Abstract
The ability to produce diacetyl from pyruvate and l-serine was studied in various strains of Pediococcus pentosaceus and Pediococcus acidilactici isolated from cheese. After being incubated on both substrates, only P. pentosaceus produced significant amounts of diacetyl. This property correlated with measurable serine dehydratase activity in cell extracts. A gene encoding the serine dehydratase (dsdA) was identified in P. pentosaceus, and strains that showed no serine dehydratase activity carried mutations that rendered the gene product inactive. A functional dsdA was cloned from P. pentosaceus FAM19132 and expressed in Escherichia coli. The purified recombinant enzyme catalyzed the formation of pyruvate from l- and d-serine and was active at low pH and elevated NaCl concentrations, environmental conditions usually present in cheese. Analysis of the amino acid profiles of culture supernatants from dsdA wild-type and dsdA mutant strains of P. pentosaceus did not show differences in serine levels. In contrast, P. acidilactici degraded serine. Moreover, this species also catabolized threonine and produced alanine and α-aminobutyrate.
INTRODUCTION
Pediococcus pentosaceus and Pediococcus acidilactici are homofermentative Gram-positive cocci that produce d- and l-lactate from carbohydrates. These bacteria are regularly found in the nonstarter population of raw milk cheeses at the end of the ripening process. Thus, Pediococcus spp. were isolated from cheddar, ewes' milk cheeses, Comté, AOC Salers, Puzzone di Moena, and various raw milk cheeses traditionally produced in Switzerland, such as Gruyère, Emmentaler, Appenzeller, and Tilsit (1–8).
Although several reports have indicated that pediococci alone or combined with lactobacilli can accelerate cheese ripening and enhance flavor development, respectively (9–13), knowledge about metabolic activities affecting the maturation of cheese is limited. Studies on the physiological and biochemical activities of Pediococcus spp. have mainly focused on carbohydrate metabolism and proteolytic and lipolytic activities (14–16). Researchers have also reported that pediococci racemize l-lactate to d-lactate under anaerobic conditions and that pediococci can oxidize lactate to acetate and carbon dioxide under aerobic conditions (17). However, information on the enzymatic degradation of amino acids, important precursors for flavor compounds, is sparse.
Diacetyl contributes to desirable flavor in fermented dairy products. A pathway well-known for producing diacetyl is through degradation of citrate (18). In this pathway, pyruvate is formed as an intermediary compound that is then converted via α-acetolactate to diacetyl. Although pediococci do not appear to utilize citrate, these species have been reported to form diacetyl (19, 20). Pyruvate, which is derived from the metabolism of carbohydrates or amino acids, is likely used for synthesizing diacetyl.
In this study, the ability of various strains of P. acidilactici and of P. pentosaceus isolated from cheese to produce diacetyl from pyruvate and serine was analyzed. Furthermore, serine dehydratase activity that releases pyruvate from serine was investigated in detail in these strains. The deamination of serine is of interest since this compound is released by proteolysis during cheese ripening and is a potential precursor for flavor compounds.
MATERIALS AND METHODS
Bacterial strain, media, and growth conditions.
The strains used in this study are listed in Table 1. Pediococci which had been identified with 16S rRNA gene sequence analyses were stored at −80°C in sterile reconstituted skim milk powder (10%, wt/vol). The bacteria were cultivated at 30°C in MRS broth (21) or in a basal broth that had been used to study serine metabolism in Lactobacillus plantarum (22). For our study, the basal broth (pH 7.0) was slightly modified and consisted of KH2PO4 at 9 g liter−1, yeast extract at 5 g liter−1, casein hydrolysate at 1 g liter−1, MgSO4 · 7H2O at 0.1 g liter−1, MnCl2 · 4H2O at 0.1 g liter−1, galactose at 2 g liter−1, and 5 mM l-serine.
Table 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Genotype, relevant properties, or sequence (5′–3′) | Source |
|---|---|---|
| Strains | ||
| E. coli BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm (DE3) | Invitrogen |
| P. pentosaceus | ||
| FAM13073 | Tilsit | |
| FAM17622 | dsdA mutant (deletion at position 107) | Gruyère |
| FAM18813 | Tête de Moine | |
| FAM19080 | dsdA mutant (amino acid substitutions T67A and P278La) | Gruyère |
| FAM19132 | Gruyère | |
| FAM19144 | dsdA mutant (deletion from positions 90 to 108) | Sbrinz |
| FAM19169 | Tilsit | |
| FAM20650 | dsdA mutant (nonsense mutation C → T at position 151) | Emmentaler |
| DSM 20336 | Type strain | German Collection of Microorganisms and Cell Cultures |
| P. acidilactici | ||
| FAM13473 | Emmentaler | |
| FAM13881 | Emmentaler | |
| FAM17411 | Gruyère | |
| FAM17418 | Gruyère | |
| FAM17422 | Gruyère | |
| FAM18098 | Gruyère | |
| FAM18987 | Tête de Moine | |
| FAM19460 | Emmentaler | |
| FAM20559 | Gruyère | |
| DSM 20284 | Type strain | German Collection of Microorganisms and Cell Cultures |
| Plasmids | ||
| pEXP5-CT/TOPO | E. coli cloning vector, Ampr | Invitrogen |
| pEXP5-CT/dsdA[19132] | Plasmid expressing dsdA from FAM19132, Ampr | This study |
| pEXP5-CT/dsdA[19080] | Plasmid expressing dsdA from FAM19080, Ampr | This study |
| Primersb | ||
| PEPE_1745F | ATGATTGATGTTGATGCTCTA | |
| PEPE_1745R | TTTAACTTGATGTCCTTCAG |
Compared to PEPE_1745 from P. pentosaceus ATCC 25745.
Primers were used for cloning and sequencing of dsdA.
Strains of Escherichia coli were grown at 37°C in LB broth (23) supplemented with ampicillin (100 μg ml−1), when necessary.
Determination of diacetyl and acetoin formation.
Pediococci were grown in 50 ml of MRS broth for 16 h at 30°C. Cells were then sedimented at 3,000 × g for 10 min, washed twice with 50 mM potassium phosphate (pH 5.5), and finally, suspended in 1 ml of the same buffer. The cell suspension (0.5 ml) was incubated with either 10 mM pyruvate or 10 mM l-serine in 70 mM potassium phosphate (pH 5.5) containing 1× RPMI 1640 vitamin solution (Sigma-Aldrich, Switzerland) and 50 μM pyridoxal 5′-phosphate (PLP) at a final volume of 5 ml in sealed tubes at 30°C for 24 h.
Diacetyl and acetoin were determined with gas chromatography (GC) with flame ionization detection (FID). Therefore, solid sodium chloride was added until the sample was saturated. After the sample was heated at 95°C for 45 min, 1 ml of the headspace was injected into the GC. The GC was equipped with an HP-ULTRA 2 capillary column with cross-linked 5% phenylmethyl silicone (50 m by 0.32 mm by 0.52 μm). The transfer to the capillary column was performed in split mode (1:8), and helium served as the carrier gas. The temperature program was the following: initially 50°C for 7 min and then a ramp at 40°C min−1 to 270°C. Additional operating conditions of the GC were an injector temperature of 150°C and a detector temperature of 320°C.
Preparation of cell extract.
The Pediococcus spp. were grown for 16 h at 30°C in 50 ml of MRS broth or basal broth. The cells were then harvested by centrifugation (3,000 × g, 10 min, 25°C), washed twice with 20 mM sodium phosphate (pH 7.4), and finally, suspended in 1 ml of the same buffer. After approximately 0.6 g of glass beads (diameter, 212 to 300 μm) was added, the cells were disrupted in an Omni Bead Ruptor 24 (Omni International) by vigorous shaking (one 45-s mixing sequence at a speed of 6 m s−1). The extract was cleared with centrifugation (17,900 × g, 10 min, 4°C).
Protein determination.
The protein content of the extracts was determined with bicinchoninic acid (BCA) using the micro-BCA reagent protocol described by Smith et al. (24). Bovine serum albumin was used to prepare a standard curve.
Enzyme assay.
Serine dehydratase activity was determined as follows: cell extract (50 μl) or recombinant enzyme (0.2 μg) was added to 25 mM sodium phosphate (pH 7.4), 50 μM PLP, and various concentrations of l-serine or d-serine in a final volume of 200 μl. The cell extract was incubated for 30 min, and the recombinant enzyme was incubated for 10 min at 30°C. The assay was stopped by adding 20 μl of 5 M perchloric acid (Fluka, Switzerland). The solution was centrifuged (17,900 × g, 10 min), and 50 μl of the supernatant was separated by high-pressure liquid chromatography using an Aminex HPX-87H column (300 by 7.8 mm) and 3.8 mM H2SO4 as the mobile phase. The operating conditions were as follows: the column temperature was 65°C, the flow rate was 0.6 ml min−1, and detection was at 210 nm. Chromatographic identification of pyruvate was performed by comparing the retention times to those of a pyruvate standard. The amount of pyruvate formed was calculated by referring to a standard curve prepared with various concentrations of sodium pyruvate and normalized for protein content.
The values of Km and Vmax of the recombinant enzyme for l-serine and d-serine were determined with various concentrations of l-serine (0.25 to 10 mM) and d-serine (0.05 to 10 mM). Km and Vmax were calculated from Hanes-Woolf plots (the ratio of the substrate and velocity as a function of the substrate) and represent the means of triplicates.
Isolation of DNA.
Genomic DNA from Pediococcus spp. was extracted by robot extraction (BioRobot EZ1; EZ1 DNA tissue kit; Qiagen, Switzerland) from 1 ml of culture grown overnight at 30°C. Before being extracted, cells were treated with 0.05 N NaOH for 15 min at room temperature, followed by treatment with TES buffer (0.1 M Tris-HCl [pH 8.0], 10 mM EDTA, 25% sucrose) supplemented with lysozyme (1 mg ml−1) for 1 h at 37°C.
E. coli plasmid DNA was isolated with a QIAprep spin miniprep kit (Qiagen) according to the manufacturer's instructions.
DNA sequencing.
The amplification products obtained from the genomic DNA of P. pentosaceus strains with the primer pair PEPE_1745F/PEPE_1745R (Table 1) were sequenced using a BigDye Terminator cycle sequencing kit and an ABI Prism 310 genetic analyzer (Applied Biosystems, Switzerland).
Construction of plasmids.
dsdA was amplified with the primer pair PEPE_1745F/PEPE_1745R from genomic DNA of P. pentosaceus FAM19132 and P. pentosaceus FAM19080 by PCR. The PCR product was cloned into the pEXP5-CT/TOPO expression vector (Invitrogen, Switzerland) according to the manufacturer's instructions. The cloning was such that the expressed protein had eight extra amino acids, including a polyhistidine tag at the C terminus (KGHHHHHH). The plasmids containing dsdA in the proper orientation were confirmed by sequencing and called, depending on the origin of the dsdA gene (i.e., P. pentosaceus FAM19132 and P. pentosaceus FAM19080), pEXP5-CT/dsdA[19132] and pEXP5-CT/dsdA[19080]. Restriction, ligation, and calcium chloride-mediated transformation of E. coli cells were performed using standard procedures (23).
Expression and purification of His-tagged proteins.
E. coli BL21(DE3) transformed with pEXP5-CT/dsdA[19132] and pEXP5-CT/dsdA[19080] was grown in 200 ml LB broth containing ampicillin (100 μg ml−1) at 37°C on a shaker. When the optical density at 600 nm reached 0.5, expression of recombinant dsdA was induced by adding 1 mM isopropyl-β-d-1-thiogalactopyranoside (final concentration). Expression was performed at 27°C. After 4 h of incubation on a shaker, the bacterial cells were harvested by centrifugation, washed twice with 20 mM sodium phosphate (pH 7.4), and then frozen at −20°C.
Cells were disrupted as described above, and the recombinant proteins were purified with Protino Ni-TED 2000 packed columns (Macherey-Nagel, Switzerland) according to the manufacturer's instructions. The elution fractions were pooled and immediately applied to PD-10 columns (GE Healthcare, Switzerland) that had been equilibrated with 20 mM sodium phosphate (pH 7.4) to remove imidazole. The eluted protein fraction was concentrated with Ultracel 30K (Millipore, Ireland) and analyzed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by colloidal Coomassie staining. The concentration of the purified protein was determined as described above.
Size-exclusion chromatography.
To determine the molecular size under nondenaturing conditions, the purified recombinant proteins were analyzed with size-exclusion chromatography on a Superdex 200 HR 10/30 column (GE Healthcare) with 25 mM sodium phosphate buffer (pH 7.4) containing 150 mM NaCl at a flow rate of 0.5 ml min−1. The column was calibrated with a mixture of proteins of known molecular weight (Gel Filtration Standard; Bio-Rad, Switzerland).
2D-TLC.
Pediococcus spp. precultured in MRS broth were grown in 50 ml of MRS broth or 50 ml of a basal broth at 30°C. To study amino acid metabolism, 1 μl of the culture supernatants was applied spotwise onto high-performance thin-layer chromatography cellulose plates (Merck, Germany) and separated with two-dimensional thin-layer chromatography (2D-TLC). The plates were developed in the first dimension with n-butanol–acetone–diethylamine–water (10 + 10 + 2 + 5 parts, vol/vol/vol/vol) and in the second dimension with 2-propanol–formic acid–water (40 + 2 + 10 parts, vol/vol/vol). To visualize the amino acids, plates were immersed into 0.1% ninhydrin solution (dissolved in acetone) and heated at 110°C for 5 min.
RESULTS
Formation of diacetyl and acetoin.
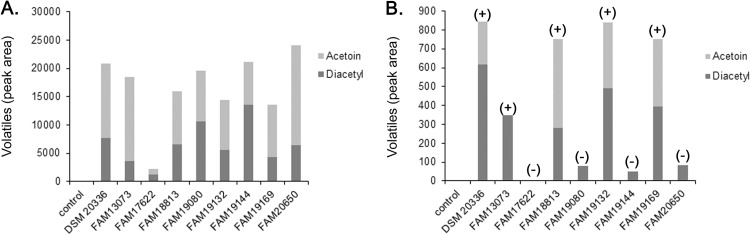
The formation of diacetyl and acetoin was studied in resting cell suspensions of 9 strains of P. pentosaceus and 10 strains of P. acidilactici (Table 1). Therefore, cells were collected 16 h after inoculation, washed, and incubated in phosphate buffer containing pyruvate at pH 5.3. After 24 h of incubation at 30°C, all studied strains of P. pentosaceus produced diacetyl and acetoin (Fig. 1A). Only very low levels of formation or no formation of these two flavor compounds was detected in the samples containing the strains of P. acidilactici (data not shown). When the Pediococcus spp. were incubated in phosphate buffer containing l-serine, diacetyl and acetoin were found in the headspace of the strains of P. pentosaceus (Fig. 1B). Four strains, namely, P. pentosaceus FAM17622, P. pentosaceus FAM19080, P. pentosaceus FAM19144, and P. pentosaceus FAM20650, did not produce diacetyl or produced only very low levels. No flavor formation from serine was observed with the strains of P. acidilactici (data not shown).
Fig 1.
Formation of diacetyl and acetoin from pyruvate (A) and l-serine (B) by various strains of P. pentosaceus. (+), the cell extracts of the strains exhibited serine dehydratase activity; (−), the cell extracts of strains lacked serine dehydratase activity.
Serine dehydratase activity.
First, serine dehydratase activity was determined in P. pentosaceus FAM19132, which produced diacetyl when incubated with serine. Therefore, cell extracts were prepared from cells that were collected at 12, 24, 48, 72, and 96 h after inoculation of MRS broth. The release of pyruvate from serine was detected only in the samples taken at 12 and 24 h. This showed the presence of a serine dehydratase whose activity was apparently downregulated in resting cells.
Then, cell extracts of all Pediococcus spp. which had been grown for 16 h in MRS broth were incubated with l-serine. All P. pentosaceus strains except P. pentosaceus FAM17622, P. pentosaceus FAM19080, P. pentosaceus FAM19144, and P. pentosaceus FAM20650 exhibited serine dehydratase activity (Fig. 1B). On the contrary, no formation of pyruvate was found in the extracts from the P. acidilactici strains (data not shown).
Sequence analysis of dsdA.
The genome data of P. pentosaceus ATCC 25745 (GenBank accession number CP000422) and the entire genome shotgun sequences of P. acidilactici DSM 20284 (GenBank accession numbers AEEG01000001 to AEEG01000012) were analyzed for the presence of genes encoding a serine dehydratase. Indeed, the gene PEPE_1745 from P. pentosaceus ATCC 25745 encodes a putative d-serine ammonia lyase. The translated sequence of this gene showed 49% identity to the well-studied PLP-dependent d-serine dehydratase (DsdA) from E. coli (GenBank accession number P00926). A homologous gene was not found in P. acidilactici DSM 20284.
On the basis of the nucleotide sequence of PEPE_1745, the primer pair PEPE_1745F/PEPE_1745R was designed and used to amplify and sequence the gene from all studied strains of P. pentosaceus. Regarding the four P. pentosaceus strains that did not exhibit serine dehydratase activity, a 1-bp deletion and a 19-bp deletion were found in P. pentosaceus FAM17622 and P. pentosaceus FAM19144, respectively. Both mutations introduced frameshift mutations (Fig. 2). In P. pentosaceus FAM20650, a transition from C to T was detected at position 151, leading to a nonsense mutation. By comparing the sequence obtained from FAM19080 with that of PEPE_1745, two missense mutations were identified that replaced a threonine residue with an alanine residue at position 67 and a proline residue with a leucine residue at position 278 in the translated sequence.
Fig 2.
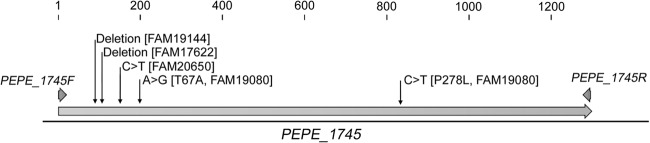
Schematic illustration of PEPE_1745 showing the mutation sites found in P. pentosaceus FAM17622, P. pentosaceus FAM19144, P. pentosaceus FAM19080, and P. pentosaceus FAM20650 that affect function. Additionally, the binding sites of the primers PEPE_1745F and PEPE_1745R used for cloning and sequencing are shown.
Purification and properties of DsdA.
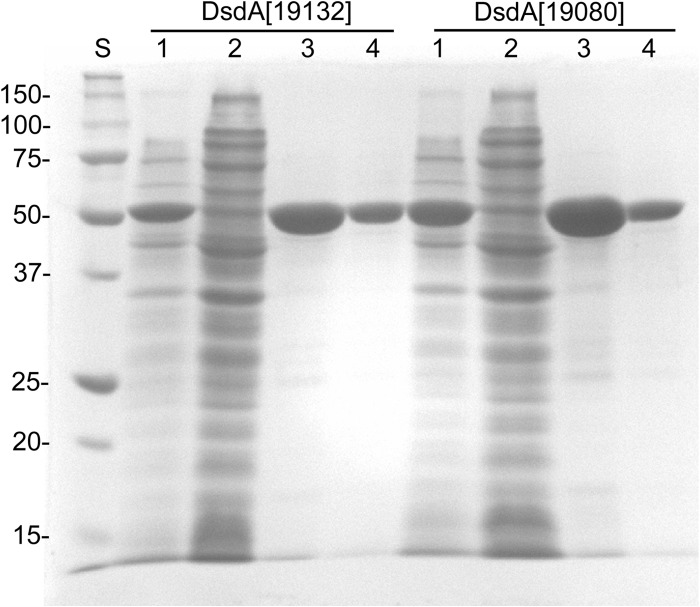
The genes homologous to PEPE_1745 were cloned from FAM19132 and FAM19080 in frame with a C-terminal polyhistidine tag and named dsdA[19132] and dsdA[19080], respectively. The proteins encoded by both genes were expressed in soluble form in E. coli BL21(DE3) and purified to apparent homogeneity with one-step nickel-affinity chromatography (Fig. 3). Both proteins migrated at 52 kDa in SDS-polyacrylamide gels (expected size, 48.5 kDa). The apparent size determined by size-exclusion chromatography under nondenaturing conditions was 120 kDa for each protein (data not shown).
Fig 3.
SDS-PAGE analysis of the purification of DsdA[19132] and DsdA[19080] by nickel-affinity chromatography. Lanes: S, protein standard; 1, cell extract of E. coli after expression; 2, unbound proteins; 3, protein fraction obtained after nickel-affinity chromatography; 4, purified protein after removal of imidazole.
The purified recombinant proteins were assayed for serine dehydratase activity. Although no enzyme activity was detected for DsdA[19080], DsdA[19132] clearly exhibited serine dehydratase activity. This protein formed pyruvate from l-serine and d-serine. No formation of a keto acid was detected when l-threonine was used as the substrate. The activity of recombinant DsdA[19132] was studied as a function of l-serine and d-serine at 30°C and pH 7.4. The enzyme followed Michaelis-Menten kinetics, and a Km value of 7.5 ± 0.3 mM (Vmax = 880.7 ± 71.3 7 μmol min−1 mg−1) for l-serine and a Km value of 0.5 ± 0.1 mM (Vmax = 71.3 ± 4.7 μmol min−1 mg−1) for d-serine were calculated.
Sodium phosphate buffer with various pHs and 15 mM l-serine were used to study the pH dependence. DsdA[19132] exhibited optimal activity between pH 6.5 and 7.4 and had about 70% residual activity at pH 5.5. Furthermore, the influence of NaCl (1%, 2%, and 4%) was studied. Whereas degradation of l-serine was slightly inhibited by increasing the amounts of NaCl, the degradation of d-serine was enhanced (data not shown). To study thermal stability, the enzyme was incubated for 1 h at 43°C, 48°C, 57°C, and 70°C. The enzyme treated at the last three temperatures showed strongly decreased serine dehydratase activity (data not shown).
Serine degradation.
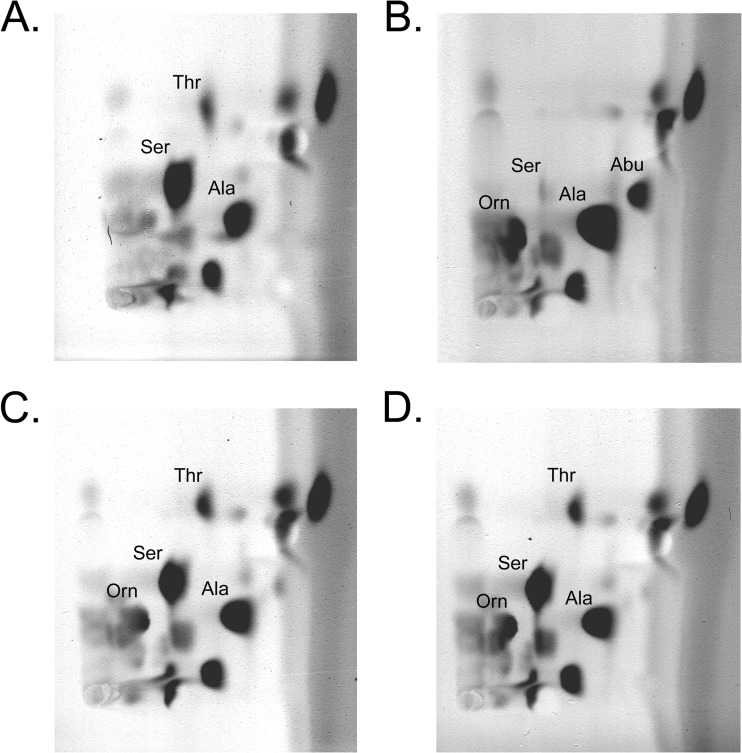
To study the bacterial metabolism of serine, the amino acids of culture supernatants were separated with 2D-TLC after 1, 2, and 3 days of fermentation. The amino acid patterns from P. pentosaceus FAM19132, P. pentosaceus FAM19144, and P. acidilactici FAM18098 were compared. No changes in the amino acid profiles were detected when the strains were grown in MRS broth (data not shown). Significant changes were found between the two strains of P. pentosaceus and P. acidilactici FAM18098 when they were cultured in basal broth. P. acidilactici FAM18098 completely metabolized serine and threonine, and increased alanine and α-aminobutyrate levels were detected (Fig. 4B). In contrast, the serine, threonine, and alanine levels in the culture supernatants of both strains of P. pentosaceus were not different from those in the basal broth, and formation of α-aminobutyrate was not observed (Fig. 4C and D).
Fig 4.
Amino acid patterns of basal broth (A) fermented with P. acidilactici FAM18098 (B), P. pentosaceus FAM19132 (C), and P. pentosaceus FAM19144 (D) for 3 days. The amino acids were separated with 2D-TLC and visualized with ninhydrin. Thr, threonine; Ser, serine; Ala, alanine; Abu, α-aminobutyrate; Orn, ornithine.
Moreover, all three pediococci increased the level of ornithine. Further analysis revealed that all studied strains of pediococci produced ornithine, but only the P. acidilactici strains degraded threonine and serine (data not shown).
DISCUSSION
Nonstarter lactic acid bacteria (NSLAB) constitute a significant part of the microbial ecosystem in cheese and influence its maturation and quality (25). Interest in using NSLAB as adjunct cultures in manufacturing cheese to accelerate and enhance flavor formation and to control the adventitious microbiota is increasing. To select appropriate strains, understanding the metabolic activities of the various species present in the NSLAB flora is crucial.
Proteolysis and degradation of amino acids are a major biochemical event during cheese ripening (26). P. acidilactici and P. pentosaceus are often detected in cheese at the end of the ripening process, and their presence suggests a role during cheese ripening. These species have proteolytic activities (14, 27, 28), but to our knowledge, reports on amino acid use are sparse. Amino acid degradation involves decarboxylases, aminotransferases, deaminases, lyases, and dehydratases (29). Most of the studies on amino acid metabolism by NSLAB have focused on transaminases and lyases. The catabolism of serine by dehydratases is of special interest since these enzymes are active in an anaerobic environment and produce pyruvate, which can be converted not only into lactic acid but also into acetic acid, ethanol, diacetyl, and acetaldehyde. These compounds are aroma active and contribute to the typical flavor of a range of fermented products.
We had observed that the studied strains of Pediococcus spp. isolated from cheese did not metabolize citrate (data not shown). Inspired by reports that Pediococcus spp. produce diacetyl, we first analyzed whether the strains can use pyruvate, an intermediary metabolite in citrate degradation and a precursor of diacetyl formation. Interestingly, all studied P. pentosaceus strains clearly produced diacetyl, a property not distinctive of the P. acidilactici strains. When serine was used as the precursor, various P. pentosaceus strains again produced diacetyl, indicating that serine dehydratase activity that converts serine to pyruvate is present. In fact, serine dehydratase activity was measured in the cell extracts of strains that produced significant amounts of diacetyl from serine. This indicated that this enzyme might be used as a marker for selecting flavor-producing strains.
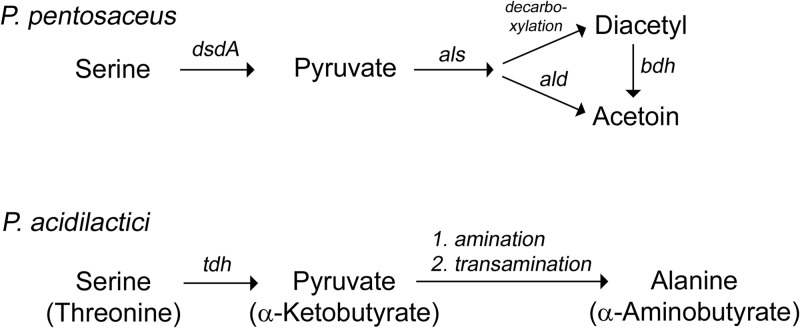
With the advent of next-generation sequencing technologies, more and more genome data on lactic acid bacteria are published. These data can be searched for genes putatively involved in amino acid degradation. In this study, we used the information to search for serine dehydratases present in pediococci and found that PEPE_1745 from P. pentosaceus ATCC 25745 encodes a putative DsdA. Interestingly, by sequencing and aligning the nucleotide sequences of the dsdA gene from the P. pentosaceus strains (see Fig. S1 in the supplemental material), mutations that clearly affect the functionality of the enzyme were identified in the strains that did not exhibit serine dehydratase activity. In P. pentosaceus FAM17622, P. pentosaceus FAM19144, and P. pentosaceus FAM20650, mutations led to truncated, nonfunctional enzymes. In P. pentosaceus FAM19080, the open reading frame of the gene seemed to be complete. However, an alignment of DsdA[19080] with all DsdA sequences from the studied P. pentosaceus strains and DsdA from E. coli showed that at position 67 a threonine residue is replaced with an alanine residue and at position 278 a proline residue is replaced with a leucine residue. The latter amino acid substitution lies next to the so-called tetraglycine loop (residues G279, G281, G282, and G283 in DsdA from E. coli), which is conserved in PLP-dependent enzymes and is involved in the binding of PLP (30). Therefore, we think that the binding of PLP is impaired in DsdA from P. pentosaceus FAM19080. This also explains why serine dehydratase activity was not detected in this strain. By cloning dsdA from P. pentosaceus FAM19080 and analyzing its gene product, we confirmed that the enzyme is actually inactive. We suggest that the serine dehydratase activity detectable in P. pentosaceus is provoked by DsdA activity and is involved in forming diacetyl from serine (Fig. 5). P. acidilactici and P. pentosaceus are difficult to separate using physiological tests (15); thus, the presence of serine dehydratase activity may be used as a marker for distinguishing the two species.
Fig 5.
Proposed pathways for the degradation of serine (and threonine) in P. pentosaceus and P. acidilactici. The synthesis of alanine from pyruvate by P. acidilactici can occur by the action of an alanine dehydrogenase (amination) or an aminotransferase (transamination). dsdA, d-serine dehydratase; als, α-acetolactate synthase; ald, α-acetolactae decarboxylase; bdh, acetoin reductase (2,3-butanediol dehydrogenase); tdh, threonine dehydratase.
To evaluate whether DsdA from P. pentosaceus is active in a cheese environment and plays a role in flavor formation, the recombinant gene product from P. pentosaceus FAM19132 was characterized. The recombinant enzyme used l-serine and d-serine but showed a higher affinity for d-serine. However, l-serine was decomposed more rapidly than d-serine. l-Threonine was not accepted as the substrate. The low pH and elevated salt concentrations present in cheese did not have a major impact on the activity. Interestingly, salt seemed to regulate the rate of dehydratation of l-serine and d-serine. That bacterial d-serine dehydratase uses d- and l-serine has been reported for E. coli, Salmonella enterica serovar Typhimurium, and Klebsiella pneumoniae (31–33). In contrast to the DsdA from P. pentosaceus, these enzymes showed more activity toward d-serine and eluted as monomers during size-exclusion chromatography. Since the recombinant DsdA[19080] and DsdA[19132] eluted with an apparent molecular mass of 120 kDa, we assume that these proteins formed a dimer. The discrepancy between apparent and calculated molecular masses is most likely explained by a nonglobular shape of the protein. The same property was observed for DsdA from Klebsiella, which showed a molecular mass of 40 and 46 kDa in SDS-polyacrylamide gels and by size-exclusion chromatography, respectively. The comparable elution behavior of both recombinant proteins indicates that the amino acid substitutions in DsdA[19080] did not affect the protein conformation.
Although the biochemical properties of DsdA can be well studied, the physiological role remains to be clarified. The strains of P. pentosaceus with and without serine dehydratase activity grew in MRS broth and the serine dehydratase activity was detectable in the cell extracts of growing dsdA+ strains, but no significant differences in serine levels were found in the culture supernatants. This observation indicates that dsdA is expressed during growth in MRS but is not required for growth. In E. coli, dsdA is involved in the detoxification of d-serine, which is toxic to strains of E. coli lacking dsdA. In these strains, l-serine and pantothenate syntheses were inhibited by d-serine (34). Moreover, dsdA may also be involved in the generation of energy in P. pentosaceus, since the protein efficiently converted l-serine to pyruvate. Pyruvate not only is a precursor for diacetyl and acetoin but also can be metabolized to acetate by the action of pyruvate oxidase and acetate kinase. The presence of this pathway in pediococci has been reported (17), and a gene encoding a pyruvate oxidase (PEPE_0639) and two genes encoding acetate kinase (PEPE_0336 and PEPE_1822) are present in the genome of P. pentosaceus ATCC 25745. We plan to perform growth studies in chemical defined medium to understand whether d-serine is a growth inhibitor and l-serine can serve as an energy source for P. pentosaceus.
Since physiological assays with MRS broth do not always reflect the metabolism taking place in a cheese environment, we also analyzed amino acid metabolism in a basal broth previously used for studying serine metabolism of L. plantarum B3089 (22). Lactose was replaced by galactose to avoid glucose repression. Amino acid analysis showed that ornithine was formed by P. acidilactici and P. pentosaceus. This is explained by an active arginine deiminase pathway that converts arginine via citrulline to ornithine and is present in both species (15). Regarding the serine and threonine levels, again, no differences were observed between the dsdA wild-type and dsdA mutant strains of P. pentosaceus. Surprisingly, P. acidilactici FAM18098 degraded serine and threonine and produced significant amounts of alanine and α-aminobutyrate. This metabolism was also observed in all of the other studied P. acidilactici strains (data not shown), indicating that this biochemical activity is generally present in this species. We propose the following pathway: first, serine and threonine are deaminated by a threonine dehydratase to pyruvate and α-ketobutyrate (Fig. 5). That threonine dehydratases catalyze the deamination of both amino acids was shown for E. coli (35). Second, pyruvate and α-ketobutyrate are either aminated by the action of an alanine dehydrogenase or transaminated by the action of an aminotransferase. Evidence for the proposed pathway is found in the genome data of P. acidilactici DSM 20284. There, two neighboring genes named HMPREF0623_0054 and HMPREF0623_0055 encode a putative alanine dehydrogenase and threonine dehydratase, respectively. The organization of these genes implies that both genes may form a transcriptional unit and may be involved in the amino acid conversion mentioned above. However, homologous genes (named PEPE_0166 and PEPE_1067) are also present in P. pentosaceus ATCC 25745. Gene expression analysis and biochemical characterization of these genes and their gene products are necessary, to understand if these genes are in fact involved in forming alanine and α-aminobutyrate and why this activity is not detected in P. pentosaceus.
The use of P. acidilactici and P. pentosaceus as adjunct cultures in cheese making opens up interesting perspectives for controlling cheese quality and flavor formation. On the one hand, these species have proteolytic activity that can accelerate proteolysis and therefore the maturation of cheese (14). On the other hand, investigations of the amino acid metabolism show the presence of the arginine deiminase pathway. This pathway produces gas and ammonia, which can affect pH and eye formation in cheese. Furthermore, the presence of an active serine dehydratase can lead to the production of flavor compounds such as diacetyl and acetate, which contribute to the desirable aroma of various dairy products. However, the role of the serine dehydratase from P. pentosaceus during cheese ripening has to be investigated in the cheese matrix. The observation that P. acidilactici produces alanine, a compound known to confer sweetness to dairy products (36), is promising since these species could be used as natural sweeteners of dairy products. In general, the acceleration of ripening by adding pediococci during cheese manufacturing has already been described (10, 12, 13). However, in these reports, the composition of amino acids and volatiles has not been analyzed in detail. We plan to perform cheese trials with the strains presented in this report to study their influence on amino acid metabolism and flavor formation in cheese. Understanding the metabolic activity of pediococci in more detail is of interest not only for the dairy industry, since these species are already used as starter cultures for various types of sausages and are known to produce undesirable flavor compounds during the manufacture of pickles, wine, and beer (37).
Supplementary Material
ACKNOWLEDGMENT
We thank Monika Haueter for providing technical assistance.
Footnotes
Published ahead of print 14 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03085-12.
REFERENCES
- 1. Bouton Y, Guyot P, Grappin R. 1998. Preliminary characterization of microflora of Comté cheese. J. Appl. Microbiol. 85:123–131 [DOI] [PubMed] [Google Scholar]
- 2. Callon C, Millet L, Montel MC. 2004. Diversity of lactic acid bacteria isolated from AOC Salers cheese. J. Dairy Res. 71:231–244 [DOI] [PubMed] [Google Scholar]
- 3. Coppola R, Nanni M, Iorizzo M, Sorrentino A, Sorrentino E, Grazia L. 1997. Survey of lactic acid bacteria isolated during the advanced stages of the ripening of Parmigiano Reggiano cheese. J. Dairy Res. 64:305–310 [Google Scholar]
- 4. Franciosi E, Settanni L, Carlin S, Cavazza A, Poznanski E. 2008. A factory-scale application of secondary adjunct cultures selected from lactic acid bacteria during Puzzone di Moena cheese ripening. J. Dairy Sci. 91:2981–2991 [DOI] [PubMed] [Google Scholar]
- 5. Fryer TF, Sharpe ME. 1966. Pediococci in cheddar cheese. J. Dairy Res. 33:325–331 [Google Scholar]
- 6. Isolini D, Fröhlich-Wyder MT. 2003. Pediokokken kommen auch in Schweizer Rohmilchkäsesorten vor. Schweiz. Milchzeitung 129:7 [Google Scholar]
- 7. Nunez M, Medina M, Gaya P. 1989. Ewes' milk cheese: technology, microbiology and chemistry. J. Dairy Res. 56:303–321 [DOI] [PubMed] [Google Scholar]
- 8. Tzanetakis N, Litopoulou-Tzanetaki E. 1992. Changes in numbers and kinds of lactic acid bacteria in feta and teleme, two Greek cheeses from ewes' milk. J. Dairy Sci. 75:1389–1393 [Google Scholar]
- 9. Barouei J, Karbassi A, Ghoddusi HB, Mortazavi A, Ramezani R, Moussavi M. 2011. Impact of native Lactobacillus paracasei subsp. paracasei and Pediococcus spp. as adjunct cultures on sensory quality of Iranian white brined cheese. Int. J. Dairy Technol. 64:526–535 [Google Scholar]
- 10. Bhowmik T, Riesterer R, van Boekel MAJS, Marth EH. 1990. Characteristics of low-fat cheddar cheese made with added Micrococcus or Pediococcus species. Milk Sci. Int. 45:230–235 [Google Scholar]
- 11. Goodwins J, Mornet A, Manoury E. November 2008. Aromatization of a milk product using at least one bacteria producing a bacteriocin and belonging to the Pediococcus genus. US patent 2008/0292749 A1
- 12. Tzanetakis N, Litopouloutzanetaki E, Vafopouloumastrojiannaki A. 1991. Effect of Pediococcus pentosaceus on microbiology and chemistry of Teleme cheese. LWT-Food Sci. Technol. 24:173–176 [Google Scholar]
- 13. Vafopoulou-Mastrojiannaki A, Litopoulou-Tzanetaki E, Tzanetakis N. 1990. Effect of Pediococcus pentosaceus on ripening changes of feta cheese. Microbiol. Alim. Nutr. 8:53–62 [Google Scholar]
- 14. Bhowmik T, Marth EH. 1990. Role of Micrococcus and Pediococcus species in cheese ripening: a review. J. Dairy Sci. 73:859–866 [Google Scholar]
- 15. Holzapfel WH, Franz CMAP, Ludwig W, Back W, Dicks LMT. 2006. The genera Pediococcus and Tetragenococcus. Prokaryotes 4:229–266 [Google Scholar]
- 16. Tzanetakis N, Litopoulou-Tzanetaki E. 1989. Biochemical activities of Pediococcus pentosaceus isolates of dairy origin. J. Dairy Sci. 72:859–863 [Google Scholar]
- 17. Thomas TD, McKay LL, Morris HA. 1985. Lactate metabolism by pediococci isolated from cheese. Appl. Environ. Microbiol. 49:908–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McSweeney PLH, Fox PF. 2004. Metabolism of residual lactose and of lactate and citrate, p 361–372 In Fox PF, McSweeney PLH, Cogan TM, Guinee TP. (ed), Cheese: chemistry, physics and microbiology. Elsevier, London, United Kingdom [Google Scholar]
- 19. Litopoulou-Tzanetaki E, Vafopoulou-Mastrojiannaki A, Tzanetakis N. 1989. Biotechnologically important metabolic activities of Pediococcus isolates from milk and cheese. Microbiol. Alim. Nutr. 7:113–122 [Google Scholar]
- 20. Pasteris SE, Strasser de Saad AM. 2005. Aerobic glycerol catabolism by Pediococcus pentosaceus isolated from wine. Food Microbiol. 22:399–407 [DOI] [PubMed] [Google Scholar]
- 21. de Man JC, Rogosa M, Sharpe ME. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 131:82–91 [Google Scholar]
- 22. Liu SQ, Holland R, McJarrow P, Crow VL. 2003. Serine metabolism in Lactobacillus plantarum. Int. J. Food Microbiol. 89:265–273 [DOI] [PubMed] [Google Scholar]
- 23. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24. Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76–85 [DOI] [PubMed] [Google Scholar]
- 25. Beresford T, Williams A. 2004. The microbiology of cheese ripening, p 287–317 In Fox PF, McSweeney PLH, Cogan TM, Guinee TP. (ed), Cheese: chemistry, physics & microbiology, vol 1 Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 26. McSweeney PLH. 2004. Biochemistry of cheese ripening. Int. J. Dairy Technol. 57:127–144 [Google Scholar]
- 27. El Soda M, Ezzat N, El Howby T, Zeiday I. 1994. The peptide hydrolase system of pediococci: cell-bound enzymatic activities. Microbiol. Alim. Nutr. 12:145–154 [Google Scholar]
- 28. Vafopoulou-Mastrojiannaki A, Litopoulou-Tzanetaki E, Tzanetakis N. 1994. Proteinase, peptidase and esterase activity of crude cell-free extracts of Pediococcus pentosaceus isolated from cheese. LWT-Food Sci. Technol. 27:342–346 [Google Scholar]
- 29. Ardö Y. 2006. Flavour formation by amino acid catabolism. Biotechnol. Adv. 24:238–242 [DOI] [PubMed] [Google Scholar]
- 30. Urusova DV, Isupov MN, Antonyuk S, Kachalova GS, Obmolova G, Vagin AA, Lebedev AA, Burenkov GP, Dauter Z, Bartunik HD, Lamzin VS, Melik-Adamyan WR, Mueller TD, Schnackerz KD. 2012. Crystal structure of d-serine dehydratase from Escherichia coli. Biochim. Biophys. Acta 1824:422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bharath SR, Shveta Bisht S, Savithri HS, Murthy MRN. 2011. Crystal structures of open and closed forms of d-serine deaminase from Salmonella typhimurium—implications on substrate specificity and catalysis. FEBS J. 278:2879–2891 [DOI] [PubMed] [Google Scholar]
- 32. Kikuchi S, Ishimoto M. 1978. A d-serine dehydratase acting also on l-serine from Klebsiella pneumoniae. J. Biochem. 84:1133–1138 [DOI] [PubMed] [Google Scholar]
- 33. Labow R, Robinson WG. 1966. Crystalline d-serine dehydrase. J. Biol. Chem. 241:1239–1243 [PubMed] [Google Scholar]
- 34. Cosloy SD, McFall E. 1973. Metabolism of d-serine in Escherichia coli K-12: mechanism of growth inhibition. J. Bacteriol. 114:685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Umbarger HE, Brown B. 1957. Threonine deamination in Escherichia coli. II. Evidence for two l-threonine deaminases. J. Bacteriol. 73:105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van der Kaaij J, Zink R, Mollet B. June 2003. Use of l-alanine as a sweetener. European patent EP1154698
- 37. Carr FJ, Chill D, Maida N. 2002. The lactic acid bacteria: a literature survey. Crit. Rev. Microbiol. 28:281–370 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.