Abstract
The pyridoxal phosphate-dependent enzyme, 1-aminocyclopropane-1-carboxylate synthase (ACS; EC 4.4.1.14), catalyzes the rate-limiting step in the ethylene biosynthetic pathway in plants. The Arabidopsis genome encodes nine ACS polypeptides that form eight functional (ACS2, ACS4 –9, ACS11) and one nonfunctional (ACS1) homodimers. Because the enzyme is a homodimer with shared active sites, the question arises whether the various polypeptides can form functional heterodimers. Intermolecular complementation experiments in Escherichia coli by coexpressing the K278A and Y92A mutants of different polypeptides show that all of them have the capacity to heterodimerize. However, functional heterodimers are formed only among gene family members that belong to one or the other of the two phylogenetic branches. ACS7 is an exception to this rule, which forms functional heterodimers with some members of both branches when it provides the wt K278 residue. ACS1, the nonfunctional polypeptide as a homodimer, can also form functional heterodimers with members of its phylogenetic branch when its partners provide the wt K278 residue. The ACS gene family products can potentially form 45 homo- and heterodimers of which 25 are functional. Bimolecular fluorescence complementation and biochemical coaffinity purification assays show that the inactivity of certain heterodimers is not due to the absence of heterodimerization but rather to structural restraint(s) that prevents the shared active sites from being functional. We propose that functional heterodimerization enhances the isozyme diversity of the ACS gene family and provides physiological versatility by being able to operate in a broad gradient of S-adenosylmethionine concentration in various cells/tissues during plant growth and development. Nonfunctional heterodimerization may also play a regulatory role during the plant life cycle.
The gas ethylene (C2H4) is used by plants as a signaling molecule for regulating a variety of developmental processes and stress responses (1). These processes include seed germination, leaf and flower senescence, fruit ripening, cell elongation, nodulation, and pathogen responses. Ethylene production is induced by a variety of external factors, including wounding, viral infection, elicitors, auxin treatment, chilling injury, drought, Cd2+ and Li+ ions, and O3, SO2, and other pollutants (2–5).
Ethylene is biosynthesized from methionine, which is converted to S-adenosylmethionine (AdoMet) by the enzyme AdoMet synthase. AdoMet is converted by the 1-aminocyclopropane-1-carboxylate synthase (ACS, EC 4.4.1.14) to methylthioadenosine and 1-aminocyclopropane-1-carboxylic acid (ACC), the precursor of ethylene (3, 6, 7). ACC is oxidized to CO2, HCN, and C2H4 by ACC oxidase (ACO). ACS is a cytosolic enzyme with a short half-life that requires pyridoxal phosphate as a cofactor (2, 3, 8). The activity of ACS is regulated at the transcriptional (3, 6, 7) and posttranscriptional levels (9, 10).
ACS is encoded by a multigene family in every plant species examined (3, 7). In Arabidopsis, the ACS gene family encodes nine polypeptides, of which eight form functional and one nonfunctional homodimers (11). The primary sequence encoded by these genes shows sequence conservation ranging from 50% to 96% amino acid sequence identity with the highest variability at the carboxylic end of the protein (11). The variable carboxyl terminus is unimportant for enzyme activity but serves as a regulatory domain responsible for posttranslational regulation of the enzyme (10, 12). ACS shares sequence similarity with other pyridoxal phosphate-dependent enzymes and is most closely related to the subgroup 1 aminotransferases, which include aspartate aminotransferase (13, 14). Complementation studies with ACS mutants have shown that the enzyme functions as a homodimer whose active site is formed from the interaction of residues from the monomeric subunits similar to aspartate aminotransferase (15, 16). In particular, the Y92 residue, which is involved in substrate recognition (17), interacts with the active-site residue K278, which forms a covalent Schiff base with the pyridoxal phosphate cofactor from the adjacent subunit. The 3D structure of ACS has confirmed this model (17) and, together with available biochemical data, explains the catalytic roles of the conserved and nonconserved active-site residues (16, 18, 19).
The Arabidopsis genome has nine ACS genes and their encoded polypeptides form eight enzymatically active and one (ACS1) inactive homodimers (11). It has been postulated that the presence of ACS isozymes may reflect tissue-specific expression that satisfies the biochemical environment of the cells or tissues in which each isozyme is expressed (13). Biochemical characterization of the eight functional homodimers supports this proposition (11). Because the isozymes are homodimers, the question arises whether their subunits can form functional heterodimers and thus further enhance isozyme diversity. Here, we have used a functional complementation assay in Escherichia coli JAde6 (19) for testing the formation of functional heterodimers by intermolecular complementation of K278A and Y92A mutants of various ACS polypeptides.
Materials and Methods
Molecular Biology. Restriction and DNA modifying enzymes were obtained from New England Biolabs and Roche Diagnostics. Acrylamide and SDS/PAGE reagents were from ICN and Bio-Rad. All other chemicals used for biochemical analysis were of analytical grade and purchased from Aldrich and Sigma. Oligonucleotides were purchased from Operon Technologies (Alameda, CA) or synthesized in house with a Polyplex oligonucleotide synthesizer (GeneMachines, San Carlos, CA). Routine DNA manipulations are described in Supporting Text, which is published as supporting information on the PNAS web site.
Bacterial Strains and Plasmids. The bacterial strain used for most transformations was E. coli DH5α; supE44, hsdR17, recA1, endA1, gyrA1, thi-1, relA1, lacU169 (80lacZ M15). E. coli JAde 6 was constructed by integrating the ACC deaminase gene into the genome of JHM544 as described by Tarun et al. (19). The vectors used for expression of ACSs were pQE-80 (Qiagen, Valencia, CA) and pQE801. pQE801 was derived from pQE80 by removing the 73-bp EcoRI–SacI fragment containing the His tag and replacing it with an oligonucleotide containing the T7 tag. The cosntruction of the various plasmids is described in Supporting Text.
Intermolecular Complementation in E. coli JAde6. For the details of this procedure, refer to Supporting Text.
Biochemical Coaffinity Purification Assay and Immunoblot Analysis. For the details of this procedure, refer to Supporting Text.
Imaging of Yellow Fluorescence in E. coli. E. coli cultures expressing various YC/YN-containing gene fusions were grown to A600 = 2.0. Cells corresponding to 1 ml of culture were collected and resuspended into 100 μl of phosphate saline buffer. The fluorescence of the E. coli cells was monitored by using an Axioplan Zeiss Fluorescence Microscopy System with spot 2.2 software. The excitation filter was HQ500/20, the emission filter was HQ520lp, and the beamsplitter was Q515lp (Chroma Technology, Rockingham, VT).
Results
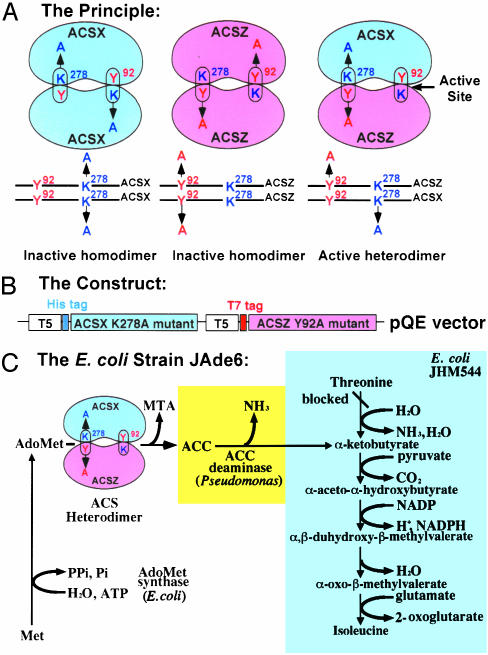
The Strategy. Fig. 1 illustrates how functional heterodimerization can be tested among the various ACS polypeptides by coexpressing mutant proteins in E. coli JAde6 (19). A prerequisite of this approach is that the mutations should be targeted on residues that are essential for enzyme activity and occupy distinct domains in the active site (Fig. 1 A). Y92 and K278 fulfill these criteria based on site-directed mutagenesis (18, 19), in vitro and in vivo intermolecular complementation (15, 16), and x-ray crystallographic data (17). According to the shared active-site model, association between two inactive proteins containing two different active-site mutations results in the formation of an active heterodimer (positive complementation). This principle can tested for functional homo- and heterodimerization among various polypeptides (Fig. 1 A). To allow hybridization to occur between two mutant ACS proteins, we have used an in vivo expression system in E. coli (Fig. 1C). This coexpression system has the advantage of using the natural efficiency of subunit assembly that occurs in the cytoplasmic environment (20). This approach has been used successfully to demonstrate shared active sites in other proteins; however, in most cases, a two-plasmid system has been used (20, 21). Because these two plasmids use two different replication origins and antibiotic markers for maintenance, they are often present in different copy members in the same cell. Thus, expression of the genes found of these two plasmids can be different and difficult to quantify. To minimize this problem, we have designed an expression vector to carry two copies of the ACS gene that may have different mutations under the control of the same promoter (Fig. 1B). To facilitate the selection of nonfunctional heterodimers, we have used a genetic screen based on the functional complementation of E. coli Ile auxotrophic cells, as shown in Fig. 1C. We have used E. coli JAde6 (19) where endogenous AdoMet synthetase catalyzes the formation of AdoMet, which can be used as a substrate by ACS to make ACC. The enzyme ACC deaminase (22) converts ACC to α-ketobutyrate, which can be used to synthesize isoleucine, thus complementing the mutation. Introduction of an active ACS in this strain will provide Ile prototrophy, whereas an inactive ACS will fail to do so.
Fig. 1.
The strategy for determining heterodimeric interactions among the various polypeptides encoded by the Arabidopsis ACS gene family members. (A) The principle of the method: schematic presentation of an active heterodimer formation by coexpressing the K278A and Y92A mutants of two ACS polypeptides X and Z, respectively. (B) The construct: a pQE double-gene construct used to coexpress the K278A and Y92A mutants of various ACSs as His- and T7-tagged proteins with the T5 promoter. (C) The E. coli strain JAde6. The biosynthetic pathway of isoleucine in the E. coli auxotroph JHM544 is shown (shaded blue). The pathway was engineered to allow Ile prototrophy by integrating ACC deaminase into the genome, yielding E. coli JAde6 (19). The strain can metabolize ACC synthesized when transformed with a functional ACS heterodimer. MTA, methylthioadenosine.
Identification of Functional Heterodimers. Fig. 6, which is published as supporting information on the PNAS web site, shows that all the His- or T7-tagged wt ACSs are enzymatically active and capable of providing Ile prototrophy in JAde6 (compare growth in LB with that in M9 medium). ACS1 is an exception because it is a nonfunctional isozyme due to the absence of the conserved tripeptide TNP (23). Furthermore, the K278A and Y92A mutations in the functional ACSs abolish ACS activity (Fig. 6) and prevent growth of JAde6 on M9 medium (compare growth of wt ACSs with the growth of the K278A and Y92A mutants). The Y92A mutation in ACS5 and ACS9 does not abolish their activity completely, allowing some growth in M9 medium (Fig. 6). Consequently, deletions of this residue (ΔY92) were constructed and used for the complementation studies (Fig. 6). The numbering of the K278 and Y92 residues is that of Le-ACS2 (13) and is different in the various ACSs, but it is used throughout this work for uniformity (see Supporting Text for the actual numbering of the K and Y residues). The absence of enzyme activity in the mutant protein is not due to a decreased level of protein expression (stability), because all mutant proteins are expressed to approximately the same level as the wt proteins (Fig. 7, which is published as supporting information on the PNAS web site). The K278A mutants inhibit enzyme activity 100%; however, the degree of inhibition by the Y92A mutation varies from 93.5% to 99.4% compared with the wt level of activity (Fig. 7).
Coexpression of K278A and Y92A mutants of different functional polypeptides allowed the identification of functional heterodimers. Functional intermolecular complementation allows Ile prototrophy in M9 medium (Fig. 2 A and B). Because the Y92A mutants of ACS5 and ACS9 have enough residual activity capable of complementing JAde6, their deletion mutation (ΔY92) was used for this assay. The growth observed on M9 medium (Fig. 2B) is supported by measurements of enzyme activity in the corresponding strains expressing double-gene constructs (Fig. 7). Furthermore, immunoblot analysis shows that the absence of enzyme activity in certain heterodimers is not due to the absence of protein expression (Fig. 7).
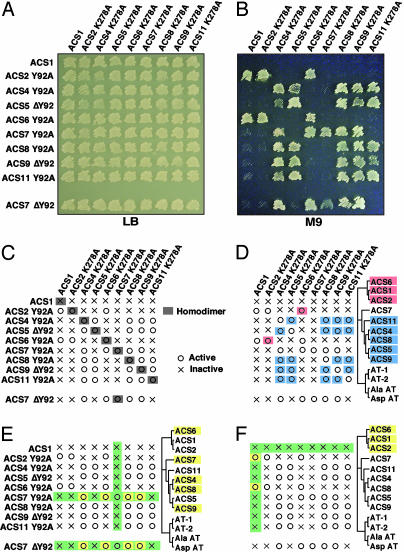
Fig. 2.
Growth of E. coli JAde6 expressing mutant ACSs. (A and B) Growth of JAde6 on rich (LB) and minimum (M9) media coexpressing the K278A and Y92A mutants of various ACSs. (C–F) Schematic presentation of the results in B regarding the formation of active and inactive heterodimers in relation to their phylogenetic location. (C) Functional homodimerization by coexpressing K278A and Y92A mutants of ACSs. (D) Functional heterodimerization among the various ACSs depending on their phylogenetic location. (E) Functional heterodimerization between the Y92 mutants of ACS7 and some members of both phylogenetic branches. (F) Functional heterodimerization between the wt ACS1 and the Y92A mutants of ACS2 and ACS6, respectively. AspAT, aspartate aminotransferase; AlaAT, alanine aminotransferase.
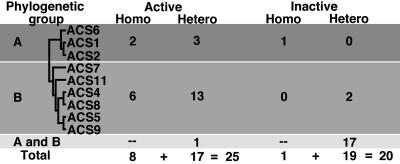
A schematic representation of the results of Fig. 2 A and B is shown in Fig. 2 C–F. First, all functional ACSs are homodimers with shared active sites (Fig. 2C). Second, functional heterodimers are formed only among members of the same phylogenetic branch (Fig. 2D). Third, ACS7 is an exemption to this rule. The ACS7 Y92A mutant can form functional heterodimers with some members of both branches (ACS6, ACS4, ACS8, and ACS9) only when the ACS7 provides the wt K278 residue (Fig. 2E). Similar results were obtained with the ACS7 ΔY92 mutant (Fig. 2E). Fourth, ACS1, the nonfunctional ACS, can form active heterodimers with ACS2 and ACS6, both members of the same branch (Fig. 2F). The ACS1 functional heterodimerization depends on which subunit provides the wt K278A residue. The ACS1 K278 residue is unable to provide functional heterodimerization with the ACS2 and ACS6 K278 mutants (Fig. 2F). Table 1 summarizes the results of the homo- and heterodimerization studies. The ACS polypeptides potentially can form 45 homo- and heterodimers. Twenty-five of them are enzymatically active (8 homodimers and 17 heterodimers). The remaining 20 are inactive (1 homodimer and 19 heterodimers).
Table 1.
Formation of enzymatically active and inactive homo- and heterodimers among the Arabidopsis ACS gene family proteins
Biochemical Coaffinity Purification Assays. The absence of functional heterodimerization among some of the polypeptides raised the question of whether it was due to the absence of protein–protein interaction among the nonhomologous subunits. We used two different approaches to address this question. First, we performed biochemical coaffinity purification assays; and second, we carried out bimolecular fluorescence complementation (BiFC) (24) experiments with the E. coli strains expressing functional and nonfunctional heterodimers.
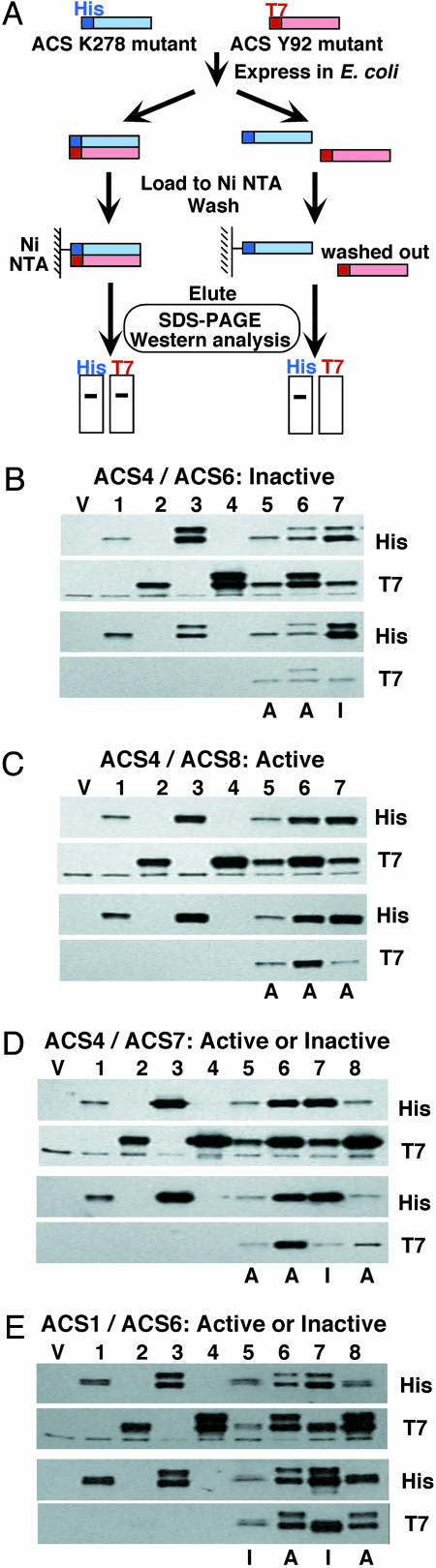
Fig. 3A shows a schematic representation of the assay that utilizes His- and T7-tagged ACS mutants coupled with affinity purification for testing their heterodimerization. Potential heterodimers are purified by Ni-nitrilotriacetic acid (Ni-NTA) affinity chromatography and analyzed by immunoblotting with His and T7 antibodies. Detection of ACS polypeptides by both antibodies is indicative of heterodimerization, whereas detection of only one subunit with the His antibody suggests the absence of heterodimerization. We first used this assay for testing heterodimerization of an inactive heterodimer such as ACS4/ACS6 (Fig. 3B). The results indicate the ACS4/ACS6 heterodimer is formed and its inactivity is not due to the absence of protein–protein interaction between the ACS4 and ACS6 mutant polypeptides (Fig. 3B, compare lane 7, an inactive heterodimer, with lanes 5 and 6, which represent the active homodimers of ACS4 and ACS6, respectively). The higher molecular size band detected by the His antibody in lanes 3, 6, and 7 (Fig. 3B) is attributed to ACS6 dimer formation. Similar analysis with the active ACS4/ACS8 heterodimer reveals that the ACS4 and ACS8 mutant polypeptides interact (Fig. 3C, compare lane 7, the active heterodimer, with lanes 5 and 6, which represent the active ACS4 and ACS8 homodimers). We also carried out similar analyses with the ACS4/ACS7 heterodimer which yielded both active and inactive heterodimers depending on which polypeptide provided the wt K278 (unidirectional/functional complementation). The results of Fig. 3D show that physical interaction occurs between the ACS4 and ACS7 subunits independent of their enzyme activity (Fig. 3D, compare lane 7, which represents the inactive heterodimer, with lane 8, which represents the active heterodimer. Lanes 5 and 6 show the interaction of the ACS4 and ACS7 homodimers.). Finally, the assay was used to test the interaction between the wt ACS1 and the K278A and Y92A mutants of ACS6 which give rise to two heterodimers, one active and the other inactive, depending on whether ACS6 provides the wt K278 residue. The results of Fig. 3E show that in both heterodimers, wt ACS1/ACS6 K278 (lane 7, inactive) and wt ACS1/ACS6 Y92A (lane 8, active), physical interaction occurs between the two polypeptides. Similar interaction is observed with the inactive wt ACS1 (Fig. 3E, lane 5) and active ACS6 homodimers (Fig. 3E, lane 6).
Fig. 3.
Determination of protein–protein interactions in active and inactive ACS heterodimers. (A) Schematic presentation of the biochemical coaffinity purification assay. Dimers of His- and T7-tagged K278A and Y92A mutants of ACS were purified by Ni-NTA affinity chromatography as described in Supporting Text and analyzed by immunoblotting with His and T7 antibodies. (B) ACS4/ACS6 heterodimers. Lane 1, H-ACS4K278A; lane 2, T-ACS4Y92A; lane 3, H-ACS6K278A; lane 4, T-ACS6Y92A; lane 5, H-ACS4K278A/T7-ACS4Y92A; lane 6, H-ACS6K278A/T7-ACS6Y92A; lane 7, H-ACS6K278A/T7-ACS4Y92A. (C) ACS4/ACS8 heterodimers. Lane 1, H-ACS4K278A; lane 2, T-ACS4Y92A; lane 3, H-ACS8K278A; lane 4, T-ACS8Y92A; lane 5, H-ACS4K278A/T-ACS4Y92A; lane 6, H-ACS8K278A/T-ACS8Y92A; lane 7, H-ACS8K278A/T-ACS4Y92A. (D) ACS4/ACS7 heterodimers. Lane 1, H-ACS4K278A; lane 2, T-ACS4Y92A; lane 3, H-ACS7K278A; lane 4, T-ACS7Y92A; lane 5, H-ACS4K278A/T-ACS4Y92A; lane 6, His-ACS7K278A/T-ACS7Y92A; lane 7, H-ACS7K278A/T-ACS4Y92A; lane 8, H-ACS4K278A/T-ACS7Y92A. (E) ACS1/ACS6 heterodimers. Lane 1, H-ACS1K278A; lane 2, T-ACS1Y92A; lane 3, H-ACS6K278A; lane 4, T-ACS6Y92A; lane 5, H-ACS1K278A/T-ACS1Y92A; lane 6, H-ACS6K278A/T-ACS6Y92A; lane 7, H-ACS6K278A/T-ACS1Y92A; lane 8, H-ACS1K278A/T-ACS6Y92A. The upper two rows in B–E are immunoblots with total E. coli extracts before Ni-NTA affinity chromatography. The lower two rows in B–E are immunoblots after Ni-NTA affinity chromatography. V, a protein sample of the E. coli DH5α with the vector alone; A and I, active and inactive homo- or heterodimers, respectively; H and T, His- and T7-tags, respectively
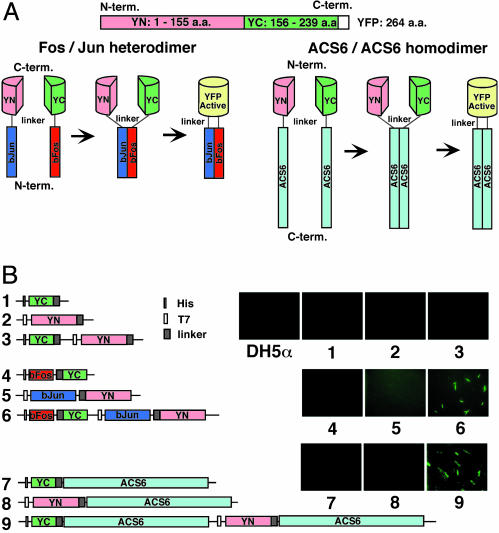
BiFC. BiFC allows detection of protein–protein interaction in vivo without disturbance of the cellular environment (24). We first used BiFC to test whether we could use this method for detecting homodimerization of the wt ACS6 subunits. Fig. 4 shows the results of the BiFC analysis demonstrating the formation of bJun/bFos heterodimer (positive control; ref. 24) and ACS6 homodimer. The N-terminal region defined as YN (amino acids 1–155) and the C-terminal region defined as YC (amino acids 156–239) of yellow fluorescence protein (YFP) were fused by a short linker to the C terminus of the bJun and bFos, respectively, or to the N terminus of the wt ACS6 polypeptide, as shown in Fig. 4A. Subsequently, the nine gene constructs (single and double) were transformed into E. coli DH5α, and the emission of yellow fluorescence by the various strains was monitored as described in Supporting Text. The results of Fig. 4B show that fluorescence emission is detected because of heterodimerization of Jun/Fos (Fig. 4B, image 6) and homodimerization of ACS6 (Fig. 4B, image 9).
Fig. 4.
BiFC. (A) Principle of the BiFC method for detecting bFos:bJun heterodimerization and ACS6:ACS6 homodimerization. The N and C termini of YFP marked in red and green, respectively, were fused to the C terminus of bJun and bFos, respectively, or to the N terminus of ACS6. (B) The gene constructs used for transforming E. coli and visualization of protein–protein interaction by YFP fluorescence imaging as described in Supporting Text.
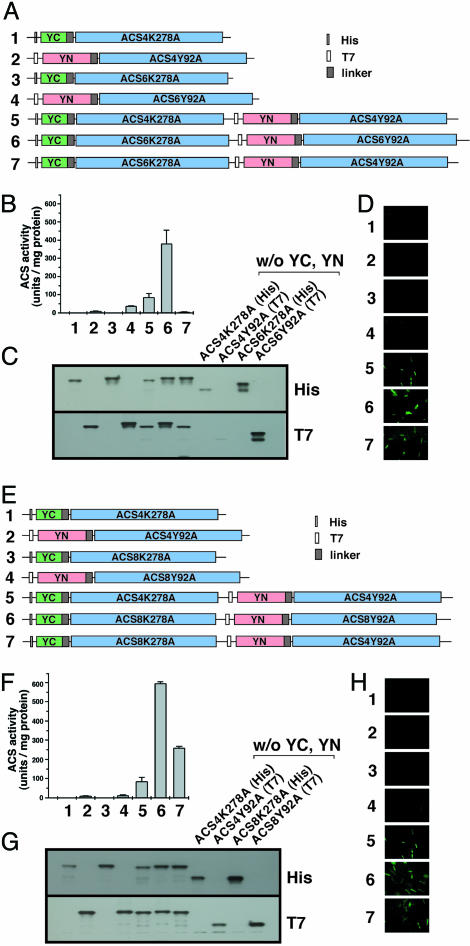
Subsequently BiFC analysis was performed for testing protein–protein interaction among some of the active and inactive ACS heterodimer pairs. We tested the same pairs as the ones examined by the biochemical coaffinity purification assay shown in Fig. 3. Fig. 5 shows that the strain expressing the inactive ACS4/ACS6 heterodimer emits fluorescence, indicating that the ACS4 and ACS6 subunits interact (Fig. 5D, image 7). In addition, the strains that express the active homodimers ACS4 and ACS6 are also fluorescing, indicating protein–protein interaction between their subunits (Fig. 5D, images 5 and 6, respectively). The YN- and YC-ACS fusions are expressed in E. coli (Fig. 5C) and are larger than the non-YN and -YC fusions (Fig. 5C, compare the protein sizes in lanes 1–7 with those in the last four lanes in the image). Furthermore, the YN and YC ACS fusions are enzymatic (Fig. 5A).
Fig. 5.
Detection of ACS heterodimer formation by BiFC. (A–D) Detection of heterodimerization between the K278A and Y92A mutants of ACS6 and ACS4 that yields inactive heterodimers. (A) The gene constructs used for detecting ACS4/ACS6 heterodimerization are numbered from 1 to 7. (B) ACS activity of E. coli strain DH5α transformed with the constructs shown above. (C) Western blot analysis of crude E. coli extracts expressing the gene constructs shown in A with use of His and T7 monoclonal antibodies. The analysis also shows immunoblotting of E. coli extracts expressing non-YN or -YC fusions of ACS4 and ACS6 K278A and Y92A mutants, respectively. (D) Visualization of protein–protein interaction by YFP imaging as described in Supporting Text. (E–H) Detection of heterodimerization between the K278A and Y92A mutants of ACS4 and ACS8 that yields active heterodimers. F, G, and H are the same as in B, C, and D, except they correspond to ACS4/ACS8 heterodimer.
A similar experiment was carried out with the active ACS4/ACS8 heterodimer and the results are shown in Fig. 5 E–H. The E. coli strain that expresses the active heterodimer and the active ACS4 and ACS8 homodimers emits fluorescence indicative of protein–protein interactions between the heterologous and homologous subunits (Fig. 5H, images 5–7). Clearly, the results of Fig. 5 show that the inactivity of the ACS4/ACS6 heterodimers is not due to the absence of heterodimerization.
BiFC was also used to test for protein–protein interactions in the unidirectional heterodimers such as ACS4/ACS7 and ACS1/ACS6. The results of the analysis are shown in Fig. 8 I and II (which is published as supporting information on the PNAS web site). The inactivity of the ACS7 K278A/ACS4 Y92A heterodimers (Fig. 8IA, lane 7) is not due to the absence of heterodimerization, because the E. coli strain emits fluorescence (Fig. 8IC, image 7), indicating that the subunits interact. Similar interactions are seen in the active homodimers (Fig. 8IC, images 5 and 6) and heterodimers (Fig. 8IC, image 8). Similar results were obtained with the active or inactive ACS1/ACS6 pair (Fig. 8II). The E. coli strain emits fluorescence independently of the enzymatic activity associated with the homodimer or heterodimer. In this case, ACS1, an inactive isozyme, forms a homodimer, but it is inactive because the conserved tripeptide TNP is missing (23). The same is true for the inactive ACS1/ACS6 heterodimer (Fig. 8IIA and C, lane and image 7, respectively).
Discussion
In this communication, we describe the potential of functional heterodimeric interactions among the ACS polypeptides encoded by the Arabidopsis gene family members (11). The polypeptides form active homodimers with shared active sites based on structural and biochemical evidence (15–17). We have used a functional complementation assay in E. coli for testing heterodimerization by intermolecular complementation of the K278A and Y92A mutants of various polypeptides. The analysis revealed that the subunits of all isozymes can form heterodimers; however, enzymatically active heterodimers are formed only among polypeptides that belong to one or the other of the two phylogenetic branches. This finding suggests that the shared active sites formed between the heterodimeric subunits of the same branch are structurally similar to those of the corresponding homodimers and dissimilar to those from different branches. However, ACS7 represents an exemption to this generalization (Fig. 2). ACS7 can form functional heterodimers with some members of both branches only when it provides the wt K278 residue, suggesting that the Y92 residue of ACS7 cannot form a functional active site with the K278 residue of all other polypeptides. ACS7 is unique among the polypeptides because it is missing the hyperviable C terminus involved in protein stability (10, 11). Phylogenetic analysis of truncated ACS polypeptides at the C terminus yields a phylogenetic tree with the ACS7 migrating to the second phylogenetic branch (compare Fig. 9, which is published as supporting information on the PNAS web site, with Fig. 2). Such “phylogenetic migration” may explain the promiscuity of ACS7.
The most surprising finding of this analysis was the recovery of enzymatically active heterodimers of ACS1 with ACS2 and ACS6, all members of the same branch. ACS1 is enzymatically inactive as a homodimer (23). It is missing the highly conserved tripeptide Thr-Asn-Pro (TNP) (23). Introduction of TNP into ACS1 restores activity (23). The ACS1 heterodimers are active only when the partners of the ACS1 heterodimer provide the wt K278 residue (Fig. 2). The TNP tripeptide is predicted to be located in a loop region that is very close to the active-site residue N209 (17, 19). This finding suggests that the individual residues of the tripeptide may be important for the proper positioning of K278 into the active site (17, 19). Consequently, the ACS1 polypeptide has an inactive K278 residue indirectly due to the absence of the TNP tripeptide. However, the heterodimers with the ACS2 or ACS6 Y92A mutants yield enzymatically active ACS heterodimers because ACS1 provides the wt Y92, and ACS2 or ACS6 provides the wt K278, resulting in the reconstruction of an active site.
Functional heterodimerization, according to the location on the phylogenetic tree, may reflect the degree of conservation of particular amino acid residues in the ACS alignment (11). The substitution map of a protein is strongly correlated with the evolution map for a number of proteins that have been mutagenized systematically, such as T4 lysozyme (25), E. coli lac repressor (26, 27), and β-lactase (28). Comparative x-ray crystallographic analysis between active and inactive ACS heterodimers has the potential to provide structural information regarding the inactivity of certain heterodimers. In addition, the functional intermolecular complementation assay can be used to isolate second-site suppression mutations for converting inactive heterodimers to active ones and vice versa. This assay will provide additional information on the structure of the heterodimers.
Protein heterodimerization is known to enhance the regulatory potential of receptors (25–31), transcription factors (32–35), transporters (36), and enzymes (37–39). Recent biochemical characterization of the ACS isozymes encoded by the Arabidopsis gene family revealed that they are biochemically distinct with respect to their Km values for AdoMet and Ki values for AVG and sinefungin (11). It has been proposed that the biochemically diverse ACS isozymes function in unique cellular environments for the biosynthesis of C2H4. This function allows the signaling molecule to exert its unique effects in a tissue-specific or cell-specific manner (11, 13). The capacity of the various isozymes to form active heterodimers further enhances the biochemical diversity of the ACS gene family. It provides an extensive repertoire of ACS isozymes capable of operating under a very broad spectrum of AdoMet concentration during the plant cycle (40). We do not yet know whether the enzymatic properties of the various heterodimers are distinct. That is a task for the future. Heterodimerization may also regulate ACS activity by posttranscriptional control mechanisms. For example, heterodimerization of MATα2 and MATa1 proteins inhibits their mutual destruction by the ubiquitin proteosome pathway (41). ACS activity is known to be regulated by the proteosome pathway (10), although whether heterodimerization participates in ubiquitin-mediated proteosome degradation of ACS remains to be determined.
Finally, we successfully used BiFC for testing in vivo protein–protein interaction in the active and inactive heterodimers. The method was recently developed for determining interactions of transcription factors in living cells (24). In planta BiFC analysis should be performed to determine whether ACS heterodimers are formed during plant growth and development. More importantly, BiFC can potentially be used for global mapping of the plant interactome networks in planta (42, 43).
Supplementary Material
Acknowledgments
We thank Drs. Kerppola and Hu (Howard Hughes Medical Institute, University of Michigan Medical School, Ann Arbor) for the generous gift of pBiFC-YC155, pBiFC-YN155, pBiFC-bJunYN155, and pBiFC-bFosYC155 plasmids. This work was supported by National Science Foundation Grants MCB-9982895 and IBN-0211421 and U.S. Department of Agriculture/Agricultural Research Service CRIS no. 5335-21430-005-00D (to A.T.).
Abbreviations: ACC, 1-aminocyclopropane-1-carboxylic acid; ACS, 1-aminocyclopropane-1-carboxylate synthase; AdoMet, S-adenosylmethionine; YFP, yellow fluorescence protein; BiFC, bimolecular fluorescence complementation; NTA, nitrilotriacetic acid.
References
- 1.Abeles, F. B., Morgan, P. W. & Saltveit, M. E., Jr. (1992) Ethylene in Plant Biology (Academic, San Diego).
- 2.Yang, S. F. & Hoffman, N. E. (1984) Annu. Rev. Plant Physiol. 35, 155–189. [Google Scholar]
- 3.Bleecker, A. B. & Kende, H. (2000) Annu. Rev. Cell Dev. Biol. 16, 1–18. [DOI] [PubMed] [Google Scholar]
- 4.Liang, X.-W., Shen, N. F. & Theologis, A. (1996) Plant J. 10, 1027–1036. [DOI] [PubMed] [Google Scholar]
- 5.Thomma, B. P. H. J., Penninckx, I. A. M. A., Broekaert, W. F. & Cammue, B. P. A. (2001) Curr. Opin. Immunol. 13, 63–68. [DOI] [PubMed] [Google Scholar]
- 6.Wang, K. L.-C., Li, H. & Ecker, J. R. (2002) Plant Cell 14, Suppl., S131–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zarembinski, T. I. & Theologis, A. (1994) Plant Mol. Biol. 26, 1579–1597. [DOI] [PubMed] [Google Scholar]
- 8.Yip, W.-K., Dong, J.-G., Kenny, J. W., Thompson, G. A. & Yang S. F. (1990) Proc. Natl. Acad. Sci. USA 87, 7930–7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woeste, K. E., Ye, C. & Kieber, J. J. (1999) Plant Physiol. 119, 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chae, H. S., Faure, F. & Kieber, J. J. (2003) Plant Cell 15, 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamagami, T., Tsuchisaka, A., Yamada, K., Haddon, W. F., Harden, L. A. & Theologis, A. (2003) J. Biol. Chem. 278, 49102–49112. [DOI] [PubMed] [Google Scholar]
- 12.Tatsuki, M. & Mori, H. (2001) J. Biol. Chem. 276, 28051–28057. [DOI] [PubMed] [Google Scholar]
- 13.Rottmann, W. E., Peter, G. F., Oeller, P. W., Keller, J. A., Shen, N. F., Nagy, B. P., Taylor, L. P., Campbell, A. D. & Theologis, A. (1991) J. Mol. Biol. 222, 937–961. [DOI] [PubMed] [Google Scholar]
- 14.Mehta, P. K., Hale, T. I. & Christen, P. (1989) Eur. J. Biochem. 186, 249–253. [DOI] [PubMed] [Google Scholar]
- 15.Li, Y., Feng, L. & Kirsch, J. F. (1997) Biochemistry 36, 15477–15488. [DOI] [PubMed] [Google Scholar]
- 16.Tarun, A. S. & Theologis, A. (1998) J. Biol. Chem. 273, 12509–12514. [DOI] [PubMed] [Google Scholar]
- 17.Capitani, G., Hohenester, E., Feng, L., Storici, P., Kirsch, J. F. & Jansonius, J. N. (1999) J. Mol. Biol. 294, 745–756. [DOI] [PubMed] [Google Scholar]
- 18.White, M. F., Vasquez, J., Yang, S. F. & Kirsch, J. F. (1994) Proc. Natl. Acad. Sci. USA 91, 12428–12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tarun, A. S., Lee, J. S. & Theologis, A. (1998) Proc. Natl. Acad. Sci. USA 94, 9796–9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larimer, F. K., Lee, E. H., Mural, R. J., Soper, T. S. & Hartman, F. C. (1987) J. Biol. Chem. 262, 15327–15329. [PubMed] [Google Scholar]
- 21.Tan, D. & Ferreira, G. C. (1996) Biochemistry 35, 8934–8941. [DOI] [PubMed] [Google Scholar]
- 22.Klee, H. J., Hayford, M. B., Kretzmer, K. A., Barry, G. F. & Kishmore, G. M. (1991) Plant Cell 3, 1187–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang, X.-W., Oono, Y., Shen, N. F., Köhler, C., Li, K., Scolnick, P. A. & Theologis, A. (1995) Gene 167, 17–24. [DOI] [PubMed] [Google Scholar]
- 24.Hu, C.-D., Chinenov, Y. & Kerppola, T. K. (2002) Mol. Cell 9, 789–798. [DOI] [PubMed] [Google Scholar]
- 25.Rennell, D., Bouvier, S. E., Hardy, L. W. & Poteete, A. R. (1991) J. Mol. Biol. 222, 67–88. [DOI] [PubMed] [Google Scholar]
- 26.Markiewicz, P., Kleina, L. G., Cruz, C., Ehret, S. & Miller, J. H. (1994) J. Mol. Biol. 240, 421–433. [DOI] [PubMed] [Google Scholar]
- 27.Kleina, L. G. & Miller, J. H. (1990) J. Mol. Biol. 212, 295–318. [DOI] [PubMed] [Google Scholar]
- 28.Huang, W., Petrosino, J., Hirsch, M., Shenkin, P. S. & Palzkill, T. (1996) J. Mol. Biol. 258, 688–703. [DOI] [PubMed] [Google Scholar]
- 29.Olayioye, M. A., Neve, R. M., Lane, H. A. & Hynes, N. E. (2000) EMBO J. 19, 3159–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thummel, C. S. (1995) Cell 83, 871–877. [DOI] [PubMed] [Google Scholar]
- 31.Neer, E. J. & Smith, T. F. (1996) Cell 84, 175–178. [DOI] [PubMed] [Google Scholar]
- 32.Jones, N. (1990) Cell 61, 9–12.2180585 [Google Scholar]
- 33.Menkens, A. E., Schindler, U. & Cashmore, A. R. (1995) Trends Biochem. Sci. 20, 506–510. [DOI] [PubMed] [Google Scholar]
- 34.Hai, T. & Curran, T. (1991) Proc. Natl. Acad. Sci. USA 88, 3720–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winter, K. U., Weiser, C., Kaufmann, K., Bohne, A., Kirchner, C., Kanno, A., Saedler, H. & Theissen, G. (2002) Mol. Biol. Evol. 19, 587–596. [DOI] [PubMed] [Google Scholar]
- 36.Reinders, A., Schulze, W., Kuhn, C., Barker, L., Schulz, A., Ward, J. M. & Frommer, W. B. (2002) Plant Cell 14, 1567–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hobbs, A. J. (1997) Trends Pharm. Sci. 18, 484–491. [DOI] [PubMed] [Google Scholar]
- 38.Muller, L., Barret, A., Etienne, E., Meidan, R., Valdenaire, O., Corvol, P. & Tougard, C. (2003) J. Biol. Chem. 278, 545–555. [DOI] [PubMed] [Google Scholar]
- 39.Culligan, K. M. & Hays, J. B. (2000) Plant Cell 12, 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graur, D. & Li, W.-H. (2000) in Fundamentals of Molecular Evolution (Sinauer, Sunderland, MA), 2nd Ed., pp. 249–322.
- 41.Johnson, P. R., Swanson, R., Rakhilina, L. & Hochstrasser, M. (1998) Cell 94, 217–227. [DOI] [PubMed] [Google Scholar]
- 42.Yamada, K., Lim, J., Dale, J. M., Chen, H., Shinn, P., Palm, C. J., Southwick, A. M., Wu, H. C., Kim, C., et al. (2003) Science 302, 842–846. [DOI] [PubMed] [Google Scholar]
- 43.Li, S., Armstrong, C. M., Berlin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P. O., Han, J. D., Chesneau, A., Hao, T., et al. (2004) Science 303, 540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.