Abstract
The nonhomologous DNA end-joining (NHEJ) pathway contains six known components, including Artemis, a nuclease mutated in a subset of human severe combined immunodeficient patients. Mice doubly deficient for the five previously analyzed NHEJ factors and p53 inevitably develop progenitor B lymphomas harboring der(12)t(12;15) translocations and immunoglobin heavy chain (IgH)/c-myc coamplification mediated by a breakage-fusion-bridge mechanism. In this report, we show that Artemis/p53-deficient mice also succumb reproducibly to progenitor B cell tumors, demonstrating that Artemis is a tumor suppressor in mice. However, the majority of Artemis/p53-deficient tumors lacked der(12)t(12;15) translocations and c-myc amplification and instead coamplified IgH and N-myc through an intra- or interchromosome 12 breakage-fusion-bridge mechanism. We discuss this finding in the context of potential implications for mechanisms that may target IgH locus translocations to particular oncogenes.
The nonhomologous end-joining (NHEJ) pathway ligates broken DNA ends irrespective of homology and is required for both general double-strand break (DSB) repair and repair of developmentally programmed DSBs introduced by the recombination-activating gene 1/2 (RAG) endonuclease (1). There are six known mammalian NHEJ factors: Ku70, Ku80, XRCC4, and Ligase 4 are evolutionarily conserved and function in all known NHEJ reactions, whereas the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and Artemis evolved more recently and together provide a nuclease activity for NHEJ reactions that require end-processing (2, 3). In this regard, RAG cleaves between V-, D-, and J-coding segments and flanking recombination signal sequence (RS) to form hairpin coding ends and blunt RS ends. Although the four conserved NHEJ factors are needed to join both coding and RS ends, DNA-PKcs and Artemis are relatively dispensable for RS joining but absolutely required for coding end-joining because of their role in hairpin opening (3, 4).
Inactivation of NHEJ results in increased ionizing radiation sensitivity, genomic instability, and severe combined immunodeficiency (SCID) (1, 3) resulting from the inability to join RAG-cleaved gene segments in progenitor (pro)-B and T lymphocytes. Despite their inability to repair DSBs, NHEJ-deficient mice show, at most, a modest predisposition to lymphomas, because cells with unrepaired breaks are eliminated by the checkpoint protein p53 (5–8). Inactivation of p53 restores pro-B lymphocyte numbers, although it does not rescue NHEJ or lymphocyte development (5, 6, 9). Combined deficiencies for p53 and all NHEJ factors except Artemis have been analyzed and lead to consistent development of early-onset pro-B lymphomas (5–12).
The molecular mechanisms underlying transformation in NHEJ/p53 pro-B lymphocytes have been elucidated (10, 13, 14). Almost all lymphomas exhibit amplification of c-myc coupled with consistent cytogenetic abnormalities, including a nonreciprocal der(12)t(12; 15) translocation, referred to as C12;15, and a complex translocation, referred to as a complicon, containing amplified immunoglobin heavy chain (IgH) and c-myc. NHEJ/p53-deficient lymphomas have been proposed to result from unrepaired DSBs persisting into S phase, where they are replicated and fused downstream of c-myc, generating the signature C12;15 and a dicentric 12;15 (15), which then leads to c-myc amplification through a breakage-fusion-bridge (BFB) mechanism (10, 13, 14). A major unresolved question is why c-myc is nearly invariably amplified. Nonmutually exclusive possibilities include strong selection for c-myc amplification or preferential targeting of the translocation to chromosome 15.
Mutations in Artemis form the basis for radiosensitive SCID in humans (16). Despite its more restricted role in NHEJ, Artemis-deficiency also leads to increased genomic instability in cultured murine cells, suggesting that it might function as a tumor suppressor (3, 17). In this study, we have asked whether Artemis has tumor suppressor functions in mice that are unmasked in the context of a p53-deficient background.
Methods
Generation of ArtN/Np53N/N Mice. 129Sv ArtN/N mice (3) were crossed to 129Sv/C57BL/6 p53N/N mice (18) to generate ArtN/+p53N/+ mice, which were then bred to generate all cohort mice. Mice were genotyped for Artemis by PCR with the following primers: ART5–3 (CAAGAGGCATTCGTGTATATGGGTGGC) and ART3–2 (CCCGTAACAGAGCTATGACAGAACCGGG), yielding a 250-bp wild-type band, or ART5–3 and neoRev (ACCGCTATCAGGACATAGCGT), yielding a 1-kb targeted band. Mice were genotyped for p53 as described (19).
Characterization of Mice/Tumors. All mice were regularly monitored and killed at first sign of illness. Mice were fixed in bouins and subjected to complete histological analysis. Lymphoid tissue samples were characterized by flow cytometry with anti-CD4, CD8, CD3, B-220, CD43 (Pharmingen) and anti-IgM (Southern Biotechnology Associates). pro-B cell lymphomas were grown in culture as described (13), and T cell lymphomas were cultured in RPMI medium 1640 supplemented with 15% FCS/25 units per ml IL-2 (BD Biosciences)/25 ng/ml IL-7 (PeproTech, Rocky Hill, NJ). Spectral karyotyping (SKY) was performed on metaphases prepared from each tumor (20). The c-myc (13) and 3′IgHRR (19) fluorescence in situ hybridization (FISH) probes have been described. The N-myc FISH probe is a bacterial artificial chromosome (RP23-246B9) overlapping the murine N-myc locus (Research Genetics, Huntsville, AL). Southern and Northern analyses were performed as described (13). Probes were as follows: JH1.1, a 1.1-kb Nae1-EcoR1 fragment downstream of JH4; mycA, a 1.6-kb Xba fragment upstream of c-myc; N-myc, a 1.6-kb ClaI-EcoR1 fragment containing exon 3 of the murine N-myc gene (21); c-myc northern probe, a 1.5-kb PstI fragment from the c-myc cDNA that hybridizes to exon 2 and 3; LR8 control probe, a 600-bp EcoR1 fragment upstream of the LR8 locus (19); and the 3′ IgH enhancer probe, a 700-bp HincII-receptor I fragment downstream of HS4 (22).
Cloning and Sequencing Translocation Junctions. All translocation junctions were cloned by circular PCR and sequenced as described (13). The non-IgH sequence was identified by using the Celera database.
Results
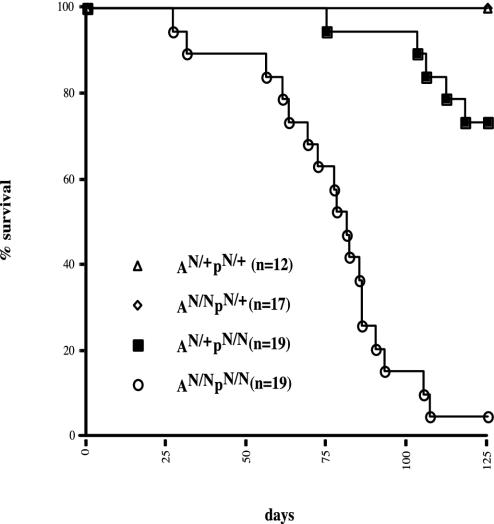
Increased Mortality in ArtN/N p53N/N Mice. To determine whether Artemis and p53 cooperate in tumor suppression, we crossed a line of Art+/N mice to p53-deficient (p53+/N) mice and then bred progeny. These breedings yielded cohorts of ArtN/N p53N/N (AN/NpN/N) mice (n = 19) and A+/Np+/N (n = 12), AN/Np+/N (n = 17), and A+/NpN/N (n = 19) mice. All genotypes were born at Mendelian ratios and appeared normal at birth. However, we observed a dramatic increase in the mortality rate of AN/NpN/N mice, compared with littermate controls (Fig. 1). The majority of AN/NpN/N mice died (or were killed because of severe morbidity) by 12 weeks of age (84 days), with 18 of 19 (95%) dead by 18 weeks (126 days). In contrast, only 5 of 19 (26%) A+/NpN/N mice died by 18 weeks of age, similar to p53N/N mice alone (18, 23, 24). Finally, all AN/Np+/N and A+/N p+/N mice lived past 18 weeks.
Fig. 1.
Increased mortality in AN/NpN/N double-deficient mice. Shown is a Kapplan–Meier curve representing the percent survival of AN/+pN/+ (n = 12), AN/NpN/+ (n = 17), AN/+pN//N(n = 19), and AN/NpN/N(n = 19) cohort mice versus age in days.
The majority of AN/NpN/N mice analyzed by histology (10/16) developed aggressive pro-B cell (B220+IgM–CD43+) lymphomas (Table 1) similarly to those reported for other NHEJ/p53 double-deficient mice (5–12). However, this was not the exclusive cause of morbidity, because other tumors were observed (Table 1). By comparison, T cell lymphomas were the most common lesion observed in AN/+pN/N mice (6/19), consistent with what has been reported for p53 N/N mice (18, 23). We did observe a pro-B cell tumor in one A+/NpN/N mouse (Table 1).
Table 1. Tumor spectrum in Art/p53 mice.
| A+/Np+/N (n = 12) | AN/Np+/N (n = 17) | A+/NpN/N (n = 19) | AN/NpN/N (n = 19) | |
|---|---|---|---|---|
| B cell lymphoma | 0 | 0 | 1 | 10 |
| T cell lymphoma | 0 | 3 | 6 | 3 |
| Teratoma | 0 | 0 | 1 | 2 |
| Colonic polyps | 0 | 0 | 0 | 1 |
| Medulloblastoma | 0 | 0 | 2 | 1 |
| Hemangiosarcoma | 0 | 0 | 1 | 0 |
| Cause of death unknown | 1 | 2 | 4 | 2 |
Numbers shown here represent all tumors observed including those appearing beyond the 18 weeks represented in Fig. 1.
Novel Chromosome 12 Aberrations in Most AN/NpN/N pro-B Cell Tumors. To analyze cytogenetic abnormalities in AN/NpN/N pro-B cell tumors, we examined metaphase spreads from cultured tumor cells by SKY. Surprisingly, of eight tumors examined, only three (AP138, AP143, and AP424) AN/NpN/N (Table 2, which is published as supporting information on the PNAS web site, and data not shown) exhibited predominant translocations involving both chromosomes 12 and 15, as observed in most other NHEJ/p53-deficient tumors. All three of these tumors had translocations involving chromosome 15, and two also had the signature C12;15. The remaining five AN/NpN/N pro-B cell tumors had predominant chromosomal aberrations that involved translocations and/or intrachromosomal gains of chromosome 12 material but did not involve chromosome 15 (Fig. 2 and Table 2). Each of these tumors (AP10, AP87, AP145, AP269, and AP270) had a chromosome 12 that by SKY appeared larger than the normal chromosome 12 (Figs. 2 and 3 and Table 2). Each large chromosome 12 did not appear to be dicentric intermediates, given that only one centromere was usually observed (Fig. 2 and data not shown). Additionally, a telomere-specific probe failed to identify any internal telomeric sequence on the enlarged chromosome 12 of tumor AP270 (Fig. 6, which is published as supporting information on the PNAS web site), suggesting that this aberration is not the result of telomeric fusions, which have been observed in ArtN/N cultured cells (17). A number of the tumors, each of which had an altered chromosome 12, also had a heterogeneous spectrum of nonclonal translocations, including C12;16 (AP10), C12;19 (AP145, AP269), C12;3 (AP269), and C12;15 (AP143) (Table 2).
Fig. 2.
A majority of AN/NpN/N pro-B cell tumors are cytogenetically distinct from other NHEJ/p53 tumors. Shown are SKY images of metaphase spreads from AP87, representative of the majority of AN/NpN/N pro-B cell tumors harboring a novel aberration, presenting as an enlarged copy of chromosome 12. SKY images are on the left, 4′,6-diamidino-2-phenylindole-stained images are in the middle, and computer-classified colors are on the right.
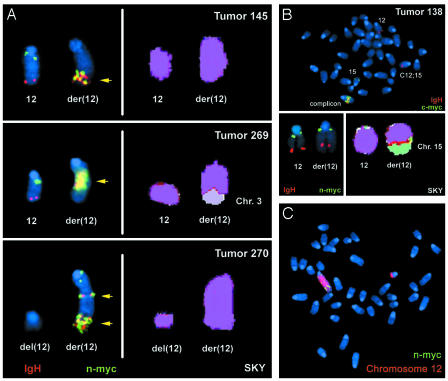
Fig. 3.
IgH and N-myc are coamplified on chromosome 12. (A) Representative FISH (Left) and SKY (Right) analyses of three tumors (AP 145, AP269, and AP270) exhibiting N-myc amplification by Southern blotting. The IgH FISH probe is red, the N-myc probe is green, and regions of coamplification are highlighted by yellow arrows. (B) Representative FISH (Upper, Lower Left) and SKY (Lower Right) analyses of tumor AP138 that exhibits c-myc amplification by Southern blotting. The IgH FISH probe is red, and the c-myc (Upper) or N-myc (Lower Left) is green. (C) Representative metaphase from AP270 analysis with a chromosome 12 paint (red) and the N-myc FISH probe (green).
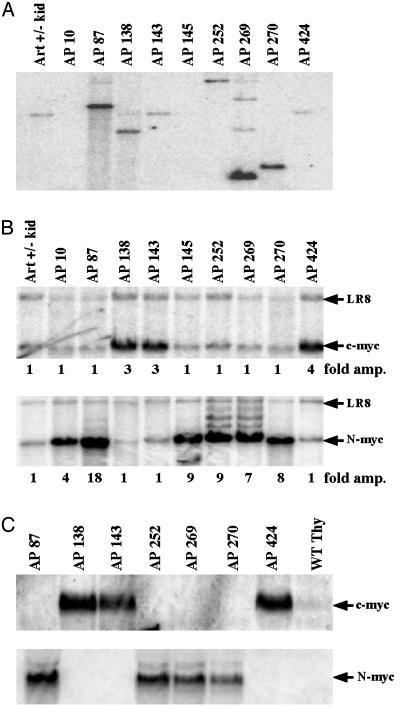
Activation of c-myc in a Minority of AN/NpN/N pro-B Tumors. We examined the amplification status of c-myc and IgH from AN/NpN/N pro-B cell tumors. Southern blot analyses with IgH-specific (JH) probes revealed that most AN/NpN/N tumors harbored either clonal rearrangements of JH or loss of the JH germline allele because of deletions extending downstream of the JH probe (Fig. 4A). A number of the tumors showed amplification of JH where it was retained (Fig. 4A), and all tumors exhibited amplification of IgH as detected by a 3′IgH regulatory region probe (Table 2 and data not shown). However, analysis with a c-myc probe revealed that only the three tumors with predominant translocations involving chromosomes 12 and 15 also contained significant amplification of c-myc (Fig. 4B and Table 2). Moreover, Northern blot analysis revealed that increased c-myc expression also was restricted to those tumors with C12;15 translocation and c-myc amplification (AP138, AP143, and AP424) (Fig. 4C), ruling out ectopic activation of c-myc as a mechanism contributing to the remaining six AN/NpN/N tumors. Metaphase spreads of one c-myc-amplified tumor (AP138) were found by FISH to be karyotypically similar to those of previously analyzed NHEJ/p53 tumors (10, 13, 14), with one normal chromosome 15, one C12;15 (lacking c-myc), one normal chromosome 12, and coamplified IgH and c-myc within a complicon (Fig. 3B). Nucleotide sequence analysis of the JH region junction from AP138 confirmed that it was fused to chromosome 15, ≈450 kb downstream of c-myc and resulted in a dicentric chromosome capable of generating c-myc amplification through BFB cycles (Fig. 5).
Fig. 4.
N-myc is amplified in a majority of AN/NpN/N pro-B cell tumors. (A and B) Southern blot analyses of tumor samples and kidney sample from a control mouse with a JH1.1 probe (A) or a N-myc probe and a c-myc probe (B). The LR8 control probe was used as a loading control to measure the amplification of relevant sequences. (C) Northern blot analysis of RNA isolated from tumors along with wild-type thymus probed with a c-myc or N-myc probe.
Fig. 5.
N-myc amplification is initiated by the translocation of IgH sequence around N-myc. (A) Schematic representation of chromosome 12 and 15 showing breakpoints of indicated tumors within the JH locus and N-myc (chr 12) or c-myc (chr 15). Breakpoint no. 4 (AP270) represents a DHJH rearrangement translocated telomeric to N-myc. (B) Sequences of the cloned translocation breakpoints from AN/NpN/N pro-B cell tumors. AP87, AP269, AP145, and AP270 have N-myc amplification, and AP138 is amplified for c-myc. JH sequences are in red, N-myc sequences are in green, and c-myc sequences are in black. Regions of homology are in blue, and nontemplated nucleotides are in purple. The locations and orientation of the translocation breakpoints are indicated to the right. TEL, telomeric; CEN, centromeric.
Amplification of N-myc in the Majority of AN/NpN/N pro-B Lymphomas. As noted, AN/NpN/N pro-B tumors that lacked c-myc amplification had unusual alterations of chromosome 12, which, in addition to the telomeric IgH, contains the Myc family member N-myc near the centromere (25). To determine whether N-myc amplification can substitute for c-myc, we analyzed these tumors by Southern blot with an N-myc exon 3 probe. All six of the AN/NpN/N pro-B tumors that lacked c-myc amplification showed substantial N-myc amplification (Fig. 4B). Furthermore, N-myc was not amplified in the three AN/NpN/N pro-B tumors having c-myc amplification. Likewise, Northern analyses of the AN/NpN/N tumors revealed that increased N-myc expression correlated with N-myc amplification (Fig. 4C), and there was no detectable N-myc expression in the three tumors with c-myc amplification and overexpression (Fig. 4C). Therefore, AN/NpN/N pro-B lymphomas can be subdivided into two distinct groups: those with translocations of IgH to chromosome 15 leading to the coamplification of c-myc and IgH sequences (similar to other NHEJ/p53 tumors) and a larger subset that has novel chromosome 12 alterations with amplification of N-myc.
Intra- or Interchromosome Translocations Lead to Coamplification of IgH and N-myc. To elucidate the mechanism of N-myc amplification in AN/NpN/N tumors, we cloned amplified IgH rearrangements from four AN/NpN/N tumors with N-myc amplification. Nucleotide sequence analyses revealed that three of the four rearrangements (AP87, AP145, and AP269) fused sequences within the JH cluster to chromosome 12 sequences ≈150–250 kb centromeric to N-myc on chromosome 12 (breakpoints 1–3 of Fig. 5). However, the orientation of these IgH/N-myc breakpoints would not have resulted in a dicentric intermediate but rather would have duplicated most of chromosome 12, with a JH/N-myc junction in the center (Fig. 3A). The rearrangement cloned from the fourth N-myc amplified tumor (AP270) involved fusion of an aberrant DJH rearrangement ≈250 kb telomeric to N-myc. This breakpoint would have generated a dicentric chromosome that could lead to subsequent BFB amplification events.
To gain additional insight into mechanisms underlying N-myc amplification, we performed FISH with probes specific for IgH and N-myc, coupled with a chromosome 12-specific paint, on several AN/NpN/N tumors with N-myc amplification. Metaphase spreads from tumors AP145, AP269, and AP270 revealed coamplification of IgH and N-myc (Fig. 3A), often at one end of the “large” chromosome 12 (Fig. 3C). However, in some metaphases, the amplified N-myc and IgH are present on an enlarged chromosome 12, which appears to contain nonchromosomal 12 sequences at its telomere, suggesting “capping” by another chromosome as has been observed in IgH/c-myc complicons. Notably, both tumors analyzed with translocation breakpoints centromeric to N-myc (AP145 and AP269) retained a normal copy of chromosome 12 by chromosome painting and by FISH (Fig. 3A) consistent with the translocations occurring between sister chromatids. Also, in many chromosome 12 derivatives containing amplified N-myc and IgH in AP145 and AP269, N-myc was not present at the centromere, suggesting complex rearrangements. Dicentric chromosomes with centrally amplified IgH and N-myc were also observed in occasional metaphases consistent with a BFB mechanism, although the amplified breakpoint junctions would not have led to a dicentric. Tumor 270, which harbored a translocation breakpoint telomeric to N-myc, lacked a normal chromosome 12 and instead contained a centromeric chromosome 12 fragment that lacked N-myc (Fig. 3 A and C), suggesting that it may have resulted from recombination between homologous chromosomes to generate a dicentric chromosome 12 plus a short centromere-containing chromosome 12 fragment. Overall, our findings support a model in which unrepaired JH DSBs in AN/NpN/N pro-B cells lead to N-myc amplification through a BFB mechanism similar to that which we have proposed for generation of c-myc complicons but involving intrachromosomal 12 rearrangements.
Discussion
Artemis Functions to Suppress Translocations and Tumors. We have shown that Artemis cooperates with p53 to suppress chromosomal translocations and tumor development in mice and therefore can be considered a tumor suppressor gene. Like other NHEJ/p53 doubly deficient mice (15), most AN/NpN/N mice succumb to pro-B cell lymphomas by 11–12 weeks of age, as opposed to T cell lymphomas that arise much later in p53-deficient mice. Despite the striking relationship between NHEJ deficiencies and tumorigenesis in mouse models, potential roles for NHEJ in tumor suppression in humans have remained unclear (15). However, inactivating mutations of Ku70, Ku80, DNA-PKcs, XRCC4, and Ligase 4 have also not been observed in the context of human immunodeficiencies, possibly because of a more severe impact of NHEJ mutations on human cells (26). In contrast, mutations in Artemis have been identified in several cohorts of human SCID patients (16, 27, 28). Therefore, our finding that Artemis functions as a tumor suppressor in mice raises the possibility of a similar function in humans. In this regard, hypomorphic alleles of Artemis have been identified in humans and have been associated with a predisposition to lymphomas (29).
The majority of AN/+pN/N control mice died from T cell lymphomas that lacked clonal translocations (data not shown) and presumably represent the T cell lymphomas that normally arise on a p53-deficient background (12, 19, 30). In contrast, of two AN/NpN/N thymic lymphomas analyzed by SKY, both had translocations involving chromosome 14, which contains the T cell antigen receptor (TCR)-α locus. In addition, two of the three AN/NpN/N T lineage tumors analyzed exhibited amplification of the TCR-α locus (data not shown). These latter findings suggest that AN/NpN/N thymic lymphomas are distinct from those that arise in p53-deficient mice. Conceivably, the AN/NpN/N thymic lymphomas may arise from a pool of pro-T cells with unresolved RAG-induced breaks that escaped elimination because of the inactivation of p53 and/or the leakiness of Artemis deficiency with respect to T cell development. In this regard, we observed T cell lymphomas in older (>8 months) ArtN/N mice (Table 1) that may be analogous to those observed in Ku70-deficient or SCID (DNA-PKcs-dead) mice (9, 11, 31–33), which are also leaky with respect to T cell development.
Activation of Endogenous N-myc or c-myc in AN/NpN/N pro-B Lymphomas. Nearly all NHEJ-p53 doubly deficient mice succumb to pro-B cell lymphomas (5–12) with amplified c-myc expression through a BFB mechanism (10, 13, 14). Therefore, it is striking that c-myc was amplified in only three of nine analyzed AN/NpN/N pro-B lymphomas and that N-myc was amplified and overexpressed in the others. N-myc was identified based on its amplification in advanced stage human neuroblastomas (34, 35). However, N-Myc can lead to pro-B lymphomas when deregulated in transgenic mice through linkage to an IgH intronic enhancer (36–38). Our current studies clearly show that translocation and amplification of endogenous N-myc can also participate in pro-B cell lymphomagenesis.
N-Myc is a putative basic region/helix-loop-helix transcription factor and shares substantial similarity to c-Myc with regards to genomic organization and protein sequence (21, 34). Although both c-Myc and N-Myc knockouts are embryonic lethal (39, 40), replacement of endogenous c-myc with N-myc allows normal development, suggesting the two proteins have largely equivalent activities and that their specificity lies in differential expression (41). We detected no c-myc expression in AN/NpN/N pro-B cell tumors with amplified N-myc, despite the fact that normal murine pro-B cells express both N-myc and c-myc (42). Conversely, the three AN/NpN/N tumors with amplified c-myc did not express detectable N-myc. Similar crossregulation of Myc family members has been reported in the context of N-myc-related transformation (36, 38, 43, 44), which further supports the role of N-myc amplification in the transformation of AN/NpN/N pro-B cells.
N-myc Complicon Formation Initiated from Intrachromosome 12 Translocations. We have proposed a molecular model for the events leading to the translocations and amplifications that underlie complicon formation in XRCC4/p53 or Ligase 4/p53-deficient mice (13). In the absence of p53, pro-B cells with unrepaired DSBs at the JH locus enter S phase, where the broken chromosome 12 is replicated. Subsequent recombination between the replicated JH DSB and a region downstream of c-myc produces a clonal C12;15 translocation and a dicentric chromosome that initiates a BFB cycle, resulting in coamplification of IgH and c-myc. A subset (3 of 9) of pro-B tumors from AN/NpN/N mice share many of the same features (c-myc amplification, C12;15, and complicon formation); thus, c-myc amplification can occur in AN/NpN/N pro-B cells by the same mechanism that predominates in other NHEJ/p53 pro-B tumors. Yet the majority of AN/NpN/N pro-B cell tumors are distinct, both karyotypically and in oncogene amplification status, from nearly all previously studied NHEJ/p53-deficient tumors, having amplified N-myc through translocations that involve IgH.
Translocation junctions characterized from AN/NpN/N pro-B tumors involved direct invasion of the N-myc flanking region by JH (or upstream DH); thus, these junctions must result from intra- or interchromosomal 12 rearrangements. Based on the locations of the junctions and cytogenetic analyses, one could envision several potential scenarios leading to N-myc amplification by BFB. We have outlined two possible mechanisms (Figs. 7 and 8, which are published as supporting information on the PNAS web site) that would lead to the different cytogenetic structures and breakpoints observed (Figs. 3 and 5). Although one cannot definitively distinguish between these and other related mechanisms, it is clear that the overall mechanism by which N-myc is amplified in AN/NpN/N pro-B tumors is highly analogous to the mechanism by which c-myc is amplified in other NHEJ/p53-deficient tumors, except that the initial V(D)J event targets the N-myc region rather than the c-myc region.
Targeting of Translocation Events. Endogenous N-myc and c-myc both can drive pro-B cell lymphomagenesis by means of a complicon-related mechanism and appear to have similar lymphomagenic potential once activated. Although still a formal possibility, it seems unlikely that N-myc versus c-myc amplification provides a specific selective growth advantage to AN/NpN/N pro-B cells as opposed to pro-B cells deficient for other NHEJ factors and p53. In the same context, the appearance of AN/NpN/N tumors that have amplified c-myc indicates that the oncogenic potential of c-myc is not limited to pro-B cells lacking other NHEJ factors.
Thus, we favor the possibility that some factor(s) must predispose c-myc versus N-myc for translocation in the different genetic settings. One such factor could be the presence of unresolved JH-coding end hairpins, which when replicated could lead to a dicentric chromosome 12 that could initiate N-myc amplification. However, all characterized AN/NpN/N junctions involve fusion of IgH to regions around N-myc; therefore, BFB appears to have been initiated by translocation rather than a reduplication of chromosome 12 (Figs. 7 and 8). Another possibility would be a unique impact of Artemis-deficiency, compared to other NHEJ deficiencies, on the availability of particular translocation acceptor sites around c-myc and/or N-myc. In this context, it also is possible that some other genetic difference between AN/NpN/N and other NHEJ/p53-deficient mouse lines, rather than the specific NHEJ deficiency per se, might affect availability of such sites. In either case, although the nature of the factor(s) that leads to preferential targeting of IgH translocations to c-myc or N-myc in certain lines will require further analysis, our current studies suggest that such factors do exist.
Supplementary Material
Acknowledgments
We thank Kevin Mills for helpful discussions and Amanda Smith and Marc Listewnik for technical assistance. This work was supported by National Institute of Health Grants AI35714 and CA92625 (to F.W.A.). F.W.A. is an Investigator of the Howard Hughes Medical Institute. J.S. is a Special Fellow of the Leukemia and Lymphoma Society. J.P.M. is supported by a Lymphoma Research Foundation Fellowship. C.L. is a Stanley Robbins Award recipient. D.O.F. is supported by National Institutes of Health Grant KO8 HL67580-02.
Abbreviations: NHEJ, nonhomologous end-joining; IgH, Ig heavy chain; DSB, double-strand break; BFB, breakage-fusion-bridge; pro, progenitor; DNA-PKcs, DNA-dependent protein kinase catalytic subunit; SKY, spectral karyotyping; FISH, fluorescence in situ hybridization; RAG, recombination-activating gene 1/2; SCID, severe combined immunodeficiency.
References
- 1.Bassing, C. H., Swat, W. & Alt, F. W. (2002) Cell 109, S45–S55. [DOI] [PubMed] [Google Scholar]
- 2.Ma, Y., Pannicke, U., Schwarz, K. & Lieber, M. R. (2002) Cell 108, 781–794. [DOI] [PubMed] [Google Scholar]
- 3.Rooney, S., Sekiguchi, J., Zhu, C., Cheng, H. L., Manis, J., Whitlow, S., DeVido, J., Foy, D., Chaudhuri, J., Lombard, D. & Alt, F. W. (2002) Mol. Cell 10, 1379–1390. [DOI] [PubMed] [Google Scholar]
- 4.Zhu, C. & Roth, D. B. (1995) Immunity 2, 101–112. [DOI] [PubMed] [Google Scholar]
- 5.Frank, K. M., Sharpless, N. E., Gao, Y., Sekiguchi, J. M., Ferguson, D. O., Zhu, C., Manis, J. P., Horner, J., DePinho, R. A. & Alt, F. W. (2000) Mol. Cell 5, 993–1002. [DOI] [PubMed] [Google Scholar]
- 6.Gao, Y., Ferguson, D. O., Xie, W., Manis, J., Sekiguchi, J., Frank, K. M., Chaudhuri, J., Horner, J., DePinho, R. A. & Alt, F. W. (2000) Nature 404, 897–900. [DOI] [PubMed] [Google Scholar]
- 7.Guidos, C. J., Williams, C. J., Grandal, I., Knowles, G., Huang, M. T. & Danska, J. S. (1996) Genes Dev. 10, 2038–2054. [DOI] [PubMed] [Google Scholar]
- 8.Nacht, M., Strasser, A., Chan, Y. R., Harris, A. W., Schlissel, M., Bronson, R. T. & Jacks, T. (1996) Genes Dev. 10, 2055–2066. [DOI] [PubMed] [Google Scholar]
- 9.Difilippantonio, M. J., Zhu, J., Chen, H. T., Meffre, E., Nussenzweig, M. C., Max, E. E., Ried, T. & Nussenzweig, A. (2000) Nature 404, 510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gladdy, R. A., Taylor, M. D., Williams, C. J., Grandal, I., Karaskova, J., Squire, J. A., Rutka, J. T., Guidos, C. J. & Danska, J. S. (2003) Cancer Cell 3, 37–50. [DOI] [PubMed] [Google Scholar]
- 11.Lim, D. S., Vogel, H., Willerford, D. M., Sands, A. T., Platt, K. A. & Hasty, P. (2000) Mol. Cell. Biol. 20, 3772–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanasse, G. J., Halbrook, J., Thomas, S., Burgess, A., Hoekstra, M. F., Disteche, C. M. & Willerford, D. M. (1999) J. Clin. Invest. 103, 1669–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu, C., Mills, K. D., Ferguson, D. O., Lee, C., Manis, J., Fleming, J., Gao, Y., Morton, C. C. & Alt, F. W. (2002) Cell 109, 811–821. [DOI] [PubMed] [Google Scholar]
- 14.Difilippantonio, M. J., Petersen, S., Chen, H. T., Johnson, R., Jasin, M., Kanaar, R., Ried, T. & Nussenzweig, A. (2002) J. Exp. Med. 196, 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mills, K. D., Ferguson, D. O. & Alt, F. W. (2003) Immunol. Rev. 194, 77–95. [DOI] [PubMed] [Google Scholar]
- 16.Moshous, D., Callebaut, I., de Chasseval, R., Corneo, B., Cavazzana-Calvo, M., Le Deist, F., Tezcan, I., Sanal, O., Bertrand, Y., Philippe, N., et al. (2001) Cell 105, 177–186. [DOI] [PubMed] [Google Scholar]
- 17.Rooney, S., Alt, F. W., Lombard, D., Whitlow, S., Eckersdorff, M., Fleming, J., Fugmann, S., Ferguson, D. O., Schatz, D. G. & Sekiguchi, J. (2003) J. Exp. Med. 197, 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donehower, L. A., Harvey, M., Slagle, B. L., McArthur, M. J., Montgomery, C. A., Jr., Butel, J. S. & Bradley, A. (1992) Nature 356, 215–221. [DOI] [PubMed] [Google Scholar]
- 19.Bassing, C. H., Suh, H., Ferguson, D. O., Chua, K. F., Manis, J., Eckersdorff, M., Gleason, M., Bronson, R., Lee, C. & Alt, F. W. (2003) Cell 114, 359–370. [DOI] [PubMed] [Google Scholar]
- 20.Ferguson, D. O., Sekiguchi, J. M., Chang, S., Frank, K. M., Gao, Y., DePinho, R. A. & Alt, F. W. (2000) Proc. Natl. Acad. Sci. USA 97, 6630–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DePinho, R. A., Legouy, E., Feldman, L. B., Kohl, N. E., Yancopoulos, G. D. & Alt, F. W. (1986) Proc. Natl. Acad. Sci. USA 83, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinaud, E., Khamlichi, A. A., Le Morvan, C., Drouet, M., Nalesso, V., Le Bert, M. & Cogne, M. (2001) Immunity 15, 187–199. [DOI] [PubMed] [Google Scholar]
- 23.Harvey, M., McArthur, M. J., Montgomery, C. A., Jr., Bradley, A. & Donehower, L. A. (1993) FASEB J. 7, 938–943. [DOI] [PubMed] [Google Scholar]
- 24.Jacks, T., Remington, L., Williams, B. O., Schmitt, E. M., Halachmi, S., Bronson, R. T. & Weinberg, R. A. (1994) Curr. Biol. 4, 1–7. [DOI] [PubMed] [Google Scholar]
- 25.Campbell, G. R., Zimmerman, K., Blank, R. D., Alt, F. W. & D'Eustachio, P. (1989) Oncogene Res. 4, 47–54. [PubMed] [Google Scholar]
- 26.Li, G., Nelsen, C. & Hendrickson, E. A. (2002) Proc. Natl. Acad. Sci. USA 99, 832–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi, N., Agematsu, K., Sugita, K., Sako, M., Nonoyama, S., Yachie, A., Kumaki, S., Tsuchiya, S., Ochs, H. D., Fukushima, Y. & Komiyama, A. (2003) Hum. Genet. 112, 348–352. [DOI] [PubMed] [Google Scholar]
- 28.Noordzij, J. G., Verkaik, N. S., van der Burg, M., van Veelen, L. R., de Bruin-Versteeg, S., Wiegant, W., Vossen, J. M., Weemaes, C. M., de Groot, R., Zdzienicka, M. Z., et al. (2003) Blood 101, 1446–1452. [DOI] [PubMed] [Google Scholar]
- 29.Moshous, D., Pannetier, C., Chasseval Rd, R., Deist Fl, F., Cavazzana-Calvo, M., Romana, S., Macintyre, E., Canioni, D., Brousse, N., Fischer, A., et al. (2003) J. Clin. Invest. 111, 381–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao, M. J., Zhang, X. X., Hill, R., Gao, J., Qumsiyeh, M. B., Nichols, W. & Van Dyke, T. (1998) Mol. Cell. Biol. 18, 3495–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Custer, R. P., Bosma, G. C. & Bosma, M. J. (1985) Am. J. Pathol. 120, 464–477. [PMC free article] [PubMed] [Google Scholar]
- 32.Gu, Y., Jin, S., Gao, Y., Weaver, D. T. & Alt, F. W. (1997) Proc. Natl. Acad. Sci. USA 94, 8076–8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, G. C., Ouyang, H., Li, X., Nagasawa, H., Little, J. B., Chen, D. J., Ling, C. C., Fuks, Z. & Cordon-Cardo, C. (1998) Mol. Cell 2, 1–8. [DOI] [PubMed] [Google Scholar]
- 34.Kohl, N. E., Kanda, N., Schreck, R. R., Bruns, G., Latt, S. A., Gilbert, F. & Alt, F. W. (1983) Cell 35, 359–367. [DOI] [PubMed] [Google Scholar]
- 35.Schwab, M., Alitalo, K., Klempnauer, K. H., Varmus, H. E., Bishop, J. M., Gilbert, F., Brodeur, G., Goldstein, M. & Trent, J. (1983) Nature 305, 245–248. [DOI] [PubMed] [Google Scholar]
- 36.Rosenbaum, H., Webb, E., Adams, J. M., Cory, S. & Harris, A. W. (1989) EMBO J. 8, 749–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheppard, R. D., Samant, S. A., Rosenberg, M., Silver, L. M. & Cole, M. D. (1998) Oncogene 17, 2073–2085. [DOI] [PubMed] [Google Scholar]
- 38.Dildrop, R., Ma, A., Zimmerman, K., Hsu, E., Tesfaye, A., DePinho, R. & Alt, F. W. (1989) EMBO J. 8, 1121–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charron, J., Malynn, B. A., Fisher, P., Stewart, V., Jeannotte, L., Goff, S. P., Robertson, E. J. & Alt, F. W. (1992) Genes Dev. 6, 2248–2257. [DOI] [PubMed] [Google Scholar]
- 40.Davis, A. C., Wims, M., Spotts, G. D., Hann, S. R. & Bradley, A. (1993) Genes Dev. 7, 671–682. [DOI] [PubMed] [Google Scholar]
- 41.Malynn, B. A., de Alboran, I. M., O'Hagan, R. C., Bronson, R., Davidson, L., DePinho, R. A. & Alt, F. W. (2000) Genes Dev. 14, 1390–1399. [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmerman, K. A., Yancopoulos, G. D., Collum, R. G., Smith, R. K., Kohl, N. E., Denis, K. A., Nau, M. M., Witte, O. N., Toran-Allerand, D., Gee, C. E., et al. (1986) Nature 319, 780–783. [DOI] [PubMed] [Google Scholar]
- 43.Nisen, P. D., Zimmerman, K. A., Cotter, S. V., Gilbert, F. & Alt, F. W. (1986) Cancer Res. 46, 6217–6222. [PubMed] [Google Scholar]
- 44.Ma, A., Smith, R. K., Tesfaye, A., Achacoso, P., Dildrop, R., Rosenberg, N. & Alt, F. W. (1991) Mol. Cell. Biol. 11, 440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.