Abstract
The fungal arginine attenuator peptide (AAP) is a regulatory peptide that controls ribosome function. As a nascent peptide within the ribosome exit tunnel, it acts to stall ribosomes in response to arginine (Arg). We used three approaches to probe the molecular basis for stalling. First, PEGylation assays revealed that the AAP did not undergo overall compaction in the tunnel in response to Arg. Second, site-specific photocrosslinking showed that Arg altered the conformation of the wild-type AAP, but not nonfunctional mutants, with respect to the tunnel. Third, using time-resolved spectral measurements with a fluorescent probe placed in the nascent AAP, we detected sequence-specific changes in the disposition of the AAP near the peptidyltransferase center in response to Arg. These data provide evidence that an Arg-induced change in AAP conformation and/or environment in the ribosome tunnel is important for stalling.
Keywords: Translational regulation, nascent peptide, photocrosslinking, upstream open reading frame, ribosome exit tunnel
Introduction
Polypeptides synthesized by the ribosome pass through the ribosome exit tunnel, which starts at the peptidyl transferase center (PTC) and spans the large ribosomal subunit. The ribosome interacts with certain nascent peptides in the tunnel, and these nascent peptides can exert regulatory functions from within the tunnel, in both prokaryotes and eukaryotes 1; 2; 3; 4. Nascent peptides known to regulate translation in this manner cause translation arrest: they stall translation at either the elongation or termination phases. How such nascent stalling peptides (NSPs) function in the ribosome tunnel is a fundamental question that needs to be answered.
One class of NSPs causes stalling in response to small molecules. In eukaryotes, the fungal arginine attenuator peptide (AAP) stalls ribosomes in response to arginine (Arg) 5.AAP is encoded by an upstream open reading frame (uORF) in the mRNA specifying the small subunit of arginine-specific carbamoyl phosphate synthetase; this enzyme catalyzes one of the initial reactions required for de novo Arg biosynthesis in fungi 6. The uORF-encoded AAP causes the ribosome to stall in response to Arg with the uORF’s termination codon in the peptidyltransferase center (PTC) 5; 7; 8. The stalled ribosome blocks additional ribosomes from scanning to the downstream initiation codon for the ORF specifying the biosynthetic enzyme, thus reducing gene expression at the level of translation 5; 9. The stall at the uORF termination codon also triggers nonsense-mediated mRNA decay 10. While the AAP naturally functions to stall fungal ribosomes at the uORF termination codon, it can also stall eukaryotic ribosomes at both termination and elongation phases of translation in response to Arg 11; 12.
The AAP resembles the bacterial TnaC and SecM NSPs in that functionally important residues are similarly positioned in their respective sequences 2; 3. For example, a region of these peptides that is crucial for stalling is located approximately 12-residues from the site at which synthesis stalls 2; 4; 13; 14; 15. Thus, Asp-12 and Tyr-13 of the 24-residue Neurospora crassa AAP (Fig. 1) are absolutely conserved in all AAPs identified so far 16; 17 and mutations at either position eliminate stalling 17. A difference between the AAP and these bacterial NSPs, however, is that there is no demonstrated requirement for the AAP to have a specific residue at the PTC when stalling occurs 4; 15; 17; 18; 19; 20. Despite conservation of length, there is considerable sequence divergence of AAPs at their C-termini 16; 17.
Fig. 1. The sequences of AAP constructs used in this study.
(a) The sequences of tape measure AAP (TmAAP) constructs. Mutations and selected restriction enzyme are shown below the sequence. The Cys-residue that is PEGylated in the polypeptide is indicated by an arrowhead. Amino acid residues matching the wild-type N. crassa AAP sequence are correspondingly numbered. (b) The sequences of AAP constructs used for incorporating crosslinking and fluorescent probes. The sequence shown begins with the 5′ boundary of the linear DNA template amplified by PCR to generate AAP-encoding mRNA and ends at AAP codon-25, which, for the wild-type uORF-encoded AAP, is a termination codon. Amino acid residues matching the wild-type N. crassa AAP sequence are correspondingly numbered. Mutations and selected restriction enzyme sites are shown below the sequence.
Nascent peptide-mediated stalling is in many cases thought to be triggered by interactions between the peptide and the ribosome. Functionally important interactions of TnaC and SecM peptides with the bacterial ribosome have been identified; specific sites in large-subunit proteins L22 and L4, and in the 23S rRNA, are important for function, as evidenced by the identification of mutations in these ribosome components that affect stalling 2; 13; 14; 21. Bacterial nascent peptides that stall ribosomes in response to antibiotics also interact with the large ribosome subunit 22. The specific sites of interaction that are functionally important differ, indicating that there are multiple ways that nascent peptides cause stalling. Less is known concerning the interactions of eukaryotic NSPs with ribosomes. Examples of these regulatory peptides include AAP, cytomegalovirus gp48 uORF2 peptide23, mammalian S-adenosyl methionine decarboxylase uORF peptide24, and a conserved internal domain encoded within plant cystathionine γ-synthase 25. Cryo-EM studies of ribosomes containing in the exit tunnel either the AAP (in the absence of Arg) or the gp48 uORF2 peptide indicate interactions between these NSPs and the ribosome that may be potentially important for function 26.
Here we investigated the interactions of AAP with eukaryotic ribosomes using accessibility assays, site-specific crosslinking, and fluorescence lifetime measurements to show that Arg, in association with its capacity to induce stalling of the ribosome, altered the relative conformation of AAP with respect to the ribosome exit tunnel. In the presence of high Arg, the AAP’s conformation shifted with respect to ribosomal proteins L17 (the eukaryotic homolog of bacterial L22) and L4. These data indicate that AAP conformation and/or environment in the ribosome, as well as the dynamic interactions between the AAP and ribosome that are altered in response to Arg, are important for stalling.
Results
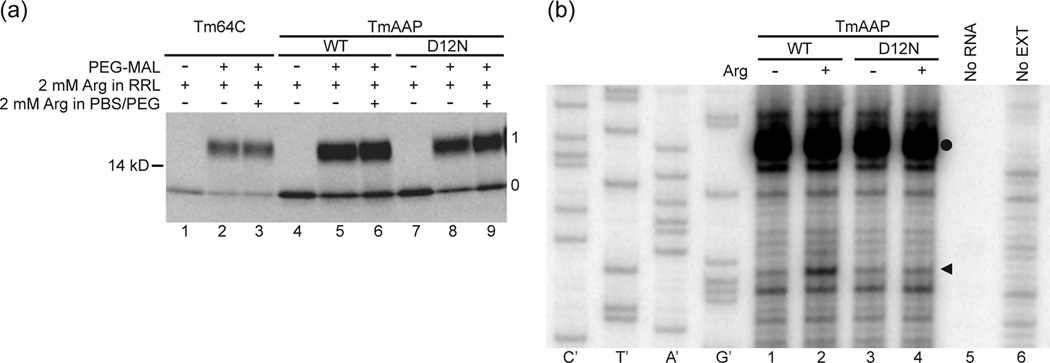
The molecular basis for AAP-mediated ribosome stalling in response to Arg is unknown. Reports have shown that a change in secondary structure (i.e., helical compaction), which alters the axial location of the nascent peptide in the tunnel, is associated with translation arrest mediated by E. coli SecM 15 and Arabidopsis CGS1 27. It is possible that the AAP similarly compacts in the presence of Arg to stall the ribosome. To evaluate this possibility, we took advantage of the observation that AAP functions in cellfree translation systems from rabbit reticulocyte11 and used a PEGylation assay previously shown to be monotonically dependent on distance from the PTC 28. We translated a truncated mRNA made from DNA that was digested with BstEII to produce a message that lacks a stop codon. In this case, the nascent peptide remains attached to the tRNA and the ribosome. A reporter cysteine, engineered to be 32 residues from the PTC in an all-extended control peptide (“tape measure” 28), will be maximally modified covalently by PEG-maleimide (PEGylated, fractionally 0.75–0.80 of the total protein) if the intervening residues are in an extended conformation. We substituted wild-type and D12N AAP sequences into the tape measure (Fig 1a) to make WT TmAAP and D12N TmAAP constructs, respectively, and determined the fraction of peptide PEGylated in high and low Arg compared to unsubstituted tape measures 28. All final samples were treated with RNase A to convert all PEGylated and unPEGylated peptidyl-tRNA species to PEGylated and unPEGylated peptides, and the peptides were analyzed by protein gel electrophoresis. This simplifies quantification of the fraction PEGylated.
As shown in Fig. 2a and Table 1, the fraction PEGylated for the all-extended tape measure is ~0.75 (Fig. 2a, lanes 2 and 3). The fraction PEGylated for both wild-type and D12N AAP substituted into this tape measure is ~0.70 in high Arg (Fig. 2a, lanes 5, 6, 8, 9). The results of replicate experiments for the wild-type and D12N AAPs in both high and low Arg are shown in Table 1. These data indicate that AAP is in a relatively extended conformation with at most one kink, that this conformation appears similar for both wild-type and D12N AAPs, and that no detectable change in the AAP’s overall length is observed in response to Arg. These results are consistent with recently published cryo-EM model of the AAP in wheat ribosomes in the absence of Arg 26. Our results do not preclude a dynamic equilibrium between compact and extended conformations. However, several lines of evidence and published controls 28; 29; 30; 31 indicate that the extent of PEGylation is a reliable and compelling measure of the axial location of a target cysteine within the ribosomal tunnel.
Fig. 2. PEGylation of AAP and toeprinting of AAP in tape-measure context.
(a) Nascent peptides were PEGylated and fractionated on polyacrylamide gels as described in the Methods. A control construct, an all-extended tape measure with a reporter position at residue 64C (Tm, lanes 1–3) was compared to AAP substituted into the tape measure two residues from the PTC (TmAAP, lanes 4–6) and D12N AAP in the same location (TmAAP D12N, lanes 7–9). Lanes 1, 4, and 7 are derived from a sample not treated with PEG-MAL. All other lanes derive from samples incubated with 1mM PEG-MAL for 3h. Gels were 4–12% NuPAGE Bis-Tris gels with MOPS running buffer. The number to the left of the gel is a molecular weight standard; numbers to the right of the gel indicate unPEGylated (0) and singly PEGylated (1) protein. (b). WT TmAAP and D12N TmAAP mRNA were used to program reticulocyte lysates as indicated. Lysates were supplemented with 10 µM (−) or 3 mM (+) Arg and 10 µM each of the other 19 amino acids After 20 min of translation, 3 µl of translation mixtures were toeprinted with primer Tm. Lanes indicated as no RNA or no EXT show toeprinting of lysate without added RNA and RNA in the absence of lysate, respectively. The Tm primer was also used for dideoxynucleotide sequencing of WT TmAAP DNA template (leftmost sequencing markers). The arrowhead indicates the position of the toeprint corresponding to ribosomes with AAP codon-25 (a CCA Pro-codon in this construct) in the A site. The toeprint signal marked with a closed circle is also observed in translation reactions programmed with these mRNAs to which cycloheximide was added at time 0 and thus does not represent ribosomes stalled during translation of this region of TmAAP.
Table 1. PEGylation of AAP-containing tape measures.
An all-extended peptide, referred to as a “tape measure”, Tm, was substituted with AAP (TmAAP) or a mutant (TmAAPD12N) and pegylated. Values are means ± SEM for triplicate samples. Significance was calculated as pairwise comparisons to give P>0.1 for experiments carried out at 2 mM or 20 µM Arg (present in both translation reactions, RRL, and in the pegylation reaction).
| 2 mM Arg | 20 µM Arg | ||||
|---|---|---|---|---|---|
| Tm | TmAAP | TmAAP D12N |
Tm | TmAAP | TmAAP D12N |
| 0.75±0.02 | 0.68±0.02 | 0.69±0.01 | 0.81±0.03 | 0.73±0.03 | 0.70±0.02 |
While the PEGylation data for the WT TmAAP and D12N TmAAP were similar, the WT TmAAP but not D12N TmAAP showed a regulatory response to Arg when translated in reticulocyte lysates as determined by toeprinting (Fig. 2b). In the presence of high Arg, a translation-dependent toeprint increased in intensity at a position corresponding to ribosomes stalled during elongation of WT TmAAP with the Pro-codon that would correspond to AAP codon-25 in the ribosome A-site (Fig. 2b, compare lanes 1 and 2) but not for D12N TmAAP (Fig. 2b, compare lanes 3 and 4). For the PEGylation experiments employing truncated RNA, the TmAAP-tRNA would thus be similarly positioned in the ribosome to be Arg-responsive.
The results from the PEGylation experiments indicate that AAP in the tape-measure context does not compact axially in response to Arg. However, this does not exclude the possibility that, in high Arg, the AAP’s conformation changes with respect to the ribosome. To examine AAP-ribosome proximity under both non-stalling low Arg and stalling high Arg conditions, we used cell-free translation systems and a photocrosslinking strategy. We incorporated a photoactivatable crosslinker at a specific residue in the AAP nascent chain (Fig. 1b) and examined the pattern of AAP crosslinking to ribosomal proteins. The photoreactive lysine analog Nε-(5 azido-2-nitrobenzoyl)-lysine (εANB-Lys) was placed in the AAP by selectively substituting AAP sense-codons with amber (UAG) stop codons (hereafter designated “*”, see Fig. 1b for example); translating the resulting mRNA in the presence of εANB-Lys-tRNAamb will incorporate εANB-Lys at those codons. To ensure that each nascent chain received only a single probe, each coding sequence contained only a single in-frame amber codon. The mRNAs that contained selectively placed amber stop codons in the AAP coding region were truncated after a C-terminal AAP sense-codon to generate stable ribosome-nascent chain complexes (RNCs) in wheat germ and N. crassa cell-free translation systems. In the absence of other constraints on translation, the final codon of the truncated RNA will be in the ribosomal P site and the amino acid corresponding to this codon will be placed in the nascent chain17; 32. To visualize these nascent peptides, additional Met-codons were added at the N-terminus of the AAP (Fig. 1b); the Met9AAP confers ribosome stalling in response to Arg and is readily detected by [35S]Met-labeling 11; 17; 33.
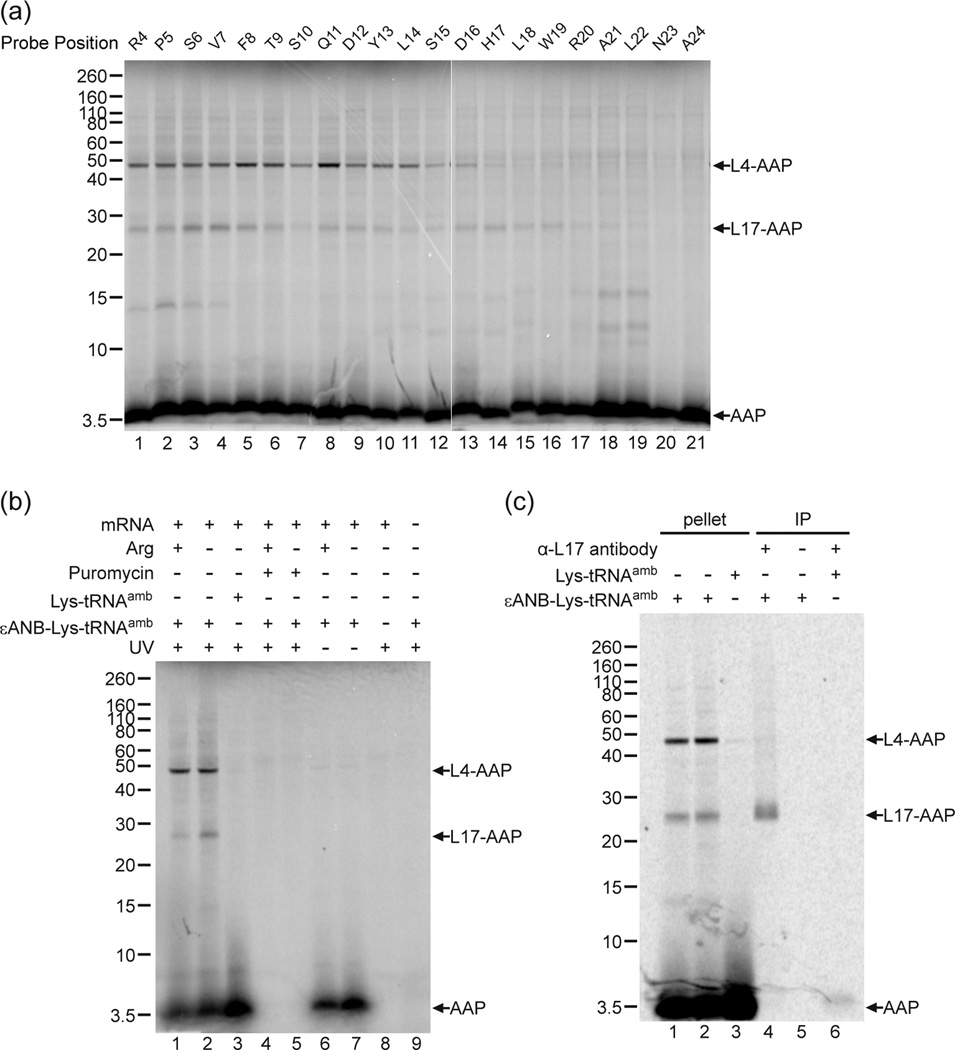
Fig. 3a shows the results of an experiment in which we examined crosslinking to ribosomal proteins by systematically placing the εANB-Lys probe at every residue of the AAP from Arg-4 to Ala-24 in an AAP construct truncated after Val-25. In this construct, codon-25, which is normally a stop codon, was replaced by a Val-codon; the AAP can be extended by a single residue and stalling activity is retained 12. When the photocrosslinker was placed at positions between Arg-4 and His-17 of the AAP, the AAP consistently formed two different photoadducts with apparent molecular masses of ~47 and ~25 kDa, which indicated crosslinking of the 4 kDa [35S]Met9AAP to ribosomal proteins L4 and L17 34; 35; 36; 37; 38; 39; 40; 41, respectively. The assignment of the ~47 kDa photoadduct to L4 is based on the knowledge that L4 has a mass of approximately 40 kDa and is the largest protein lining the ribosome’s exit tunnel 42; 43; no antibodies against eukaryotic L4 are available to confirm the assignment. The assignment of the ~25 kDa photoadduct to L17 was confirmed by immunoprecipitation with antibodies directed against L17 (see below). Nascent AAP photocrosslinking to L4 diminished when the probe was placed C-terminal to AAP His-17 (Fig. 3a, lane 14), and photocrosslinking to L17 began to diminish when the probe was C-terminal to AAP Arg-20 (lane 17). When the probe was placed close to the PTC (Asn-23 and Ala-24, lanes 20 and 21), nascent AAP photocrosslinking to ribosomal proteins was minimal. This is consistent with the region of the ribosome near the PTC being composed mainly of rRNA 34. Additional photoadducts to ribosomal proteins migrating with an estimated mass of 15-kDa or less were not considered further.
Fig. 3. Photocrosslinking of the nascent AAP to ribosomal proteins.
(a) Free RNCs with a single photoreactive probe εANB-Lys placed at the indicated positions in the nascent chain of AAP were photolyzed and analyzed on 12% NuPAGE gels. Nascent chain photoadducts to ribosomal proteins identified as L4 and L17 are indicated by arrows, as is the uncrosslinked AAP at the bottom of the gel. The increased signal from lower molecular weight bands between 10–15 kDa in lanes 18 and 19 were not reproducibly seen. (b) AAP nascent chain photoadducts to the ribosome depended on the photoreactive probe, UV irradiation, and the in vitro translation of AAP-encoding mRNAs. Lanes 1 and 2, AAP nascent chains containing a single εANB-Lys at position Val-7 were synthesized in the presence of 2 mM (+) or 30 µM (−) Arg, and photolyzed. Lane 3, AAP nascent chain containing a normal Lys at position Val-7 was synthesized and photolyzed. Lanes 4 and 5, parallel to lanes 1 and 2, except 1µM puromycin was added at the start of translation. Lanes 6 and 7, parallel to lanes 1 and 2, but RNCs were not irradiated by UV before being analyzed on gel. Lane 8, as lane 2, but neither εANB-Lys-tRNAamb nor Lys-tRNAamb was included in the translation reaction. Lane 9, as lane 2, except that the translation reaction was not programmed with mRNA. (c) Identification of the crosslinked product as wheat germ L17 by immunoprecipitation. Lanes 1–3, aliquots of material in ribosome pellets used for immunoprecipitation in lanes 4–6, respectively. Lanes 1 and 2 are from duplicate reactions containing εANB-Lys-tRNAamb and lane 3 is from a reaction containing Lys-tRNAamb; lanes 4 and 6, immunoprecipitation with L17 antibody; lane 5, mock immunoprecipitation (no antibody added). All of the cross-linking data shown in this and other figures are representative of multiple experiments.
AAP crosslinking to L4 and L17 required both the εANB-Lys probe (Fig. 3b, compare lane 3 with lanes 1 and 2) and UV irradiation (Fig. 3b, compare lanes 6 and 7 with lanes 1 and 2). AAP-L4 and AAPL17 photoadducts were associated with pelleted ribosomes following velocity sedimentation (Fig. 3c, lanes 1 and 2), but when the reaction mixtures contained Lys-tRNAamb (to produce AAP devoid of crosslinker) instead of εANB-Lys-tRNAamb, only [35S]Met-labeled AAP and no crosslinked ribosomal proteins were detected in the pelleted ribosome fraction (Fig. 3c lane 3). The appearance of all strongly radiolabeled species was dependent on both addition of AAP-encoding mRNA to the extracts (Fig. 3b, lane 9 and Fig. 5b, lane 9) and translation, as evidenced by their absence when the translation inhibitor puromycin was added to reactions prior to incubation (Fig. 3b, compare lanes 4 and 5 to lanes l and 2).
Fig. 5. Effects of Arg on photocrosslinking of the nascent AAP to ribosomal proteins.
(a) Wild-type (WT) and non-functional D12N AAP nascent chains with a single εANB-Lys probe at position Val-7 (lanes 1, 3, 5 and 7) were synthesized in wheat germ extract with 2 mM Arg (+) or 30 µM Arg (−). As controls, parallel reactions were performed to produce AAPs containing Lys instead of εANB-Lys (lanes, 2, 4, 6 and 8). RNCs were analyzed as in Fig. 1. (b) Effects of Arg on photocrosslinking of the AAP to N. crassa ribosomal proteins. The experiment was identical to panel a except that N. crassa extracts were used. The extracts contained 2 mM Arg (+) or 10 µM Arg (−) and the RNCs were pelleted by centrifugation through a sucrose cushion prior to analysis. (c) AAP-tRNA is the predominant form of the AAP in RNCs and is the form that photocrosslinked to ribosomal proteins. WT and non-functional D12N AAP nascent chains with a single εANB-Lys probe at position Val-7 were synthesized in wheat germ extract with 2 mM Arg (+) (lane 1) or 30 µM Arg (−) (lane 2). Parallel control translation was performed with Lys incorporated in place of photoreactive εANB-Lys probe (Lane 3). RNCs were pelleted after photolysis and analyzed without RNase treatment. Under each gel image, quantitative analyses of the crosslink products show the relative amount of radiolabel in bands representing AAP crosslinked to L4 and L17 indicated as the fraction of the sum of both crosslink products. Complete quantification data are provided in Supplementary Table 3.
The identity of L17 was verified by immunoprecipitation using rabbit antiserum against the N-terminus of eukaryotic L17 39, generously provided by Martin Pool (Fig. 3c, lane 4). Mock-immunoprecipitation of photoadducts in the absence of antibody (Fig. 3c, lane 5), or immunoprecipitation of photolyzed RNCs lacking the εANB-Lys probe (Fig. 3c, lane 6), did not yield significant amounts of radiolabeled products.
Crosslinking to L4 and L17 occurred with probes placed at different sites within a relatively long stretch of the AAP. This would seem surprising because the positions of L4 and L17 are fixed in the tunnel. An explanation for this is that there is motion of the nascent AAP in the ribosome and that these results arise because the nascent AAP rotates and/or moves back and forth in the tunnel, as has been proposed to explain results obtained with εANB-Lys-tRNAamb in studies of the interactions of nascent transmembrane segments with the ribosome tunnel 44.
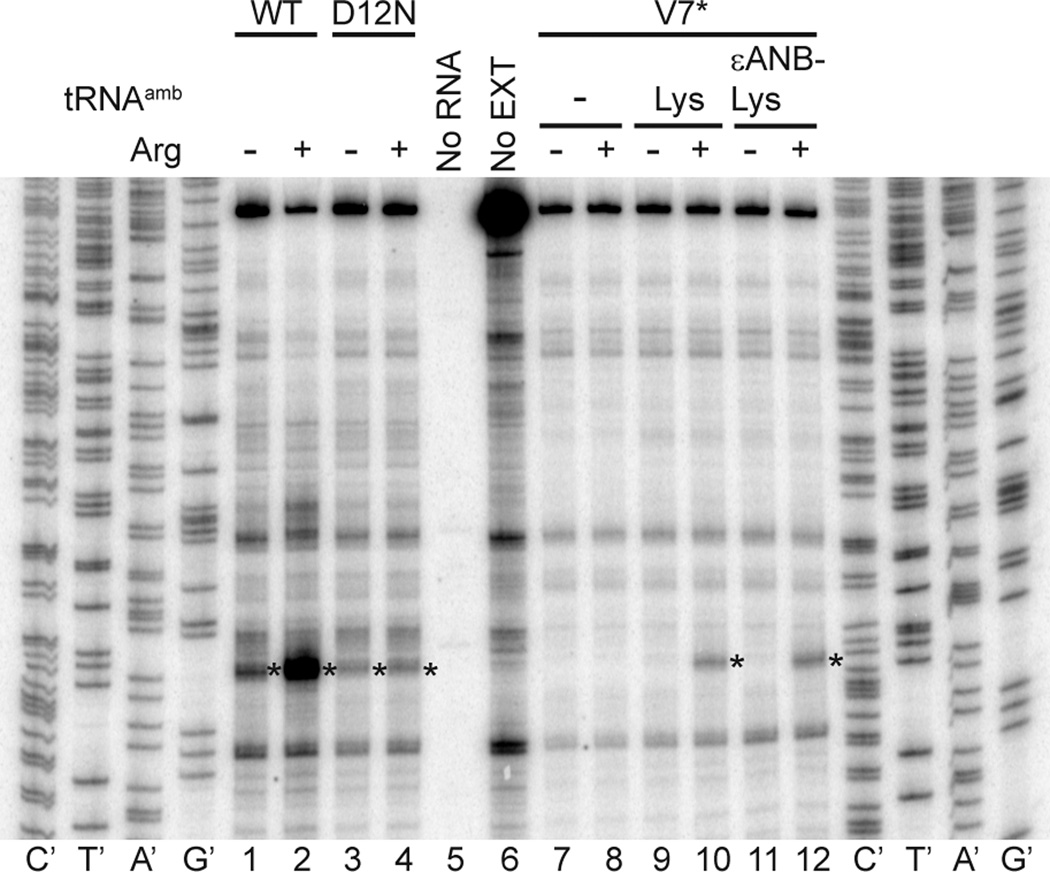
Using the suppressor tRNA approach, a photoreactive probe could in principle be placed anywhere within the AAP to detect changes in AAP proximity to ribosomal tunnel components in response to Arg. However, systematic mutagenesis of the N. crassa AAP and analysis of its capacity to cause regulated stalling of ribosomes indicated that residues Thr9 to Arg20 comprised the primary functional region and were relatively intolerant to substitution 17. We examined the effects of placing εANB-Lys at V7* and observed that Arg-specific regulation was retained based on the toeprinting assay (Fig. 4). The wild-type AAP, when encoded as a uORF, causes the ribosomes that are synthesizing it to stall with the uORF termination codon-25 in the ribosome A-site in the presence of high Arg 17 as shown in Fig. 4, lanes 1 and 2. The D12N mutation abolishes Arg-specific stalling at the termination codon (Fig. 4, lanes 3 and 4). The AAP containing the V7* mutation, which specifies an amber stop codon at AAP codon-7, does not enable ribosome-stalling at codon-25 in the absence of amber stop-codon suppression (Fig. 4 lanes 7 and 8) because the ribosomes do not translate past the amber stop codon 33. However, when either Lys-tRNAamb or εANB-Lys-tRNAamb were added to N. crassa extracts programmed with V7* mRNA, the signal corresponding to ribosomes at codon-25 was observed, and ribosome-stalling in response to Arg at this codon was retained (Fig 4, lanes 9–12).. Therefore we focused on using εANB-Lys at V7* to assess the impact of Arg on the proximity of nascent AAP to ribosomal components.
Fig. 4. Toeprint analyses demonstrate that AAP containing ANB-Lys at the position corresponding to Val-7 stalls ribosomes in response to high Arg.
The different mRNAs used to program N. crassa extracts are indicated on the top. Either no tRNAamb, Lys-tRNAamb, or εANB-Lys- tRNAamb were added to extracts programmed with V7* as indicated. Reaction mixtures contained 10 µM (−) or 2 mM (+) Arg and 10 µM each of the other 19 amino acids After 20 min of translation, 3 µl of translation mixtures were toeprinted with primer ZW4. Lanes indicated as no RNA or no EXT show toeprinting of extract without added RNA, and of pR7013-derived RNA in the absence of extract, respectively. This primer was also used for dideoxynucleotide sequencing of pR101 (leftmost sequencing markers) and pR7013 (rightmost sequencing markers). The asterisk indicates the position of the toeprint corresponding to ribosomes with the AAP uORF codon-25 termination codon in the A-site.
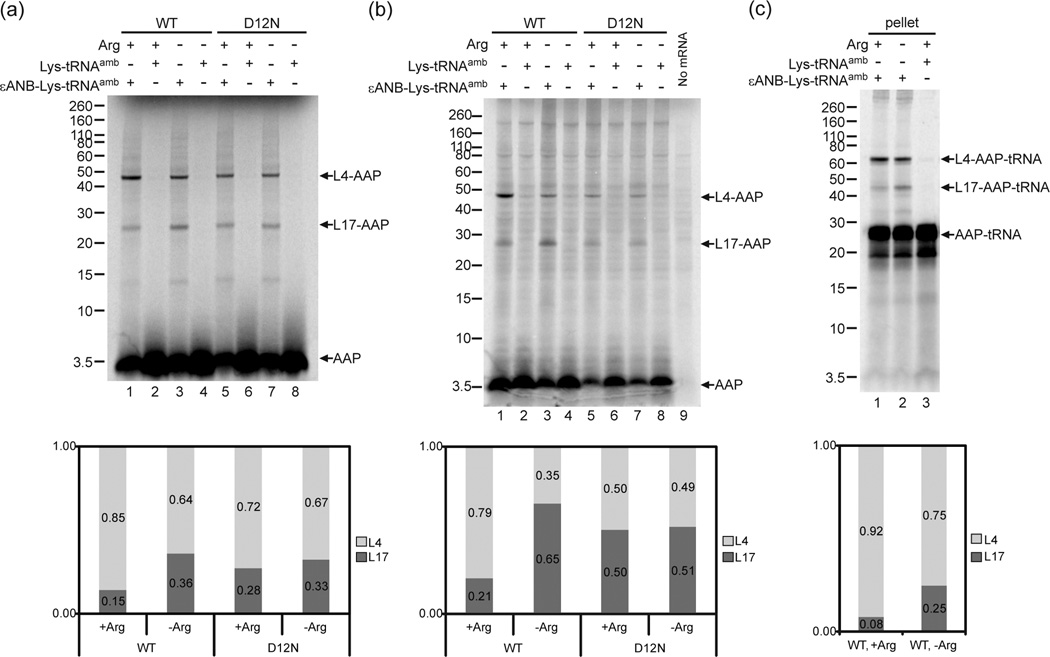
Fig. 5a and b show the results of crosslinking the V7*AAP to wheat germ and N. crassa ribosomes, respectively. The nascent AAP was synthesized in translation reactions that contained either high Arg [2 mM (+)] or low Arg [30 µM (−)], and either εANB-Lys-tRNAamb or Lys-tRNAamb. The crosslinking of wild-type AAP was compared to AAP containing the D12N mutation; this mutation eliminates Arg-specific ribosome stalling 5; 45. Crosslinking of AAP to wheat germ and N. crassa ribosomal proteins L4 and L17 was εANB-Lys-dependent (compare lanes 1, 3, 5, and 7 to lanes 2, 4, 6, and 8 in Fig. 5a and b). Importantly, in both wheat germ and N. crassa extracts, the relative intensities of crosslinking of nascent wild-type AAP to L4 and L17 changed in response to high Arg (compare lanes 1 and 3 in Fig. 5a and b; full quantification data are given in Supplementary Table 3): the relative level of crosslinking to L4 increased, and the relative level of crosslinking to L17 decreased. The change in the relative levels of crosslinking in high Arg did not occur with the nonfunctional D12N AAP (compare lanes 5 and 7 in Fig. 5a and b). All of the data presented in this and other figures to compare the effects of Arg on the relative crosslinking of the AAP to L4 and L17 are representative of multiple experiments and an overall summary of the results is given in Supplementary Table 4.
These data indicated that the wild-type AAP, but not the D12N AAP, underwent a change in conformation and/or environment in response to high Arg. We next determined that this change in AAP occurred while AAP was linked to tRNA, as would be expected if the nascent peptide was at the PTC. When the translation products were not treated with RNase prior to gel-analysis, both uncrosslinked AAP and crosslinked AAP increased in apparent mass by an additional ~22 kDa due to the covalently linked tRNA (Fig. 5c), and the Arg-specific effect on the relative crosslinking to L4 and L17 was maintained (Fig. 5c compare lanes 1 and 2). AAP lacking εANB-Lys did not crosslink to ribosomal proteins, but was present predominantly as AAP-tRNA and not free AAP (Fig. 5c, lane 3). Thus it was AAP-tRNA that underwent a change in location within the tunnel in response to Arg.
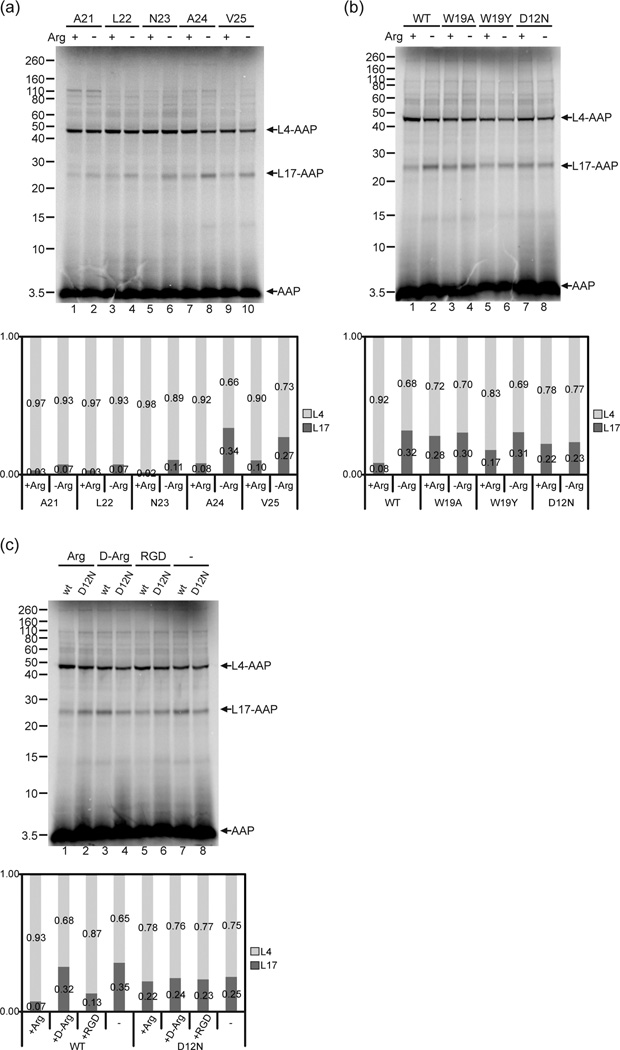
At what stage during AAP synthesis does AAP change its conformation and/or environment in response to Arg? To examine this, we used V7* constructs with truncations of the AAP coding region after Ala-21, Leu-22, Asn-23, Ala-24, or Val-25. In all cases (Fig. 6a), Arg caused the level of crosslinking to L17 to decrease relative to the level of crosslinking to L4 in wheat germ extracts. As the AAP was progressively shortened at its C-terminus, the efficiency of crosslinking to L17 decreased overall, likely reflecting that V7* was dynamically in a different set of positions in the ribosome tunnel. That an Arg-dependent change in the AAP-ribosome conformation was observed with C-terminal truncated AAPs indicated that the full-length AAP need not be synthesized before Arg could affect the AAP’s configuration with respect to the ribosome.
Fig. 6. The change in AAP’s relative conformation detected by photocrosslinking is specifically induced by Arg and requires an AAP sequence that is functional for stalling.
(a) The Arg-induced change in relative conformation occurred when the AAP was shortened at its C-terminus. AAP chains of different lengths with a single εANB-Lys probe at position Val-7 were synthesized in wheat germ extracts in 2 mM Arg (+) or 30 µM Arg (−). Truncation of mRNA was after Ala-21, Leu-22, Asn-23, Ala-24 or Val-25. RNCs were photolyzed and analyzed as in Fig. 2. (b) The Arg-dependent conformational change is related to the AAP’s ability to stall the ribosome. Wild-type (WT), W19A, W19Y, and D12N AAP nascent chains truncated after Ala-24 with a single εANB-Lys probe at position Val-7 were synthesized in 2 mM Arg (+) or 30 µM Arg (−). RNCs were photolyzed and analyzed as in Fig. 2. (c) The Arg-dependent change in photocrosslinking is not a nonspecific electrostatic effect. WT and D12N AAP nascent chains truncated after Ala-24 with a single εANB-Lys probe at position Val-7 were synthesized in the presence of 2 mM L-Arg, D-Arg, or RGD (Arg-Gly-Asp tripeptide), or 30 µM Arg (−). RNCs were photolyzed and analyzed as in Fig. 2. Under each gel image, quantitative analyses of the crosslink products show the relative amount of radiolabel in bands representing AAP crosslinked to L4 and L17 indicated as the fraction of the sum of both crosslink products. Complete quantification data are provided in Supplementary Table 3.
It is possible that the D12N mutation eliminated the AAP conformational change detected by the V7* probe for reasons not associated with its causing a stalling defect (e.g., proximity to the probe). Since changing the more distal and also highly conserved Trp-19 residue can either eliminate (W19A) or reduce (W19Y) stalling 17, we tested the effects of W19A and W19Y mutations for their impact on crosslinking in wheat germ extracts. The W19A mutation eliminated Arg-dependent changes in crosslinking, and thus was similar to the D12N mutation in its inability to stall ribosomes and to affect the AAP-ribosome conformation in response to Arg (Fig. 6b, lanes 3, 4 and 7, 8). The W19Y AAP showed an Arg-dependent 2-fold reduction in the fraction of AAP-L17 photoadducts, significantly less than the 4-fold decrease in the fraction of AAP-L17 photoadducts observed with wild-type AAP in the presence of high Arg (Fig. 6b, lanes 1, 2 and 5, 6), consistent with the effect of W19Y to reduce but not eliminate stalling. Thus, the extent to which the mutations affected AAP conformation and/or environment in the tunnel in response to Arg correlates with its capacity to stall ribosomes.
To eliminate the possibility that the Arg-dependent change in AAP arose from a nonspecific electrostatic effect of Arg, we tested whether D-Arg, an optical isomer that does not elicit AAP-mediated ribosome stalling46, affected crosslinking. D-Arg had no impact on the relative levels of wild-type AAPL4 and AAP-L17 photoadducts (Fig. 6c, compare lanes 3 and 7), while in parallel experiments, Arg altered the relative levels of L4 and L17 crosslinking (Fig. 6c, compare lanes 1 and 7). Addition of the tripeptide Arg-Gly-Asp (RGD), which is commercially available at high purity, and, like Arg, induces stalling of ribosomes through the function of the AAP 17, produced crosslinking results similar to those obtained with Arg. Relative crosslinking to L4 increased in its presence (Fig. 6c, compare lanes 5 and 7), although the increase was not as pronounced as with Arg. In parallel experiments with the D12N AAP, no change in relative crosslinking was observed in the presence of Arg, RGD, or D-Arg (Fig. 6c, lanes 2, 4, 6, and 8), consistent with the D12N mutant’s inability to manifest a regulatory response to Arg. Thus, only those small molecules that caused stalling also affected the AAP-ribosome conformation in the ribosome.
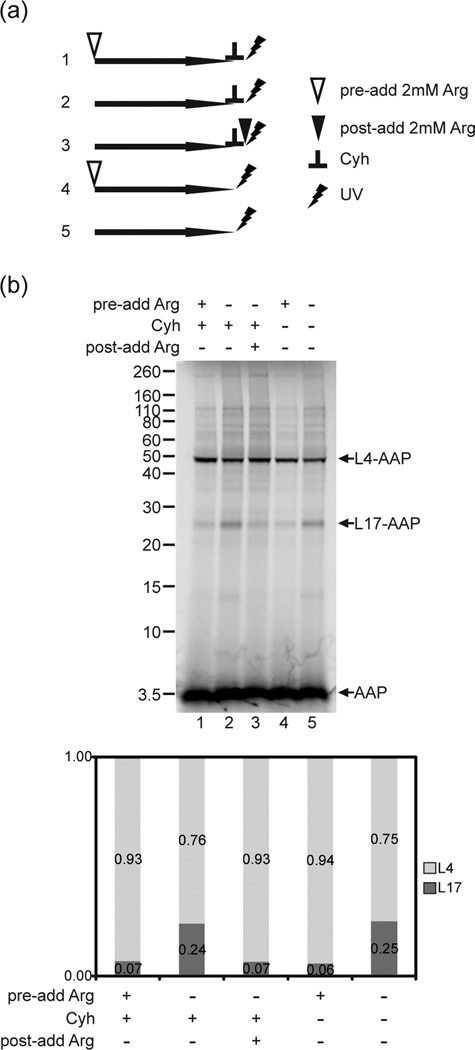
A possible explanation for the observed Arg-dependent change in AAP is that the wild-type AAP was not fully synthesized in the presence of high Arg and was thus in a different location in the tunnel relative to the location of the full-length AAP that was synthesized in low Arg. We tested this using the strategy depicted in Fig. 7a, which used cycloheximide (Cyh) to block translation elongation. We synthesized the AAP for 15 min in translation reactions containing low Arg and then added Cyh to stop translation elongation. Any possible effects of high Arg on AAP synthesis were thus eliminated. We then added Arg to this reaction and observed increased crosslinking to L4 and decreased crosslinking to L17 (Fig. 7b, lane 3). This indicated that the change in crosslinking in response to the level of Arg cannot be attributed simply to differences in AAP synthesis that occurred in low versus high Arg. Cyh did not impact UV-crosslinking, since reactions in which translation occurred in high Arg versus low Arg, and to which Cyh was added before UV irradiation, were indistinguishable from parallel reactions to which Cyh was not added (Fig. 7b, compare lanes 1 and 2 to lanes 4 and 5).
Fig. 7. The Arg-dependent change in crosslinking arises from a change in the relative conformation of AAP in the ribosome tunnel.
(a) Schematic experimental design indicating the relative order of addition of 2 mM Arg and/or cycloheximide (Cyh) to reactions. Numbers correspond to the sample lanes in panel b. (b) Wild-type (WT) and D12N nascent chains truncated after Ala-24 with a single εANB-Lys probe at position Val-7 were synthesized in 2 mM (+) or 30 µM (−) Arg (indicated by the row, “pre-add Arg”. Then Cyh was added to some reactions to a final concentration of 0.5 mg/ml to stop nascent peptide elongation (lanes 1–3) (indicated by + in the row “Cyh”). To a reaction containing 30 µM Arg that was stopped with Cyh, 2 mM Arg (+) or no Arg (−) was subsequently added (lane 3) (indicated by the row “post-add Arg”). RNCs from these reactions and parallel reactions without added Cyh (lanes 4–5) were photolyzed and examined as in Fig. 2. Under each gel image, quantitative analyses of the crosslink products show the relative amount of radiolabel in bands representing AAP crosslinked to L4 and L17 indicated as the fraction of the sum of both crosslink products. Complete quantification data are provided in Supplementary Table 3.
AAP-mediated ribosome stalling in response to Arg is likely to be mediated by altered activity of the PTC, and this could be caused by a change in AAP conformation and/or environment at the PTC. To detect changes in the disposition of the nascent peptide at a position near the PTC, we used Nε-6-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)aminohexanyl-Lys tRNALys (εNBD-Lys-tRNALys) to incorporate a fluorescent NBD dye into the nascent peptide at position 2032. The residue corresponding to N. crassa AAP Arg-20 is commonly substituted with Lys in other AAPs. The spectral properties of NBD vary with the polarity and hydrogen bonding capacity of its environment, and the average fluorescence lifetime (<τ>) is lower in an aqueous environment than in a nonpolar environment 32; 44; 47. Thus, changes in fluorescence lifetime will reveal changes in the microenvironment of an NBD placed near the C-terminus of ribosome-bound AAP. When the NBD <τ> was measured in nascent wild-type and D12N AAPs in high and low Arg in two independent experiments, the increase in Arg concentration had opposite effects on NBD lifetime: <τ> increased for NBD in wild-type AAP, while <τ> decreased for NBD in D12N AAP (Table 2). The differing spectral changes in response to Arg reveal that NBD environment differs for the wild-type and mutant AAPs in RNCs, most likely dynamically 41; 44, and that the addition of Arg accentuates the difference between the two AAPs in the tunnel.
Table 2. NBD fluorescence lifetimes of WT and D12N AAP in the presence and absence of 2 mM Arg.
| Expt | AAP | average τ (ns) | average τ (ns) | lifetime (ns) difference (high Arg-low Arg) |
||
|---|---|---|---|---|---|---|
| high Arg | chi-square | low Arg | chi-square | |||
| 1 | WT | 6.6 | 1.5 | 5.3 | 2.0 | 1.3 |
| 2 | WT | 5.7 | 1.0 | 4.6 | 0.8 | 1.1 |
| 1 | D12N | 4.9 | 0.3 | 5.7 | 0.4 | −0.8 |
| 2 | D12N | 5.2 | 1.0 | 5.7 | 1.1 | −0.5 |
Discussion
Ribosome stalling mediated by the nascent AAP in response to Arg is an example of a genetic regulatory mechanism in which a small molecule effector and a nascent peptide regulate protein synthesis. Here we obtained direct evidence that AAP-mediated ribosome stalling is accompanied by an Arg-dependent change in the conformation of the nascent AAP and/or its environment in the ribosome tunnel. Eukaryotic cell-free translation systems were used for PEGylation studies, crosslinking analyses, and fluorescence lifetime measurements to examine these Arg-induced changes. A PEGylation assay, which sensitively measures the axial distance of an N-terminal cysteine relative to the ribosome’s PTC, indicated that the AAP was in a relatively extended conformation in the ribosome. Because PEGylation efficiency was not affected in response to Arg, it appears that the Arg-induced changes in AAP do not involve a substantial change in the overall level of compaction of the nascent peptide. Data obtained by incorporating a photo-reactive crosslinker at a specific site in the nascent AAP and monitoring the photoadducts to ribosomal proteins produced in the presence of low and high Arg showed that the crosslinker at this site reacted relatively more with ribosomal protein L4 and less with L17 when the level of Arg was high. Mutations at highly conserved AAP residues that are essential for stalling reduced the Arg-mediated changes in AAP photocrosslinking, indicating that relative arrangement of AAP and ribosome tunnel components is important for stalling. These Arg-induced changes were not due to nonspecific electrostatic interactions caused by the positive charge of Arg since they only occurred with l-Arg and not d-Arg. Finally, fluorescence lifetime measurements indicate that changes in AAP environment close to the PTC also occur in response to Arg.
Crosslinking analyses using C-terminal truncations of the wild-type AAP show that shortened peptides still underwent a change in AAP-ribosome conformation in response to Arg. This indicates that the last two or three residues at the AAP C-terminus are not necessary for the Arg-induced changes in the ribosome. However, based on primer-extension inhibition (toeprint) assays that can detect ribosomes at mRNA positions where translation is rate-limited, such C-terminally shortened AAPs are less able to stall ribosomes 5; 12, suggesting that the nascent AAP must be fully synthesized before it can stall ribosomes efficiently. These data are consistent with a model for AAP function in which the Arg-induced change in AAP can occur prior to the synthesis of the full-length AAP, but this change is insufficient to cause ribosome stalling until AAP is fully synthesized and optimally positioned within the ribosome. Consistent with the idea that AAP must be optimally positioned in the ribosome to cause stalling, even if its relative conformation in the ribosome changes in response to Arg before it is fully synthesized, the evolutionarily conserved AAPs so far identified are at least as long as the N. crassa AAP when their conserved residues are aligned 17, despite considerable divergence in the specific identities of residues at their C-termini. Thus, while the length of the AAP appears important, the specific C-terminal residues encoded are not crucial for its function, which is different from what has been reported for many other peptides that cause stalling 2; 17.
Cryo-EM studies (in low Arg) reveal a reconstructed density for the C-terminus of AAP adjacent to the PTC that can be fit with a single helical turn model 26. Compaction of the nascent regulatory peptide accompanying translation arrest has been indicated for E. coli SecM peptide 15, although the peptide appears mostly in an extended conformation 48, and for the Arabidopsis CGS1 nascent peptide, which stalls ribosomes in response to S-adenosyl methionine 27. In our study, no helical compaction of AAP in response to Arg was detected by the PEGylation assay. AAP appears to be in an extended configuration with at most one α-helical turn in either low or high Arg. Recently, we showed that a Pro can be tolerated, albeit with reduced regulatory activity, at many positions in the AAP’s C-terminal region17. Thus, while an α-helical structure could exist at the C-terminus of wild-type AAP as suggested by secondary structure predictions for AAP in solution49 and cryo-EM data for AAP in the tunnel26, our data suggest the presence of, at most, a kink or partial turn in AAP in the tunnel. Although such minimalist structures may contribute to the compaction observed by cryo-EM, they are detected equally by PEGylation in both wild-type and mutant (no stalling) AAP and therefore suggest that helical secondary structure alone does not modulate AAP regulatory function.
The interactions between critical residues of nascent peptides and the ribosome exit tunnel have been reported for many of the bacterial NSPs. At the PTC, E. coli 23S rRNA nucleotides U2585, U2609, A2062, and A2602 interact with essential C-terminal residues of TnaC, SecM, and ErmCL 48; 50; 51; 52. Mutations in both NSPs and ribosomal RNA at these sites have been shown to abolish translation arrest 13; 52; 53. In the narrowest region of the exit tunnel, nascent peptides interact with bacterial ribosomal proteins L4 and L22, as well as 23S rRNA nucleotide A751 48; 50; 51. Mutations at the tip of the β-hairpin of L22 and insertions at A751 relieve the translation arrest caused by TnaC and SecM 13; 14, indicating this site in the tunnel has a crucial role in translation arrest mediated by these two peptides. It is proposed that the interaction of nascent peptide with the exit tunnel helps certain critical residues to be precisely positioned so that they would trigger the conserved rRNA nucleotides at the PTC to adopt a conformation that “silences” the PTC 48. Similar interactions of AAP with components in the exit tunnel have been predicted by recent cryo-EM structural data 26. The critical Asp-12 residue of AAP is sandwiched between L17 and 26S rRNA nt A751 (E. coli numbering) on one side of the tunnel and L4 on the other side of the tunnel. The C-terminus of AAP also contacts rRNA nucleotides U2585, A2062, and A2058 26. An additional contact is observed between the region near AAP Ser6 and Val7 (where the photo-reactive crosslinker we used is located) and L4. Since an increase in the lifetime of an NBD probe in the C-terminal region of AAP near the PTC correlated with the Arg-dependent change in AAP crosslinking to L4 and L17, a change near the PTC could be responsible for inactivation of the PTC in the Arg-arrested state.
The experiments described here that examined the AAP conformation and environment in the ribosome using cross-linking are to our knowledge the first to show that a small molecule effector has the ability to change the contacts between a nascent peptide and components of the eukaryotic ribosome exit tunnel. These changes are associated with the capacity of the nascent peptide to stall ribosomes. It is possible that Arg interacts solely with the ribosome to cause AAP-induced stalling. Alternatively, Arg might interact directly with AAP to alter its conformation and/or environment to induce stalling. A third possibility, which resembles the situation for erythromycin-induced stalling triggered by the ErmCL peptide 54, is that Arg interacts with both AAP and the ribosome to create a combined structural arrangement that leads to stalling.
Materials and Methods
Plasmids, mRNA and tRNA
For PEGylation assays, capped truncated mRNAs coding for nascent peptides of defined length in the tape-measure context were transcribed in vitro by SP6 RNA polymerase using BstEII-linearized plasmids 28. For toeprinting ribosomes translating these coding regions, plasmids were linearized at a downstream NotI site to use as templates for capped mRNA synthesis. For photocrosslinking and time-resolved spectral measurement assays, plasmids containing the coding sequences for Met9AAP were derived from pKL20233, which encodes Met9AAPTAA-LUC. A poly(A)30 sequence was introduced into the 5′ untranslated region to increase the translation of capped mRNAs lacking poly(A) tails in N. crassa extracts. An amber stop codon and/or other mutations were introduced into the AAP-coding sequence by replacing a restriction fragment with synthetic oligonucleotides (Supplementary Table 1) or by PCR (primers listed in Supplementary Table 2). Each construct was confirmed by DNA sequencing (Fig 1). Truncated mRNAs coding for nascent peptides of defined length were transcribed in vitro by T7 RNA polymerase 55 using PCR-produced DNA fragments (Supplementary Table 2 lists PCR-primers). The yield of RNA was quantified using ImageQuantTL by comparison to standard markers with known masses using ethidium bromide-stained agarose gels imaged with a GE Typhoon Trio phosphorimager. [14C]Lys-tRNAamb, εANB-[14C]Lys-tRNAamb, [14C]Lys-tRNALys, and εNBD-[14C]Lys-tRNALys were prepared as before 56; 57; 58 (tRNA Probes, LLC, College Station, TX). For toeprinting ribosomes synthesizing wild-type, D12N, or V7* AAP, plasmids pR101, pS101, and pR7013 5; 33 were linearized with EcoRI and RNA transcribed using T7 RNA polymerase 55.
Preparation of cell-free translation extracts
Wheat germ extract was prepared as described 57; 59. Neurospora extract was prepared as described 55 except cells were ground with frozen Buffer A beads (1 ml/g of cell weight) by three 2-min grinding cycles (rate set at 10) with 1-min cooling between each cycle in a SPEX SamplePrep 6850 Freezer/Mill.
Translation and photocrosslinking
Wheat germ in vitro translation reaction mixtures (50 µl, programmed with 240 ng of truncated RNA) were incubated in the dark at 26°C for 5 min 60; 61. Reaction mixtures contained 110 mM KOAc, 30 mM HEPES-KOH (pH 7.5), 3.5 mM Mg(OAc)2, 30 µM of each amino acid except methionine, 20 µCi of [35S]Met (MP Biomedical, > 1000 Ci/mmol), 60 pmol of εANB-Lys-tRNAamb or Lys-tRNAamb, other components as described 32, and (where indicated) 2 mM arginine or Arg analogs. Samples were photolyzed on ice for 15 min using a 500W mercury arc lamp 60. After photolysis, samples were incubated at 22°C for 20 min in the presence of 10 mM EDTA and 10 µg/ml RNase A. Photocrosslinked material was analyzed by 12% NuPAGE gels (Invitrogen) in MES running buffer and visualized using a GE Typhoon Trio phosphorimager.
Neurospora in vitro translation reaction mixtures (50 µl, programmed with 240 ng of truncated mRNA) were performed as described11 in the dark at 26°C for 5 min. Reaction mixtures contained in the presence of 120 mM KOAc, 35 mM HEPES-KOH (pH 7.6), 2.7 mM Mg(OAc)2, 20 µM of each amino acid except methionine, 20 µCi of [35S]Met, 60 pmol of εANB-Lys-tRNAamb or Lys-tRNAamb, and (where indicated) 2 mM arginine. Samples were photolyzed as described above. After photolysis, samples were sedimented through a 110 µl sucrose cushion [0.5 M sucrose, 25 mM HEPES (pH 7.5), 130 mM KOAc, 3 mM Mg(OAc)2] in a Beckman TLA-100 rotor (100,000 rpm, 10 min, 4°C). Pellets containing RNC complexes were resuspended in 50 µl of TE containing 10 µg/ml RNase A and analyzed by NuPAGE as described above.
Primer-extension inhibition (toeprint) assays
For toeprinting in reticulocyte lysates (Promega) or N. crassa extracts, translation reaction mixtures (10 µl were programmed with 60 ng of mRNA and incubated at 26°C for 20 min. Toeprint assays were performed by adding 3 µl of translation reaction mixtures (or pure mRNA) to tubes containing 5.5 µl of ice-cold reverse transcription buffer 7; 55. The tubes were heated at 55°C for 3 min and then immediately placed on ice again. 32P-labeled primer ZW4 (TCCAG GAACC AGGGC GTA) or Tm (AAGTA ATTGA AGTTG GAAAC AATCA C) (1 µl; approximately 2 × 106 cpm of Cerenkov) was added to each tube and annealed to the template by placing tubes in a 37°C water bath for 5 min. Then 0.5 µl (100 U) of Superscript III reverse transcriptase (Invitrogen) was added, and the mixtures were incubated for 30 min at 37°C. The reactions were terminated by extraction with 10 µl of phenol/chloroform. The aqueous phase was removed and mixed with an equal volume of DNA sequencing stop solution. Samples were heated at 85°C for 5 min, cooled on ice, and loaded on 6% polyacrylamide sequencing gels (7 µl of sample per lane). The gels were dried and visualized using a GE Typhoon Trio phosphorimager.
Immunoprecipitation
Following translation in wheat germ extract, photolysis and pelleting of RNC complexes, the RNC pellets were resuspended in 50 µl of 3% (w/v) SDS, 50 mM Tris-HCl (pH 7.5) and placed at 55°C for 30 min. The samples were adjusted to 500 µl with buffer B [150 mM NaCl, 50 mM Tris-HCl (pH 7.6), 1% (v/v) Triton X-100]. Samples were then pre-cleared by rocking with 30 µl of protein A-Sepharose (Sigma) at 22°C for 1 hr before the Sepharose beads were removed by sedimentation. Rabbit antiserum against the N-terminus of L17 was added to each supernatant, and the samples were rocked overnight at 4°C, after which protein A-Sepharose (40 µl) was added and incubated for 2 hr at 4°C. The immunoprecipitate was recovered by sedimentation, washed three times with buffer B, and then washed a final time with the same buffer containing no detergent. Immunoprecipitated material was incubated at 22°C for 20 min in the presence of 10 µg/ml RNase A, and then analyzed by NuPAGE as described above.
PEGylation
As described previously 28, translation reaction mixture (40 µl) was added to 500 µl phosphate-buffered saline (PBS, Ca-free, containing 4 mM MgCl2 (pH 7.3)) with 2 mM DTT. Either 2 mM or 20 µM Arg, depending on the experiment, was present in the translation reaction and the PBS buffer. The suspension was centrifuged (Beckman Optima TLX Ultracentrifuge, Beckman TLA 100.3 rotor) through a sucrose cushion (100 µl; 0.5 M sucrose, 100 mM KCl, 5 mM MgCl2, 50 mM HEPES-KOH, and 1 mM DTT (pH 7.5)) for 20 min at 70,000rpm (~245,000xg) at 4° C to isolate RNC complexes. The pellet was resuspended on ice in 90 µl of PBS Ca-free buffer containing 4 mM Mg2+ and 100 µM DTT (pH 7.3) with either no Arg or 2 mM Arg. 10µl buffer containing 10 mM PEG-MAL (5 kDa, SunBio Co.) was added (final PEG-MAL concentration of 1 mM) and incubated at 0°C for 3–5h. PEGylation reactions were terminated by addition of 100 mM DTT, followed by incubation of the mixture at room temperature for 10–15 min. Samples were collected either by centrifugation at 70,000 rpm at 4°C for 20 min and/or by precipitation with a 90% final volume of cold acid-acetone (900 µl; a stock acid-acetone solution was made by adding 10 µl of 12N HCl to 120 ml acetone). The final sample was analyzed by NuPAGE gel as described above. The fraction of protein PEGylated for each residue was calculated as the ratio of counts per minute in the PEGylated band (band 1) to the sum of the counts per minute in the PEGylated and unPEGylated (band 0) bands.
Time-resolved spectral measurements
Wheat germ in vitro translations (500 µl) were performed as described in the dark at 26°C for 20 min. Reaction mixtures contained 20 mM HEPES (pH 7.5), 3.2 mM Mg(OAc)2, 110 mM KOAc (pH 7.5), 30 µM of each amino acid except lysine, 300 pmol of εNBD-Lys- tRNALys or Lys-tRNALys, other components as described 32, 2 mM arginine where indicated, and 4.8 µg truncated mRNA. Before the addition of mRNA and tRNA, each reaction was incubated at 26°C for 5 min to complete the translation of any residual endogenous mRNA fragments. At the end of a the incubation period, each sample was adjusted to 500 mM KOAc and incubated on ice for 10 min. RNC complexes were then purified at 4°C on Sepharose CL-4B columns (Sigma) equilibrated with RNC buffer [50 mM HEPES (pH 7.5), 5 mM Mg(OAc)2, 40 mM KOAc] or RNC buffer with 2 mM Arg, and peak fractions were equalized based on background light scattering 41; 44. Fluorescence lifetimes were measured in RNC Buffer at 4°C with an ISS K2-002 spectrofluorometer with a laser diode (λex = 470 nm) for excitation 62. Fluorescence lifetimes were measured in RNC Buffer at 4°C with an ISS K2-002 spectrofluorometer with a laser diode (λex = 470 nm) for excitation 62. The sample chamber was maintained at 4°C and flushed with N2 to prevent condensation. NBD emission was collected using a 495 nm cut-on filter, and lifetimes were measured in the frequency domain (2–54 MHz) against a fluorescein reference standard (τ = 4.05 ns in 0.1 M NaOH; fluorescein concentration was adjusted to an emission intensity similar to that of the biochemical samples being examined). An equivalent sample lacking NBD was prepared in parallel to subtract background phase and modulation data 44; 63 Background-subtracted data were combined and fit to several different models to determine which model provided the simplest fit while still yielding a low χ2 value using Vinci multidimensional fluorescence spectroscopy analysis software (ISS, Champaign, Illinois). The best fit was almost always obtained by assuming two discrete exponential decay components. The fit of the data was not significantly improved by assuming the samples contained three components with distinguishable lifetimes, or by using a Lorentzian fit instead of a discrete fit. When NBD was incorporated into nascent AAP, the lifetime data were best-fit by a combination of two discrete NBD lifetimes. The relative mole fractions of the different lifetimes were calculated from the pre-exponential factors. In aqueous solution, an NBD dye has a short lifetime (τ~ 1 ns), whereas in a nonpolar solvent, its τ is ~8 ns 32. The variation in the mole fraction of long lifetimes in different samples indicates the dynamic distribution of NBD in distinct environments of different polarity within the tunnel,. and the average lifetime reflects this distribution 41; 44.
Supplementary Material
Acknowledgments
We thank Randolph Addison for assistance with the PEGylation experiments. This work was supported by National Institutes of Health grants to CD (R01 GM 52302), AEJ (R01 GM 26494) and MSS (R01 GM 47498) and a Robert A. Welch Foundation Chair grant (BE-0017) to AEJ.
Abbreviations used
- AAP
Arginine attenuator peptide
- Cyh
cycloheximide
- εANB-Lys
Nε-(5 azido-2-nitrobenzoyl)-lysine
- εNBD-Lys-tRNALys
Nε-6-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)aminohexanyl-Lys tRNALys
- NSP
nascent stalling peptide
- PEG
polyethylene glycol
- PTC
peptidyl transferase center
- RNC
ribosome nascent chain
- RGD
Arg-Gly Asp tripeptide
- uORF
upstream open reading frame
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mankin AS. Nascent peptide in the "birth canal" of the ribosome. Trends Biochem. Sci. 2006;31:11–13. doi: 10.1016/j.tibs.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Ito K, Chiba S, Pogliano K. Divergent stalling sequences sense and control cellular physiology. Biochem. Biophys. Res. Commun. 2010;393:1–5. doi: 10.1016/j.bbrc.2010.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cruz-Vera LR, Sachs MS, Squires CL, Yanofsky C. Nascent polypeptide sequences that influence ribosome function. Curr. Opin. Microbiol. 2011;14:160–166. doi: 10.1016/j.mib.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 4.Beringer M. Modulating the activity of the peptidyl transferase center of the ribosome. RNA. 2008;14:795–801. doi: 10.1261/rna.980308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, Sachs MS. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol. Cell. Biol. 1997;17:4904–4913. doi: 10.1128/mcb.17.9.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis RH. Compartmental and regulatory mechanisms in the arginine pathways of Neurospora crassa and Saccharomyces cerevisiae. Microbiol. Rev. 1986;50:280–313. doi: 10.1128/mr.50.3.280-313.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Fang P, Sachs MS. The evolutionarily conserved eukaryotic arginine attenuator peptide regulates the movement of ribosomes that have translated it. Mol. Cell. Biol. 1998;18:7528–7536. doi: 10.1128/mcb.18.12.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Gaba A, Sachs MS. A highly conserved mechanism of regulated ribosome stalling mediated by fungal arginine attenuator peptides that appears independent of the charging status of arginyl-tRNAs. J. Biol. Chem. 1999;274:37565–37574. doi: 10.1074/jbc.274.53.37565. [DOI] [PubMed] [Google Scholar]
- 9.Gaba A, Wang Z, Krishnamoorthy T, Hinnebusch AG, Sachs MS. Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J. 2001;20:6453–6463. doi: 10.1093/emboj/20.22.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaba A, Jacobson A, Sachs MS. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell. 2005;20:449–460. doi: 10.1016/j.molcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 11.Fang P, Spevak CC, Wu C, Sachs MS. A nascent polypeptide domain that can regulate translation elongation. Proc. Natl. Acad. Sci. USA. 2004;101:4059–4064. doi: 10.1073/pnas.0400554101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang P, Wang Z, Sachs MS. Evolutionarily conserved features of the arginine attenuator peptide provide the necessary requirements for its function in translational regulation. J. Biol. Chem. 2000;275:26710–26719. doi: 10.1074/jbc.M003175200. [DOI] [PubMed] [Google Scholar]
- 13.Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol. Cell. 2005;19:333–343. doi: 10.1016/j.molcel.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 15.Woolhead CA, Johnson AE, Bernstein HD. Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol. Cell. 2006;22:587–598. doi: 10.1016/j.molcel.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 16.Hood HM, Spevak CC, Sachs MS. Evolutionary changes in the fungal carbamoyl-phosphate synthetase small subunit gene and its associated upstream open reading frame. Fungal Genet. Biol. 2007;44:93–104. doi: 10.1016/j.fgb.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Spevak CC, Ivanov IP, Sachs MS. Sequence requirements for ribosome stalling by the arginine attenuator peptide. J. Biol. Chem. 2010;285:40933–40942. doi: 10.1074/jbc.M110.164152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muto H, Nakatogawa H, Ito K. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol. Cell. 2006;22:545–552. doi: 10.1016/j.molcel.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 19.Ivanov IP, Loughran G, Sachs MS, Atkins JF. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1) Proc Natl Acad Sci U S A. 2010;107:18056–18060. doi: 10.1073/pnas.1009269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cruz-Vera LR, Gong M, Yanofsky C. Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide. Proc. Natl. Acad. Sci. USA. 2006;103:3598–3603. doi: 10.1073/pnas.0600082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence MG, Lindahl L, Zengel JM. Effects on translation pausing of alterations in protein and RNA components of the ribosome exit tunnel. J. Bacteriol. 2008;190:5862–5869. doi: 10.1128/JB.00632-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramu H, Mankin A, Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol. Microbiol. 2009;71:811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- 23.Janzen DM, Frolova L, Geballe AP. Inhibition of translation termination mediated by an interaction of eukaryotic release factor 1 with a nascent peptidyl-tRNA. Mol. Cell. Biol. 2002;22:8562–8570. doi: 10.1128/MCB.22.24.8562-8570.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raney A, Law GL, Mize GJ, Morris DR. Regulated translation termination at the upstream open reading frame in S-adenosylmethionine decarboxylase mRNA. J. Biol. Chem. 2002;277:5988–5994. doi: 10.1074/jbc.M108375200. [DOI] [PubMed] [Google Scholar]
- 25.Onouchi H, Nagami Y, Haraguchi Y, Nakamoto M, Nishimura Y, Sakurai R, Nagao N, Kawasaki D, Kadokura Y, Naito S. Nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in the CGS1 gene of Arabidopsis. Genes Dev. 2005 doi: 10.1101/gad.1317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhushan S, Meyer H, Starosta AL, Becker T, Mielke T, Berninghausen O, Sattler M, Wilson DN, Beckmann R. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol. Cell. 2010;40:138–146. doi: 10.1016/j.molcel.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Onoue N, Yamashita Y, Nagao N, Goto DB, Onouchi H, Naito S. S-Adenosyl-L-methionine induces compaction of nascent peptide chain inside the ribosomal exit tunnel upon translation arrest in the Arabidopsis CGS1 gene. J. Biol. Chem. 2011;286:14903–14912. doi: 10.1074/jbc.M110.211656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu J, Deutsch C. Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry. 2005;44:8230–8243. doi: 10.1021/bi050372q. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, Deutsch C. Folding zones inside the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2005;12:1123–1129. doi: 10.1038/nsmb1021. [DOI] [PubMed] [Google Scholar]
- 30.Tu L, Wang J, Deutsch C. Biogenesis of the T1-S1 linker of voltage-gated K+ channels. Biochemistry. 2007;46:8075–8084. doi: 10.1021/bi700319f. [DOI] [PubMed] [Google Scholar]
- 31.Tu LW, Deutsch C. A folding zone in the ribosomal exit tunnel for Kv1.3 helix formation. J. Mol. Biol. 2010;396:1346–1360. doi: 10.1016/j.jmb.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crowley KS, Reinhart GD, Johnson AE. The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation. Cell. 1993;73:1101–1115. doi: 10.1016/0092-8674(93)90640-c. [DOI] [PubMed] [Google Scholar]
- 33.Fang P, Wu C, Sachs MS. Neurospora crassa supersuppressor mutants are amber codon-specific. Fungal Genet. Biol. 2002;36:167–175. doi: 10.1016/s1087-1845(02)00014-2. [DOI] [PubMed] [Google Scholar]
- 34.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 35.Berisio R, Schluenzen F, Harms J, Bashan A, Auerbach T, Baram D, Yonath A. Structural insight into the role of the ribosomal tunnel in cellular regulation. Nat. Struct. Biol. 2003;10:366–370. doi: 10.1038/nsb915. [DOI] [PubMed] [Google Scholar]
- 36.Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for cotranslational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- 37.Weil JE, Hadjithomas M, Beemon KL. Structural characterization of the Rous sarcoma virus RNA stability element. J. Virol. 2009;83:2119–2129. doi: 10.1128/JVI.02113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunkle JA, Xiong L, Mankin AS, Cate JH. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. USA. 2010;107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pool MR. A trans-membrane segment inside the ribosome exit tunnel triggers RAMP4 recruitment to the Sec61p translocase. J Cell Biol. 2009;185:889–902. doi: 10.1083/jcb.200807066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- 41.Lin P-J, Jongsma CG, Liao S, Johnson AE. Transmembrane segments of nascent polytopic membrane proteins control cytosol/ER targeting during membrane intergration. J. Cell Biol. doi: 10.1083/jcb.201103117. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, Marquez V, Mielke T, Thomm M, Berninghausen O, Beatrix B, Soding J, Westhof E, Wilson DN, Beckmann R. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc. Natl. Acad. Sci. USA. 2010 doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 44.Lin P-J, Jongsma CG, Pool MR, Johnson AE. Polytopic membrane protein folding at L17 in the ribosome tunnel initiates cyclical changes at the translocon. J. Cell Biol. doi: 10.1083/jcb.201103118. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freitag M, Dighde N, Sachs MS. A UV-induced mutation in Neurospora that affects translational regulation in response to arginine. Genetics. 1996;142:117–127. doi: 10.1093/genetics/142.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Sachs MS. Arginine-specific regulation mediated by the Neurospora crassa arg-2 upstream open reading frame in a homologous, cell-free in vitro translation system. J. Biol. Chem. 1997;272:255–261. doi: 10.1074/jbc.272.1.255. [DOI] [PubMed] [Google Scholar]
- 47.Johnson AE. Fluorescence approaches for determining protein conformations, interactions and mechanisms at membranes. Traffic. 2005;6:1078–1092. doi: 10.1111/j.1600-0854.2005.00340.x. [DOI] [PubMed] [Google Scholar]
- 48.Bhushan S, Hoffmann T, Seidelt B, Frauenfeld J, Mielke T, Berninghausen O, Wilson DN, Beckmann R. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1000581. e1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hood HM, Neafsey DE, Galagan J, Sachs MS. Evolutionary roles of upstream open reading frames in mediating gene regulation in fungi. Annu Rev Microbiol. 2009;63:385–409. doi: 10.1146/annurev.micro.62.081307.162835. [DOI] [PubMed] [Google Scholar]
- 50.Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache JP, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, Steitz TA, Beckmann R. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trabuco LG, Harrison CB, Schreiner E, Schulten K. Recognition of the regulatory nascent chain TnaC by the ribosome. Structure. 2010;18:627–637. doi: 10.1016/j.str.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vazquez-Laslop N, Ramu H, Klepacki D, Kannan K, Mankin AS. The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 2010 doi: 10.1038/emboj.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang L, Huang K, Yang X. Dielectric Properties of N,N-Dimethylformamide Aqueous Solutions in External Electromagnetic Fields by Molecular Dynamics Simulation. J Phys Chem A. 2009 doi: 10.1021/jp909802c. [DOI] [PubMed] [Google Scholar]
- 54.Vazquez-Laslop N, Klepacki D, Mulhearn DC, Ramu H, Krasnykh O, Franzblau S, Mankin AS. Role of antibiotic ligand in nascent peptide-dependent ribosome stalling. Proc. Natl. Acad. Sci. USA. 2011;108:10496–10501. doi: 10.1073/pnas.1103474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu C, Amrani N, Jacobson A, Sachs MS. The use of fungal in vitro systems for studying translational regulation. In: Lorsch J, editor. Translation initiation: extract systems and molecular genetics. Vol. 429. San Diego: Elsevier; 2007. pp. 203–225. [DOI] [PubMed] [Google Scholar]
- 56.Johnson AE, Woodward WR, Herbert E, Menninger JR. Nepsilon-acetyllysine transfer ribonucleic acid: a biologically active analogue of aminoacyl transfer ribonucleic acids. Biochemistry. 1976;15:569–575. doi: 10.1021/bi00648a018. [DOI] [PubMed] [Google Scholar]
- 57.Krieg UC, Walter P, Johnson AE. Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc. Natl. Acad. Sci. USA. 1986;83:8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flanagan JJ, Chen JC, Miao Y, Shao Y, Lin J, Bock PE, Johnson AE. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detectedsubnanomolar affinity that does not diminish as the nascent chain lengthens. J. Biol. Chem. 2003;278:18628–18637. doi: 10.1074/jbc.M300173200. [DOI] [PubMed] [Google Scholar]
- 59.Erickson AH, Blobel G. Cell-free translation of messenger RNA in a wheat germ system. Methods Enzymol. 1983;96:38–50. doi: 10.1016/s0076-6879(83)96007-x. [DOI] [PubMed] [Google Scholar]
- 60.Do H, Falcone D, Lin J, Andrews DW, Johnson AE. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell. 1996;85:369–378. doi: 10.1016/s0092-8674(00)81115-0. [DOI] [PubMed] [Google Scholar]
- 61.Liao S, Lin J, Do H, Johnson AE. Both lumenal and cytosolic gating of the aqueous ER translocon pore are regulated from inside the ribosome during membrane protein integration. Cell. 1997;90:31–41. doi: 10.1016/s0092-8674(00)80311-6. [DOI] [PubMed] [Google Scholar]
- 62.Alder NN, Jensen RE, Johnson AE. Fluorescence mapping of mitochondrial TIM23 complex reveals a water-facing, substrate-interacting helix surface. Cell. 2008;134:439–450. doi: 10.1016/j.cell.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 63.Reinhart GD, Marzola P, Jameson DM, Gratton E. A method for on-line background subtraction in frequency domain fluorometry. J. Fluoresc. 1991;1:153–162. doi: 10.1007/BF00865362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.