Abstract
The malaria parasite lives within erythrocytes and depends on the binding of parasite ligands to host cell surface receptors for invasion. The most virulent human malaria parasite, Plasmodium falciparum, uses multiple ligands, including EBA-175, BAEBL, and JESEBL of the Duffy-binding-like (DBL) family of erythrocyte-binding proteins, for invasion of human erythrocytes. Region II of these parasite ligands is the erythrocyte-binding domain. Previously, we had shown that polymorphism in region II of BAEBL leads to different erythrocyte-binding specificities. We have now identified and characterized the binding specificity of six JESEBL variants. We sequenced region II of JESEBL from 20 P. falciparum clones collected from various parts of the world where malaria is endemic. We observed eight JESEBL variants that contained amino acid polymorphisms at five positions among all clones. Seven of the eight variants could be connected by a single base change that led to an amino acid change. We investigated the functional significance of these polymorphisms by transiently expressing region II from six of JESEBL variants on the surface of Chinese hamster ovary cells. We observed four erythrocyte-binding patterns to enzyme-treated erythrocytes. Thus, P. falciparum DBL ligands JESEBL and BAEBL can recognize multiple receptors on the erythrocyte surface. In contrast to Plasmodium vivax, which has disappeared from West Africa because of the Duffy-negative blood group, P. falciparum may have been successful in endemic areas because it has mutated the ligands of the DBL family to create multiple pathways of invasion, thus making selection of refractory erythrocytes unlikely.
Keywords: parasite polymorphism
All of the clinical manifestations associated with human malaria infection are due to the asexual erythrocytic phase of the Plasmodium life cycle. Although the process by which the parasite enters erythrocytes is complex and not well understood, invasion of erythrocytes by Plasmodium can be divided into multiple steps. Initial attachment of the parasite to the erythrocyte is followed by reorientation to bring the apical end of the parasite into close contact with the surface of the erythrocyte (1). The parasite forms a junction between its apical end and the erythrocyte surface. Junction formation is irreversible and is followed by entry through invagination of the erythrocyte membrane (2). The process of junction formation is mediated by interaction between specific erythrocyte receptors and specific parasite ligands, as indicated by the inability of Plasmodium knowlesi, a simian malaria parasite that can infect humans, to form a junction with human erythrocytes lacking the Duffy blood group antigen and Plasmodium vivax to invade erythrocytes and infect Duffy-negative individuals (3–5).
Unlike P. vivax, which is restricted to the erythrocytes expressing the Duffy blood group antigen and to reticulocytes, invasion of human erythrocytes by Plasmodium falciparum involves a number of apparently redundant receptor–ligand interactions between the merozoite and the erythrocyte surface (6). The first identified P. falciparum ligand was EBA-175 (7). It is orthologous to the P. vivax Duffy-binding protein (DBP), which is involved in the formation of the junction during invasion. The erythrocyte receptor for EBA-175 is glycophorin A (8). Five paralogues of EBA-175 have been identified from the P. falciparum genome. They are all characterized by a 5′ cysteine-rich motif, also called Duffy binding-like domain, or region II, which defines the erythrocyte-binding domain, a 3′ cysteine motif, a transmembrane domain, and a short cytoplasmic tail (6).
Studies aiming at the identification of erythrocyte receptors used by P. falciparum merozoites have made use of mutant erythrocytes deficient in specific surface proteins, as well as enzymes that modify the erythrocyte surface. These studies have revealed that distinct isolates of P. falciparum have varying abilities to use alternative invasion pathways, although many of the erythrocyte receptors are yet to be defined (9–13). On the erythrocyte side, several invasion pathways have been described involving the erythrocyte glycophorins, glycophorin A, and glycophorin C/D, receptor X, receptor Y, and, potentially, glycolipids (8, 14–19).
The diversity of P. falciparum invasion pathways does not rely solely on the number of parasite ligands but also relies on polymorphisms in a single parasite ligand, BAEBL (EBA-140), each of which can recognize a different erythrocyte receptor (20). The polymorphic sites that we described in the F1 region of BAEBL by using our laboratory-maintained isolates were also found in field isolates from Ibadan, Nigeria (21). In addition to these sites, a fifth site was found in the F2 region of BAEBL from those field isolates (21). Previous characterization of P. falciparum ligand JESEBL (also called EBA-181), a paralogue of EBA-175 and BAEBL, demonstrated its erythrocyte-binding specificity to depend on a trypsin-resistant, chymotrypsin-sensitive molecule different from glycophorin B. Furthermore, targeted disruption of the eba-181 gene had no effect on the invasion phenotype of the parasite (22). In this report, we demonstrate that variations in the erythrocyte-binding domain of JESEBL/EBA-181 also change its erythrocyte-binding specificity. We have sequenced the erythrocyte-binding domain of JESEBL from numerous P. falciparum clones from various parts of the world. We have identified a distinct set of nonsynonomous changes in region II of JESEBL and have investigated the functional significance of these nonsynonomous substitutions.
Materials and Methods
Antisera. Antisera to JESEBL region II and region VI of Dd2/Nm were generated by immunization of rats with a DNA vaccine by using the vector VR1050 (kindly supplied by S. Hoffman, Naval Medical Research Institute, Silver Spring, MD) that contains the T cell epitopes P2P30 from tetanus toxin. Regions II and VI gene fragments were amplified from P. falciparum clone Dd2/Nm and were cloned into VR1050 vector, previously described as VR1012tPAP2P30 by Becker et al. (23), but now renamed VR1050. The inserts for regions II and VI of JESEBL/EBA-181 from Dd2/Nm spanned from amino acids Y112-S726 and A1363-T1448, respectively (GenBank accession no. AF332918). Rats were immunized intradermally in the abdomen with 500 μg of DNA at 3-week intervals for a total of four immunizations. Sera were obtained from the rats 1 week after the fourth immunization.
Rabbit anti-region II of EBA-175 was a kind gift of A. Stowers (Malaria Vaccine Development Unit, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda). Mouse anti-RAP-1 monoclonal antibody 7H8/50 [MRA-79, Malaria Research and Reference Reagent Resource (MR4) Center (American Type Culture Collection; Manassas, VA)] was a kind gift of A. Saul (Queensland Institute of Medical Research, Brisbane QLD, Australia).
Immunolocalization of JESEBL/EBA-181. The methods for immunolocalization of JESEBL/EBA-181 by confocal microscopy were performed as described (15), with the following modifications. The blocking buffer consisted of PBS, pH 7.4, containing 0.1% Triton X-100 (Bio-Rad) and 2.5 mg/ml normal goat serum (Jackson ImmunoResearch). The secondary antisera consisted of Alexa 488-conjugated goat anti-rat IgG, Alexa 488-conjugated goat anti-mouse, or Alexa 594-conjugated goat anti-rabbit IgG (Molecular Probes), which were all diluted 1:500 in blocking buffer. For antiquenching, we mounted labeled parasites in Prolong Antifade (Molecular Probes).
Parasite Clones. Parasites used in this study are itemized in Table 1 (24). Genotypes of the parasite lines were confirmed by microsatellite analysis (25). Parasite culture and DNA isolation were performed as described (20).
Table 1. Position of amino acid polymorphisms in JESEBL and location of point mutations.
| Region II
|
|||||
|---|---|---|---|---|---|
| Clones (origin) | 359 | 363 | 414 | 443 | 637 |
| PNG3 (PNG) | K(aag) | D(gat) | I(att) | Q(caa) | K(aaa) |
| ↕ | |||||
| Indochina (Vietnam) | K(aag) | D(gat) | I(att) | Q(caa) | N(aat) |
| ↕ | |||||
| Pc/26 (Peru) | K(aag) | V(gtt) | I(att) | Q(caa) | N(aat) |
| PNG2, PNG5 (PNG) | ↕ | ||||
| DIV30 (Brazil) | |||||
| Dd2, Dd2/Nm (Vietnam) | R(agg) | V(gtt) | I(att) | Q(caa) | N(aat) |
| MT/S-1 | ↕ | ||||
| HB3 (Honduras) | R(agg) | V(gtt) | N(aat) | Q(caa) | N(aat) |
| M24 (Kenya) | ↕ | ||||
| 7G8 (Brazil) | |||||
| T2/C6 (Thailand) | |||||
| Fab9 (Swaziland) | |||||
| PNG4 (PNG) | |||||
| 3D7 (Africa) | R(agg) | V(gtt) | N(aat) | K(aaa) | N(aat) |
| Camp (Malaysia) | ↕ | ||||
| Sc-D6 (Sierra Leone) | |||||
| PC49 (Peru) | R(agg) | V(gtt) | N(aat) | K(aaa) | K(aaa) |
| D10 (PNG) | K(aag) | H(cac) | I(att) | Q(caa) | N(aat) |
Sequence Analysis of jesebl/eba-181 Genes. ORF sequences of jesebl/eba-181 were amplified from genomic DNA. Oligonucleotide primer sequences were TACATAGATATCCAGTTAGT and TCACAGAATTGTGATTTACA. After treatment with shrimp alkaline phosphatase/ExoI (United States Biochemical, Cleveland), PCR products were directly sequenced by using Big Dye terminator chemistry on an ABI3100 sequencer (Applied Biosystems; ref. 15). DNA sequences were aligned by using sequencer 3.1 (Gene Codes, Ann Arbor, MI) or assemblylign software (Oxford Molecular, Madison, WI).
Expression of Region II of JESEBL/EBA-181 on CHO-K1 and COS7 Cells. Region II of JESEBL/EBA-181 was cloned into the pRE4 vector, which contains the gene encoding herpes simplex virus glycoprotein D, under the control of the Rous sarcoma virus LTR promoter in a mammalian expression vector, has been described (26, 27). The 5′ primer 5′-TATTTGAATAGAAATAGTTTTGTTCAAAG-3′ and downstream 3′ primer 5′-AACAAAGAAATGTTATCTATAGACTCA-3′ were used to generate region II from HB3 (RVNQN), D2/Nm (RVIQN), 3D7 (RVNKN), DIV30 (KVIQN), PC49 (RVNKK), and PNG3 (KDIQK) by using the PvuII and ApaI sites, respectively. PCR conditions were 94°C for 2 min, and 30 cycles at 94°C for 50 sec, 55°C for 30 sec, and 72°C for 3 min.
Cell Culture and Transfection of CHO-K1 and COS7 Cells. CHO-K1 cells (American Type Culture Collection) were cultured in RPMI medium1640 with 10% FCS/2% glutamine/1% penicillin-streptomycin/1% Hepes, pH 7.5 (Invitrogen) in a humidified 5% CO2 incubator at 37°C. Cells (1 × 106) were seeded (90% confluency) in 3.5-cm diameter wells and were transfected with 4 μg of plasmid DNA by using the cationic lipid LipofectAMINE 2000, following the manufacturer's protocol (Invitrogen). The transfected cells were used for immunofluorescence or erythrocyte-binding assays 24 h after transfection. COS7 cells (American Type Culture Collection) were cultured in DMEM with 10% FCS/2% glutamine/1% penicillin-streptomycin/1% Hepes (Invitrogen) in 5% CO2 incubator at 37°C. COS7 cells were transfected by CaPO4 as described (27).
Enzyme Treatment of Erythrocytes. Erythrocytes were collected in 10% citrate phosphate dextrose and stored at 4°C. Erythrocytes were washed three times in RPMI medium 1640, pH 7.4, containing 0.05% hypoxanthine (Invitrogen) before use. Erythrocytes at 5% hematocrit were treated twice in 25 units of neuraminidase (Vibrio cholerae; Calbiochem) in RPMI medium 1640 for 2 h at 37°C. Erythrocytes were treated twice with 1 mg/ml trypsin-N-p-tosyl-l-phenylalanine chloromethyl ketone or 1 mg/ml chymotrypsin-Nα-p-tosyl-l-lysine chloromethyl ketone (Sigma) for 2 h at 37°C. The enzymes were removed by washing twice with RPMI medium 1640. Trypsin was inhibited with 1 mg/ml soybean trypsin inhibitor (Sigma) by incubation at room temperature for 15 min. Chymotrypsin was inhibited with 1 mg/ml trypsin-chymotrypsin inhibitor by incubation at room temperature for 15 min (Sigma). Erythrocytes were resuspended to an hematocrit of 10% in complete RPMI medium 1640.
Genetically Deficient Human Erythrocytes. Glycophorin A and glycophorin B null erythrocytes [En(a–) and S–s–U–, respectively] had been frozen and thawed by the Red Cross method (28).
Erythrocyte-Binding Assay on CHO-K1 and COS7 Cells. Fourty microliters of a 10% erythrocyte suspension was added to transfected CHO-K1 cells grown on coverslips. The erythrocytes were allowed to settle for 2 h at 37°C. The cells were washed by inversion on glass support for 20 min in PBS. Transfected CHO-K1 cells were scored by using an inverted microscope. Transfected cells with five or more bound erythrocytes were scored as positive. Transfection efficiency was assayed by an immunofluorescence assay as described below. Binding of untreated and enzyme-treated erythrocytes to untransfected CHO-K1 cells, and to cells expressing CIDR1 in pRE4 (29), a PfEMP1 domain for endothelial cytoadherence, was tested as a negative control. For the immunofluorescence assay, transfected cells were washed in PBS and fixed in 3.7% formaldehyde for 5 min at room temperature. This procedure was followed by three washes in PBS for 1 min each at room temperature. The cells expressing the fusion protein in pRE4 were stained with mouse monoclonal antibodies DL6 and ID3 directed against the herpes simplex glycoprotein D sequences as described (refs. 26 and 27; gifts from R. Eisenberg and G. Cohen, University of Pennsylvania, Philadelphia), and rat polyclonal antisera directed against region II of JESEBL, respectively.
Erythrocyte-Binding Assay Using Radiolabeled Culture Supernatant. Metabolically labeled supernatant from P. falciparum clone HB3 and 3D7 were used in erythrocyte absorption and elution assays as described (15). JESEBEL/EBA-181 was immunoprecipitated by using rat polyclonal antisera directed against region VI as described (15).
Results
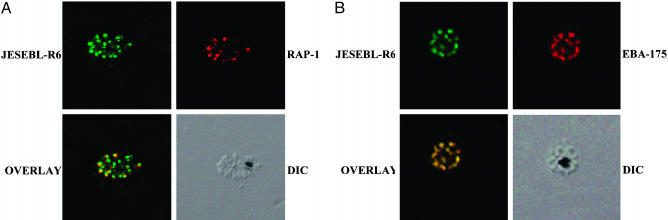
Expression and Subcellular Localization of JESEBL/EBA-181. The apical end of merozoites contains several organelles that are involved in erythrocyte invasion. Molecular markers for two organelles, EBA-175 for micronemes and RAP-1 for rhoptries, were used in this study. Antibodies to the two cysteine-rich domains (regions II and VI) for JESEBL/EBA-181 of P. falciparum 3D7 were used to determine localization and expression of the protein. Immunolocalization of a rhoptry protein, RAP-1, with the two antibodies against regions II (data not shown) and VI show that JESEBL/EBA-181 is adjacent to, but not overlapping with, the rhoptry marker (Fig. 1A). Furthermore, antibodies to regions II (data not shown) and VI localize to the same organelle as does EBA-175 (Fig. 1B). This distribution confirms the localization of JESEBL within micronemes (22), a distribution identical to EBA-175, BAEBL, and the P. knowlesi DBP (15, 19, 30, 31).
Fig. 1.
JESEBL/EBA-181 localizes to the micronemes. (A) Schizonts from P. falciparum 3D7 were double-labeled with anti-JESEBL region VI (JESEBL-R6) and anti-RAP-1, a rhoptry marker. The schizont immunolabeled with anti-JESEBL region VI (JESEBL-R6) has been stained with Alexa 488 (green) secondary antibody. Schizonts labeled with anti-RAP1 have been stained with Alexa 594 (red) secondary antibody. (B) Schizonts from P. falciparum 3D7 were double-labeled with anti-JESEBL region VI (JESEBL-R6) and anti-EBA-175, a microneme marker. The schizonts immunolabeled with anti-JESEBL region VI (JESEBL-R6) have been stained with Alexa 488 (green) secondary antibody. The schizonts labeled with anti-EBA-175 have been stained with Alexa 594 (red) secondary antibody. The unstained schizont is shown by differential interference contrast microscopy. The two labeled proteins are shown in the overlay.
Furthermore, antiserum to region VI immunoprecipitated a protein >175 kDa from radiolabeled culture supernatant that has been immunodepleted of EBA-175 (data not shown), confirming that the antiserum was not cross-reactive with EBA-175, because EBA-175 was not immunoprecipitated from the immunodepleted supernatant.
Sequences of Region II of JESEBL/EBA-175 From Various P. falciparum Clones. JESEBL/EBA-181, like other members of the Duffy-binding-like-erythrocyte-binding protein (DBL-EBP) family, has two cysteine-rich regions: one at the amino terminus and the second at the carboxyl terminus of the molecule. In addition to the signal sequence, it is predicted to have a transmembrane domain and a short cytoplasmic tail. Its exon boundaries are similar to that of BAEBL and EBA-175. Region II of P. falciparum DBL genes, the domain of the protein that binds to the erythrocyte surface, is duplicated, forming the F1 and F2 domains. Region II of BAEBL is more similar to region II of EBA-175 than to JESEBL/EBA-181 (Fig. 2). They both contain a total of 26 cysteine residues: 12 in the F1 domain and 14 in the F2 domain. In contrast, region II of JESEBL/EBA-181 contains five extra cysteine residues. BAEBL and EBA-175 are closer to each other in lineage than they are to JESEBL/EBA-181 (Fig. 2). The deletions and insertions of bases are the same for BAEBL and EBA-175, as compared with JESEBL/EBA-181. We sequenced region II of JESEBL from 20 clones from different parts of the world (GenBank accession nos. AY495335, AY495336, AY496955, AY496956, and AY495679–AY495693). We identified eight polymorphisms in the nucleotide sequence of region II, all leading to changes in amino acids in five positions (Table 1). No synonymous mutations were observed. Three base substitutions in region II of JESEBL/EBA-181 occurred in the F1 domain, and two occurred in the F2 domain (Fig. 2). With the exception of D10, all polymorphisms in JESEBL/EBA-181 can be connected through single-point mutations from one sequence to the next (Table 1). This finding suggests linkage between the sequences and the possibility that the mutations can change the sequence in either direction. The mutation that changes K to R (amino acid 359; Table 1) uses infrequent codons for K (aag; 10.1% codon usage) and R (agg; 12.6% codon usage). This use of infrequent codons again suggests a link between the polymorphisms. The positions of the variations in JESEBL/EBA-181 differ from that of BAEBL and EBA-175. The mutations of JESEBL/EBA-181 start at the carboxyl-terminal region of F1 and go into the F2 region (Fig. 2), whereas the polymorphisms in BAEBL are mostly restricted to the amino-terminal section of the F1 region.
Fig. 2.
Alignment and location of polymorphisms in JESEBL/EBA-181, BAEBL, and EBA-175. Positions of polymorphism in JESEBL/EBA-181 (present study), BAEBL, and EBA-175 are highlighted in black. The F1 and F2 domains are indicated by arrows. Identical amino acids are shown with asterisks, the positions of conserved substitutions are shown with colons, the positions of semiconserved substitutions are shown by periods, and the positions of insertions/deletions are shown with hyphens. The sequence of region II of BAEBL is from the 3D7 clone (GenBank accession no. AF425236). The sequence of region II of JESBEL is from the 3D7 clone (GenBank accession no. AY496955). The sequence of region II of EBA-175 is from the Camp clone (GenBank accession no. X52524).
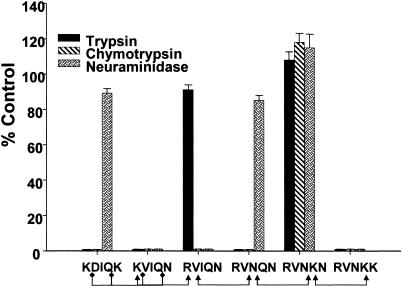
JESEBL Variants Recognize Different Erythrocyte Receptors. We investigated the functional significance of the polymorphisms in the erythrocyte-binding domain (region II) of JESEBL/EBA-181. Region II of six variants of JESEBL/EBA-181 was expressed transiently on the surface of CHO-K1 cells in the pRE4 vector. For all transfected constructs, positive surface expression was confirmed by immunofluorescence with three antibodies: two were directed against the epitope on the amino terminus and carboxyl terminus of the heterologous fusion protein in the vector pRE4 and the third was directed against region II of JESEBL/EBA-181. Binding to CHO-K1 cells was performed with untreated and enzyme-treated (trypsin, chymotrypsin, and neuraminidase) erythrocytes. Four erythrocyte-binding patterns were observed, indicating at least four different erythrocyte receptors. Identical erythrocyte-binding patterns do not indicate recognition of identical erythrocyte receptors. Region II from Dd2 (RVIQN) was sensitive to neuraminidase and chymotrypsin but was resistant to trypsin as described (22). This variant bound to S–s–U–human erythrocytes (ref. 22 and this study), indicating that its erythrocyte receptor was not glycophorin B. At least two other patterns of binding specificity were observed: clone 3D7 (RVNKN) bound trypsin-treated, neuraminidase-treated, and chymotrypsin-treated erythrocytes; and DIV30 (KVIQN) and PC49 (RVNKK) bound to neither neuraminidase-, trypsin-, nor chymotrypsin-treated erythrocytes. Both KVIQN and RVNKK bound to En (a–) indicating glycophorin A was not the erythrocyte receptor for these two variants.
HB3 (RVNQN) expressed on CHO-K1 cells did not bind to untreated or enzyme-treated erythrocytes. HB3 (RVNQN) variant expressed on the surface of CHO-K1 did not react with antiserum against region II of JESEBL. Region II failed to react, despite the fact that the heterologous fusion protein was expressed at high levels on the surface of these cells, as determined by the two antibodies to the amino and carboxyl termini of the heterologous fusion protein. Sequence analysis of the pRE4 plasmid confirmed that there were no mutations in region II. In contrast to the studies with the RVNQN variant, the anti-JESEBL region II antiserum reacted to the surface of CHO-K1 cells transfected with all other constructs described above.
To understand the absence of reactivity of antiserum to region II or binding of erythrocytes to the HB3 (RVNQN) variant in CHO-K1 cells, we expressed the variant in COS7 cells and probed for surface expression by using the antiserum directed against region II of JESEBL/EBA-181. Unlike the expression on CHO-K1 cells, the RVNQN variant was detected on the surface of COS7 cells by using the antiserum against region II of JESEBL/EBA-181. The RVNQN variant expressed in COS7 cells bound human erythrocytes. This variant bound to neuraminidase-treated, but not to trypsin- or chymotrypsin-treated erythrocytes (Fig. 3). Two possible reasons for the variation in expression of region II of the RVNQN variant between COS7 and CHO-K1 cells could be due to differences in the folding or other posttranslational modifications.
Fig. 3.
JESEBL/EBA-181 variants recognize different erythrocyte receptors. Region II of JESEBL/EBA-181 expressed in CHO-K1 cells or COS7 (RVNQN) cells contains the mutations shown at the positions delineated in Table 1. Transfected cells with five or more attached erythrocytes were counted, and the total per coverslip was recorded. The transfection efficiency of all constructs into CHO-K1 cells was 80–90%. Data are shown as the mean of three independent experiments, and the error bar denotes the SE. Data from enzyme-treated erythrocytes are expressed as the percentage of binding to untreated erythrocytes. The control normal untreated erythrocyte samples contained between 100 and 198 cells with bound erythrocytes. Arrowheads and diamonds represent point mutations (see Table 1).
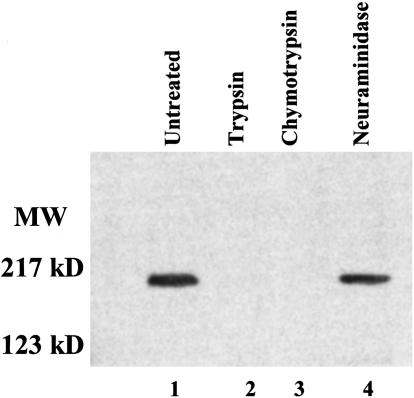
We next determined whether the specificity of binding of the RVNQN variant of JESEBL/EBA-181 observed on COS7 cells mirrored the binding of the native protein in culture supernatants. JESEBL/EBA-181 from culture supernatants of HB3 bound to human erythrocytes with the identical specificity as that described above for region II of RVNQN expressed on COS7 cells, namely, binding to neuraminidase-treated erythrocytes but not to trypsin- or chymotrypsin-treated erythrocytes (Fig. 4). This finding indicates that the two assays are measuring the same specificity. Thus, like Duffy of P. knowlesi and P. vivax (27), and BAEBL and EBA-175 of P. falciparum (8, 15), region II of JESEBL/EBA-181 defines the erythrocyte-binding specificity of the native protein [RVNQN-HB3 and RVNKN-3D7 (CHO-K1 cells and data not shown for binding of native protein)].
Fig. 4.
Erythrocyte-binding specificity of RVNQN variant from JESEBL. Lane 1, immunoprecipitate of eluate of untreated human erythrocytes. Lane 2, immunoprecipitate of eluate of trypsin-treated human erythrocytes. Lane 3, immunoprecipitate of eluate of chymotrypsin-treated human erythrocytes. Lane 4, immunoprecipitate of eluate of neuraminidase-treated human erythrocytes.
We showed that a single base change leading to a single amino acid substitution in each of the five positions in region II of JESEBL led to recognition of different molecules on human erythrocytes (KDIQK to KVIQN to RVIQN to RVNQN to RVNKN and RVNKK).
Discussion
Region II of JESEBL/EBA-181 from numerous P. falciparum clones isolated worldwide contained distinct nonsynonymous substitutions at five positions. These substitutions led to changes in amino acids that gave rise to eight JESEBL/EBA-181 polymorphisms. With one exception, the variants seemed to have arisen by single-point mutations that can be linked (Table 1). As in the case of BAEBL, a single-point mutation in each of the five positions of JESEBL/EBA-181 modifies its erythrocyte-binding specificity. Therefore, in addition to the multiple copies of DBL genes, P. falciparum has evolved another mechanism for recognition of different molecules on the erythrocytes by creating various forms of a single P. falciparum erythrocyte-binding ligand.
Region II of P. falciparum is duplicated and is referred to as F1 and F2 (Fig. 2). The location of the changes in the polymorphic sites of JESEBL/EBA-181 does not overlap with those of BAEBL, which are mostly restricted to the amino terminus of region II. Polymorphisms in JESEBL/EBA-181 are located at three positions in the carboxyl terminus of the F1 domain and at two positions in the F2 domain. The amino acid changes that we have noted in JESEBL/EBA-181 all led to changes in receptor specificity and may be located in the receptor pocket. However, the structure of region II of any of the DBL family of EBPs (DBL-EBP) has yet to be solved.
The DBL-EBP members are assumed to have the same function, based on primary structure (including intron/exon structure), location in merozoites, and the capability to bind to human erythrocytes. The inability of P. vivax and P. knowlesi to invade human Duffy-negative erythrocytes correlated with the failure of the DBP to bind to Duffy-negative human erythrocytes. The P. knowlesi merozoites attach to Duffy-negative erythrocytes and reorient apically, but no junction is formed with Duffy-negative human erythrocytes and invasion fails to proceed (4). P. knowlesi DBP was shown to localize to micronemes at the apical end of the merozoite (31). Likewise, EBA-175 of P. falciparum localizes to the same organelle (30). By confocal microscopy, BAEBL and JESEBL also appear to localize in the micronemes. These three P. falciparum molecules bind to human erythrocytes, as in the case of P. knowlesi and P. vivax. By analogy with P. knowlesi, they are assumed to be involved in junction formation during erythrocyte invasion.
Multiple nonsynonymous mutations were identified throughout region II in the DBP of P. vivax (32) and in EBA-175 of P. falciparum (21, 33). These polymorphisms did not change the erythrocyte-binding specificity (32, 33). In contrast, each point mutation in BAEBL and JESEBL led to the recognition of different receptors on the erythrocyte. Why are there fewer mutations in region II of BAEBL and JESEBL and why do they all affect their erythrocyte-binding specificities? It is possible that neither JESEBL nor BAEBL play a critical role in erythrocyte invasion. We also speculate that both JESEBL and BAEBL are less exposed to the immune system, or that numerous mutations in region II of both molecules would lead to defective parasite ligands.
Why has P. falciparum evolved so many pathways for invasion? The erythrocyte surface molecules are highly polymorphic within the human population (34–38). It has been reported that the fastest evolving genes in the human lineage are the glycophorins A and B (39). EBA-175 that binds glycophorin A may have driven this polymorphism. There are at least 40 blood types that are caused by variations in glycophorin A and B (40), many of which occur in Africa. Gerbich-negative polymorphism of glycophorin C occurs in 50% of the population of a malaria endemic region of Papua, New Guinea (41, 42) and is the erythrocyte receptor for one variant of BAEBL (15, 20). The erythrocyte receptors for JESEBL/EBA-181 have not been identified. Polymorphisms in the erythrocyte-binding domain of JESEBL/EBA-181 may provide the parasite population with an advantage in human populations.
There are no known mutations that change the erythrocyte receptor specificity of P. falciparum EBA-175 or P. vivax DBP, and yet, for BAEBL and JESEBL/EBA-181, each variant bound to a different erythrocyte receptor. P. vivax did not change its receptor specificity and disappeared from West Africa, where the population became almost 100% Duffy-negative. A mutation in P. vivax that would have changed its receptor specificity has yet to be identified. Similarly, there are no known mutations in EBA-175 of P. falciparum that alter its specificity for sialic acid. Both orthologues require the peptide backbone of their respective erythrocyte receptors for binding, although there is a posttranslational modification involved in the binding site for both (e.g., sialic acid for glycophorin A). Their specificity for peptides may have limited their ability to switch receptors.
The best example of a point mutation that led to a novel receptor was the influenza hemagglutinin, where a mutation in the hemagglutinin receptor pocket led to recognition of a new sugar linkage (sialic acid α2,3Gal to sialic acid α2,6Gal) (43). The receptor for one of the BAEBL variants is glycophorin C/D, based on its inability to bind Gerbich-negative (15, 16) and glycophorin C/D-negative erythrocytes (17). On human erythrocytes carrying the Gerbich-negative blood group antigen, glycophorin C is missing part of its peptide backbone. In addition, it contains an abnormal N-linked glycan (44). It is possible that the N-linked glycan on glycophorin C is the receptor for BAEBL, because a point mutation in BAEBL is unlikely to recognize an unrelated protein. Although speculative, variation in recognition of N-linked sugar glycans or other carbohydrate residues by BAEBL will better describe some of its variants and those of JESEBL/EBA-181. This conclusion could explain why point mutations in P. vivax DBP, which recognizes a peptide in the Duffy blood group protein could not adjust to the growing frequency of absence of the Duffy blood group protein in West Africa.
Acknowledgments
We thank Drs. Susan K. Pierce and Karl B. Seydel for scientific discussion, Drs. Gary Cohen and Rosalind Eisenberg for the gift of monoclonal antibodies ID3 and DL6, and Drs. Owen Schwartz and Juraj Kabat for assistance with confocal microscopy.
Abbreviations: DBP, Duffy-binding protein; DBL, Duffy-binding-like; JESEBL/EBA-181 and BAEBL, Plasmodium falciparum DBL ligands; EBP, erythrocyte-binding protein.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY495679–AY495693, AY495335, AY495336, AY496955, and AY496956).
References
- 1.Dvorak, J. A., Miller, L. H., Whitehouse, W. C. & Shiroishi, T. (1975) Science 187, 748-749. [DOI] [PubMed] [Google Scholar]
- 2.Aikawa, M., Miller, L. H., Johnson, J. & Rabbege, J. (1978) J. Cell Biol. 77, 72-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller, L. H., Mason, S. J., Clyde, D. F. & McGinniss, M. H. (1976) N. Engl. J. Med. 295, 302-304. [DOI] [PubMed] [Google Scholar]
- 4.Miller, L. H., Aikawa, M., Johnson, J. G. & Shiroishi, T. (1979) J. Exp. Med. 149, 172-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnwell, J. W., Nichols, M. E. & Rubinstein, P. (1989) J. Exp. Med. 169, 1795-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams, J. H., Kaneko, O., Blair, P. L. & Peterson, D. S. (2001) Trends Parasitol. 17, 297-299. [DOI] [PubMed] [Google Scholar]
- 7.Camus, D. & Hadley, T. H. (1985) Science 230, 553-556. [DOI] [PubMed] [Google Scholar]
- 8.Sim, B. K. L., Chitnis, C. E., Wasniowska, K., Hadley, T. J. & Miller, L. H. (1994) Science 264, 1941-1944. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell, G. H., Hadley, T. J., McGinnis M. H., Klotz F. W. & Miller L. H. (1986) Blood 67, 1519-1521. [PubMed] [Google Scholar]
- 10.Dolan, S. A., Miller, L. H. & Wellems, T. E. (1990) J. Exp. Med. 86, 618-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolan, S. A., Proctor J. L., Alling, D. W., Okubo, Y., Wellems T. E. & Miller L. H. (1994) Mol. Biochem. Parasitol. 64, 55-63. [DOI] [PubMed] [Google Scholar]
- 12.Okoyeh, J. N., Pillai, C. R. & Chitnis, C. E. (1999) Infect. Immun. 67, 5784-5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baum J., Pinder, M. & Conway, D. J. (2003) Infect. Immun. 71, 1856-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pasvol, G. J., Wainscoat, J. S & Weatherall, D. J. (1982) Nature 297, 64-66. [DOI] [PubMed] [Google Scholar]
- 15.Mayer, D. C. G., Kaneko, O., Hudson-Taylor, D. E., Reid, M. E. & Miller, L. H. (2001) Proc. Natl. Acad. Sci. USA 98, 5222-5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maier, A. G., Duraisingh, M. T., Reeder, J. C., Patel, S. S., Kazura, J. W., Zimmerman P. A. & Cowman, A. F. (2003) Nat. Med. 9, 87-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lobo C. A., Rodriguez, M., Reid, M. E. & Lustigman, S. (2003) Blood 101, 4628-4631. [DOI] [PubMed] [Google Scholar]
- 18.Rayner, J. C., Vargas-Serrato, E., Huber, C. S., Galinski, M. R. & Barnwell, J. W. (2001) J. Exp. Med. 194, 1571-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson, J. K., Triglia, T., Reed, M. B. & Cowman, A. F. (2001) Mol. Microbiol. 41, 47-58. [DOI] [PubMed] [Google Scholar]
- 20.Mayer, D. C. G., Mu, J. B., Feng, X., Su, X. & Miller, L. H. (2002) J. Exp. Med. 196, 1523-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baum, J., Thomas, A. W. & Conway, D. J. (2003) Genetics 163, 1327-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilberger, T. W., Thompson J. K., Triglia, T., Good, R. T., Duraisingh, M. T. & Cowman, A. F. (2003) J. Biol. Chem. 278, 14480-14486. [DOI] [PubMed] [Google Scholar]
- 23.Becker, S. I., Wang, R., Hedstrom, R. C., Aguiar, J. C., Jones, T. R., Hoffman, S. L. & Gardner, M. J. (1998) Infect. Immun. 66, 3457-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mu, J., Ferdig, M. T., Feng, X., Joy, D. A., Duan, J., Furuya, T., Subramanian, G., Aravind, L., Cooper, R. A., Wooton, J. C., et al. (2003) Mol. Microbiol. 49, 977-989. [DOI] [PubMed] [Google Scholar]
- 25.Su, X. Z., Carucci, D. J. & Wellems, T. E. (1998) Exp. Parasitol. 89, 262-265. [DOI] [PubMed] [Google Scholar]
- 26.Cohen, G. H., Wilcox, W. C., Sodora, D. L., Long, D., Levin, J. Z. & Eisenberg, R. J. (1988) J. Virol. 62, 1932-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chitnis, C. E. & Miller, L. H. (1994) J. Exp. Med. 180, 497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mallory, D., ed. (1993) in Immunohematology Methods and Procedures (American Red Cross, National Reference Laboratory, Rockville, MD), pp. 125.1-125.2.
- 29.Baruch, D. I., Gamain, B. & Miller, L. H. (2003) Infect. Immun. 71, 4536-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sim, B. K., Toyoshima, T., Haynes, J. D. & Aikawa, M. (1992) Mol. Biochem. Parasitol. 51, 157-159. [DOI] [PubMed] [Google Scholar]
- 31.Adams, J. H., Hudson, D. H., Torii, M., Ward, G. E., Wellems, T. E., Aikawa, M. & Miller, L. H. (1990) Cell, 63, 142-153. [DOI] [PubMed] [Google Scholar]
- 32.Xainli, J., Adams, J. H. & King, C. L. (2000) Mol. Biochem. Parasitol. 111, 253-256. [DOI] [PubMed] [Google Scholar]
- 33.Liang, H. & Sim, B. K. (1997) Mol. Biochem. Parasitol. 84, 241-245. [DOI] [PubMed] [Google Scholar]
- 34.Mourant, A. E., Tills, D., Kopec, A. C., Warlow A., Teesdale, P., Booth, P. B. & Hornabrook, R. W. (1981) Hum. Genet. 59, 77-80. [DOI] [PubMed] [Google Scholar]
- 35.Tills, D., Warlow, A., Kopec, A. C., Fridriksson, S. & Mourant, A. E. (1982) Ann. Hum. Biol. 9, 507-520. [DOI] [PubMed] [Google Scholar]
- 36.Tills, D., Kopec, A. C., Warlow, A., Barnicot, N. A., Tills, D., Kopec, A. C., Marin, A., Bennet, F. J. & Woodburn, J. C. (1982) Hum. Genet. 61, 52-59. [DOI] [PubMed] [Google Scholar]
- 37.Tills, D., Kopec, A. C., Fox, R. F. & Mourant, A. E. (1979) Hum. Hered. 29, 172-176. [DOI] [PubMed] [Google Scholar]
- 38.Tills, D., Teesdale, P. & Mourant, A. E. (1977) Ann. Hum. Biol. 4, 23-24. [DOI] [PubMed] [Google Scholar]
- 39.Wang, H. Y., Tang, H., Shen, C. K. J. & Wu, C. I. (2003) Mol. Biol. Evol. 20, 1795-1804. [DOI] [PubMed] [Google Scholar]
- 40.Blumenfeld, O. O. & Huang, C. H. (1995) Hum. Mutat. 6, 199-209. [DOI] [PubMed] [Google Scholar]
- 41.Booth, P. B., Tills, D., Warlow, A., Kopec, A. C., Mourant, A. E., Teesdale, P. & Hornabrook, R. W. (1982) Hum. Hered. 32, 385-403. [DOI] [PubMed] [Google Scholar]
- 42.Patel, S. S., Mehlotra, R. K., Kastens, W., Mgone, C. S., Kazura, J. W. & Zimmerman, P. A. (2001) Blood 98, 3489-3491. [DOI] [PubMed] [Google Scholar]
- 43.Rogers, G. N., Paulson, J. C., Daniels, R. S., Shekel, J. J., Wilson, I. A. & Wiley, D. C. (1983) Nature 304, 76-78. [DOI] [PubMed] [Google Scholar]
- 44.Reid, M. E. & Spring, F. A. (1994) Trans. Med. 4, 139-146. [DOI] [PubMed] [Google Scholar]