Abstract
Blue light receptors in Arabidopsis include two types of proteins, cryptochromes and phototropins. Previous studies have suggested that the cryptochromes cry1 and cry2 function mainly in photomorphogenic responses and that the phototropins phot1 and phot2 mainly regulate photo-induced movements. Receptors in the same family have redundant functions, although their responses to the fluence rate of blue light differ. To uncover functions of blue light receptors that may be concealed by their functional redundancy, we conducted analyses of combinatorial multiple mutants of blue light receptors. Comparison of the responses of the quadruple mutant cry1 cry2 phot1 phot2 to blue light with those of related triple mutants revealed that cryptochromes function in blue light-dependent, random hypocotyl-bending and that phototropins function in one photomorphogenic response, cotyledon expansion. Microarray analysis suggested that cry1 and cry2 independently function as key regulators of early blue light-induced genes, whereas phot1 and phot2 play subsidiary roles in transcriptional regulation by blue light.
Blue light induces many adaptive responses in plants (1–5). These responses are classified into two types by their reversibility and speed and by the kind of photoreceptor: photo-induced movements or photomorphogenic responses. Phototropism, chloroplast movement, and stomatal opening are observed immediately and occur reversibly; therefore, these are photo-induced movements. The photoreceptors regulating photo-induced movements are the phototropins phot1 and phot2 (6). Previous research with a phot1 phot2 double mutant revealed that these phototropins show functional redundancy in phototropic response, chloroplast movement, and stomatal opening in a fluence-rate-dependent manner (7, 8). On the other hand, a photomorphogenic response comprises relatively slow and irreversible responses, such as inhibition of hypocotyl elongation, cotyledon expansion, and cotyledon opening and induction of anthocyanin accumulation. These responses are regulated by the cryptochromes cry1 and cry2. Analyses of blue light responses of a cry1 cry2 double mutant and of plants overexpressing CRY1 or CRY2 support the hypothesis that these two cryptochromes play redundant roles in photomorphogenic responses in response to blue light (9, 10).
Cryptochromes show many functional similarities to the red/far-red light receptors known as phytochromes (11, 12). However, the functional relationship between the two types of blue light receptors, phototropins and cryptochromes, has scarcely been investigated. They show no similarities in terms of subcellular localization or molecular function. Little evidence is available that they share either the same blue light responses or common signal-transduction factors downstream from them. The only function of phototropins in photomorphogenic responses known so far is that of phot1 in the initial rapid inhibition of hypocotyl elongation by blue light irradiation (13). Similarly, the possible functions of cryptochromes in photo-induced movements have not yet been fully investigated. cry1 cry2 double mutant shows a normal phototropic response and normal chloroplast movement under blue light (14, 15), suggesting that cryptochromes do not contribute to these responses. However, a slight contribution of other blue light receptors to the phototropic response was predicted in an analysis of the phot1 phot2 double mutant (7). Whether phototropins regulate photo-induced movements independently of cryptochromes remains unknown.
We attempted to investigate the functional relationship between cryptochromes and phototropins by analyzing the functions of each photoreceptor in blue light responses. Each blue light receptor in the same family functions redundantly in response to the fluence rate of blue light, and such redundancy makes analysis of the function of individual photoreceptors difficult. To solve this problem, we conducted an analysis of combinatorial multiple mutants of blue light receptors. By comparing a cry1 cry2 phot1 phot2 quadruple mutant with related triple mutants, we performed physiological and microarray analyses of the blue light-signaling pathways. Our results indicate additional functions of cryptochromes in a blue light response (blue light-dependent, random hypocotyl-bending) and of phototropins in one type of photomorphogenic response (cotyledon expansion). Each blue light receptor functions independently of the other three receptors in most cases. The microarray analysis suggested that cryptochromes play major roles and phototropins play minor roles in the transcriptional regulation of blue-lightresponsive genes.
Materials and Methods
Plant Materials and Light Sources. The quadruple mutant and each triple mutant line were generated by crossing a cry1 cry2 mutant (hy4-2.23N fha-1) with a phot1 phot2 mutant (phot1-101/nph1-101 phot2-5/cav1-5/npl1-1). Genotypes of those mutants were confirmed with PCR-based DNA markers (data not shown). Genomic regions of CRY1, CRY2, PHOT1, and PHOT2 were obtained from an EcoRI-digested ≈7.6-kb fragment of BAC clone T3H13, a SalI-digested ≈6.9-kb fragment of BAC clone F19P19, a HindIII-digested ≈11.6-kb fragment of BAC clone T6D9, and a MscI-digested ≈11.2-kb fragment of P1 clone MCK7, respectively. Transformation of the quadruple mutant with each genomic fragment was performed by the vacuum infiltration method of ref. 16. Homozygous T3 plants were used in the experiments.
Light irradiation for all experiments was obtained with light-emitting diode blue (470 ± 30 nm) light lamps (LED-mB or LED-B, Eyela, Tokyo) or red (660 ± 20 nm) light lamps (LED-mR or LED-R, Eyela). The fluence rate of blue light was controlled with filters (film no. 72, Tokyo Butai Shoumei, Tokyo). Fluence rates were determined by using a quantum meter (model Li-250, Li-Cor, Lincoln, NE).
Hypocotyl-Bending Measurement. Germinating seedlings were grown on Okada and Shimura (OS) 1.5% agar medium under continuous blue light irradiation from above at indicated fluence rates or in the dark for 3 days (17). Degree of deviation of hypocotyl growth from phototropic vector (antigravitropic vector) was measured. At least 25 seedlings were counted per histogram.
Cotyledon Expansion Measurement. Germinating seedlings were grown on Murashige and Skoog (MS) 1.5% agar medium in plates under continuous blue light irradiation or in the dark for 6 days. The area of the cotyledon of each plant was measured with image-pro plus 4.0 software (Media Cybernetics, Silver Spring, MD). Experiments were repeated independently three times, and >20 cotyledons were measured each time.
Microarray Analysis. Eleven-day-old seedlings grown on MS 1.0% agar medium with 10% sucrose under constant white light were irradiated with red light (40 μmol·m–2·sec–1) for 3 days and then with blue light (10 μmol·m–2·sec–1) plus red light (40 μmol·m–2·sec–1) for 1 h. As the control, plants grown under the same conditions were irradiated with red light for 3 days and then with only red light (40 μmol·m–2·sec–1) for 1 h. The blue light-irradiated seedlings were used for the experimental array, and the control seedlings were used for the baseline array. Poly(A)+ RNA was isolated from ≈1,000 seedlings and used for cRNA synthesis as described (18). Biotin-labeled cRNA probes were hybridized to oligonucleotide microarrays (Arabidopsis GeneChip, Affymetrix, Santa Clara, CA) containing 8,300 probe sets. Further protocols were performed according to instructions provided by Affymetrix. To ensure the reproducibility of the results, we performed three independent biological experiments.
We applied two separate statistical analyses to eliminate false-positive genes. Initial analyses were performed with microarray suite 5.0 software (mas 5.0, Affymetrix), and subsequent analyses were carried out with genespring 4.1.1 software (Silicon Genetics, Redwood City, CA) (for further details, see Supporting Text, which is published as supporting information on the PNAS web site).
Classification of genes and clustering of each gene were performed with genespring 4.1.1 software.
Prediction of Subcellular Localization. Prediction of subcellular localization of proteins encoded by genes identified by the microarray analysis was performed with targetp server 1.01 software (www.cbs.dtu.dk/services/TargetP).
Northern Blot Analysis. Total RNA was prepared from the seedlings as described (19). RNA samples (4 μg) were separated on a 1.25% agarose gel containing formaldehyde, transferred to a Hybond-N blotting membrane (Amersham Pharmacia Bioscience), and UV cross-linked. The membrane was hybridized to a randomly primed 32P-labeled DNA probe in Perfect-Hyb hybridization buffer (Toyobo, Osaka), washed, and exposed according to the manufacturer's instructions. The regions used as probes in each gene were as follows (the position of the first nucleotide of each coding sequence was arbitrarily set as 1): At3g55800, 1–526; At2g30390, 123–790; At5g05580, 26–526; At35090, 154–703; At2g21330, 8–757; and At29450, 69–558. Autoradiograms were scanned with a Typhoon 8600 scanner (Amersham Pharmacia Bioscience).
Results and Discussion
Making of the Quadruple Mutant, Triple Mutants, and Transgenic Quadruple Mutants. To clarify the precise function of each of the blue light photoreceptors, cry1, cry2, phot1, and phot2, by an analysis of combinatorial multiple mutants, we made a quadruple mutant, cry1 cry2 phot1 phot2 (quad), and the four triple mutants, CRY1+/+ cry2 phot1 phot2 (called +cry1 for short), CRY2+/+ cry1 phot1 phot2 (+cry2), PHOT1+/+ cry1 cry2 phot2 (+phot1), and PHOT2+/+ cry1 cry2 phot1 (+phot2), by crossing the cry1 cry2 mutant with the phot1 phot2 mutant. The background ecotype of the phot2 mutant was Wassilewskija (WS), and those of the others were Landsberg erecta (Ler). The individual isolates of the triple mutants and the quadruple mutant were heterogeneous with respect to the background ecotype. It has been reported that WS and Ler show similar phototropic responses and chloroplast movements (7, 15), and we confirmed that WS and Ler responded to blue light in a similar manner for all the responses examined (see below). However, in case a crossed-background ecotype has an effect on blue light responses not manifested in the parental lines, the results were reconfirmed with another respective line of the triple mutant and the quadruple mutant, which were isolated in an independent manner (see below). Furthermore, to be completely sure, we made artificial triple mutants (transgenic quadruple mutants), CRY1 cry1 cry2 phot1 phot2 (called +cry1T for short), CRY2 cry1 cry2 phot1 phot2 (+cry2T), PHOT1 cry1 cry2 phot1 phot2 (+phot1T), and PHOT2 cry1 cry2 phot1 phot2 (+phot2T), by transforming the quadruple mutant with plasmids containing the genomic regions of CRY1, CRY2, PHOT1, or PHOT2.
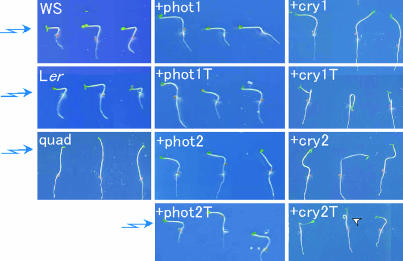
cry1 and cry2 Regulate a Photo-Induced Movement: Blue Light-Dependent, Random Hypocotyl-Bending. The function of cryptochromes in photo-induced movement remained unsolved in previous analyses because of their possible functional redundancy with phototropins. To clarify the contribution of cryptochromes to the phototropic response, we compared hypocotyl curvature of the +cry1, +cry1T, +cry2, and +cry2T mutants with that of the quadruple mutant under blue light. The hypocotyls of the quadruple mutant showed hardly any curvature at all of the fluence rates examined (Fig. 1). Unexpectedly, hypocotyl curvature in the +cry1, +cry1T, +cry2, and +cry2T mutants was not related to the direction of the light source, and sometimes a knot even formed (Fig. 1). Because this seems to be a blue light response that differs from phototropism, we further analyzed the response in detail. We exposed seedlings to blue light irradiation from above and measured degrees deviation of the direction of hypocotyl growth from the phototropic vector. Blue light irradiation (1 μmol·m–2·sec–1 or 100 μmol·m–2·sec–1) induced the random hypocotyl bending of 60° to 130° in the +cry1, +cry1T, +cry2, and +cry2T mutants, whereas the quadruple mutant under blue light scarcely showed any random hypocotyl-bending, deviating by 5–15°, just as it did in the dark (Fig. 2). Similar results were obtained with all independent lines of the +cry1, +cry2, +cry1T, and +cry2T mutants and of the quadruple mutant (data not shown). The hypocotyls of wild-type and +phot1, +phot1T, +phot2, and +phot2T seedlings showed similar straight-upward growth under all conditions with deviations from 5° to 15° (Fig. 2), indicating that this new response is promoted only by cryptochromes. Although it has been reported that phytochromes induce a similar random hypocotyl-bending (change in growth orientation) by red light irradiation (20, 21), the results that the quadruple mutant that has a normal complement of phytochromes scarcely show the random hypocotyl-bending, indicating that blue light activation of phytochrome was minimal. Therefore, we concluded that the blue light-dependent, random hypocotyl-bending in the absence of both phototropins is induced by cry1 and cry2.
Fig. 1.
The +cry1, +cry1T, +cry2, and +cry2T mutants show hypocotyl curvatures that are irrelevant to the phototropic vector. Shown are phototropic responses of the wild types (WS and Ler), the quadruple mutant (quad), and the +phot1, +phot1T, +phot2, +phot2T, +cry1, +cry1T, +cry2, and +cry2T mutants. Each plant was exposed to blue light at a fluence rate of 100 μmol·m–2·sec–1 for 12 h. (For further details, see Supporting Text.) The light source was to the left in the pictures. The arrowhead indicates a knot in one hypocotyl.
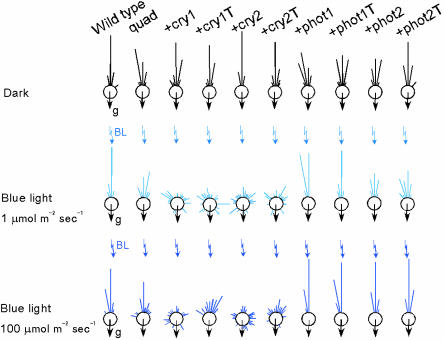
Fig. 2.
Blue light-dependent, random hypocotyl-bending is observed only in the +cry1, +cry1T, +cry2, and +cry2T mutants. Seedlings of wild type, the quadruple mutant, and the +cry1, +cry1T, +cry2, +cry2T, +phot1, +phot1T, +phot2, and +phot2T mutants were grown in the dark or under blue light irradiation for 3 days. The degree of deviation of hypocotyl growth from phototropic vector was measured.
The bending induced by blue light irradiation, which was identified in this study, is similar to that induced by red light irradiation. It has been reported that the function of cryptochromes in photomorphogenic responses overlapped almost entirely with the function of phytochromes and that they share the regulation of photomorphogenic signaling pathways (11, 12), suggesting a common mechanisms in cryptochrome- and phytochrome-dependent bending.
One hypothesis to explain the random hypocotyl-bending by red light irradiation is that phytochromes suppress the negative hypocotyl gravitropism (20, 21). Therefore, it is possible that cryptochromes also suppress the negative hypocotyl gravitropism, at least in the absence of both phototropins. However, on the other hand, this hypothesis could not explain the blue light-dependent, random hypocotyl-bending in etiolated seedlings of the +cry1, +cry1T, +cry2, and +cry2T mutants as observed in Fig. 1. In this condition, the random hypocotyl-bending seems to occur by active effects of cryptochromes to induce the bending rather than by inhibitory effects of cryptochromes to the negative gravitropism in hypocotyl. Further analyses are required to understand how cryptochromes and phytochromes induce the random hypocotyl-bending.
Our previous analysis indicated the following: phot1 functions in the phototropic response under blue light irradiation at fluence rates ranging from 0.01 to 100 μmol·m–2·sec–1; phot2 functions only at high fluence rates; and phot1 and phot2 act redundantly (7). We confirmed those observations by comparing hypocotyl curvatures in the +phot1, +phot1T, +phot2, and +phot2T mutants with those in the quadruple mutant (Fig. 5A, which is published as supporting information on the PNAS web site). Furthermore, our results suggest that each phototropin functions independently of the other three blue light receptors.
Function of Each Blue Light Receptor in Other Photo-Induced Movements. We also examined the involvement of each photoreceptor in other photo-induced movements, stomatal opening and chloroplast movement. There was little difference in stomatal aperture between seedlings under blue plus red light and those under red light only in the quadruple mutant or the +cry1, +cry1T, +cry2, or +cry2T mutants, whereas blue light-induced stomatal opening in +phot1, +phot1T, +phot2, and +phot2T mutants, although to a lesser extent than that in WS or Ler (Fig. 5B). These results indicate that phot1 and phot2 partly regulate blue light-activated stomatal opening by themselves and that neither cry1 nor cry2 does so in the absence of the other three blue light receptors. Chloroplast relocation was also observed in each mutant. As predicted (7), the quadruple mutant and the +cry1, +cry1T, +cry2, and +cry2T mutants showed no response, the +phot1 and +phot1T mutants showed an accumulation response at all fluences of blue light (2 and 40 μmol·m–2·sec–1), and the +phot2 and +phot2T mutants showed an accumulation response at low fluence and an avoidance response at high fluence rates of blue light (Fig. 5C). These results indicate that phot1 induces a chloroplast accumulation response at high and low fluence rates and that phot2 induces an accumulation response at low fluence rates and an avoidance response at high fluence rates, independently of the other blue light receptors.
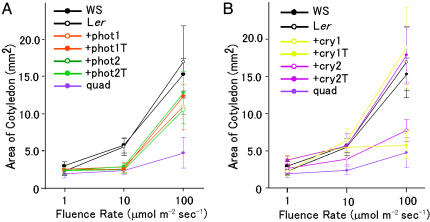
Phototropins Regulate a Photomorphogenic Response: Cotyledon Expansion. We examined the contribution of phototropins to four photomorphogenic responses, inhibition of hypocotyl elongation, anthocyanin accumulation, cotyledon expansion, and cotyledon opening, in the same way. The quadruple mutant showed hardly any response at any fluence rate of blue light examined, except for a slight response at the highest fluence rate of 100 μmol·m–2·sec–1, indicating that photoreceptor(s) other than cryptochromes and phototropins play little, if any, role in those photomorphogenic responses (Fig. 3 and Fig. 6, which is published as supporting information on the PNAS web site). The +phot1, +phot2, +phot1T, and +phot2T mutants showed little, if any, inhibition of hypocotyl elongation, anthocyanin accumulation, or cotyledon opening at all fluence rates of blue light examined, indicating that phototropins do not contribute to those responses (Figs. 6 D, F, and G). However, the cotyledons of the +phot1, +phot1T, +phot2, and +phot2T mutants expanded more than those of the quadruple mutant at a high fluence rate of 100 μmol·m–2·sec–1 (Fig. 3A). Similar results were obtained with all independent lines of the +phot1, +phot1T, +phot2 and +phot2T mutants and of the quadruple mutant (data not shown). These results indicate that both phototropins induce cotyledon expansion independently of the other blue light receptors at high fluence rates. The phot1 phot2 mutant is known to have small curly leaves when grown under white light (7, 8, 22). These results suggest that phot1 and phot2 regulate the photomorphogenic response of leaf tissue.
Fig. 3.
Cotyledon expansion in the wild type, the quadruple mutant, and the +phot1, +phot1T, +phot2, and +phot2T mutants (A) or in the wild type, the quadruple mutant, and the +cry1, +cry1T, +cry2, and +cry2T mutants (B) grown under blue light at the indicated fluence rates for 6 days. Data points and error bars represent the means ± SD of the average value of the cotyledon area in three independent experiments.
Previous reports suggested that cry1 regulates those photomorphogenic responses mainly at high fluence rates and regulates cry2 mainly at low fluence rates. Those reports are reconfirmed by our analysis. The difference between the +cry1 (+cry1T) mutant and the quadruple mutant for all those responses was almost as much as the difference between wild type and the quadruple mutant, indicating that cry1 functions in those photomorphogenic responses independently of the other three blue light receptors (Figs. 3B and 6 C, E, and G). The difference between the +cry2 (or +cry2T) mutant and the quadruple mutant in those photomorphogenic responses indicated that cry2 regulates these kinds of photomorphogenic responses in part at fluence rates of 10 and 100 μmol·m–2·sec–1 and fully at 1 μmol·m–2·sec–1 or lower, independently of the other blue light receptors.
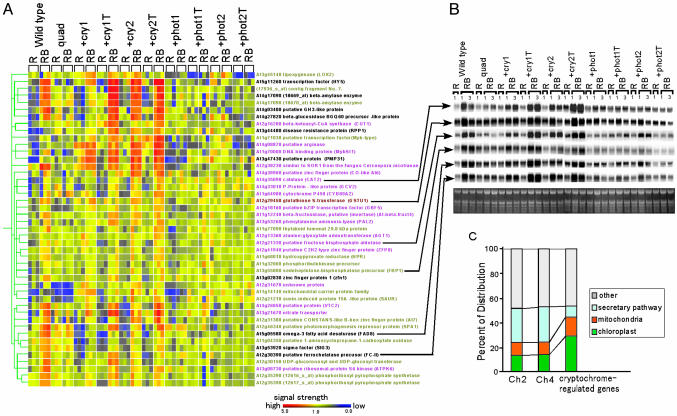
cry1 and cry2 Independently Play a Major Role in Regulating Blue Light-Induced Gene Expression. Our physiological results indicated that phototropins and cryptochromes shared very little function in most blue light responses. To clarify whether they shared a common regulation of blue light-induced genes, we applied the combinatorial loss-of-function approach to a microarray analysis with an Affymetrix GeneChip array that included ≈8,300 Arabidopsis genes. To diminish the effects of phytochromes that also absorb blue light, seedlings were first irradiated with red light and then simultaneously with red and blue light. Of 8,300 genes, 369 were isolated as induced by blue light in the +cry1 mutant by using the mas algorithm, and among these, 37 met the condition of at least a 1.3-fold induction, and the difference was shown to be significant by t test. Similarly, 250, 73, 116, 5, 8, 2, and 6 genes were isolated as being induced by blue light in the +cry1T, +cry2, +cry2T, +phot1, +phot1T, +phot2, and +phot2T mutants, respectively (Fig. 7 A–D, which is published as supporting information on the PNAS web site). After eliminating genes that might be induced by blue light in the quadruple mutant, 26 genes were induced in both the +cry1 and +cry1T mutants; these genes were considered to be regulated by cry1. Similarly, 28, 1, and 0 genes were identified as genes regulated by cry2, phot1, and phot2, respectively (Table 1, which is published as supporting information on the PNAS web site). All genes regulated by each photoreceptor were clustered into a hierarchical gene by using a Pearson correlation (Fig. 4A). Of the total of 45 genes, 44 (98%) were induced >1.3-fold by blue light in the wild type, indicating that each blue light receptor functions in regulating those genes in wild-type seedlings. Furthermore, 35 genes (84%) were induced in both the +cry1 and +cry1T mutants, 44 genes (98%) were induced in both the +cry2 and +cry2T mutants, 1 (2%) was induced in both the +phot1 and +phot1T mutants, and no gene was induced in both the +phot2 and +phot2T mutants. These results indicate that most genes regulated by blue light receptors are regulated by cry1 and cry2 independently. Several genes were randomly selected from these genes, and all of them, including one gene regulated by phot1, were confirmed by a Northern blot analysis to be induced by blue light in wild type and in the +cry1, +cry1T, +cry2, and +cry2T mutants but not in the quadruple mutant (Fig. 4B). In Northern blot analysis, we confirmed that one gene regulated by phot1 was induced in the +phot1 and +phot1T mutants but not in the quadruple mutant, although their induction in the triple mutants was mild. These results indicated that cry1 and cry2 independently play a major role in regulating gene expression early in the blue light signal-transduction pathway and that the effects of phototropins are much weaker than those of cryptochromes. Those results are consistent with the subcellular localization and functions of those apoproteins. Cryptochromes can localize in the nucleus and, in general, regulate photomorphogenic responses that are relatively slow, taking at least several hours to detect. On the other hand, phototropins are kinases that localize to the plasma membrane region (22, 23) and mainly regulate photo-induced movements that are relatively fast responses observable within ≈1 h.
Fig. 4.
Gene profiling of blue light-induced genes in the wild type and in each mutant. (A) Clustergram of the 45 genes regulated by each photoreceptor. Signal intensities of the probe for the 45 blue light-induced genes on a microarray from the wild type, the quadruple mutant, and the +cry1, +cry1T, +cry2, +cry2T, +phot1, +phot1T, +phot2, and +phot2T mutant seedlings that were illuminated with control irradiation (R) or with blue light irradiation (RB) in three replicate assays are shown. The color of each signal in the clustergram represents the signal intensity (red, high; yellow, medium; and blue, low). The color of each gene represents regulation by each photoreceptor (ocher, cry1; pink, cry2; brown, photo1; and black, both cry1 and cry2). (B) Northern blot analysis of blue light-induced genes. An 11-day-old wild-type, quadruple mutant, and +cry1, +cry1T, +cry2, +cry2T, +phot1, +phot1T, +phot2, and +phot2T mutant seedlings were irradiated with red light (R) or blue plus red light (RB) for 1 or 3 h before preparation of total RNA. The bottom gel shows the rRNA in each total RNA sample as a control. (C) Predicted distribution of subcellular localization of proteins encoded by the 45 blue light-induced genes identified as being regulated by cry1 and/or cry2. Subcellular localization was determined with targetp server 1.01 software. Localization of all annotated proteins on chromosomes 2 and 4 of Arabidopsis are shown for comparison.
Involvement of Cryptochromes in the Regulation of Photosynthetic Reaction. To detect a peculiarity in the 45 blue light-induced genes regulated by cryptochromes, we examined whether some of them shared a common pathway, regulatory sequence, or protein localization. First, we queried the subcellular localization of the 45 genes with targetp server 1.01 which predicts the localization from the protein sequence. The results were synthesized and compared with the localization of all annotated proteins on chromosomes 2 and 4 of Arabidopsis that were officially predicted by targetp server and that shows a general trend. The percentages of proteins encoded by blue light-induced genes localizing in chloroplasts (28.9%) were much higher than those of the annotated proteins on chromosomes 2 and 4 (13.1% and 13.9%; Fig. 4C). These results indicate that the proteins encoded by blue light-inducible genes regulated by cryptochromes tend to be located in chloroplasts. In chloroplasts, two photoreactions (photosynthetic light reaction and Calvin cycle) occur. cry1 and cry2 mediate the induction of psbD, which is a major component of PS II of photosynthetic light reaction under blue light (24). In this context, one of our cryptochrome-regulated genes [At3g53920 (SIG3)] encodes a transcriptional cofactor, sigma factor, that functions in the transcription of psbD (25), suggesting that both cryptochromes are involved in the transcriptional regulation of psbD by SIG3. On the other hand, three of the genes in the list of genes regulated by cryptochromes function in the other photoreaction (the Calvin cycle): phosphoribulokinase (At1g32060), sedoheptulose-bisphosphatase (At3g55800), and fructose bisphosphate aldolase (At2g21330) (Fig. 7). To clarify whether cryptochromes regulate genes in the Calvin cycle, the expression patterns of six other genes functioning in the pathway were examined. Two genes [ribose-5-phosphate isomerase (At2g01290) and fructose-bisphosphatase (At3g54050)] were induced more than 1.3-fold by blue light in the wild type and in most +cry1, +cry1T, +cry2, and +cry2T mutants, but not in the quadruple mutant. These results indicate that cryptochromes regulate some part of gene expression in the Calvin cycle. Those microarray data may support the hypothesis that cryptochromes control both photosynthetic reactions under blue light.
Conclusion
Analysis of each blue light receptor by using the combinatorial multiple mutant enabled us to discover blue light-dependent, random hypocotyl-bending regulated by cry1 or cry2. Similarly, the involvement of phototropin in one photomorphogenic response, cotyledon expansion, was clarified. Our microarray analyses revealed the principal roles played by cryptochromes in blue light-induced gene expression. Furthermore, in most of those responses, including regulation of blue light-induced genes, each photoreceptor functions independently of the other three blue light receptors.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing the BAC clones T3H13, F19P19, and T6D9; the Kazusa DNA Research Institute for the P1 clone MCK7; Drs. Hideki Goda, Yukihisa Shimada, and Shigeo Yoshida (RIKEN Plant Science Center) for technical assistance and discussion of the microarray analysis; and our laboratory colleagues for discussion of the physiological analysis.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ahmad, M. & Cashmore, A. R. (1996) Plant Mol. Biol. 30, 851–861. [DOI] [PubMed] [Google Scholar]
- 2.Fankhauser, C. & Chory, J. (1997) Annu. Rev. Cell. Dev. Biol. 13, 203–229. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad, M. (1999) Curr. Opin. Plant Biol. 2, 230–235. [DOI] [PubMed] [Google Scholar]
- 4.Briggs, W. R. & Huala, E. (1999) Annu. Rev. Cell. Dev. Biol. 15, 33–62. [DOI] [PubMed] [Google Scholar]
- 5.Lin, C. (2002) Plant Cell 14, Suppl., S207–S225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briggs, W. R. & Christie, J. M. (2002) Trends Plant Sci. 7, 204–210. [DOI] [PubMed] [Google Scholar]
- 7.Sakai, T., Kagawa, T., Kasahara, M., Swartz, T. E., Christie, J. M., Briggs, W. R., Wada, M. & Okada, K. (2001) Proc. Natl. Acad. Sci. USA 98, 6969–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinoshita, T., Doi, M., Suetsugu, N., Kagawa, T., Wada, M. & Shimazaki, K. (2001) Nature 414, 656–660. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad, M., Jarillo, J. A. & Cashmore, A. R. (1998) Plant Cell 10, 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin, C., Yang, H., Guo, H., Mockler, T., Chen, J. & Cashmore, A. R. (1998) Proc. Natl. Acad. Sci. USA 95, 2686–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casal, J. J. (2000) Photochem. Photobiol. 71, 1–11. [DOI] [PubMed] [Google Scholar]
- 12.Frankhauser, C. & Stainger, D. (2002) Planta 216, 1–16. [DOI] [PubMed] [Google Scholar]
- 13.Folta, K. M. & Spalding, E. P. (2001) Plant J. 26, 471–478. [DOI] [PubMed] [Google Scholar]
- 14.Lasceve, G., Leymarie, J., Olney, M. A., Liscum, E., Christie, J. M., Vavasseur A. & Briggs W. R. (1999) Plant Physiol. 120, 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kagawa, T. & Wada, M. (2000) Plant Cell Physiol. 41, 84–93. [DOI] [PubMed] [Google Scholar]
- 16.Bechtold, N. & Pelletier, G. (1998) Methods Mol. Biol. 82, 259–266. [DOI] [PubMed] [Google Scholar]
- 17.Okada, K. & Shimura, Y. (1992) Aust. J. Plant Physiol. 19, 439–448. [Google Scholar]
- 18.Goda, H., Shimada, Y., Asami, T., Fujioka, S. & Yoshida, S. (2002) Plant Physiol. 130, 1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagy, F., Kay, S. A. & Chua, N.-H. (1988) in Plant Molecular Biology Manual, eds. Given, S. & Schilperoort, R. (Kluwer, Dordrecht, The Netherlands), pp. 1–12.
- 20.Liscum, E. & Hangarter, R. P. (1993) Plant Physiol. 103, 15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robson, P. R. H. & Smith, H. (1996) Plant Physiol. 110, 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakamoto, K. & Briggs, W. R. (2002) Plant Cell 14, 1723–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harada, A., Sakai, T. & Okada K. (2003) Proc. Natl. Acad. Sci. USA 100, 8583–8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thum, K. E., Kim, M., Christopher, D. A. & Mullet, J. E. (2001) Plant Cell 13, 2747–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsunoyama, Y., Morikawa, K., Shiina, T. & Toyoshima, Y. (2002) FEBS Lett. 516, 225–228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.