Abstract
Iron-sulphur cluster biogenesis requires coordinated delivery of iron and sulphur to scaffold proteins, followed by transfer of the assembled clusters from scaffold proteins to target proteins. This complex process is accomplished by a group of dedicated iron-sulphur cluster assembly proteins that are conserved from bacteria to humans. While sulphur in iron-sulphur clusters is provided by L-cysteine via cysteine desulfurase, the iron donor(s) for iron-sulphur cluster assembly remains largely elusive. Here we report that among the primary iron-sulphur cluster assembly proteins, IscA has a unique and strong binding activity for mononuclear iron in vitro and in vivo. Furthermore, the ferric iron centre tightly bound in IscA can be readily extruded by L-cysteine, followed by reduction to ferrous iron for iron-sulphur cluster biogenesis. Substitution of the highly conserved residue tyrosine 40 with phenylalanine (Y40F) in IscA results in a mutant protein that has a diminished iron binding affinity but retains the iron-sulphur cluster binding activity. Genetic complementation studies show that the IscA Y40F mutant is inactive in vivo, suggesting that the iron binding activity is essential for the function of IscA in iron-sulphur cluster biogenesis.
Introduction
Throughout evolution, iron-sulphur clusters have become integral parts of diverse physiological processes including photosynthesis, nitrogen fixation, sugar metabolism, co-factor biogenesis, RNA modification and translation, DNA replication and repair, and gene expression regulation.1-4 While iron-sulphur clusters can be assembled in proteins in vitro with ferrous iron and sulphide, iron-sulphur cluster assembly in vivo requires a group of dedicated proteins that are conserved from bacteria to humans.5 In Escherichia coli, the proteins encoded by a gene cluster iscSUA-hscBA-fdx are primarily responsible for iron-sulphur cluster assembly under normal growth conditions.5 The homologues of the six proteins encoded by iscSUA-hscBA-fdx have been identified in eukaryotic organisms.2 Among them, IscS is a cysteine desulfurase that catalyzes desulfurization of L-cysteine6-8 and provides sulphur for iron-sulphur cluster assembly in a scaffold protein IscU.9-11 IscU in turn transfers the assembled clusters to target proteins.12, 13 IscA has been characterized as an alternative scaffold or intermediate carrier for iron-sulphur cluster assembly.14, 15 A comprehensive characterization of iron-sulphur cluster binding in an IscA homolog NifIscA from Azotobacter vinelandii has recently been reported.16 Two heat shock cognate proteins, HscB and HscA, have specific interactions with IscU17 and promote the transfer of assembled clusters from IscU to target proteins in an ATP-dependent reaction.18, 19 Ferredoxin, which contains a stable [2Fe-2S] cluster,20 is likely involved in the reductive formation of a [4Fe-4S] cluster in scaffold protein IscU 21, 22.
While the sulphur donor for iron-sulphur cluster assembly has been well established,6-8 the iron donor(s) remains largely elusive. Frataxin, a mitochondrial protein that is linked to neurodegenerative disease Friedreich’s ataxia,23 has previously been characterized as a likely iron donor for iron-sulphur cluster assembly.24, 25 Deficiency of frataxin results in diminished activity of iron-sulphur enzymes in yeast cells,26 and frataxin has physical interactions with iron-sulphur cluster assembly protein IscU,27-29 the IscU/IscS complex,30, 31 and with the iron-sulphur enzyme aconitase in mitochondria.32 However, frataxin has a weak iron binding activity with an iron binding constant of 4 to 55 μM.24, 33, 34 Such a low iron binding affinity may preclude frataxin from binding any significant amounts of iron in mitochondria under normal physiological conditions. Furthermore, deletion of frataxin homologue CyaY in E. coli does not affect iron-sulphur cluster biogenesis,35 and frataxin-deficient yeast cells can be rescued by either expressing ferritin36 or vacuolar iron transporter CCC1.37 In fact, scavenging H2O2 can effectively restore iron-sulphur enzyme activities in a Drosophila model of Friedreich’s ataxia.38 Thus, the primary function of frataxin could be to maintain iron homeostasis under oxidative stress39, 40 or to modulate overall iron-sulphur cluster assembly in cells.30, 31
In previous studies, we reported that unlike other iron-sulfur cluster assembly scaffold proteins such as IscU, IscA has a strong iron binding activity with an apparent iron association constant of 1.0 × 1019 M−1.41-45 A similar strong iron binding activity has been observed for IscA homologues from humans,46 yeast cells,47 and A. vinelandii NifIscA.48 In this study, we present new evidence showing that among the primary iron-sulphur cluster assembly proteins, IscA is unique in binding iron, and that the tightly bound ferric iron in IscA can be readily extruded by L-cysteine, followed by reduction to ferrous iron for iron-sulphur cluster assembly. Site-directed mutagenesis studies show that the iron-binding activity is crucial for the physiological function of IscA in iron-sulphur cluster biogenesis.
Experimental
Protein preparation
The iron-sulphur cluster assembly proteins IscS, IscU, IscA, HscB, and HscA from E. coli were prepared as described previously.43 The IscA mutant in which tyrosine 40 was substituted with phenylalanine (Y40F) was constructed using the site-directed mutagenesis kit (Stratagene co). The mutation of gene iscA in the cloned plasmid was confirmed by direct sequencing. Human frataxin was subcloned from plasmid pETHF249 to expression plasmid pET28b+ and prepared as described in.46 The E. coli frataxin homologue CyaY was prepared as described in.44 The protein concentration was determined from the absorption peak at 260 nm or 280 nm using the previously published extinction coefficient for each protein.
In vitro iron binding and iron-sulphur cluster assembly in IscA
For the iron binding experiments, each of purified iron-sulphur cluster assembly proteins (50 μM in monomer) was incubated with Fe(NH4)2(SO4)2 (50 μM) and sodium citrate (5 mM) in buffer containing NaCl (200 mM), Tris (20 mM, pH 8.0) in the presence of dithiothreitol (2 mM) at room temperature for 20 min, followed by passing through a High-trap Desalting column. Total iron contents in re-purified protein samples were determined using an iron indicator FerroZine as described in.50 For iron-sulphur cluster assembly, purified IscA was incubated with IscS (0.5 μM), Tris (20 mM, pH 8.0), NaCl (200 mM), Fe(NH4)2(SO4)2 (50 μM), and dithiothreitol (2 mM) at 37°C for 5 min under anaerobic conditions. L-cysteine (1 mM) was then added to initiate the iron-sulphur cluster assembly reaction. The amount of iron-sulphur clusters assembled in protein was monitored in a Beckman DU-640 UV-Visible spectrophotometer.
EPR measurements
EPR (electron paramagnetic resonance) spectra were recorded at X-band on a Bruker ESP-300 EPR spectrometer using an Oxford Instruments ESR-9 flow cryostat. The routine EPR conditions were: microwave frequency, 9.45 GHz; microwave power, 10 mW; modulation frequency, 100 kHz; modulation amplitude, 2.0 mT; temperature, 10 K; receive gain, 1.0×105.
In vivo activity assay of the IscA Y40F mutant
For the in vivo activity of the IscA Y40F mutant, gene encoding Y40F was cloned into a plasmid pBAD (Invitrogen co.). The constructed plasmid pBAD/Y40F was introduced into an E. coli mutant in which gene iscA and its paralogue sufA were in-frame deleted.51 The E. coli iscA/sufA double mutant was viable in rich LB medium, but had a null-growth phenotype in M9 minimal medium under aerobic conditions.51 The E. coli iscA/sufA double mutant cells containing pBAD/iscA and pBAD served as positive and negative control, respectively.
Results and discussion
IscA has a unique iron binding activity among the iron-sulphur cluster assembly proteins
To explore the iron binding activity of the primary iron-sulphur cluster assembly proteins encoded by the gene cluster iscSUA-hscBA-fdx from E. coli,5 we expressed each protein in wild-type E. coli cells grown in LB media. Each protein was purified to a single band on the SDS-PAGE gel.
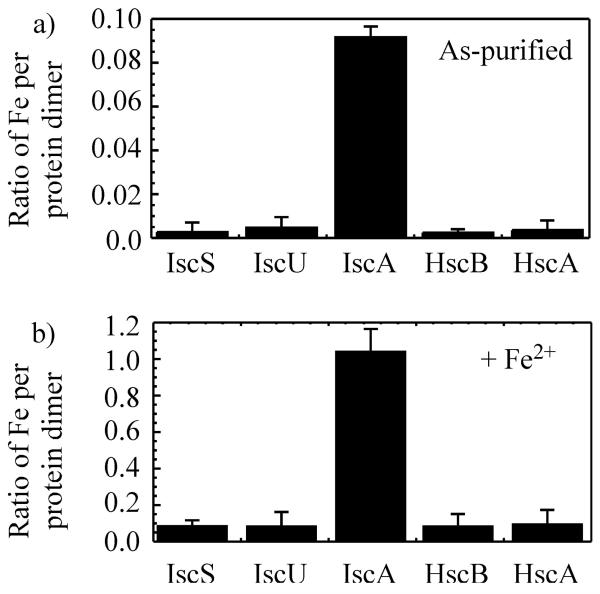
Because ferredoxin contained a stable [2Fe-2S] cluster,20 ferredoxin was not included for the iron binding analysis. Among the other iron-sulphur cluster assembly proteins encoded by iscSUA-hscBA-fdx, only purified IscA contains 0.09±0.02 iron atoms per IscA dimer (Fig. 1a). No acid-labile sulphur is detectable in purified IscA, indicating that purified IscA does not contain any significant amounts of iron-sulphur clusters.
Fig. 1.
Iron binding of primary iron-sulphur cluster assembly proteins. Iron-sulphur cluster assembly proteins IscS, IscU, IscA, HscB and HscA were purified from E. coli cells grown in LB media. a), iron content of purified iron-sulphur cluster assembly proteins. Iron content was presented as the ratio of iron atoms per protein dimer. b), iron content of iron-sulphur cluster assembly proteins (50 μM in monomer) after incubation with Fe(NH4)2(SO4)2 (50 μM), sodium citrate (5 mM) and dithiothreitol (2 mM). Proteins were re-purified from incubation solutions. Iron content was presented as the ratio of iron atoms per protein dimer. The data are averages plus standard deviations from three independent experiments.
The iron binding activity of purified iron-sulphur cluster assembly proteins is also analyzed in vitro. Each purified protein is incubated with freshly prepared ferrous iron in the presence of dithiothreitol under aerobic conditions, followed by re-purification of protein Fig. 1b) shows that while the iron content in IscA is increased to 1.05±0.10 iron atoms per IscA dimer after incubation with an equivalent amount of ferrous iron (relative to protein monomer), the iron content in other proteins is less than 0.1 iron atoms per protein dimer, demonstrating that IscA has a unique iron binding activity.
The only other candidate proposed for the iron donor to iron-sulphur cluster assembly in scaffold protein IscU is frataxin and its bacterial homologue CyaY.24, 25 To explore the iron binding activity of frataxin/CyaY in vitro under the same experimental conditions, purified E. coli CyaY and human frataxin are incubated with an equivalent amount of ferrous iron in the presence of dithiothreitol. Very little or no iron binding is found in re-purified CyaY and human frataxin (data not shown), suggesting that unlike IscA from E. coli 41 and humans,46 CyaY/frataxin has a very weak iron binding activity in the presence of dithiothreitol.
The ferric iron centre in IscA can be extruded b y L-cysteine, followed by reduction to ferrous iron
To access the iron in IscA for iron-sulphur cluster biogenesis, the iron stored in the protein must be quickly released when there is such a demand. Previously, we reported that L-cysteine, but not other related biological thiols including N-acetyl-L-cysteine or reduced glutathione, can efficiently release iron from the iron-bound IscA.52 However, the mechanism underlying the L-cysteine-mediated iron release from IscA was not clear.
The iron-bound IscA has an unusual EPR signal at g = 4.3 and 6.0.41 The signal is completely eliminated upon removal of iron from the protein, and fully restored after reconstitution with iron, indicating that the EPR signal represents the iron binding in IscA.41 Interestingly, similar EPR signal was previously reported for the S = 3/2, reduced [4Fe-4S] cluster in proteins such as nitrogenase53 and 2-hydroxyglutaryl-CoA dehydratase.54 While further biophysical studies will be important to determine the spin state of the iron center in E. coli IscA, recent optical and magnetic spectroscopic characterization of NifIscA, an IscA homologue from A. vinelandii, provided comprehensive and convincing evidence for the assignment of the iron site in NifIscA to a ferric iron in an unusual rhombic spin S = 3/2 state.48 The EPR spectra of the iron-bound NifIscA from A. vinelandii48 and the iron-bound IscA from E. coli41 are essentially the same. Furthermore, the iron center in both E. coli IscA and NifIscA is redox active: the EPR signal at g = 4.3 and 6.0 disappears when the iron centre in IscA is reduced by sodium dithionite.41, 48
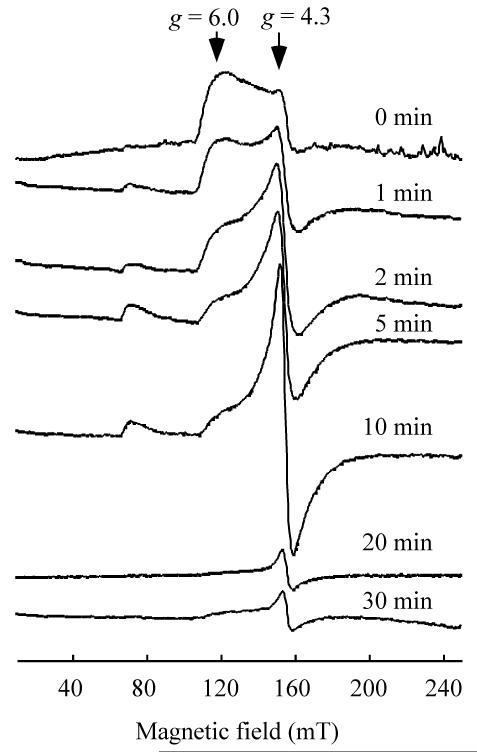
To follow the process of the L-cysteine-mediated iron release from IscA, we used EPR to monitor the redox state of the iron centre in IscA when the iron-bound IscA was incubated with L-cysteine under aerobic conditions. At different time points, aliquots were taken from the incubation solution after addition of L-cysteine for EPR measurements.
Fig. 2 shows that upon addition of L-cysteine, the EPR signal at g = 4.3 and 6.0 of the iron-bound IscA is quickly replaced with a new EPR signal at g = 4.3, representing “free” ferric iron in solution.55 After further incubation (over 20 min), the newly formed EPR signal at g = 4.3 is gradually decreased, indicating that the “free” ferric iron is reduced to the EPR-silent ferrous iron. As a control, incubation of the iron-bound IscA with N-acetyl-L-cysteine or reduced glutathione does not affect the EPR signal at g = 4.3 and 6.0 of the iron-bound IscA (data not shown). On the other hand, D-cysteine has the same activity as L-cysteine in releasing iron from IscA,52 suggesting that the redox property, but not stereochemistry, of L-cysteine is responsible for the iron release from IscA.
Fig. 2.
L-cysteine-mediated iron release from IscA. Iron-bound IscA dimer (300 μM) was incubated with 2 mM L-cysteine under aerobic conditions. Aliquots were taken at indicated time and frozen immediately for the liquid helium-temperature EPR measurements.
The transition of the EPR signal of the iron-bound IscA from g = 4.3 and 6.0 to g = 4.3 upon addition of L-cysteine indicates that the L-cysteine-mediated iron release from IscA may have two distinctive steps. In the first step, L-cysteine extrudes ferric iron from IscA via ligand exchange to form the L-Cys-Fe3+-IscA and/or the L-cys-Fe3+ complex which attributes to the observed EPR signal at g = 4.3 (Fig. 2). In the second step, the L-Cys-Fe3+-IscA and/or L-cys-Fe3+ complex is reduced by L-cysteine to the EPR-silent ferrous iron which will be accessible for iron-sulphur cluster biogenesis.43
Mutation at residue tyrosine 40 to phenylalanine diminishes the iron binding activity of IscA
In crystal structure, E. coli IscA is a homodimer with the three conserved cysteine residues (Cys-35, Cys-99 and Cys-101) projected to form a “cysteine pocket” between two monomers.56, 57 Mutation of any of the three conserved cysteine residues to serine produced IscA mutant proteins that have a diminished iron binding activity 58 and are inactive in E. coli cells.51 However, because the cysteine residues may accommodate either a mononuclear iron or iron-sulphur cluster in IscA, mutation of cysteine residues would have affected both the iron and iron-sulphur cluster binding in the protein.
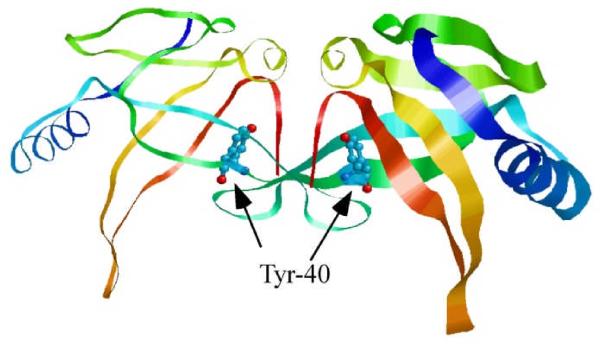
Among the amino acid residues that are in the vicinity of the putative iron binding site in IscA dimer, tyrosine 40 is highly conserved and located in the interface between two IscA monomers (Fig. 3). Unlike other nearby conserved residues that form a hydrophobic packing between IscA monomers,56 the hydroxyl group of tyrosine 40 may act as a possible oxygenic ligand for iron binding in IscA dimer.48
Fig. 3.
Location of Tyr-40 in E. coli IscA dimer. Tyr-40 is located in the interface between two IscA monomers, and is in the vicinity of the putative iron binding site (adapted from 56). The conserved iron binding residues (Cys-99 and Cys-101) of IscA are not visible in the electron density map, likely because of the high flexibility.
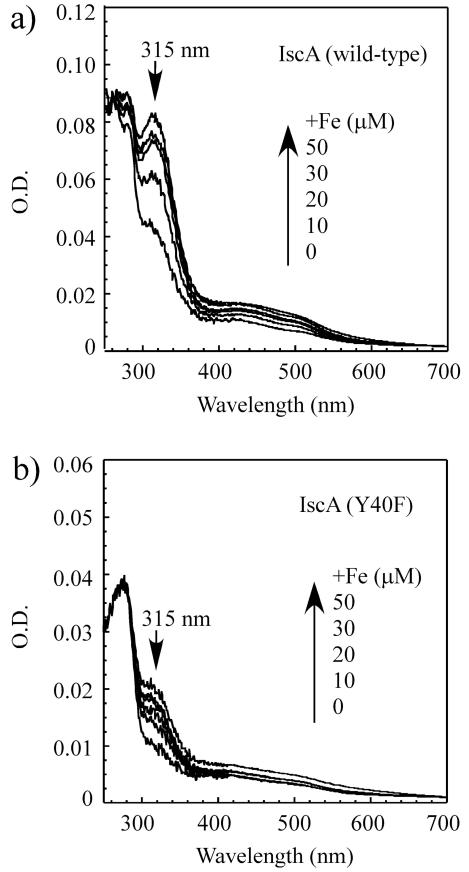
To explore the role of tyrosine 40 in the iron binding and iron-sulphur cluster binding of IscA, we constructed an IscA mutant in which tyrosine 40 was substituted with phenylalanine (Y40F). The IscA mutant Y40F protein was purified (Supplementary Figure A), and incubated with increasing concentrations of ferrous iron, followed by re-purification of the protein. Fig. 4 shows that unlike wild-type IscA, the absorption peak at 315 nm of the iron binding in Y40F is only slightly increased after incubation with ferrous iron in the presence of dithiothreitol. The iron content analyses revealed that after incubation with two-fold excess of iron per protein dimer, the ratio of iron to the wild-type IscA dimer is 1.05±0.10, while the ratio of iron to the IscA mutant Y40F dimer is less than 0.15 (Supplementary Figure B). The wild-type IscA and Y40F mutant proteins after reconstitution with two fold excess of iron were re-purified concentrated, and subjected to the EPR measurements. While wild-type IscA has a typical EPR spectrum of the iron-bound protein with g = 4.3 and 6.0, Y40F only has a small EPR signal at g = 4.3 region (data not shown). In the light of recent publication on the iron binding property of NifIscA,48 it would be interesting to determine whether tyrosine 40 actually provides the oxygenic ligand for the iron binding in IscA. Regardless, the results shown in Figure 4 and Supplementary Figure clearly demonstrate that mutation at Y40F severely diminishes the iron binding activity of E. coli IscA in vitro.
Fig. 4.
IscA mutant Y40F has a diminished iron binding activity. a), purified wild-type IscA dimer (25 μM) was incubated with indicated concentrations of Fe(NH4)2(SO4)2 in the presence of dithiothreitol (2 mM) at room temperature for 20 min, followed by re-purification of protein. Spectra were calibrated to the same amplitude of the absorption peak at 260 nm of the IscA sample after reconstitution with two-fold excess of iron. b), same as in a), except IscA mutant Y40F dimer (25 μM) was used. Spectra were calibrated to the same amplitude of the absorption peak at 260 nm of the Y40F sample after reconstitution with two-fold excess of iron.
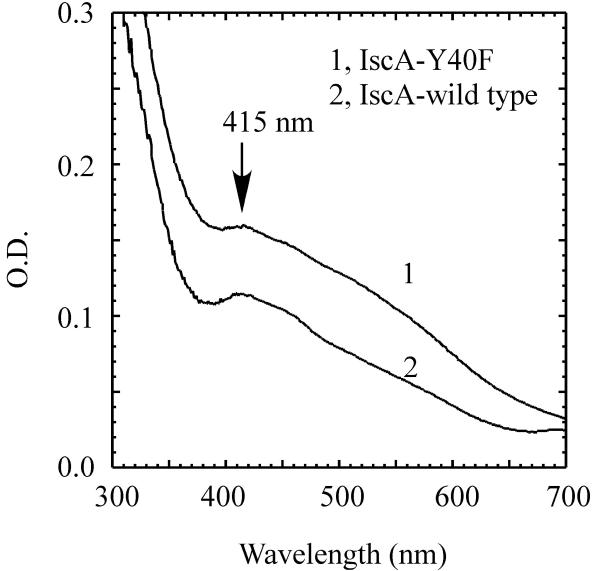
Next, we explored the iron-sulphur cluster binding activity of the wild-type IscA and IscA mutant Y40F. Purified IscA and the IscA mutant Y40F proteins were incubated with ferrous iron, L-cysteine, and a catalytic amount of IscS in the presence of dithiothreitol under anaerobic conditions. After 20 min incubation, the iron-sulphur cluster assembly in IscA was analyzed. Fig. 5 shows that like wild-type IscA, the IscA mutant Y40F retains the iron-sulphur cluster binding activity, suggesting that mutation of Y40F does not significantly affect the iron-sulphur cluster assembly in IscA in vitro.
Fig. 5.
In vitro iron-sulphur cluster assembly in IscA and IscA mutant Y40F. Purified IscA mutant Y40F (spectrum 1) or wild-type IscA dimer (spectrum 2) (25 μM) was incubated with IscS (0.5 μM), Fe(NH4)2(SO4)2 (100 μM), dithiothreitol (2 mM) in buffer containing Tris (20 mM, pH 8.0) and NaCl (200 mM) at 37°C under anaerobic conditions. L-cysteine (1 mM) was then added to the incubation solutions to initiate the iron-sulphur cluster assembly reaction. Spectra were taken 20 min after addition of L-cysteine. The absorption peak at 415 nm reflects the iron-sulphur cluster formation in IscA.
IscA mutant Y40F is inactive in E. coli cells
E. coli has at least two IscA paralogs: ErpA and SufA. ErpA, which maps at a distance from any iron-sulphur cluster assembly-related genes, has been characterized as a dedicated scaffold for maturation of the key iron-sulphur enzymes in the isoprenoids biosynthesis pathway.59 SufA, on the other hand, is a member of second iron-sulphur cluster assembly gene cluster sufABCDSE in E. coli.60 Purified SufA, like IscA, has a strong iron binding activity and provides iron for iron-sulphur cluster assembly in IscU in vitro.51 While deletion of IscA or SufA in E. coli cells only has a mild effect on cell growth, deletion of both IscA and SufA results in a null-growth phenotype in M9 minimal media under aerobic conditions.51, 61 Re-introducing IscA or SufA restores the cell growth of the E. coli iscA/sufA double mutant,51, 62 suggesting that IscA and SufA are complementary to each other.
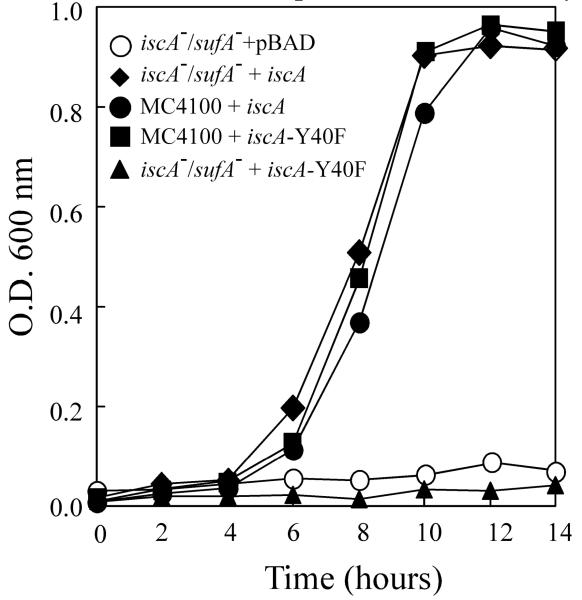
To determine the in vivo activity of the IscA mutant Y40F, a plasmid expressing Y40F was introduced into the E. coli iscA/sufA double mutant cells. The cells were inoculated in M9 minimal media and grown under aerobic conditions. Fig. 6 shows that unlike wild-type IscA, Y40F failed to restore the cell growth of the E. coli iscA/sufA double mutant in M9 minimal media under aerobic conditions. Since the IscA mutant Y40F has a diminished iron binding activity but retains the iron-sulphur cluster binding activity, we propose that the iron binding activity is essential for the physiological function of IscA in iron-sulphur cluster biogenesis.
Fig. 6.
Complementary activity of the IscA mutant Y40F in E. coli mutant with deletion of IscA and its paralog SufA. The E. coli wild-type (MC4100) and the mutant cells with deletion of IscA/SufA (iscA−/sufA−) containing expression plasmids were grown in M9 minimal media at 37°C under aerobic conditions. Cell growth was monitored at O.D. at 600 nm for 14 hours after inoculation of 1:100 dilutions.
Phylogenomic analyses revealed that IscA is highly conserved from prokaryotic to eukaryotic organisms.63 In A. vinelandii, depletion of IscA produces a null-growth phenotype in modified Burks minimal medium under elevated oxygen conditions.64 In E. coli, deletion of IscA and its paralog SufA results in a mutant that fails to grow in M9 minimal media under aerobic conditions.51, 61 In S. cerevisiae, depletion of IscA homologues leads to iron accumulation in mitochondria and dependency on lysine and glutamate in media.65 In human Hela cells, RNAi knockdown of IscA homologue results in decreased activities of iron-sulphur enzymes in both mitochondria and cytosol.66 Evidently, IscA has a crucial role for iron-sulphur cluster biogenesis, especially under aerobic conditions. However, the physiological function of IscA remains controversial. One hypothesis stated that IscA acts as an alternative scaffold protein or intermediate carrier for iron-sulphur cluster biogenesis, as IscA is able to bind an iron-sulphur cluster and transfer the cluster to target proteins.14-16 However, unlike other scaffold proteins such as IscU,9 IscA has a strong iron binding activity with a maximum binding of one iron per IscA dimer.41-45 Recent spectroscopic characterization of NifIscA from A. vinelandii confirms that NifIscA is able to bind one iron per dimer, and that the the iron center in IscA is in an unusual intermediate S = 3/2 spin state.48 The results presented in this study provide new evidence for the hypothesis that IscA is a bona fide iron binding protein. Among the primary iron-sulphur cluster assembly proteins encoded by the gene cluster iscSUA-hscBA-fdx5 and the putative iron donor frataxin/CyaY,24, 25 only IscA has a strong binding activity for mononuclear iron in the presence of dithiothreitol. Furthermore, the ferric iron centre tightly bound in IscA can be readily extruded by L-cysteine, followed by reduction to ferrous iron. Additional studies further reveal that mutation at tyrosine 40, a possible oxygenic ligand for the iron binding in IscA,48 diminishes iron binding activity but retains iron-sulphur cluster binding activity of IscA. Genetic complementation studies show that Y40F is inactive in vivo, suggesting that the iron binding activity is essential for the physiological function of IscA. Collectively, the results support the hypothesis that IscA may act as an iron donor for iron-sulphur cluster biogenesis. Nevertheless, since IscA can also bind iron-sulphur clusters 14, 15 and transfer the assembled clusters to target proteins,16 the role of IscA as an alternative scaffold protein cannot be excluded. Perhaps, the iron binding in IscA could be the initial step for iron-sulphur cluster assembly either in IscA or in other scaffold proteins such as IscU.43 Evidently, additional experiments are required to illustrate the role of IscA in iron-sulphur cluster biogenesis.
It is worth pointing out that IscA/SufA is dispensable for cell growth of E. coli in M9 minimal media under anaerobic conditions.45 Interestingly, most anaerobic organisms contain no IscA homologues and almost all aerobic organisms contain at least one copy of IscA in their genomes.63 It may be envisioned that under anaerobic conditions intracellular “free” iron is more freely available for iron-sulphur cluster biogenesis, and IscA is unnecessary. In aerobic organisms, a functional IscA will be required to recruit intracellular iron and deliver iron or iron-sulphur clusters for iron-sulphur cluster biogenesis under aerobic conditions.
Conclusions
Among the iron-sulphur cluster assembly proteins, IscA is unique in binding mononuclear iron. The tightly-bound iron centre in IscA can be readily extruded by L-cysteine, followed by reduction to ferrous iron. An IscA mutant that has a diminished iron binding activity but retains iron-sulphur cluster binding in vitro is inactive in vivo. The results suggest that IscA is a bona fide iron binding protein and that the iron binding activity is essential for physiological function of IscA in iron-sulphur cluster biogenesis.
Supplementary Material
Acknowledgements
We thank Hao Huang for some preliminary experiments. This work was supported by the National Cancer Institute of the National Institutes of Health under award number R01CA107494.
References
- 1.Beinert H, Holm RH, Munck E. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 2.Lill R. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 3.White MF, Dillingham MS. Current Opinion in Structural Biology. 2012;22:94–100. doi: 10.1016/j.sbi.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Crack JC, Green J, Thomson AJ, Le Brun NE. Curr Opin Chem Biol. 2012;16:35–44. doi: 10.1016/j.cbpa.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Zheng L, Cash VL, Flint DH, Dean DR. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 6.Zheng L, White RH, Cash VL, Jack RF, Dean DR. Proc Natl Acad Sci U S A. 1993;90:2754–2758. doi: 10.1073/pnas.90.7.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flint DH. J Biol Chem. 1996;271:16068–16074. [PubMed] [Google Scholar]
- 8.Shi Y, Ghosh MC, Tong WH, Rouault TA. Hum Mol Genet. 2009;18:3014–3025. doi: 10.1093/hmg/ddp239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agar JN, Krebs C, Frazzon J, Huynh BH, Dean DR, Johnson MK. Biochemistry. 2000;39:7856–7862. doi: 10.1021/bi000931n. [DOI] [PubMed] [Google Scholar]
- 10.Smith AD, Agar JN, Johnson KA, Frazzon J, Amster IJ, Dean DR, Johnson MK. J Am Chem Soc. 2001;123:11103–11104. doi: 10.1021/ja016757n. [DOI] [PubMed] [Google Scholar]
- 11.Urbina HD, Silberg JJ, Hoff KG, Vickery LE. J Biol Chem. 2001;276:44521–44526. doi: 10.1074/jbc.M106907200. [DOI] [PubMed] [Google Scholar]
- 12.Unciuleac MC, Chandramouli K, Naik S, Mayer S, Huynh BH, Johnson MK, Dean DR. Biochemistry. 2007;46:6812–6821. doi: 10.1021/bi6026665. [DOI] [PubMed] [Google Scholar]
- 13.Raulfs EC, O’Carroll IP, Dos Santos PC, Unciuleac MC, Dean DR. Proc Natl Acad Sci U S A. 2008;105:8591–8596. doi: 10.1073/pnas.0803173105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ollagnier-De-Choudens S, Sanakis Y, Fontecave M. J Biol Inorg Chem. 2004;9:828–838. doi: 10.1007/s00775-004-0581-9. [DOI] [PubMed] [Google Scholar]
- 15.Gupta V, Sendra M, Naik SG, Chahal HK, Huynh BH, Outten FW, Fontecave M, Ollagnier de Choudens S. J Am Chem Soc. 2009;131:6149–6153. doi: 10.1021/ja807551e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mapolelo DT, Zhang B, Naik SG, Huynh BH, Johnson MK. Biochemistry. 2012 [Google Scholar]
- 17.Bonomi F, Iametti S, Morleo A, Ta D, Vickery LE. Biochemistry. 2011;50:9641–9650. doi: 10.1021/bi201123z. [DOI] [PubMed] [Google Scholar]
- 18.Chandramouli K, Johnson MK. Biochemistry. 2006;45:11087–11095. doi: 10.1021/bi061237w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonomi F, Iametti S, Morleo A, Ta D, Vickery LE. Biochemistry. 2008;47:12795–12801. doi: 10.1021/bi801565j. [DOI] [PubMed] [Google Scholar]
- 20.Ta DT, Vickery LE. J Biol Chem. 1992;267:11120–11125. [PubMed] [Google Scholar]
- 21.Chandramouli K, Unciuleac MC, Naik S, Dean DR, Huynh BH, Johnson MK. Biochemistry. 2007;46:6804–6811. doi: 10.1021/bi6026659. [DOI] [PubMed] [Google Scholar]
- 22.Shi Y, Ghosh M, Kovtunovych G, Crooks DR, Rouault TA. Biochim Biophys Acta. 2012;1823:484–492. doi: 10.1016/j.bbamcr.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Canizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, Pandolfo M. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 24.Yoon T, Cowan JA. J Am Chem Soc. 2003;125:6078–6084. doi: 10.1021/ja027967i. [DOI] [PubMed] [Google Scholar]
- 25.Layer G, Ollagnier-de Choudens S, Sanakis Y, Fontecave M. J Biol Chem. 2006;281:16256–16263. doi: 10.1074/jbc.M513569200. [DOI] [PubMed] [Google Scholar]
- 26.Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- 27.Wang T, Craig EA. J Biol Chem. 2008;283:12674–12679. doi: 10.1074/jbc.M800399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerber J, Muhlenhoff U, Lill R. EMBO Rep. 2003;4:906–911. doi: 10.1038/sj.embor.embor918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cook JD, Kondapalli KC, Rawat S, Childs WC, Murugesan Y, Dancis A, Stemmler TL. Biochemistry. 2010;49:8756–8765. doi: 10.1021/bi1008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai CL, Barondeau DP. Biochemistry. 2010;49:9132–9139. doi: 10.1021/bi1013062. [DOI] [PubMed] [Google Scholar]
- 31.Prischi F, Konarev PV, Iannuzzi C, Pastore C, Adinolfi S, Martin SR, Svergun DI, Pastore A. Nat Commun. 2010;1:95. doi: 10.1038/ncomms1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bulteau AL, O’Neill HA, Kennedy MC, Ikeda-Saito M, Isaya G, Szweda LI. Science. 2004;305:242–245. doi: 10.1126/science.1098991. [DOI] [PubMed] [Google Scholar]
- 33.Bou-Abdallah F, Adinolfi S, Pastore A, Laue TM, Dennis Chasteen N. J Mol Biol. 2004;341:605–615. doi: 10.1016/j.jmb.2004.05.072. [DOI] [PubMed] [Google Scholar]
- 34.Kondapalli KC, Kok NM, Dancis A, Stemmler TL. Biochemistry. 2008;47:6917–6927. doi: 10.1021/bi800366d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li DS, Ohshima K, Jiralerspong S, Bojanowski MW, Pandolfo M. FEBS Lett. 1999;456:13–16. doi: 10.1016/s0014-5793(99)00896-0. [DOI] [PubMed] [Google Scholar]
- 36.Campanella A, Isaya G, O’Neill HA, Santambrogio P, Cozzi A, Arosio P, Levi S. Hum Mol Genet. 2004;13:2279–2288. doi: 10.1093/hmg/ddh232. [DOI] [PubMed] [Google Scholar]
- 37.Chen OS, Kaplan J. J Biol Chem. 2000;275:7626–7632. doi: 10.1074/jbc.275.11.7626. [DOI] [PubMed] [Google Scholar]
- 38.Anderson PR, Kirby K, Orr WC, Hilliker AJ, Phillips JP. Proc Natl Acad Sci U S A. 2008;105:611–616. doi: 10.1073/pnas.0709691105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreno-Cermeno A, Obis E, Belli G, Cabiscol E, Ros J, Tamarit J. J Biol Chem. 2010;285:41653–41664. doi: 10.1074/jbc.M110.149443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irazusta V, Obis E, Moreno-Cermeno A, Cabiscol E, Ros J, Tamarit J. Free Radic Biol Med. 2010;48:411–420. doi: 10.1016/j.freeradbiomed.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 41.Ding H, Clark RJ. Biochem J. 2004;379:433–440. doi: 10.1042/BJ20031702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding H, Harrison K, Lu J. J Biol Chem. 2005;280:30432–30437. doi: 10.1074/jbc.M504638200. [DOI] [PubMed] [Google Scholar]
- 43.Yang J, Bitoun JP, Ding H. J Biol Chem. 2006;281:27956–27963. doi: 10.1074/jbc.M601356200. [DOI] [PubMed] [Google Scholar]
- 44.Ding H, Yang J, Coleman LC, Yeung S. J Biol Chem. 2007;282:7997–8004. doi: 10.1074/jbc.M609665200. [DOI] [PubMed] [Google Scholar]
- 45.Wang W, Huang H, Tan G, Si F, Liu M, Landry AP, Lu J, Ding H. Biochem J. 2010;432:429–436. doi: 10.1042/BJ20101507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu J, Bitoun JP, Tan G, Wang W, Min W, Ding H. Biochem J. 2010;428:125–131. doi: 10.1042/BJ20100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muhlenhoff U, Richter N, Pines O, Pierik AJ, Lill R. J Biol Chem. 2011;286:41205–41216. doi: 10.1074/jbc.M111.296152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mapolelo DT, Zhang B, Naik SG, Huynh BH, Johnson MK. Biochemistry. 2012 [Google Scholar]
- 49.Cavadini P, Adamec J, Taroni F, Gakh O, Isaya G. J Biol Chem. 2000;275:41469–41475. doi: 10.1074/jbc.M006539200. [DOI] [PubMed] [Google Scholar]
- 50.Cowart RE, Singleton FL, Hind JS. Anal Biochem. 1993;211:151–155. doi: 10.1006/abio.1993.1246. [DOI] [PubMed] [Google Scholar]
- 51.Lu J, Yang J, Tan G, Ding H. Biochem J. 2008;409:535–543. doi: 10.1042/BJ20071166. [DOI] [PubMed] [Google Scholar]
- 52.Ding B, Smith ES, Ding H. Biochem J. 2005;389:797–802. doi: 10.1042/BJ20050405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindahl PA, Day EP, Kent TA, Orme-Johnson WH, Münck E. Journal of Biological Chemistry. 1985;260:11160–11173. [PubMed] [Google Scholar]
- 54.Hans M, Buckel W, Bill E. European Journal of Biochemistry. 2000;267:7082–7093. doi: 10.1046/j.1432-1327.2000.01809.x. [DOI] [PubMed] [Google Scholar]
- 55.Woodmansee AN, Imlay JA. Methods Enzymol. 2002;349:3–9. doi: 10.1016/s0076-6879(02)49316-0. [DOI] [PubMed] [Google Scholar]
- 56.Bilder PW, Ding H, Newcomer ME. Biochemistry. 2004;43:133–139. doi: 10.1021/bi035440s. [DOI] [PubMed] [Google Scholar]
- 57.Cupp-Vickery JR, Silberg JJ, Ta DT, Vickery LE. J Mol Biol. 2004;338:127–137. doi: 10.1016/j.jmb.2004.02.027. [DOI] [PubMed] [Google Scholar]
- 58.Ding H, Clark RJ, Ding B. J Biol Chem. 2004;279:37499–37504. doi: 10.1074/jbc.M404533200. [DOI] [PubMed] [Google Scholar]
- 59.Loiseau L, Gerez C, Bekker M, Ollagnier-de Choudens S, Py B, Sanakis Y, Teixeira de Mattos J, Fontecave M, Barras F. Proc Natl Acad Sci U S A. 2007;104:13626–13631. doi: 10.1073/pnas.0705829104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takahashi Y, Tokumoto U. J Biol Chem. 2002;277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 61.Mettert EL, Outten FW, Wanta B, Kiley PJ. J Mol Biol. 2008;384:798–811. doi: 10.1016/j.jmb.2008.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan G, Lu J, Bitoun JP, Huang H, Ding H. Biochem J. 2009;420:463–472. doi: 10.1042/BJ20090206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vinella D, Brochier-Armanet C, Loiseau L, Talla E, Barras F. PLoS Genet. 2009;5:e1000497. doi: 10.1371/journal.pgen.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson DC, Unciuleac MC, Dean DR. J Bacteriol. 2006;188:7551–7561. doi: 10.1128/JB.00596-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jensen LT, Culotta VC. Mol Cell Biol. 2000;20:3918–3927. doi: 10.1128/mcb.20.11.3918-3927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song D, Tu Z, Lee FS. J Biol Chem. 2009;284:35297–35307. doi: 10.1074/jbc.M109.040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.