase. The genes encoding these two enzymes are absent in S. cerevisiae. (3) Methionine salvage pathway. Genes highlighted in gray are repressed under high-methionine conditions. Genes in black boxes and highlighted in gray are overexpressed under high-methionine conditions. The reactions carried out by unidentified or assortments of enzymes are indicated with dotted arrows. Histograms are utilized only for compounds with significant differences. Values and error bars in histograms represent means with extreme values.
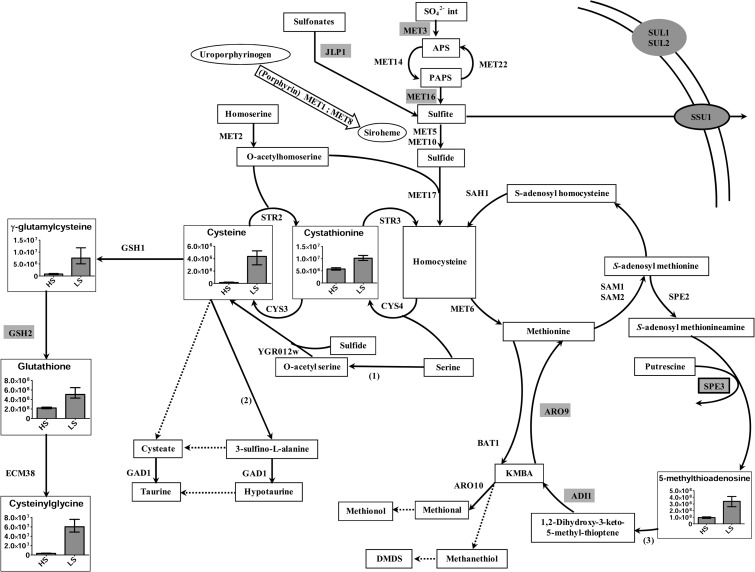
(iii) Drastic decrease in glutathione intermediates when sulfate is supplied.
Under conditions of high sulfate supply (Fig. 3), we observed a repression of genes involved in sulfite production, as well as the overexpression of a sulfite exporter. Sulfite probably provokes a large amount of stress, since all pools of the intermediates of the glutathione pathway were decreased under high-sulfate conditions. It has been demonstrated that the exporter of sulfite is of major importance for sulfite resistance in S. cerevisiae (27).
Fig 3.
Overview of sulfur metabolism in Y. lipolytica under high- versus low-sulfate conditions. (1) Serine-O-acetyltransferase. (2) Cysteine dioxygenase. The genes encoding these two enzymes are absent in S. cerevisiae. (3) Methionine salvage pathway. Genes highlighted in gray are repressed under high-methionine conditions. Genes in black boxes and highlighted in gray are overexpressed under high-methionine conditions. The reactions carried out by unidentified or assortments of enzymes are indicated with dotted arrows. Histograms are utilized only for compounds with significant differences. Values and error bars in histograms represent means with extreme values.
VSC production. (i) New insights into methionine catabolism.
VSC production was observed mainly with a high methionine supply. Concerning aminotransferase-encoding genes possibly involved in methionine catabolism (ARO8 and ARO9 aromatic aminotransferase genes and BAT1 and BAT2 branched-chain aminotransferase genes), only BAT1 was overexpressed under high-methionine conditions. This gene could therefore be a good candidate for methionine catabolism. Bondar et al. (6) demonstrated that the overexpression of BAT1 in Y. lipolytica is correlated with an increase in VSC production. BAT1 was also overexpressed under high-cystine conditions, with no increase in VSC production. However, this gene, which is not mainly involved in sulfur metabolism, could be overexpressed due to other metabolic regulations. Since the intracellular methionine pool was increased only under high-methionine conditions, it could result in an overexpression of BAT1 and VSC production. Furthermore, since we did not observe an intracellular pool of KMBA, we hypothesized its active excretion due to its toxic potential. We propose that the transporter gene YALI0C15488g, which is overexpressed under high-methionine conditions, could be responsible for KMBA export. This hypothesis is supported by the previous observation of extracellular KMBA accumulation (28). KMBA can then be converted to various VSCs, depending on the physicochemical properties of the extracellular medium (pH, redox status, and ionic strength).
(ii) Coexistence of two mechanisms for H2S production.
We observed the production of H2S only under high-methionine and high-sulfate conditions. We suppose that this production with a high methionine supply was due to cysteine catabolism, as previously described (29), resulting in an increase in the cysteine pool, whereas the production with a high sulfate supply may have resulted from the sulfate assimilation pathway. H2S production from sulfate could be the consequence of a high sulfur flux in the sulfur assimilation pathway, leading to an H2S leak before its incorporation into a carbon chain. These two phenomena have already been described for cheese and wine, respectively. We confirm here that H2S production strongly depends on sulfur supply, not on differences in the biosynthesis pathway. In wine, where sulfate is predominant, H2S is produced through the sulfate assimilation pathway (30, 31). In contrast, in cheese, where sulfur is retrieved mainly in methionine and cysteine, the production of H2S could arise from cysteine catabolism (9, 32).
Finally, the absence of H2S production under high-cystine conditions was quite surprising, because the addition of cysteine is strictly correlated with H2S production in several yeasts (29). As a consequence, we postulate that cystine assimilation and/or catabolism is different from that of its reduced form, cysteine.
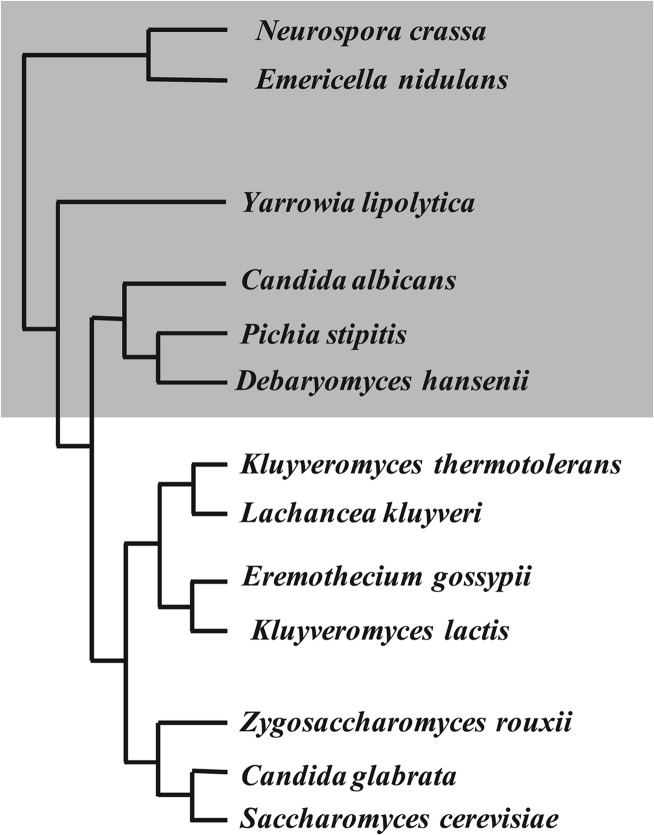
New insights into sulfur metabolism. (i) Identification of taurine metabolism in yeasts.
For the first time, to our knowledge, our metabolomic analysis has proved the existence of a taurine/hypotaurine pool in a yeast species. We performed an in silico study of a limited set of ascomycete species representative of this clade, which led to the identification of the genes involved in taurine/hypotaurine biosynthesis in the part of the evolutionary tree including Yarrowia, the CTG clade members (e.g., Candida albicans and Debaryomyces hansenii), and ascomycetous fungi. This pathway appeared to be absent from the other part of the tree, which included the Saccharomyces clade and the Kluyveromyces clade (Fig. 4). Our previous results confirmed the absence of hypotaurine and taurine pools in Kluyveromyces lactis (17). The hypotaurine and taurine pools, which are increased under high-methionine conditions, could serve as a sulfur reservoir. Furthermore, the slight decrease of γ-glutamylcysteine under high methionine supply could be due to a redirection of the sulfur flux toward taurine and hypotaurine.
Fig 4.
Phylogeny of ascomycetes. The portion in gray shows yeasts and fungi with a complete taurine and hypotaurine pathway.
Taurine has already been found in mammals (33) and in some fish, shellfish, and plants living in extreme environments (34). It is thought to function as an osmoprotectant, antioxidant, and membrane stabilizer. Furthermore, the antistress functions of taurine have led to its use as an antifreezing and antioxidative agent for the preservation of cells, including dried yeasts (35). Meanwhile, its physiological role remains unclear. Our finding that Yarrowia lipolytica naturally synthesizes taurine will help to elucidate its biological role by studying specific mutants thanks to the availability of efficient genetic engineering tools for this yeast. Since the glutathione pool is not modified under the two conditions studied here, this mechanism could also maintain glutathione homeostasis.
(ii) Sulfonates: an important sulfur source for Y. lipolytica.
In S. cerevisiae, JLP1 encodes a sulfonate/alpha-ketoglutarate dioxygenase (36), involved in sulfonate catabolism, which is overexpressed under sulfur starvation (37). In Y. lipolytica, six orthologs of the JLP1 gene product belonging to the TauD protein family (Pfam number Pf02668) have been identified. All of them are repressed under high-sulfur conditions (methionine, cystine, and sulfate). This multiplicity of genes and their strong regulation by the sulfur supply suggest that Y. lipolytica could assimilate sulfonates as a sulfur source.
(iii) Genes related to sulfur metabolism.
In this transcriptomic study, we retrieved numerous genes of unknown function that have already been linked to sulfur metabolism. FMO1 and MXR1 are related to the folding of disulfide-bonding proteins (38) and the response to oxidative stress (39). PRX1, TSA1, and TRX1 code for thioredoxin peroxidases that are repressed under high-methionine conditions. Genes encoding glutathione transferases that are orthologs of fungal genes are repressed at high sulfur concentrations. To our knowledge, orthologs of these genes are absent in other hemiascomycetous yeasts.
Another gene of unknown function (YALI0C22088g) that is repressed at high sulfur concentrations has sequence similarities with four genes involved in sulfur metabolism (16). The repression of YALI0C22088g and its high conservation in hemiascomycetous yeasts confirm its involvement in sulfur metabolism.
YIL166C and SEO1 encode putative transporters that belong to the complex allantoate transporter family, including the recently defined high-affinity cysteine transporter (encoded by YCT1) (40, 41). Boer et al. (42) identified YIL166c in S. cerevisiae as a gene induced under sulfur limitation. All of these observations strongly suggest that these transporters could be linked to sulfur metabolism. The phylogenetic divergences and multiplicity of transporters in Y. lipolytica provide an invaluable opportunity for studying transporter evolution and specificity.
To conclude, Yarrowia lipolytica's sulfur metabolism confirms the particular status of this yeast, located at the frontier of hemiascomycetes and fungi. As a consequence, this yeast is an excellent candidate for evolutionary metabolism studies. In addition, the great interest in Yarrowia lipolytica for biotechnology makes this yeast a major element in fundamental research as well as applied research.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the EcoMet program (grant ANR-06-PNRA-014), funded by the French National Research Agency (ANR). A.H. and M.-P.F.-G. are grateful to the ANR for Ph.D. scholarships.
We thank Armelle Delile, Nour Elhayate Badazzi, Roselyne Tâche, and Emmanuelle Rebours for their helpful technical assistance.
Footnotes
Published ahead of print 7 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03259-12.
REFERENCES
- 1. Bankar AV, Kumar AR, Zinjarde SS. 2009. Environmental and industrial applications of Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 84:847–865 [DOI] [PubMed] [Google Scholar]
- 2. Corsetti A, Rossi J, Gobbetti M. 2001. Interactions between yeasts and bacteria in the smear surface-ripened cheeses. Int. J. Food Microbiol. 69:1–10 [DOI] [PubMed] [Google Scholar]
- 3. Gonzalez-Lopez CI, Szabo R, Blanchin-Roland S, Gaillardin C. 2002. Genetic control of extracellular protease synthesis in the yeast Yarrowia lipolytica. Genetics 160:417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guerzoni ME, Lanciotti R, Vannini L, Galgano F, Favati F, Gardini F, Suzzi G. 2001. Variability of the lipolytic activity in Yarrowia lipolytica and its dependence on environmental conditions. Int. J. Food Microbiol. 69:79–89 [DOI] [PubMed] [Google Scholar]
- 5. Suzzi G, Lanorte MT, Galgano F, Andrighetto C, Lombardi A, Lanciotti R, Guerzoni ME. 2001. Proteolytic, lipolytic and molecular characterisation of Yarrowia lipolytica isolated from cheese. Int. J. Food Microbiol. 69:69–77 [DOI] [PubMed] [Google Scholar]
- 6. Bondar DC, Beckerich JM, Bonnarme P. 2005. Involvement of a branched-chain aminotransferase in production of volatile sulfur compounds in Yarrowia lipolytica. Appl. Environ. Microbiol. 71:4585–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cholet O, Hénaut A, Casaregola S, Bonnarme P. 2007. Gene expression and biochemical analysis of cheese-ripening yeasts: focus on catabolism of l-methionine, lactate, and lactose. Appl. Environ. Microbiol. 73:2561–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hébert A, Beckerich JM, Landaud S, Bonnarme P. Sulphur metabolism of the cheese-ripening yeast Yarrowia lipolytica. Microbiol. Monogr., in press [Google Scholar]
- 9. Landaud S, Helinck S, Bonnarme P. 2008. Formation of volatile sulfur compounds and metabolism of methionine and other sulfur compounds in fermented food. Appl. Microbiol. Biotechnol. 77:1191–1205 [DOI] [PubMed] [Google Scholar]
- 10. Bonnarme P, Lapadatescu C, Yvon M, Spinnler HE. 2001. l-Methionine degradation potentialities of cheese-ripening microorganisms. J. Dairy Res. 68:663–674 [DOI] [PubMed] [Google Scholar]
- 11. Bonnarme P, Psoni L, Spinnler HE. 2000. Diversity of l-methionine catabolism pathways in cheese-ripening bacteria. Appl. Environ. Microbiol. 66:5514–5517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arfi K, Landaud S, Bonnarme P. 2006. Evidence for distinct l-methionine catabolic pathways in the yeast Geotrichum candidum and the bacterium Brevibacterium linens. Appl. Environ. Microbiol. 72:2155–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lopez del Castillo Lozano M, Tâche R, Bonnarme P, Landaud S. 2007. Evaluation of a quantitative screening method for hydrogen sulfide production by cheese-ripening microorganisms: the first step towards l-cysteine catabolism. J. Microbiol. Methods 69:70–77 [DOI] [PubMed] [Google Scholar]
- 14. Mansour S, Beckerich JM, Bonnarme P. 2008. Lactate and amino acid catabolism in the cheese-ripening yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 74:6505–6512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dujon B. 2006. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 22:375–387 [DOI] [PubMed] [Google Scholar]
- 16. Hébert A, Casaregola S, Beckerich J-M. 2011. Biodiversity in the sulfur metabolism in hemiascomycetous yeasts. FEMS Yeast Res. 11:366–378 [DOI] [PubMed] [Google Scholar]
- 17. Hébert A, Forquin-Gomez Roux M-PA, Aubert J, Junot C, Loux V, Heilier J-F, Bonnarme P, Beckerich J-M, Landaud S. 2011. Exploration of sulfur metabolism in the yeast Kluyveromyces lactis. Appl. Microbiol. Biotechnol. 91:1409–1423 [DOI] [PubMed] [Google Scholar]
- 18. Godard P, Urrestarazu A, Vissers S, Kontos K, Bontempi G, van Helden J, André B. 2007. Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 27:3065–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15 doi:10.1093/nar/30.4.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aubert J. 2010. anapuce: tools for microarray data analysis. R package, version 2.2. http://CRAN.R-project.org/package=anapuce
- 21. Delmar P, Robin S, Daudin JJ. 2005. VarMixt: efficient variance modelling for the differential analysis of replicated gene expression data. Bioinformatics 21:502–508 [DOI] [PubMed] [Google Scholar]
- 22. Forquin M-P, Hébert A, Roux A, Aubert J, Proux C, Heilier J-F, Landaud S, Junot C, Bonnarme P, Martin-Verstraete I. 2011. Global regulation of the response to sulfur availability in the cheese-related bacterium Brevibacterium aurantiacum. Appl. Environ. Microbiol. 77:1449–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. 2006. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78:779–787 [DOI] [PubMed] [Google Scholar]
- 24. Kraidlova L, Van Zeebroeck G, Van Dijck P, Sychrová H. 2011. The Candida albicans GAP gene family encodes permeases involved in general and specific amino acid uptake and sensing. Eukaryot. Cell 10:1219–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Knijnenburg TA, Daran JMG, van den Broek MA, Daran-Lapujade PA, de Winde JH, Pronk JT, Reinders MJT, Wessels LFA. 2009. Combinatorial effects of environmental parameters on transcriptional regulation in Saccharomyces cerevisiae: a quantitative analysis of a compendium of chemostat-based transcriptome data. BMC Genomics 10:53 doi:10.1186/1471-2164-10-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hansen J, Johannesen PF. 2000. Cysteine is essential for transcriptional regulation of the sulfur assimilation genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 263:535–542 [DOI] [PubMed] [Google Scholar]
- 27. Avram D, Bakalinsky AT. 1997. SSU1 encodes a plasma membrane protein with a central role in a network of proteins conferring sulfite tolerance in Saccharomyces cerevisiae. J. Bacteriol. 179:5971–5974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kagkli DM, Tâche R, Cogan TM, Hill C, Casaregola S, Bonnarme P. 2006. Kluyveromyces lactis and Saccharomyces cerevisiae, two potent deacidifying and volatile-sulphur-aroma-producing microorganisms of the cheese ecosystem. Appl. Microbiol. Biotechnol. 73:434–442 [DOI] [PubMed] [Google Scholar]
- 29. Lopez del Castillo-Lozano M, Delile A, Spinnler HE, Bonnarme P, Landaud S. 2007. Comparison of volatile sulphur compound production by cheese-ripening yeasts from methionine and methionine-cysteine mixtures. Appl. Microbiol. Biotechnol. 75:1447–1454 [DOI] [PubMed] [Google Scholar]
- 30. Cordente AG, Heinrich A, Pretorius IS, Swiegers JH. 2009. Isolation of sulfite reductase variants of a commercial wine yeast with significantly reduced hydrogen sulfide production. FEMS Yeast Res. 9:446–459 [DOI] [PubMed] [Google Scholar]
- 31. Linderholm AL, Findleton CL, Kumar G, Hong Y, Bisson LF. 2008. Identification of genes affecting hydrogen sulfide formation in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 74:1418–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lopez del Castillo-Lozano M, Mansour S, Tâche R, Bonnarme P, Landaud S. 2008. The effect of cysteine on production of volatile sulphur compounds by cheese-ripening bacteria. Int. J. Food Microbiol. 122:321–327 [DOI] [PubMed] [Google Scholar]
- 33. Dominy J, Eller S, Dawson R., Jr 2004. Building biosynthetic schools: reviewing compartmentation of CNS taurine synthesis. Neurochem. Res. 29:97–103 [DOI] [PubMed] [Google Scholar]
- 34. Stintzing FC, Schieber A, Carle R. 2001. Phytochemical and nutritional significance of cactus pear. Eur. Food Res. Technol. 212:396–407 [Google Scholar]
- 35. Honjoh K, Machida T, Nishi K, Matsuura K, Soli KW, Sakai T, Ishikawa H, Matsumoto K, Miyamoto T, Iio M. 2007. Improvement of freezing and oxidative stress tolerance in Saccharomyces cerevisiae by taurine. Food Sci. Technol. Res. 13:145–154 [Google Scholar]
- 36. Hogan DA, Auchtung TA, Hausinger RP. 1999. Cloning and characterization of a sulfonate/alpha-ketoglutarate dioxygenase from Saccharomyces cerevisiae. J. Bacteriol. 181:5876–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang N, Merlotti C, Wu J, Ismail T, El-Moghazy AN, Khan SA, Butt A, Gardner DC, Sims PF, Oliver SG. 2001. Functional analysis of six novel ORFs on the left arm of chromosome XII of Saccharomyces cerevisiae reveals three of them responding to S-starvation. Yeast 18:325–334 [DOI] [PubMed] [Google Scholar]
- 38. Suh JK, Poulsen LL, Ziegler DM, Robertus JD. 1999. Yeast flavin-containing monooxygenase generates oxidizing equivalents that control protein folding in the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 96:2687–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Luikenhuis S, Perrone G, Dawes IW, Grant CM. 1998. The yeast Saccharomyces cerevisiae contains two glutaredoxin genes that are required for protection against reactive oxygen species. Mol. Biol. Cell 9:1081–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hellborg L, Woolfit M, Arthursson-Hellborg M, Piskur J. 2008. Complex evolution of the DAL5 transporter family. BMC Genomics 9:164 doi:10.1186/1471-2164-9-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kaur J, Bachhawat AK. 2007. Yct1p, a novel, high-affinity, cysteine-specific transporter from the yeast Saccharomyces cerevisiae. Genetics 176:877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boer VM, de Winde JH, Pronk JT, Piper MDW. 2003. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 278:3265–3274 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.