Abstract
The human activator-recruited cofactor (ARC), a family of large transcriptional coactivator complexes related to the yeast Mediator, was recently identified based on functional association with the activation domains of multiple cellular and viral transcriptional activators, including the herpes simplex viral activator VP16, sterol regulatory element binding protein, and NF-κB. Here we describe the biochemical purification and cloning of the 92-kDa ARC/Mediator subunit, ARC92, that is specifically targeted by the activation domain of the VP16 transactivator. Affinity chromatography using the VP16 activation domain followed by peptide microsequencing led to the identification of ARC92 as a specific cellular interaction partner of the VP16 activation domain. ARC92 associates with the VP16 activation domain in vitro and in vivo, and the VP16 binding domain of ARC92 is a strong competitive inhibitor of Gal4-VP16 in vivo. Moreover, small interfering RNA-mediated knockdown of ARC92 in human cells results in selective inhibition of Gal4-VP16 gene activation. Taken together, our results suggest that ARC92 is a direct and specific target of the VP16 transactivator that serves in the context of the ARC/Mediator coactivator as an important transducer of transcription activating signals from the VP16 activation domain to the RNA polymerase II transcriptional machinery.
Transcriptional activation in metazoans is exquisitely regulated in a spatio-temporal fashion to ensure accurate developmental programming as well as appropriate responses to environmental and homeostatic cues (1). Gene-selective transcription requires the action of sequence-specific activators that bind to cognate enhancer and promoter DNA elements and communicate the activating signals to RNA polymerase II (Pol II) and the general transcription factors (TFIIA, -B, -D, -E, -F, and -H) that together compose the transcriptional machinery. Transcriptional activators generally contain modular domains that allow sequence-specific DNA binding and multimerization, as well as activation domains that are required to stimulate gene transcription. Activation domains are protein–protein interaction modules that directly recruit diverse types of cellular activities collectively referred to as coactivators, some of which facilitate chromatin remodeling, whereas others assist activators in the recruitment of the transcriptional machinery (2).
One class of coactivators, termed the Mediator, was originally identified in yeast as a cofactor that appears to bridge between activators and Pol II by interacting with activation domains and the C-terminal domain (CTD) on the large subunit of Pol II (3). Recent studies have revealed that metazoan organisms also harbor a family of coactivators that appears to be homologous to the yeast Mediator. For example, the activator-recruited cofactor (ARC) mediates gene activation in cell-free transcription reactions by several different classes of gene regulators, including the activation domain of the potent herpes simplex transactivator VP16, sterol regulatory element binding protein, and NF-κB (4, 5). Other groups have identified similar or identical co-factors, referred to as TRAP, DRIP, CRSP, PC-2, NAT, SMCC, and mammalian Srb/Mediator, suggesting that ARC is part of a family of closely related mammalian transcriptional coactivator complexes (3). The human ARC/Mediator was recently found to be composed of two highly related cofactor complexes sharing many subunits (6). The smaller of the two complexes (ARC-S) appears to be identical to the CRSP coactivator complex and is the active component, whereas the larger ARC/Mediator complex (referred to as ARC-L) is inactive in biochemical transcription assays. Like the yeast Mediator, the smaller ARC-S/CRSP complex interacts strongly with the Pol II CTD (7), suggesting that Pol II recruitment is an important function of this conserved class of coactivator. Several studies have suggested that individual subunits of the mammalian ARC/Mediator family of coactivators may play specialized roles in mediating specific gene regulatory pathways. For example, the 205/220-kDa subunit is a target of ligand-bound nuclear receptors (8–12), whereas the 130-kDa subunit (also known as Sur-2) binds to and is required for transactivation by the adenoviral E1A13S protein and the Ras/mitogen-activated protein kinase-regulated activator Elk-1 (13, 14). Additionally, a recent study uncovered the 105-kDa subunit of ARC/Mediator (ARC105) as a critical mediator of TGFβ/Activin/Nodal signaling via the SMAD2/3 activators in metazoans (15).
The function of the VP16 activator has been extensively investigated over the last two decades, and it has consequently gained importance as a benchmark activator. VP16 stimulates transcription of immediate-early (IE) viral genes by nucleating the formation of a complex with the cellular DNA binding factors Oct-1 and host cell factor on IE viral gene promoters and then serving to recruit activities that are required for transcription initiation (16). The latter function is accomplished by a discrete C-terminal activation domain that is unusually potent when fused to heterologous DNA binding domains (17). A number of functional targets of the VP16 activation domain have been proposed, including general transcription factors like the TBP and TAF9 components of TFIID, as well as TFIIA, TFIIB, and TFIIH (18). Other studies in yeast and mammalian cells have suggested that the VP16 activation domain can also recruit chromatin-directed activities, such as SAGA, NuA4, SWI/SNF, and CBP/p300 (5, 19–23). Our recent finding that a VP16 activation domain-affinity column can be used to purify the human ARC/Mediator to near homogeneity in one step from human nuclear extracts (5) suggests that the ARC/Mediator likely represents one of the most important cellular targets of the VP16 activation domain in human cells. Moreover, a chimeric activator composed of the VP16 activation domain fused to the yeast Gal4 DNA-binding domain requires ARC/Mediator for full transcriptional activation in biochemical transcription reactions reconstituted with chromatin templates (5). Similar findings by other groups support the view that the ARC/Mediator family of coactivators plays a critical role in transmitting the activating signal by the VP16 activation domain (9, 13). As VP16 is an archetypal activator used in many studies of gene activation mechanisms in a variety of organisms, cells and in vitro systems, identification of the ARC/Mediator subunit that interacts physically and functionally with the VP16 activation domain could lead to a better understanding of general mechanistic principles that govern gene activation in eukaryotes.
We have now taken an unbiased biochemical approach based on VP16 activation domain affinity chromatography to identify VP16-targeted proteins from fractionated human nuclear extract. This strategy has resulted in the identification and cloning of a 92-kDa ARC/Mediator subunit (ARC92) that is specifically targeted by the VP16 activation domain in vitro and in vivo. The functional importance of ARC92 in VP16 transactivation was demonstrated by both dominant negative experiments and small interfering RNA (siRNA)-mediated depletion of ARC92 in mammalian cells. Together, our findings suggest that ARC92 serves as a functionally important target within the ARC/Mediator coactivator of transcriptional activators.
Materials and Methods
Purification of ARC92. HeLa cell nuclear extract (100 ml) was applied to a 50-ml phosphocellulose (PC) column preequilibrated with 0.1 M KCl HEG (20 mM Hepes, pH 7.6/0.1 mM EDTA/10% glycerol/1 mM DTT/1 mM benzamidine/0.25 mM PMSF/2 μg/ml aprotinin). The column was step-eluted by using two column volumes of 0.2 M, 0.5 M, and 1.0 M KCl HEG. The fractions were then dialyzed to 0.1 M KCl. Large-scale purification of ARC92 was achieved by incubating 50 ml of the PC 0.1 M KCl (flow through) fraction (precleared with GST beads twice for 1 h) with 1 ml of GST-VP16-coupled beads for 3 h nutating at 4°C. The beads were washed seven times in 15 ml of 0.5 M KCl HEG plus 0.1% Nonidet P-40 (HEGN). Bound proteins were eluted by incubating with 1 ml of 0.3% Sarkosyl in 0.1 M KCl HEGN for 1 h. GST-VP16-bound proteins were separated by SDS/PAGE, and the band corresponding to the 92-kDa polypeptide was excised and subjected to trypsin digestion followed by HPLC separation of peptides. The purified peptides were analyzed by microsequencing.
Cloning of Human ARC92 cDNA. First-strand cDNA synthesis was performed with mRNA prepared from HepG2 cells. According to the peptide microsequence data of ARC92 and EST information from the GenBank database, we designed PCR primer pairs: 5′-ATGGTCCCCGGGTCCGAGGGCCC and 5′-CATCCACTGAGGCAGGTTTGGGTT for base pairs 1–1,255 (here, the A in the first ATG was designated as base pair 1), 5′-GGCTCCACGGCACAGCCCGGGG and 5′-GCAACTGGACTTGTAAATCCCCCG for base pairs 1,068–2,720. The PCR products were subcloned into pGEM-T-Easy vector (Promega). The NcoI site in the overlapping region of the PCR products was used to ligate them to generate full-length ARC92 cDNA.
Northern Blotting. Human tissue mRNA blots were purchased from Clontech and were blotted according to the manufacturers protocol. The cDNA probes used for blotting correspond to the first 1,200 nt of human ARC92 coding sequence and nucleotides 591–1,159 for human β-actin (BC014861).
Plasmids. The ARC92 cDNA was subcloned into the pCDNA3 vector with or without an N-terminal Flag-epitope tag. All other derivatives of ARC92 were generated by PCR using ARC92 cDNA as template. The PTOV1 cDNA was subcloned into pCDNA3 with an N-terminal Flag-tag. The VP16 (amino acids 413–490) and Myb (amino acids 241–325) activation domains were fused to the Gal4 DNA binding domain (amino acids 1–94) and subcloned into pCDNA3 with a hemagglutinin (HA)-tag at the N terminus. G4BE-luc and RN-luc were generous gifts from Nick Dyson (Massachusetts General Hospital Cancer Center). The siRNA-resistant pCDNA3-Flag-ARC92 was generated by mutating 5′-AAGAAGATGCGCGAGCAGATT (base pairs 387–405) to 5′-AAGAAaATGCGgGAaCAGATT by using PCR-based mutagenesis.
GST-Pulldown Assays. GST-fusion proteins were expressed in Escherichia coli BL21 cells and purified by glutathione Sepharose (Amersham Pharmacia). Nuclear extracts (NE) from human cell lines were prepared as described (24). In vitro translated (Promega) and 35S-labeled proteins (3 μl) were diluted 100-fold with 0.15 M NaCl HEGN. Flag-ARC92, or Flag-hSRB4 were expressed in U2OS cells and extracted into binding buffer with 0.5% Nonidet P-40 to disrupt the ARC/Mediator complex. Whole cell extracts were diluted 4-fold with binding buffer without detergent. For E1A binding experiments, EDTA was excluded and 100 μM ZnCl2 was added. NE, in vitro translated proteins, or whole cell extracts was applied to 25 μl of GST-fusion protein beads and incubated at 4°C for 3 h. Beads were washed seven times in 1 ml with 0.25 M KCl HEGN. Bound proteins were eluted with 0.3% sarkosyl.
Antibodies. Anti-Flag was purchased from Sigma. Anti-HA was purchased from Covance Research Products (Princeton). Anti-TRAP220/ARC205, CRSP150, CRSP70, Anti-CDK8, Anti-Med6, and Anti-Lamin A/C were purchased from Santa Cruz Biotechnology. Anti-ARC105 monoclonal antibodies were generated as described (15). Rabbit antisera were generated against the first 175 aa of human ARC92 by Strategic BioSolution.
Immunoprecipitation and Immunoblotting. For immunoprecipitation, 5 μl of anti-Flag (or anti-HA), 60 μl of anti-Med6 (or anti-TRAP220/ARC205, or anti-CDK8), 10 μl of goat serum, or 1 ml of anti-ARC105 hybridoma supernatant were bound to 20 μl of protein A/G beads (Amersham Pharmacia) and, after washing, 1 ml of HeLa or U2OS nuclear extract or whole cell lysate was added and incubated for 3 h nutating at 4°C. After washing five times with 1 ml of 0.25 M KCl HEGN, bound proteins were eluted with 0.3% sarkosyl or 10 mM Flag peptide. For immunoblotting, protein samples were processed as described (4).
Transfection and Luciferase Assay. To overexpress tagged proteins in U2OS cells for whole cell lysates, we transfected 2 μg of plasmids by Lipofectamine 2000 (Invitrogen) into each well (2 × 106 cells) of six-well plates. Whole cell extracts were prepared after 24 h. For luciferase assays, we plated 5 × 105 cells per well into 24-well plates and cotransfected with 100 ng of 2× G4BE-luciferase reporter, 5 ng of Renilla luciferase plasmid, and 2 ng of HA-Gal4-VP16 [or HA-Gal4-Myb, or HA-Gal4-E1A(CR3)]. Transfected cells were lysed after 24 h and analyzed by using the Dual Luciferase System (Promega).
siRNA. Double-stranded siRNA oligos were generated by Dharmacon Research. The sense sequences are 5′-GAAGAUGCGCGAGCAGAUU(TT)-3′ for human ARC92 and 5′-UCCUGGCCAACAUCCGCUC(TT)-3′ for human ARC105. siRNA oligos (2 μg per well) were cotransfected with pBABE (100 ng per well) into HeLa cells in six-well plates by Lipofectamine 2000. Cells were cultured overnight and then selected by 1 mg/liter puromycin (Sigma) for 24 h. After washing with PBS, the surviving cells were replated into 24-well plates and transfected with reporters and Gal4-fusion vectors. Cells were harvested for luciferase assays and immunodetection 65 h after the initial siRNA transfection.
Results
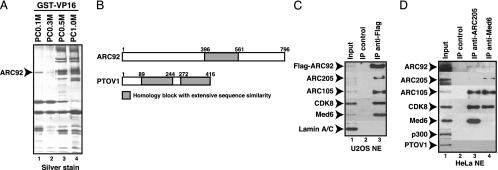
Identification of ARC92 as a Specific Target of the VP16 Activation Domain. A biochemical approach was used to identify the VP16-targeted ARC/Mediator subunit. Separation of HeLa cell nuclear extract over a phosphocellulose column followed by GST-VP16 affinity chromatography resulted in the purification of the ARC/Mediator complexes from the 0.5 M and 1 M KCl eluted fractions, as previously reported (Fig. 1A, lane 3 and 4) (4, 5). Interestingly, we also found strong association of the VP16 activation domain with a single polypeptide with a molecular mass of ≈92 kDa present in the phosphocellulose flow through fraction, PC0.1M (Fig. 1 A, lane 1). Peptide microsequencing revealed that this polypeptide is identical to a previously identified ARC/Mediator subunit, ARC92 (or DRIP97) (5, 10). ARC92 present in the PC0.1M fraction does not appear to associate with the ARC/Mediator complexes or other subunits; hence, our data suggest that this subunit may represent the VP16 activation domain target in the ARC/Mediator coactivator.
Fig. 1.
ARC92 is a VP16-targeted subunit of the ARC/Mediator coactivator complex. (A) ARC92 was purified as a VP16-interacting protein. HeLa cell nuclear proteins were first bound to the phosphocellulose (PC) column and were then successively eluted by the indicated concentration of KCl. The PC fractions were incubated with GST-VP16 beads, and specifically bound proteins were visualized by SDS/PAGE and silver stain. The band indicated by the arrowhead was identified as human ARC92 by peptide microsequencing. (B) Schematic representation of ARC92 and the related PTOV1 proteins. The amino acid sequence of one portion in ARC92 (gray box) is highly similar to two segments of PTOV1, a protein overexpressed in prostate tumors. (C) The ARC/Mediator complex is coimmunoprecipitated with the overexpressed Flag-ARC92. Flag-tagged ARC92 was expressed in U2OS cells and immunoprecipitated from nuclear extract by anti-Flag or anti-HA (IP control) antibodies and analyzed by immunoblotting for the indicated proteins. (D) Endogenous ARC92 is coimmunoprecipitated with the ARC/Mediator complex. Anti-TRAP220/ARC205 and anti-Med6 specifically coimmunoprecipitated endogenous ARC92 along with other components of the ARC/Mediator complexes from HeLa cell nuclear extracts. Other nuclear proteins, such as p300 and PTOV1, were not coprecipitated.
Because the ARC92 subunit had not been cloned, we then set out to isolate the ARC92 cDNA. Based on the peptide amino acid sequences, we searched the GenBank human EST database and found a series of overlapping ESTs that, when assembled, appear to represent the longest transcript in human cells and tissues that would be consistent with the 92-kDa molecular mass of ARC92. The N terminus of ARC92 was confirmed by N-terminal microsequencing and corresponds to the most 5′ EST with a preceding in-frame stop codon. The putative EST-based cDNA assembly served as a template for the design of PCR oligos that were then used in RT-PCR reactions to isolate the full-length human ARC92 cDNA from HepG2 cells (see Fig. 5A, which is published as supporting information on the PNAS web site, for protein sequence). Subsequent to our cloning of the ARC92 cDNA, another group reported the isolation of a cDNA encoding a 78-kDa protein of unknown function that appears to represent an alternatively spliced form of ARC92 (25). Indeed, based on further examination of the ESTs in the GenBank database, there are several splice variants of ARC92 that differ in the 3′ end. The relative expression of these alternative splice forms in different tissues and cells and their functional relevance is not known. ARC92 exhibits homologs in higher eukaryotes, including frog, zebrafish, fruit fly, and plants; however, we have been unable to identify homologs in Caenorhabditis elegans or yeast, suggesting that ARC92 may either have diverged during evolution or that it has emerged as a subunit in higher eukaryotes in response to the demands imposed by an increasingly complex gene regulatory apparatus (data not shown). A homology search also revealed that a segment of ARC92 appears to be highly related to a repeated sequence in a protein of unknown function that was identified based on its overexpression in prostate tumors (PTOV1) (26) (Fig. 1B). The ARC92 and PTOV1 genes are located next to each other on chromosome 19, suggesting a recent gene duplication event. Northern blotting revealed that ARC92 is ubiquitously expressed in human tissues, with two major transcripts of 2.5 and 4.4 kb (Fig. 5B).
To investigate whether the cloned ARC92 was capable of incorporating into the human ARC/Mediator complexes, we initially performed coimmunoprecipitation experiments with the Flag-tagged ARC92 expressed in 293 and U2OS cells. These studies showed that Flag-ARC92 can coprecipitate several endogenous ARC/Mediator subunits, including CDK8, Med6, ARC105, and ARC205/TRAP220, from U2OS nuclear extracts (Fig. 1C and data not shown). In contrast, nuclear Lamin A/C, which is not a component of ARC/Mediator, was not coprecipitated (Fig. 1C). Reciprocal immunoprecipitation experiments showed that antibodies directed against Med6 and ARC205/TRAP220 also coprecipitate endogenous ARC92 from HeLa cell nuclear extract, although with reduced efficiency that may indicate a pool of free ARC92 or that ARC92 dissociates during the washing steps (Fig. 1D). The unrelated coactivator p300 was not coprecipitated (Fig. 1D). Additionally, the ARC92-related PTOV1 protein did not coprecipitate with the human ARC/Mediator components, consistent with previous studies by others and us that did not identify PTOV1 as a component of the ARC/Mediator family of coactivators (Fig. 1D). Together, these results demonstrate that ARC92 is a bona fide subunit of the human ARC/Mediator and that the cloned ARC92 protein can specifically incorporate into the human ARC/Mediator complexes.
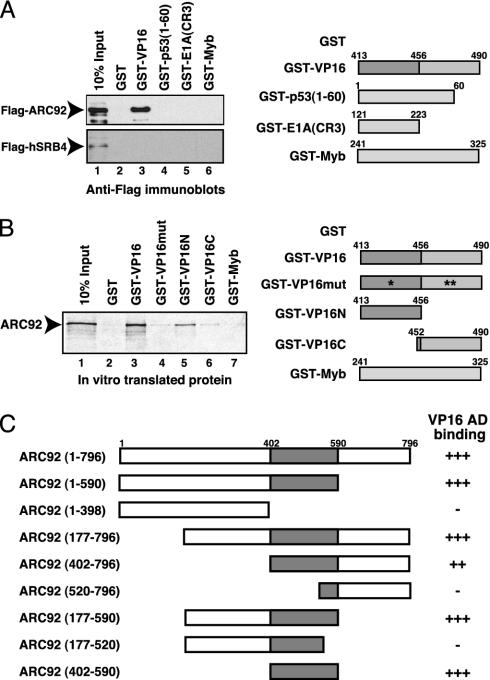
ARC92 Specifically Associates Via a Discrete Region with the Acidic Activation Domain of VP16 but Not with the Acidic Activation Domain of p53. Next, we examined the ability of the cloned ARC92 to interact with the activation domains of VP16 and other activators. Previous studies had classified the VP16 and p53 tumor suppressor activation domains as acidic. Interestingly, Flag-tagged ARC92 overexpressed in human U2OS osteosarcoma cells bound strongly to the GST-VP16 activation domain resin; however, the p53 activation domain did not interact with ARC92, although it bound strongly to the ARC/Mediator (Fig. 2A Upper, lanes 3 and 4 and data not shown). These findings would then suggest that VP16 and p53 might have distinct targets in the ARC/Mediator, despite their classification as acidic. No significant binding of ARC92 was observed to the E1A13S activation domain (which targets the 130 kDa ARC/Mediator subunit; ref. 13), nor the activation domain of the c-Myb transactivator, which does not interact with the ARC/Mediator, but which binds to the CBP coactivator (27) (Fig. 2 A Upper, lanes 5 and 6). These results suggest that the ARC92 ARC/Mediator subunit is a specific target of the VP16 activation domain. The 80-kDa subunit of the ARC/Mediator, a putative homolog of the yeast SRB4 Mediator subunit (28), was previously shown to interact with the activation domains of both VP16 and p53 when translated in vitro (9). We have attempted to address whether the human SRB4 homolog overexpressed in human cells can indeed bind to activators. Flag-tagged hSRB4/ARC/DRIP/TRAP80 (labeled hSRB4 for simplicity) was expressed in U2OS cells, and binding experiments identical to and in parallel with the Flag-ARC92 pulldown experiments with GST-activation domain fusions were performed. Interestingly, none of the activation domains were able to bind specifically to the hSRB4 subunit under the same conditions as where ARC92 associates strongly with the VP16 activation domain (Fig. 2 A Lower). These findings imply that, in contrast with previous reports, the hSRB4 ARC/Mediator subunit does not specifically bind to the VP16 activation domain. We have attempted to confirm and further characterize the interaction of the VP16 activation domain with ARC92 by performing GST pulldown studies with recombinant and in vitro translated proteins. Binding experiments with 35S-labeled in vitro translated ARC92 showed that ARC92 interacted strongly with the VP16 activation domain fused to GST, but not with GST alone (Fig. 2B, lanes 1–3). In contrast, ARC92 did not interact significantly with a mutated VP16 activation domain where three functionally critical phenylalanines were changed to alanines (29, 30) (Fig. 2B, compare lanes 3 and 4). These mutations also disrupt VP16 association with the ARC/Mediator complexes (5). Our results then demonstrate that ARC92 binds effectively only to a functional VP16 activation domain. The activation domain of VP16 is composed of two regions, each of which contribute to the transactivation ability of VP16, but which may also function independently (31). We find that the N-terminal portion of the VP16 activation domain (amino acids 413–456) is primarily responsible for the association with ARC92, whereas the C-terminal region (amino acids 452–490) binds less well to ARC92 (Fig. 2B, lanes 5 and 6). In keeping with its lack of binding to the ARC/Mediator and ARC92 expressed in cells, the GST-Myb activation domain fusion protein is incapable of specific binding to the in vitro-translated ARC92 subunit, demonstrating the specificity of the interaction between the VP16 activation domain and ARC92 (Fig. 2B, lane 7). To identify the VP16-binding domain (VBD) in ARC92, a series of deletion mutants of ARC92 were generated, translated in vitro, and tested for interaction with the GST-VP16 activation domain resin as above. These binding experiments revealed that the ARC92VBD resides in a discrete region between amino acids 402 and 590 (Fig. 2C and Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 2.
ARC92 specifically interacts with the VP16 activation domain in vitro.(A) Overexpressed ARC92 specifically associates with the VP16 activation domain. Flag-tagged ARC92 and hSRB4 were expressed in U2OS cells and incubated with the indicated GST-fusion proteins. Specifically bound proteins were analyzed by immunoblotting with anti-Flag antibodies. (B) In vitro translated human ARC92 specifically interacts with the VP16 activation domain. Human ARC92 was translated in vitro to generate 35S-labeled protein, which was incubated with the indicated GST-fusion proteins. The bound proteins were resolved by SDS/PAGE and visualized by autoradiography. (C) Identification of the VP16 binding domain in ARC92 (ARC92VBD). Fragments of human ARC92 were expressed in vitro and incubated with GST-VP16 as in B. The VP16 binding domain (VBD) resides in a region between amino acids 402 and 590 of ARC92 (gray box).
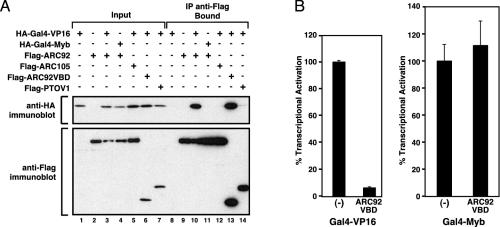
ARC92 Interacts with and Is Required for Full Transactivation by the VP16 Activation Domain in Mammalian Cells. As a prelude to investigation of the in vivo functional relevance of the ARC92 interaction with the VP16 activation domain, we have initially performed coimmunoprecipitation to ascertain that ARC92 and the VP16 activation domain can specifically associate in vivo. Flag-epitope-tagged full-length ARC92 or ARC92VBD coexpressed in human cells with HA-epitope-tagged Gal4-VP16 strongly coprecipitated in an anti-Flag immunoprecipitation experiment (Fig. 3A, lanes 10 and 13). In contrast, Flag-tagged ARC105, which had previously been found to serve as a specific target and mediator of transcriptional signaling by SMAD2/3 (15), was unable to associate with the Gal4-VP16 activation domain (Fig. 3A, lane 12). Additionally, HA-tagged Gal4-Myb did not coprecipitate with Flag-tagged ARC92 (Fig. 3A, lane 11), and Flag-tagged PTOV1 was not found in association with HA-tagged Gal4-VP16 (Fig. 3A, lane 14). Together these results demonstrate that the ARC92 ARC/Mediator subunit is a specific target of the VP16 activation domain in vivo.
Fig. 3.
ARC92 interacts with the VP16 activation domain in human cells. (A) Gal4-VP16 specifically coimmunoprecipitates with ARC92 and ARC92VBD in U2OS cells. The indicated Flag- and HA-tagged proteins were coexpressed in U2OS cells, immunoprecipitated by anti-Flag antibodies, and eluted by incubation with Flag peptide. Bound proteins were detected by immunoblotting. (B) ARC92VBD inhibits Gal4-VP16 transcriptional activation. Transient expression of Flag-ARC92VBD in HeLa cells inhibits HA-Gal4-VP16 transactivation but not activation by HA-Gal4-Myb.
Based on the strong association of Gal4-VP16 with the ARC92VBD fragment in human cells (Fig. 3A, lane 13), we hypothesized that the ARC92VBD would also compete effectively for VP16 binding to endogenous ARC92 present in the ARC/Mediator and thus function as a potent dominant negative inhibitor. Indeed, expression of the ARC92VBD fragment strongly inhibits Gal4-VP16 gene activation, without affecting transactivation by the Gal4-Myb activator (Fig. 3B).
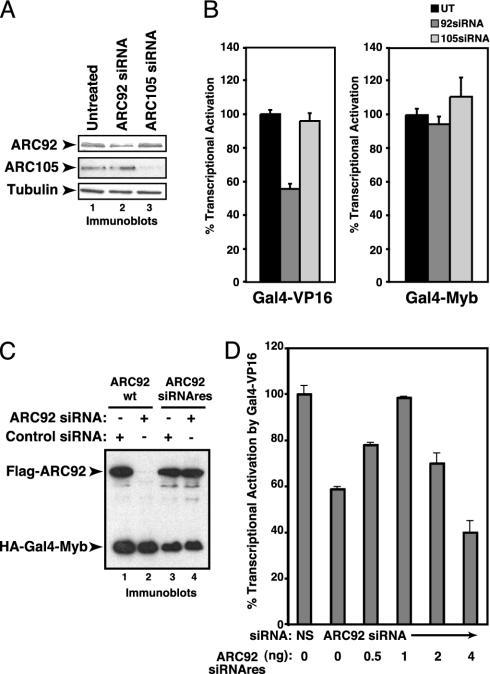
To further examine the functional role of ARC92 in VP16 transactivation, we have used siRNA-mediated knockdown of ARC92 in cultured mammalian cells. Transient transfection of ARC92-specific siRNA into HeLa cells resulted in a 50–60% depletion of ARC92 (Fig. 4A). Concomitant with a decrease in ARC92 protein levels, we observed a reproducible and significant (40–60%) reduction in Gal4-VP16 transactivation, whereas gene activation by the Gal4-Myb activator was unaffected (Fig. 4B). In contrast, siRNA-mediated depletion of the SMAD2/3-targeted ARC105 subunit exerted no significant influence on transactivation by Gal4-VP16 and Gal4-Myb (Fig. 4B). Our results then demonstrate that ARC92 is specifically required for full transactivation by Gal4-VP16.
Fig. 4.
ARC92 is required for full transcriptional activation by Gal4-VP16. (A) ARC92 siRNA specifically depleted endogenous ARC92 protein. siRNA oligos were transfected into HeLa cells, and ARC92, ARC105, and β-tubulin (loading control) were detected by immunoblotting. (B) Depletion of ARC92 specifically inhibits Gal4-VP16 transcriptional activation. siRNA-depleted cells were transfected with HA-Gal4-VP16, HA-Gal4-Myb, and reporters and analyzed for luciferase activity (represented as percentage of transactivation by the Gal4-activation domain fusion). (C) Generation of siRNA-resistant ARC92. Silent point mutations at the core of the siRNA-targeted sequence of ARC92 were introduced. The expressed, mutated ARC92 was resistant to degradation induced by cotransfection of siRNA against wild-type ARC92. Flag-tagged wild-type ARC92 (WT) or mutated ARC92 (siRNAres) was introduced with siRNA directed against ARC92 or ARC105 (control) into HeLa cells. The cells were analyzed by anti-Flag and -HA immunoblotting. (D) Small amounts of siRNA-resistant ARC92 (Flag-ARC92siRNAres) can rescue Gal4-VP16 transcriptional activation inhibited by ARC92 depletion by ARC92 siRNA. After siRNA treatments as in A, HeLa cells were cotransfected with the indicated amount of point-mutated ARC92, HA-Gal4-VP16, and reporter and analyzed as above.
To further confirm that the effect on Gal4-VP16 by the ARC92-directed siRNA is caused by knockdown of ARC92 rather than a nonspecific effect on other cellular proteins/processes, we generated a siRNA-resistant ARC92 by changing three base pairs in the sequence targeted by the siRNA that preserves the amino acid sequence (Fig. 4C). Cotransfection of the siRNA-resistant ARC92 vector with the ARC92-directed siRNA increased Gal4-VP16 gene activation at low concentrations of the siRNA-resistant ARC92 vector, but inhibited activation at higher levels, indicating that the siRNA exerts its inhibitory effects on Gal4-VP16 transactivation by specifically targeting ARC92 present in the ARC/Mediator coactivator complex (Fig. 4D). Together, these results confirm the functional requirement of ARC92 for full transcriptional activation by the VP16 activation domain in human cells.
Discussion
The transcriptional function of the herpes simplex activator VP16 has been extensively studied, and early biochemical experiments showed that the VP16 activation domain can interact with a number of cellular factors, including general transcription factors (GTFs). Studies of the functional relevance of the VP16 activation domain interactions with GTFs have largely relied on in vitro reconstituted transcription reactions. However, biochemical transcription reaction reconstituted with purified TBP, TFIIA, -B, -E, -F, -H, and Pol II cannot support transactivation by Gal4-VP16 (32), suggesting that additional nuclear factors may represent more functionally relevant targets of the VP16 activation domain. We previously used the activation domain of VP16 as an affinity resin to purify to near homogeneity the human ARC/Mediator coactivator and showed that this coactivator complex is required for gene activation by Gal4-VP16 in a reconstituted, chromatin-based transcription reaction (5). In this study, we describe the purification, cloning, and initial characterization of an ARC/Mediator subunit, ARC92, that is specifically targeted by the VP16 activation domain. A previous biochemical study has suggested that the TRAP80/SRB4 subunit of the ARC/Mediator is a specific target of the VP16 activation domain (9). However, this study did not examine the functional requirement for SRB4 in VP16 transactivation in vivo. Our findings, based on both dominant interference by the ARC92 VP16 binding domain as well as knockdown of ARC92 by siRNA, suggest that the ARC92 subunit, as part of the ARC/Mediator coactivator complex, is an important transducer of transactivating signals elicited by the VP16 activation domain in human cells.
Early studies noted that the VP16 activation domain carries a net negative charge, similar to the activating region found in the yeast Gal4 and GCN4 activators, and it was therefore labeled “acidic” (33–36). Other activators, including the human p53 tumor suppressor, have subsequently been labeled as acidic, both based on amino acid composition as well as data in support of functional similarities (37–39). Our results showing that the acidic activation domain of VP16, but not of p53, interacts with the ARC92 subunit would argue that the categorization of activation domains based on charge may potentially be misleading, at least concerning the targeting of the ARC/Mediator family of coactivators. Future studies aimed at identifying the p53 target in the ARC/Mediator together with comparative structural studies of the interaction between the VP16 and p53 activation domains and their cognate coactivators will help to further elucidate the molecular and functional similarities and differences of “acidic” activators.
VP16 is a tegument protein of the herpes simplex virus that also functions to direct transcriptional activation of several immediate-early viral genes. It will be interesting to probe whether host cell ARC92 is also required for the transactivating function of VP16 in the natural viral gene expression program. Viral proteins often exploit cellular functional pathways and molecular mechanisms and it is therefore possible that ARC92 also serves as a specific target and mediator of cellular activators. Indeed, our preliminary observations suggest that the VP16 binding domain in ARC92 can interact with several cellular transactivators (F.Y. and A.M.N, unpublished data). Efforts are needed to confirm and extend these intriguing findings.
Supplementary Material
Acknowledgments
We are grateful for the generous support and advice from Robert Tjian at University of California, Berkeley, during the initial part of these studies. We also thank members of the Massachusetts General Hospital Cancer Center and the Department of Cell Biology at Harvard Medical School for stimulating discussions and advice. We thank Timothy M. Thompson (Institut de Bìologìa Molecular, Consejo Superior de Investigaciones Científicas, Barcelona) for the generous gift of the PTOV1 antibody and cDNA. These studies were supported by the Bertucci Foundation at the Massachusetts General Hospital Cancer Center. F.Y. was supported by the Fund for Medical Discovery. A.M.N. is a Dammerman Scholar of the Damon Runyon Cancer Research Foundation (DRS-36-03).
Abbreviations: ARC, activator-recruited cofactor; Pol II, RNA polymerase II; siRNA, small interfering RNA; HA, hemagglutinin.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY533507).
References
- 1.Lemon, B. D. & Tjian, R. (2000) Genes Dev. 14, 2551–2569. [DOI] [PubMed] [Google Scholar]
- 2.Näär, A. M., Lemon, B. D. & Tjian, R. (2001) Annu. Rev. Biochem. 70, 475–501. [DOI] [PubMed] [Google Scholar]
- 3.Lewis, B. A. & Reinberg, D. (2003) J. Cell Sci. 116, 3667–3675. [DOI] [PubMed] [Google Scholar]
- 4.Näär, A. M., Beaurang, P. A., Robinson, K. M., Oliner, J. D., Avizonis, D., Scheek, S., Zwicker, J., Kadonaga, J. T. & Tjian, R. (1998) Genes Dev. 12, 3020–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Näär, A. M., Beaurang, P. A., Zhou, S., Abrahams, A., Solomon, W. & Tjian, R. (1999) Nature 398, 828–832. [DOI] [PubMed] [Google Scholar]
- 6.Taatjes, D. J., Näär, A. M., Andel, F., III, Nogales, E. & Tjian, R. (2002) Science 295, 1058–1062. [DOI] [PubMed] [Google Scholar]
- 7.Näär, A. M., Taatjes, D. J., Zhai, W., Nogales, E. & Tjian, R. (2002) Genes Dev. 16, 1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu, Y., Qi, C., Jain, S., Rao, M. S. & Reddy, J. K. (1997) J. Biol. Chem. 272, 25500–25506. [DOI] [PubMed] [Google Scholar]
- 9.Ito, M., Yuan, C., Malik, S., Gu, W., Fondell, J. D., Yamamura, S., Fu, Z., Zhang, X., Qin, J. & Roeder, R. G. (1999) Mol. Cell 3, 361–370. [DOI] [PubMed] [Google Scholar]
- 10.Rachez, C., Lemon, B. D., Suldan, Z., Bromleigh, V., Gamble, M., Näär, A.M., Erdjument-Bromage, H., Tempst, P. & Freedman, L. P. (1999) Nature 398, 824–828. [DOI] [PubMed] [Google Scholar]
- 11.Ito, M., Yuan, C. X., Okano, H. J., Darnell, R. B. & Roeder, R. G. (2000) Mol. Cell 5, 683–693. [DOI] [PubMed] [Google Scholar]
- 12.Ge, K., Guermah, M., Yuan, C. X., Ito, M., Wallberg, A. E., Spiegelman, B. M. & Roeder, R. G. (2002) Nature 417, 563–567. [DOI] [PubMed] [Google Scholar]
- 13.Boyer, T. G., Martin, M. E. D., Lees, E., Riccardi, R. P. & Berk, A. J. (1999) Nature 399, 276–279. [DOI] [PubMed] [Google Scholar]
- 14.Stevens, J. L., Cantin, G. T., Wang, G., Shevchenko, A. & Berk, A. J. (2002) Science 296, 755–758. [DOI] [PubMed] [Google Scholar]
- 15.Kato, Y., Habas, R., Katsuyama, Y., Näär, A. M. & He, X. (2002) Nature 418, 641–646. [DOI] [PubMed] [Google Scholar]
- 16.Wysocka, J. & Herr, W. (2003) Trends Biochem. Sci. 28, 294–304. [DOI] [PubMed] [Google Scholar]
- 17.Sadowski, I., Ma, J., Triezenberg, S. & Ptashne, M. (1988) Nature 335, 563–564. [DOI] [PubMed] [Google Scholar]
- 18.Zawel, L. & Reinberg, D. (1995) Annu. Rev. Biochem. 64, 533–561. [DOI] [PubMed] [Google Scholar]
- 19.Utley, R. T., Ikeda, K., Grant, P. A., Côté, J., Steger, D. J., Eberharter, A., John, S. & Workman, J. L. (1998) Nature 394, 498–502. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda, K., Steger, D. J., Eberharter, A. & Workman, J. L. (1999) Mol. Cell. Biol. 19, 855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neely, K. E., Hassan, A. H., Wallberg, A. E., Steger, D. J., Cairns, B. R., Wright, A. P. & Workman, J. L. (1999) Mol. Cell 4, 649–655. [DOI] [PubMed] [Google Scholar]
- 22.Kundu, T. K., Palhan, V. B., Wang, Z., An, W., Cole, P. A. & Roeder, R. G. (2000) Mol. Cell 6, 551–561. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda, K., Stuehler, T. & Meisterernst, M. (2002) Genes Cells 7, 49–58. [DOI] [PubMed] [Google Scholar]
- 24.Dignam, J. D., Martin, P. L., Shastry, B. S. & Roeder, R. G. (1983) Methods Enzymol. 101, 582–598. [DOI] [PubMed] [Google Scholar]
- 25.Wang, C., McCarty, I. M., Balazs, L., Li, Y. & Steiner, M. S. (2002) Biochem. Biophys. Res. Commun. 296, 281–287. [DOI] [PubMed] [Google Scholar]
- 26.Benedit, P., Paciucci, R., Thomson, T. M., Valeri, M., Nadal, M., Caceres, C., de Torres, I., Estivill, X., Lozano, J. J., Morote, J. & Reventos, J. (2001) Oncogene 20, 1455–1464. [DOI] [PubMed] [Google Scholar]
- 27.Dai, P., Akimaru, H., Tanaka, Y., Hou, D. X., Yasukawa, T., Kanei-Ishii, C., Takahashi, T. & Ishii, S. (1996) Genes Dev. 10, 528–540. [DOI] [PubMed] [Google Scholar]
- 28.Boube, M., Joulia, L., Cribbs, D. L. & Bourbon, H. M. (2002) Cell 110, 143–151. [DOI] [PubMed] [Google Scholar]
- 29.Shen, F., Triezenberg, S. J., Hensley, P., Porter, D. & Knutson, J. R. (1996) J. Biol. Chem. 271, 4819–4826. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan, S. M., Horn, P. J., Olson, V. A., Koop, A. H., Niu, W., Ebright, R. H. & Triezenberg, S. J. (1998) Nucleic Acids Res. 26, 4487–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker, S., Greaves, R. & O'Hare, P. (1993) Mol. Cell. Biol. 13, 5233–5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orphanides, G., Lagrange, T. & Reinberg, D. (1996) Genes Dev. 10, 2657–2683. [DOI] [PubMed] [Google Scholar]
- 33.Hope, I. A. & Struhl, K. (1986) Cell 46, 885–894. [DOI] [PubMed] [Google Scholar]
- 34.Ma, J. & Ptashne, M. (1987) Cell 51, 113–120. [DOI] [PubMed] [Google Scholar]
- 35.Ma, J. & Ptashne, M. (1987) Cell 48, 847–853. [DOI] [PubMed] [Google Scholar]
- 36.Triezenberg, S. J., Kingsbury, R. C. & McKnight, S. L. (1988) Genes Dev. 2, 718–729. [DOI] [PubMed] [Google Scholar]
- 37.Truant, R., Xiao, H., Ingles, C. J. & Greenblatt, J. (1993) J. Biol. Chem. 268, 2284–2287. [PubMed] [Google Scholar]
- 38.Liu, X., Miller, C. W., Koeffler, P. H. & Berk, A. J. (1993) Mol. Cell. Biol. 13, 3291–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao, H., Pearson, A., Coulombe, B., Truant, R., Zhang, S., Regier, J. L., Triezenberg, S. J., Reinberg, D., Flores, O., Ingles, C. J., et al. (1994) Mol. Cell. Biol. 14, 7013–7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.