Abstract
Mutations of the polycystic kidney and hepatic disease 1 (PKHD1) gene have been shown to cause autosomal recessive polycystic kidney disease (ARPKD), but the cellular functions of the gene product (PKHD1) remain uncharacterized. To illuminate its properties, the spatial and temporal expression patterns of PKHD1 were determined in mouse, rat, and human tissues by using polyclonal Abs and mAbs recognizing various specific regions of the gene product. During embryogenesis, PKHD1 is widely expressed in epithelial derivatives, including neural tubules, gut, pulmonary bronchi, and hepatic cells. In the kidneys of the pck rats, the rat model of which is genetically homologous to human ARPKD, the level of PKHD1 was significantly reduced but not completely absent. In cultured renal cells, the PKHD1 gene product colocalized with polycystin-2, the gene product of autosomal dominant polycystic disease type 2, at the basal bodies of primary cilia. Immunoreactive PKHD1 localized predominantly at the apical domain of polarized epithelial cells, suggesting it may be involved in the tubulogenesis and/or maintenance of duct–lumen architecture. Reduced PKHD1 levels in pck rat kidneys and its colocalization with polycystins may underlie the pathogenic basis for cystogenesis in polycystic kidney diseases.
Autosomal recessive polycystic kidney disease (ARPKD) is a hereditary renal cystic disease typically affecting infants and children (1, 2). Its major clinical manifestations include ectasia of renal collecting and hepatic biliary ducts and fibrosis of both the liver and kidney (3, 4). The estimated incidence of ARPKD ranges from 1 in 20,000 to 1 in 40,000 live births (5). The clinical spectrum of ARPKD is heterogeneous with significant inter- and intrafamilial phenotypic variability (5, 6). In ≈30–50% of ARPKD cases, disease onset is during gestation, and patients are born with symmetrically large kidneys, showing ectasia of 60–90% of the renal collecting ducts. Some patients (<30%) die during early childhood because of respiratory and/or renal dysfunction (5, 7). Fibrosis of the liver and kidneys is also commonly seen (2, 8, 9), and chronic lung disease can be observed in 11% of surviving ARPKD patients (7).
Several groups have recently identified the gene responsible for ARPKD [polycystic kidney and hepatic disease 1 (PKHD1)] (10–12). Ward et al. (10) used syntenic mapping between human and pck rats, which exhibit the ARPKD phenotypes, to identify a novel protein they called fibrocystin. The gene encoding fibrocystin is composed of 67 exons, including a 12,222-bp ORF within an ≈472-kb genomic span on human chromosome 6p. PKHD1 encodes a predicted protein of 4,074 amino acids that appears to be membrane-associated. Only one putative transmembrane domain was predicted, near the COOH-terminal portion. The last 192 residues of fibrocystin are predicted to lie in the cytoplasm. Another group (11) independently reported the same gene product, which they named polyductin. They also identified diverse alternatively spliced variants of PKHD1 that could putatively generate isoforms of the PKHD1 product.
Using library screening and exon–exon cloning strategies, we identified one of the PKHD1 isoforms, named PKHD1-tentative (PKHD1-T) (12). PKHD1-T encodes a 3,396-aa protein we named tigmin; its first 60 exons are nearly identical to PKHD1-fibrocystin/polyductin (PKHD1-FP). This smaller variant (PKHD1-T) lacks the transmembrane domain predicted for fibrocystin/polyductin (PKHD1) (12).
In situ hybridization studies showed that the mouse Pkhd1 is expressed in ductal/tubular structures during mouse kidney, liver, lung, pancreas, and vascular development (12, 13). Recently, a PKHD1-related gene (PKHD1L) was also isolated and found to be 25% identical and 41% similar to PKHD1. Its expression pattern seems to be restricted to the hematopoietic system (14). Unfortunately, because the function of PKHD1L is also unknown, its discovery did not help explain the biological role of PKHD1.
In an attempt to elucidate the functions of fibrocystin/polyductin (PKHD1), we identified the full-length amino acid sequence of mouse Pkhd1 and used a panel of poly- and mAbs to examine the distribution patterns of this previously undescribed protein and its subcellular localization. In addition to demonstrating that PKHD1 is expressed in epithelial cells at critical stages of kidney, lung, liver, and CNS morphogenesis, our findings reveal it acts as a membrane-associated protein affiliated with the basal bodies/primary cilia in renal epithelial cells. The reduction or loss of this protein correlates with the phenotypes of ARPKD.
Materials and Methods
cDNA Cloning and in Situ cRNA Hybridization for Pkhd1. A 321-bp RT-PCR product that corresponds to exon 58 of Pkhd1 was amplified by predicted primers (forward, 5′-TGC TTT GGT CTG ATA ATG TGG TCC-3′; reverse, 5′-ACT GAG GAG CCT TTC TGT CG-3′) based on homology analysis between humans and mice. This PCR product was used as probe 1 (Fig. 6A, which is published as supporting information on the PNAS web site) to screen a mouse kidney cDNA library (Clontech). Cloning strategies and approaches were similar to our previous report (12). The probe for cRNA in situ hybridization was also the same as our previous study, and detailed hybridization procedures are described (12).
Cell Culture and Transfection. All cell lines, including Madin–Darby canine kidney (MDCK) epithelia, mouse cortex collecting duct (M1), mouse inner medullar collecting duct (IMCD3), and human embryonic kidney (HEK 293) were cultured under the conditions described in the American Type Culture Collection manual. For the cell lines used to establish cell polarity and primary cilia, cells were plated to confluence for at least 5 days on either 12-mm transwell plates or 6-mm culture plates (Costar). Transient transfection assays for pCMV-Tag4hARF1–2 fusion construct (pCMV-Tag4 vector, Stratagene) were performed in six-well culture plates (Costar) with lipofectamine 2000 (Invitrogen). The detailed procedures followed the manufacturer's protocol.
Preservation and Fixation of Mouse, Rat, and Human Tissue. Tissues obtained at necropsy from a human fetus of 20-wk gestation and from tumor resections of three adult kidneys were immersed in formalin-periodate-acetate-salt (FPAS), an acidified aldehyde fixative that enhances preservation for immunohistochemistry (15), and paraffin-embedded. The mice and rats were anesthetized with i.p. Nembutal (50 mg/kg), given Heparin (1,000 units/kg, i.p.) to minimize coagulation, and perfused with FPAS through the aortic trunk cannulated by means of the left ventricle. After fixation, the selected tissues were dehydrated and paraffin-embedded.
Abs. A pGEX3 GST expression vector (Amersham Pharmacia) was used as a backbone for producing PKHD1 GST-fusion proteins. The amino acid sequences of these fusion portions have been underlined in Fig. 7, which is published as supporting information on the PNAS web site. cDNA inserts for hAR-N, -C, and -C2 were fused in-frame into the vector (Fig. 6C). The PKHD1-fused plasmids were transformed to Rossetta host cells (Novagen) to produce GST-fusion antigens.
The antigens were s.c. injected into New Zealand White rabbits with 0.6 mg of each antigen per injection. mAbs against hAR-N, -C, and -C2 were generated by hybridization of myeloma derived from mouse spleen B cells, which was performed by A&G Pharmaceuticals. A PKD2 mAb (PKD2A11) against the entire COOH-terminal cytoplasmic region was also generated by A&G Pharmaceuticals, as reported in our previous studies (16). All polyclonal antisera were affinity-purified, as described (17).
Immunoprecipitation and Western Blotting. Cultured cell proteins were extracted in lysis buffer [0.5% Nonidet P-40/5% sodium deoxycholate/50 μM NaCl/10 μM Tris·HCl (pH 7.5)/1% BSA], sonicated, and centrifuged. The supernatant was incubated with hAR-Np polyclonal Ab and rProtein G agarose (Invitrogen) at 4°C overnight. Antibody–protein-bound agarose was washed with cell-lysis buffer six times and resuspended in protein loading buffer (Bio-Rad).
Protein samples were solubilized in protein-loading buffer and denatured by boiling. The samples were electrophoresed in 12%, 10%, 7.5%, or 4–20% gradient SDS/PAGE gels (Bio-Rad). The membranes were incubated with 1% BSA at room temperature for 1 h, blotted with the particular Ab at room temperature for 4 h, incubated with peroxidase-conjugate secondary Ab (Sigma), and detected with enhanced chemiluminescence (Amersham Pharmacia).
Immunofluorescence/Immunohistochemistry Staining, Confocal Microscopy, and Immunoelectron Microscopy. For immunofluorescence staining, cultured cells were first washed with 1× PBS twice and fixed with 4% paraformaldehyde at 4°C for 30 min. Then the cells were incubated with primary Abs for 2 h and washed again with 1× PBS three times. Washed cells were treated with fluorescence-conjugated secondary Abs for 1 h. For paraffin-embedded tissue sections, the slides were deparaffinized, rehydrated, and stained, as described (15). For confocal microscopy, the images of antibody staining were collected as Z series sections by using a Zeiss LSM 510 confocal microscope system with a ×40 or ×63 oil objectives. Multiple sections (0.3 μm in thickness) were projected onto one plane for presentation. For immunoelectron microscopy, cells or tissues were fixed in 4% paraformaldehyde plus 0.01% glutaraldehyde for 30 min, stained with immunogold before being embedded (5 nm, Sigma) or hAR-Np antiserum, dehydrated, embedded in epoxy, and viewed at 80 kV on a Philips/FEI CM-12 (Eindhoven, The Netherlands) electron microscope.
Results
Identification and cDNA Cloning of the Mouse Pkhd1 Transcript Corresponding to Human PKHD1-FP. The full-length cDNA transcript Pkhd1 represents a mouse orthologue of the human PKHD1-FP and is ≈12.8 kb long (GenBank accession no. AY438374). This gene spans ≈550 kb on mouse chromosome 1 and is composed of 67 exons with a 103-bp 5′UTR region (Fig. 6B). Like its human orthologue, the mouse initiation codon lies in exon 2. However, Pkhd1 utilizes two ATGs in tandem as its initiation codon. An in-frame stop codon and a consensus polyadenylation signal (AATAAA) lie in the last exon of Pkhd1. The mouse gene product PKHD1 encodes for a protein of 4,060 amino acids with ≈72% identity and ≈82% similarity to the human PKHD1 (Fig. 7). A highly hydrophobic region present at the NH-terminal end (M1-A18) represents a signal sequence (Fig. 7) (18).
Characterization of PKHD1 Ab. Three GST-fusion proteins, encoding residues D481–F700 (hAR-N), N3191–A3385 (hAR-C), and L3872–L4074 (hAR-C2) of PKHD1, were used to produce rabbit polyclonal antisera and/or mouse mAbs (Table 1, Fig. 6C). Antiserum against the hAR-Np GST-fusion protein was generated and affinity-purified. In parallel, we obtained at least three positive clones for mAbs against the hAR-N, -C, and -C2 regions and one polyclonal antiserum against the hAR-C2 region of PKHD1 (Table 1 and Figs. 6C and 7). The mAbs hAR-Nm3G12, -Cm3G6, and -C2m3C10, and the polyclonal antiserum hAR-C2p were used to confirm the specificity of polyclonal antiserum hAR-Np to PKHD1.
Table 1. Abs against PKHD1.
| Antigen and its location
|
|||
|---|---|---|---|
| hAR-N (D481-F700) | hAR-C (N3191-A3385) | hAR-C2 (L3872-L4074) | |
| Polyclonal antiserum | hAR-Np | * | hAR-C2p |
| mAbs | hAR-Nm3G12 | hAR-Cm3G6 | hAR-C2m3C10 |
| Amino acid similarity to mice, % | ≈70 | ≈81.5 | ≈63† |
| Amino acid identity to mice, % | ≈63 | ≈78 | ≈57† |
Abs have not yet been produced.
Including an 8-aa deletion in the mouse sequence.
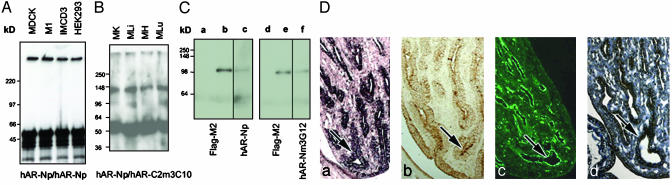
Standard Western analysis using hAR-Np failed to detect the expected ≈400-kDa bands for PKHD1, although some small-size bands appeared that were competed away by hAR-Np antigen (data not shown). However, hAR-Np immunoprecipitation (IP) Western blots obtained the expected bands at ≈400 kDa in all tested renal cell lines (Fig. 1A). To eliminate the possibility that the positive ≈400-kDa band was due to nonspecific immunoreactivity, we also tested adult mouse tissues (kidney, liver, heart, and lung) by using an additional set of poly- and mAbs hAR-C2m3C10 and -C2p, which were raised against the COOH terminus of PKHD1. The ≈400-kDa band was again obtained (Fig. 1B). However, a band at ≈135 kDa was also detected with monoclonal hAR-C2m3C10 detecting polyclonal hAR-C2p IP product. This band was not seen when hAR-Np was used as the IP product (data not shown), suggesting the ≈135-kDa protein may serve as an isoform that associates with the PKHD1 COOH terminus (Fig. 1B).
Fig. 1.
Specificity of the hAR-Np antiserum. (A) IP Western blots of a 4–20% SDS/PAGE gradient gel using the hAR-Np were detected at an ≈400-kDa band from extracts of MDCK, M1, IMCD3, and HEK293 cells. (B) Western blots using the hAR-C2p antiserum as IP and detected by hAR-C2m3C10. Bands were detected at ≈135 and ≈400 kDa from mouse tissues. MK, kidney; MLi, liver; MH, heart; MLu, lung. (C) Western blots of recombinant fusion proteins pCMV-Tag4hARF1–2. Lane a shows no band was detected by Flag-M2 in HEK293 cells without transfection (lanes a and d). Flag-M2 was reacted with an ≈105-kDa protein (lanes b and e) from the HEK293 cell with pCMV-Tag4hARF1–2 transfection. In lanes c and f, the same 105-kDa protein was also detected in extracts from the same transient transfected HEK293 cells by using hAR-Np and -Nm3G12 (lanes c and f). (D) Histological comparison of PKHD1 expression. (a) Pkhd1 antisense cRNA staining in the papillary collecting duct cells of the postnatal mouse kidney. (b) The PKHD1 protein expression is seen by using hAR-Np antiserum, which exhibits a similar staining pattern to the Pkhd1 mRNA pattern in the age-matched mouse kidney. The hAR-C2p (c) and -Cm3G6 (d) staining patterns in rat papillary tips are almost identical to those in mouse papillary staining (arrows) (b) (width, 25 μm in a–d).
To confirm the specificity of hAR-Np antiserum for PKHD1, we constructed a mammalian expression vector (pCMV-Tag4) with a Flag-tag fused in-frame with the 828-aa NH terminus of PKHD1, which includes the hAR-Np region (Figs. 6C and 7). The molecular mass of this recombinant protein was predicted to be at ≈100 kDa. Transient transfection of HEK293 cells with the same Flag-tag construct yielded immunoreactive bands at the expected size using the hAR-Np antiserum, the hAR-Nm3G12 mAb, and the Flag-M2 antibody (Table 1 and Fig. 1C). The bands at ≈100 kDa were competed away by the hAR-N antigen; however, the antigen did not block Flag-M2 binding to the Flag epitope (data not shown). This result supports the specificity of the hAR-Np and hAR-Nm3G12 Ab for PKHD1.
To further validate this specificity, we performed a histological comparison between cRNA in situ hybridization and immunohistochemistry/fluorescence by using Abs hAR-Np, -C2p, and -Cm3G6 against varying regions of PKHD1 (Table 1 and Fig. 1D). That the in situ cRNA hybridization and the hAR-Np antiserum gave similar staining patterns in mouse kidneys indicates the hAR-Np antiserum also exhibits specificity for PKHD1 on tissue sections. This concordance extends to the other polyclonal (hAR-C2p) and mAbs (hAR-Cm3G6) when examined in mouse or rat kidneys (Fig. 1D). Immunoreactive staining with affinity-purified hAR-Np in mouse and rat paraffin-embedded tissue sections was efficiently competed by preincubation with the hAR-N antigen.
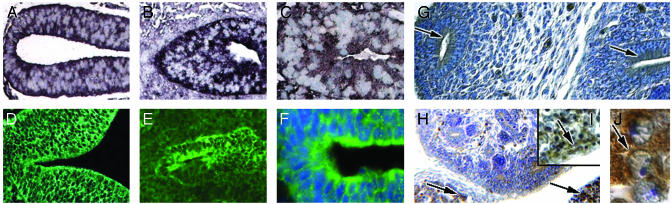
Expression of PKHD1 During Mouse Development. To elucidate the pattern of PKHD1 expression during primary organogenesis, polyclonal antibodies and mAbs (Table 1) were used for immunohistochemistry in developing mouse tissues. cRNA in situ hybridization for these particular tissues was compared with antibody staining. Clear-cut positive staining was first detected in epithelial cells of the neural tube at embryonic day (E) 9.5 (Fig. 2 A and D). Positive staining appeared in the main bronchi (Fig. 2 B and E) and in the primordial gut at E10.5 (Fig. 2 C and F). By E11.5, the epithelia of the early ureteric bud and mesonephric tubules expressed PKHD1 at their apical surfaces, as the bud penetrated into the metanephrogenic mesenchyme (Fig. 2G). The apical surfaces of epithelia from the ureteric buds continued to display PKHD1 immunoreactivity as they branched and differentiated into collecting ducts at E15.5 (Fig. 2H). Significant staining was not detected in the condensing mesenchyme, the comma-shaped body, the S shaped body, or developing glomerular vesicles of the nephrogenic zone (Fig. 2 G and H). At this stage, strong PKHD1 expression was also apparent in fasciculated cells in the developing adrenal cortex and in immature hepatocytes (Fig. 2 H–J).
Fig. 2.
PKHD1 tissue expression during mouse development. cRNA in situ hybridization of mouse embryos indicates that PKHD1 can be detected in the neural tube at E9.5 (A) and in bronchial buds (B) and epithelia of the midgut (C) at E10.5. Protein expression was also detected in each of the above organs, respectively, by using the hAR-Np (D–F). (F)4′,6-Diamidino-2-phenylindole (Sigma) for cell nuclei staining. At E11.5, PKHD1-positive staining is seen as the uretic bud projects into the metanephric primordium (right arrow, G) and was also observed in the mesonephric tubules (left arrows, G). At E15.5, PKHD1 appeared in cords of cells in the adrenal cortex (left arrow, H) and the liver (right arrow, H). Low and high powers of liver (I and J) show that PKHD1 is present in the cytoplasm of immature hepatic cells (arrows, I and J) (width, 1.5 μm, J; 10 μm, C, F, H, and I; 20 μm, A, B, D, and E; 30 μm, G).
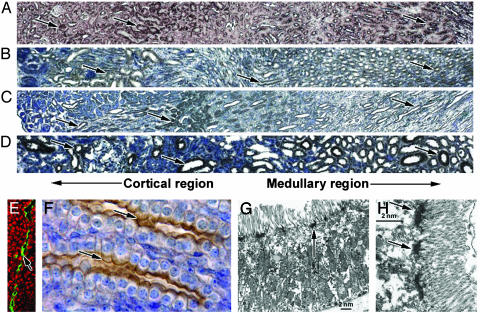
Expression of PKHD1 in Adult Renal Tubules. In the adult kidney, PKHD1 was widely expressed in the epithelia of collecting ducts, proximal convoluted tubules, and thick ascending limbs of Henle's loop (Fig. 3 A–D). The segments exhibiting hAR-Np staining were identified by double-staining of human kidneys with DBA (Dolichos biflorus agglutinin) for the cortical collecting ducts, NHE3 (Na+/H+ exchanger 3) Abs for the proximal tubules, AQP2 (Aquaporin-2) Abs for collecting ducts, and Tamm–Horsfall antiserum for the medullary thick ascending limb and distal convoluted tubules. Apical surfaces of epithelia in the collecting ducts and thick ascending limbs of Henle's loop were significantly labeled (Fig. 3 E and F), whereas basolateral membranes were weakly labeled (Fig. 3F). Cytoplasmic labeling was generally observed in proximal convoluted tubule cells (Fig. 3 B–D). Similar distribution patterns were also observed using the hAR-C2p and -Cm3G6 Abs (data not shown). Electron microscopy showed PKHD1 expression in the terminal web below the microvilli at the apical surface of the tubule epithelial cells (Fig. 3 G and H).
Fig. 3.
PKHD1 expression in the kidney. (A) Mouse adult kidneys were labeled by the same cRNA probe we used previously (12). Positive staining was observed in both the cortex and medulla regions (arrows, A). Corresponding regions of the kidney also showed positive staining (arrows) when the hAR-Np was used in mice (B), rats (C), and humans (D). Confocal microscopy using the hAR-Np reveals PKHD1 in an apical expression pattern in renal tubules (arrow, E); ethidium bromide (Sigma) was used for nuclei staining (red). The same pattern was observed in renal collecting ducts by using immunohistochemistry (arrows, F), but weak staining was also seen on the basolateral membrane. Electron microscopy using hAR-Np labeled with ImmunoGold indicates the expression of PKHD1 is localized to the terminal web below the microvilli along proximal tubule epithelial cells (arrows, G and H) (width, 5 μm, E; 10 μm, F; 150 μm, A–D).
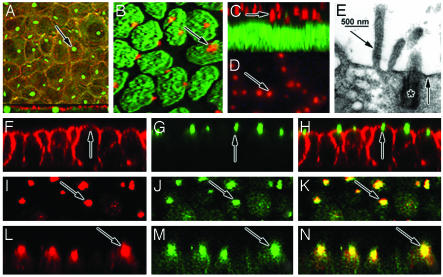
PKHD1 Localizes to the Vicinity of Basal Bodies/Primary Cilia of Renal Epithelia. In the human kidney cell line HEK293, hAR-Nm3G12 mAbs revealed highly localized spots of positive staining (Fig. 4A). Analysis of confocal lateral view showed these spots lie just beneath the cytoplasmic membrane likely associated with the basal bodies of primary cilia (Fig. 4A) (19, 20), raising the possibility that PKHD1 might physically associate with the primary cilia. Because the HEK293 cells do not fully polarize in culture, we reassessed the subcellular localization of PKHD1 in cultured MDCK cells, which establish polarity and generate cilia (21). PKHD1 immunoreactivity was restricted to one or two spots per cell when viewed en face (Fig. 4B). In the lateral view, the stained bodies were localized to the supranuclear region of the cells (Fig. 4C), but their relationship to the plasma membrane was indeterminate. Using monoclonal hAR-Nm3G12, similar staining patterns were also obtained in cultured IMCD3 cells (Fig. 4D). To elucidate cell profiles, phalloidin was used to label cytoplasmic actin (Fig. 4F). Anti-PKHD1 stained primary cilia (Fig. 4G), apparently extending through the apical actin network in merged confocal image (Fig. 4H). Immunoelectron microscopy analysis confirmed that PKHD1 is highly agglomerated at the basal body. Some positive immunoreactivity was also seen in the ciliary shaft, microvilli and plasma membrane in cultured MDCK cells (Fig. 4E).
Fig. 4.
PKHD1 is associated with the basal bodies/primary cilia in cultured renal cells. (A) In the confocal top view, costaining of cultured HEK293 cells using the hAR-Nm3G12 (green) and Rhodamine–phalloidin for F-actin (red) displayed positive staining. The hAR-Nm3G12 labeling shows centralized spots of antibody labeling one for each cell (arrow, A). The confocal lateral view (lower section, A) shows the green spots of antibody labeling to be just below the apical membrane (red), indicating that PKHD1 is localized to the basal bodies of the cells. Cultured MDCK cells were stained with the hAR-Np (red) and YO-PRO (Molecular Probes), a dye for nucleic acids (green). Positive stained cilia-shaped structures (red) can be seen clearly in each cell (arrows, B and C). In addition, hAR-N3G12 can also detect a similar result in cultured IMCD3 cells (arrow, D). In electron micrographs (E), PKHD1 immunoreactivity is concentrated at the basal body (*), the cytoplasmic surface of the apical plasmalemma (right arrow, E), and the cores of microvilli (left arrow, E). The merged confocal image (H) clearly shows PKHD1 (green) (G) localized at ciliary transition zone across the apical surface (F) stained by Rhodamine–phalloidin (red) in cultured MDCK cells. The hAR-Np (I and L, red) and Pkd2A11 mAbs (J and M, green) were used to costain cultured MDCK cells, resulting in a pair of merged confocal images (I–K, top view; L–N, lateral view), clearly indicating that the Abs against PKHD1 and polycystin-2, respectively, are colocalized at the basal bodies/primary cilia (arrows, K and N) (width, 2.5 μm, B and C; 3.5 μm, A; 4.5 μm, F–N; 10 μm, D).
Polycystin-2, a gene product associated with autosomal dominant polycystic disease, has been well-documented to localize at the primary cilium of renal cultured cell lines and tissues (22, 23). Costaining of MDCK cells with mAbs PKD2A11 (16) and hAR-Np antiserum clearly shows that PKHD1 was colocalized with polycystin-2 at the basal bodies/primary cilia (Fig. 4 I–N). This result was subsequently confirmed with the hAR-Nm3G12 and -Cm3G6 Abs (data not shown). Preincubation with the hAR-N antigens of the hAR-Np and -Nm3G12 Abs, respectively, blocked the staining of the primary cilium.
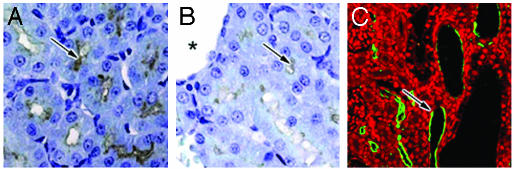
PKHD1 Exhibits Abnormal Kidney Expression in an ARPKD-Genetic Rat Model. The pck rat has been demonstrated to be a true genetic ARPKD rat model (10). To determine whether PKHD1 was abnormally expressed in the pck rat, PKHD1 Ab staining was performed to compare 16 pck rats with the same number of WT rats, all identical in age and strain. The pck rats exhibited significantly less immunoreactivity of PKHD1 than did WT rats (Fig. 5 A and B). In particular, some collecting duct epithelia completely lacked detectable PKHD1 expression at the cellular level. Only ≈5% of the pck renal cysts retained PKHD1 labeling, and most were at the early stage of cystogenesis (cystic lumen more than three times the normal tubule diameter; Fig. 5C). We did not observe any PKHD1 labeling in large cysts (>10 times of the normal renal tubule diameter) of the pck rat kidneys. These findings indicate that significant reduction or the loss of PKHD1 expression is associated with cystogenesis in ARPKD (Fig. 5).
Fig. 5.
PKHD1 expression in the pck rat kidneys. (A) The cortex region of rat kidney from 4-wk-old WT animals was stained with the hAR-C2m3C10, resulting in positive labeling in cortical proximal tubules and collecting ducts. (B) A dramatic reduction in PKHD1 labeling was seen in pck rat kidneys (arrows, A and B). Asterisk marker B indicates a renal cyst. Confocal microscopy using the hAR-Np clearly shows that PKHD1 remains in pck rat kidneys, and some were seen in epithelia of renal cysts or dilated tubules (arrow, C) (width, 20 μm, A and B; 70 μm, C).
Discussion
The availability of the mouse full-length Pkhd1 cDNA allowed us to compare conserved structural features between the orthologues of humans and mice (Fig. 7). Moreover, the amino acid sequencing result also reveals the intron–exon boundaries in Pkhd1, facilitating the identification of alternative splice isoforms in mice. However, we did not find evidence for a mouse Pkhd1-T homolog to the human PKHD1-T isoform (12). Taken together, the intracellular COOH termini were only 43% identical between humans and mice, differing from the 55% given in previously report (13), consistent with the possibility of alternative C-terminal splice variants.
Analysis of PKHD1 protein during development in mice reveals it is expressed in other tubuloepithelial derivatives before appearance of the metanephric primordia. It is widely expressed in cells of the neural tube, gut, bronchi, and vascular system. The functional role of the PKHD1 expression in these tissues is uncertain; however, its early expression suggests it may be involved in primary tubule morphogenesis (13).
After the initial submission of this work, cellular and subcellular localizations of PKHD1 were reported in cultured renal (24) and hepatic cells (25). Our results are consistent with these findings that PKHD1 associates with the primary cilia in renal epithelia. However, our studies add considerable resolution by demonstrating that PKHD1 predominantly localizes to the basal bodies of the primary cilia in cultured renal cells and to the roots of microvilli in the terminal web of renal tubular epithelium.
Primary cilia are found on diverse cell types ranging from fibroblasts to epithelia (26). PKHD1 was expressed in primary ductal epithelial cells of primordial tubules of the CNS, pulmonary, and gastrointestinal systems (Fig. 4 A–F). Cells within these specified regions generally contain motile cilia (27). It is therefore conceivable that PKHD1 expression is not limited to primary cilia but is also expressed in motile cilia.
In mammals, renal epithelia cilia are nonmotile (28). These primary cilia are present throughout nephron and collecting duct. Polycystin-1 and -2 appear to form a complex (29) colocalizing at the primary cilium (23), where they may contribute to signals initiating epithelial differentiation and proliferation by cation channel activity (30, 31). The overlapping spatial distribution of PKHD1 and the polycystins implies that PKHD1 may associate with a common pathway that alters ciliary function and cation channel activity. Recently, a kidney-specific KIF3A-deficient mouse model was reported (32). This tissue-specific mouse model exhibits multiple cysts in the kidney and fails to establish primary cilia in renal epithelia. These findings strongly suggest that dysfunction of the primary cilia associates with cystogenesis in the kidney. The presence of PKHD1 at the vicinity of the basal body raises the possibility this protein may act as an intraflagellar motor-binding protein (22, 33).
The pck rat is the rat orthologue for human ARPKD. Unexpectedly, antibody staining detected reduced but not absent PKHD1 labeling. This was the case with all of Abs tested (Table 1), including hAR-C2p and -C2m3C10, whose antigens were generated from the last COOH-terminal 192 amino acids of PKHD1 (Fig. 5 A–C). Similar findings were also obtained in Western analysis (data not shown) (25). These findings lead us to believe the mutation in the pck rats may not result in premature truncation due to nonsense mutations or an ORF shift. Rather the dramatic reduction of PKHD1 levels as compared to age-matched WT is consistent with a haploinsufficiency mechanism contributing to cystogenesis in ARPKD, which does not exclude the possibilities that absence of PKHD1 and/or functional domain deletions in the protein could cause cystic phenotypes in the kidney.
Additionally, we identified at least one previously undescribed isoform at ≈135 kDa associated with the COOH terminus of PKHD1 (Fig. 1B). A recent study using mAbs raised against the COOH terminus of PKHD1 (24) postulated several candidate isoforms. Even though isoform sizes detected by our methods are different from theirs, the results are complementary in supporting the existence of PKHD1 isoforms (11, 12). It may be of interest that we used whole-cell lysates, whereas the other study (24) used membrane lysates; therefore, the differences in isoform sizes may derive from their respective subcellular origins. The different isoforms may produce a different expression pattern in certain nephronic tubular segments and subcellular localization. Further identification and characterization of multiple PKHD1 isoforms will be required before any definitive conclusions can be reached.
In summary, the present work demonstrates that PKHD1 is a membrane-associated protein, exhibits a tissue-specific expression pattern, and appears to be developmentally regulated. In addition, PKHD1 is widely expressed in a diverse ductal structure from several organs and tissues, consistent with its involvement in primary duct formation. An analysis of the pck rat, a true ARPKD-genetic model, using a combination of Abs, indicates that cystic formation in these rats may result from a haploinsufficiency mechanism. PKHD1 was localized to epithelial basal bodies/cilia and, like several other polycystic kidney disease proteins, is a cilium-associated protein. Its coexpression with polycystin-2 suggests that PKHD1 acts in concert with polycystins and may be directly or indirectly involved in a common cystogenic pathway.
Supplementary Material
Acknowledgments
We thank Dan Liang, Heping Yan, and Yongxiong Chen for technical assistance and Drs. Alfred George and James Goldenring for critical reading of the manuscript and suggestions. This study was supported by grants from the Polycystic Kidney Research Foundation, the American Heart Association, and the National Institutes of Health (DK62373) to G.W.
Abbreviations: ARPKD, autosomal recessive polycystic kidney disease; IP, immunoprecipitation; En, embryonic day n; MDCK, Madin–Darby canine kidney; HEK, human embryonic kidney.
References
- 1.Guay-Woodford, L. M. (1996) in Polysystic Kidney Disease, eds. Torres V. & Watson, M. (Oxford Univ. Press, Oxford), pp. 237–267.
- 2.Zerres, K., Rudnik-Schoneborn, S., Steinkamm, C., Becker, J. & Mucher, G. (1998) J. Mol. Med. 76, 303–309. [DOI] [PubMed] [Google Scholar]
- 3.Potter, E. L. (1972) Normal and Abnormal Development of the Kidney (Year Book, Chicago).
- 4.Bosniak, M. A. & Ambos, M. A. (1975) Semin. Roentgenol. 10, 133–143. [DOI] [PubMed] [Google Scholar]
- 5.Zerres, K., Rudnik-Schoneborn, S., Senderek, J., Eggermann, T. & Bergmann, C. (2003) J. Nephrol. 16, 453–458. [PubMed] [Google Scholar]
- 6.Deget, F., Rudnik-Schoneborn, S. & Zerres, K. (1995) Clin. Genet. 47, 248–253. [DOI] [PubMed] [Google Scholar]
- 7.Guay-Woodford, L. M. & Desmond, R. A. (2003) Pediatrics 111, 1072–1080. [DOI] [PubMed] [Google Scholar]
- 8.Zerres, K., Mucher, G., Becker, J., Steinkamm, C., Rudnik-Schoneborn, S., Heikkila, P., Rapola, J., Salonen, R., Germino, G. G., Onuchic, L., et al. (1998) Am. J. Med. Genet. 76, 137–144. [PubMed] [Google Scholar]
- 9.Lonergan, G. J., Rice, R. R. & Suarez, E. S. (2000) Radiographics 20, 837–855. [DOI] [PubMed] [Google Scholar]
- 10.Ward, C. J., Hogan, M. C., Rossetti, S., Walker, D., Sneddon, T., Wang, X., Kubly, V., Cunningham, J. M., Bacallao, R., Ishibashi, M., et al. (2002) Nat. Genet. 30, 259–269. [DOI] [PubMed] [Google Scholar]
- 11.Onuchic, L. F., Furu, L., Nagasawa, Y., Hou, X., Eggermann, T., Ren, Z., Bergmann, C., Senderek, J., Esquivel, E., Zeltner, R., et al. (2002) Am. J. Hum. Genet. 70, 1305–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong, H., Chen, Y., Yi, Y., Tsuchiya, K., Moeckel, G., Cheung, J., Liang, D., Tham, K., Xu, X., Chen, X. Z., et al. (2002) Genomics 80, 96–104. [DOI] [PubMed] [Google Scholar]
- 13.Nagasawa, Y., Matthiesen, S., Onuchic, L. F., Hou, X., Bergmann, C., Esquivel, E., Senderek, J., Ren, Z., Zeltner, R., Furu, L., et al. (2002) J. Am. Soc. Nephrol. 13, 2246–2258. [DOI] [PubMed] [Google Scholar]
- 14.Hogan, M. C., Griffin, M. D., Rossetti, S., Torres, V. E., Ward, C. J. & Harris, P. C. (2003) Hum. Mol. Genet. 12, 685–698. [PubMed] [Google Scholar]
- 15.McKanna, J. A. & Zhang, M. Z. (1997) J. Histochem. Cytochem. 45, 527–538. [DOI] [PubMed] [Google Scholar]
- 16.Li, Q., Shen, P. Y., Wu, G. & Chen, X. Z. (2003) Biochemistry 42, 450–457. [DOI] [PubMed] [Google Scholar]
- 17.Xu, M. J., Sui, X., Zhao, R., Dai, C., Krantz, S. B. & Zhao, Z. J. (2003) Blood 102, 4354–4360. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen, H., Engelbrecht, J., Brunak, S. & von Heijne, G. (1997) Protein Eng. 10, 1–6. [DOI] [PubMed] [Google Scholar]
- 19.Taulman, P. D., Haycraft, C. J., Balkovetz, D. F. & Yoder, B. K. (2001) Mol. Biol. Cell 12, 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pazour, G. J. & Witman, G. B. (2003) Curr. Opin. Cell Biol. 15, 105–110. [DOI] [PubMed] [Google Scholar]
- 21.Praetorius, H. A. & Spring, K. R. (2001) J. Membr. Biol. 184, 71–79. [DOI] [PubMed] [Google Scholar]
- 22.Pazour, G. J., San Agustin, J. T., Follit, J. A., Rosenbaum, J. L. & Witman, G. B. (2002) Curr. Biol. 12, R378–R380. [DOI] [PubMed] [Google Scholar]
- 23.Yoder, B. K., Hou, X. & Guay-Woodford, L. M. (2002) J. Am. Soc. Nephrol. 13, 2508–2516. [DOI] [PubMed] [Google Scholar]
- 24.Ward, C. J., Yuan, D., Masyuk, T. V., Wang, X., Punyashthiti, R., Whelan, S., Bacallao, R., Torra, R., LaRusso, N. F., Torres, V. E., et al. (2003) Hum. Mol. Genet. 12, 2703–2710. [DOI] [PubMed] [Google Scholar]
- 25.Masyuk, T. V., Huang, B. Q., Ward, C. J., Masyuk, A. I., Yuan, D., Splinter, P. L., Punyashthiti, R., Ritman, E. L., Torres, V. E., Harris, P. C., et al. (2003) Gastroenterology 125, 1303–1310. [DOI] [PubMed] [Google Scholar]
- 26.Wheatley, D. N. (1995) Pathobiology 63, 222–238. [DOI] [PubMed] [Google Scholar]
- 27.O'Callaghan, C., Sikand, K. & Rutman, A. (1999) Pediatr. Res. 46, 704–707. [DOI] [PubMed] [Google Scholar]
- 28.Wheatley, D. N., Wang, A. M. & Strugnell, G. E. (1996) Cell Biol. Int. 20, 73–81. [DOI] [PubMed] [Google Scholar]
- 29.Qian, F., Germino, F. J., Cai, Y., Zhang, X., Somlo, S. & Germino, G. G. (1997) Nat. Genet. 16, 179–183. [DOI] [PubMed] [Google Scholar]
- 30.Koulen, P., Cai, Y., Geng, L., Maeda, Y., Nishimura, S., Witzgall, R., Ehrlich, B. E. & Somlo, S. (2002) Nat. Cell Biol. 4, 191–197. [DOI] [PubMed] [Google Scholar]
- 31.Nauli, S. M., Alenghat, F. J., Luo, Y., Williams, E., Vassilev, P., Li, X., Elia, A. E., Lu, W., Brown, E. M., Quinn, S. J., et al. (2003) Nat. Genet. 33, 129–137. [DOI] [PubMed] [Google Scholar]
- 32.Lin, F., Hiesberger, T., Cordes, K., Sinclair, A. M., Goldstein, L. S., Somlo, S. & Igarashi, P. (2003) Proc. Natl. Acad. Sci. USA 100, 5286–5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pazour, G. J., Dickert, B. L., Vucica, Y., Seeley, E. S., Rosenbaum, J. L., Witman, G. B. & Cole, D. G. (2000) J. Cell Biol. 151, 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pendleton, A., Pope, B., Weeds, A. & Koffer, A. (2003) J. Biol. Chem. 278, 14394–14400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.