Abstract
The ubiquitous bacterium Pseudomonas aeruginosa is the quintessential opportunistic pathogen. Certain isolates infect a broad range of host organisms, from plants to humans. The pathogenic promiscuity of particular variants may reflect an increased virulence gene repertoire beyond the core P. aeruginosa genome. We have identified and characterized two P. aeruginosa pathogenicity islands (PAPI-1 and PAPI-2) in the genome of PA14, a highly virulent clinical isolate. The 108-kb PAPI-1 and 11-kb PAPI-2, which are absent from the less virulent reference strain PAO1, exhibit highly modular structures, revealing their complex derivations from a wide array of bacterial species and mobile elements. Most of the genes within these islands that are homologous to known genes occur in other human and plant bacterial pathogens. For example, PAPI-1 carries a complete gene cluster predicted to encode a type IV group B pilus, a well known adhesin absent from strain PAO1. However, >80% of the PAPI-1 DNA sequence is unique, and 75 of its 115 predicted ORF products are unrelated to any known proteins or functional domains. Significantly, many PAPI-1 ORFs also occur in several P. aeruginosa cystic fibrosis isolates. Twenty-three PAPI ORFs were mutated, and 19 were found to be necessary for full plant or animal virulence, with 11 required for both. The large set of “extra” virulence functions encoded by both PAPIs may contribute to the increased promiscuity of highly virulent P. aeruginosa strains, by directing additional pathogenic functions.
The versatile and ubiquitous bacterium Pseudomonas aeruginosa is the quintessential opportunistic pathogen, because it can infect a broad range of hosts, from amoeba to humans (1, 2), where it is found associated with severe burns, cystic fibrosis (CF), AIDS, and cancer (3). This pathogen produces an arsenal of virulence factors (4), and displays a remarkable range of virulence, from weakly virulent isolates, to isolates that infect just a few organisms, to highly virulent broad spectrum strains, exemplified by the clinical isolate PA14 (2, 5, 6). We are interested in identifying the genomic basis of this promiscuity to better understand both the P. aeruginosa mechanisms of pathogenesis and defense and the origin, evolution, and utilization of these mechanisms by other infectious microorganisms.
Many bacterial genomes are arranged in blocks of core sequences and genomic islands (7–9). Genomic islands typically greatly differ in their G+C content, and encode a variety of accessory activities that underlie specializations such as symbiotic and pathogenesis functions. Different genes carried by a single island often have diverse origins, and blocks are built piecemeal through insertion and deletion events. Genomic islands are acquired and exchanged by lateral gene transfer, and can be found in widely divergent species, making it difficult to ascribe their initial origins. Pathogenicity islands are specialized genomic islands that encode virulence factors. Their recent characterization in a wide range of pathogenic bacteria has led to the identification of virulence factors used by these species to infect their respective hosts (8, 10–13).
P. aeruginosa possesses a large and diverse genome that is both highly conserved and plastic (14–17). Although clinical and environmental isolates have a remarkably conserved genome (14–19), distinct P. aeruginosa strains carry additional specific sequences, interspersed as genomic blocks in the conserved core genome, that account for 10% or more of their DNA (14, 18). To date, no distinct P. aeruginosa genomic island that carries virulence genes has been identified, with the exception of a recently identified genomic island that harbors the exoU virulence gene (14–17, 19, 20). Here we present the identification and characterization of two P. aeruginosa pathogenicity islands (PAPI-1 and PAPI-2) in the genome of the broad host range and highly virulent strain PA14. The large PAPI-1 island is absent from the reference strain PAO1, whereas only part of the smaller PAPI-2 element is found in this isolate. Although PA14 and PAO1 share the same host range, PA14 is considerably more virulent in several model organisms (2, 6). Most of the predicted proteins encoded by the PAPI genes exhibit no homology with any proteins of known function, and mutation of several of these genes reveals their relevance in both plant and animal pathogenicity. We speculate that PAPI-1 and PAPI-2 virulence functions promote the broad host promiscuity of highly virulent P. aeruginosa strains, such as PA14.
Materials and Methods
Strains, Plasmids, and Media. All P. aeruginosa strains are human isolates (6, 15, 16). The TnphoA mutant 33A9 has been described (21). The PA14 genomic cosmid library, constructed in pJSR (6), was grown in Escherichia coli VCS257, and subcloned in DH5α. pRK2013 and pEX18Ap (22) served as the P. aeruginosa conjugation helper plasmid and marker exchange suicide vector, respectively. Bacteria were grown in LB plus 100 μg/ml ampicillin (E. coli), 100 μg/ml rifampicin (PA14), and 250 μg/ml carbenicillin (PA14 transconjugants).
DNA Methods and Library Construction. Probes were labeled with [32P]dCTP (NEN) by using Rediprime II (Amersham Pharmacia). Genomic, cosmid, and plasmid DNA extractions followed standard procedures (23). To construct the PA14 cosmid library, a 30- to 50-kb partial Sau3AI digest of total PA14 DNA was size-fractionated in a 10–40% sucrose gradient, cloned into the BamHI site of pJSR, and packaged by using gigapack iii xl (Stratagene).
PA14 Mutants. In-frame deletions in 10 PAPI-1 ORFs and one PAPI-2 ORF were generated by PCR: 1.0- to 1.6-kb 5′ and 3′ segments were amplified from target PA14 genomic or cosmid DNA, and each amplicon, which included the first or last 10–20 aa of the target ORF plus an engineered restriction site, was ligated into pEX18Ap to produce replacement plasmids. Inframe deletion mutants were generated in PA14 via homologous recombination by sucrose resistance selection, and confirmed by hybridization.
Twelve TnphoA transposon insertion mutants of PA14 were obtained from a library that includes transposon–insertion mutants of all nonessential PA14 ORFs. Access to mutants and information about this library is currently available via a web interface (Massachusetts General Hospital Parabiosys: National Heart, Lung, and Blood Institute Program for Genomic applications, Massachusetts General Hospital and Harvard Medical School; http://pga.mgh.harvard.edu/cgi-bin/pa14/mutants/retrieve.cgi).
To evaluate pyocyanin production, bacteria were cultivated in King's A broth and the relative phenazine concentration was measured at 695 nm. Pyoverdine concentrations in culture supernatants grown in King's B medium were measured at 380 nm. Total proteolytic activity was estimated by spotting the bacteria onto skim milk agar plates (24) and measuring the zone of hydrolysis produced after 24-h incubation at 37°C. Swimming, twitching, and swarming motilities were assessed as described (25). Biofilm formation was tested by using crystal violet assay with modifications (25).
Plant and Mouse Pathogenicity Studies. Mouse mortality and Arabidopsis thaliana (ecotype Col-1) plant infection studies were as described (6, 21).
DNA Sequencing and Annotation. The nucleotide sequences of the PA14 pI48, pG22, pSK91, pSK24, pF62, and pH44 cosmids were determined by shotgun sequencing. Cosmid fragments subcloned into pBluescript SK(-) and pDN19 were sequenced by primer walking to cover gaps. Individual reads were aligned and assembled by using DNAstar and CAP (http://pbil.univlyon1.fr/cap3.html). The sequence was compared to the PAO1 annotated genome (www.pseudomonas.com) (26). tRNA genes were identified by using trna-sacn-se (www.genetics.wustl.edu/eddy/tRNAscan-SE). ORFs were predicted by using genemark.hmm (http://opal.biology.gatech.edu/GeneMark/gmhmm2_prok.cgi) (27) and ORF finder (www.ncbi.nlm.nih.gov/gorf/gorf.html). Each predicted ORF of >200 bp was analyzed for homologies and conserved motifs by using blastn, blastp, and blastx. A full array of parameters was used. psort (http://psort.nibb.ac.jp/form.html) and tmpred (www.ch.embnet.org/software/TMPRED_form.html) were used to predict cellular localization and transmembrane regions, respectively.
Sequence Accession. GenBank accession numbers for PAPI-1 and PAPI-2 are AY273869 and AY273870, respectively.
Results
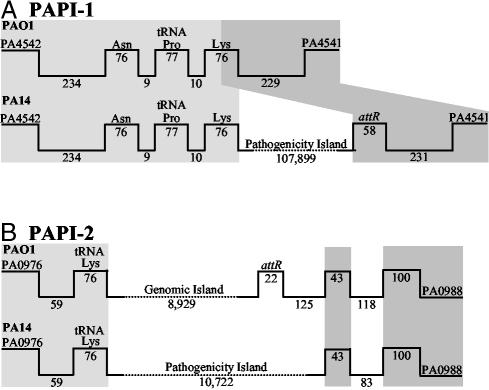
RL003 Defines Two Genomic Islands Present in PA14 and Absent from PAO1. The PA14 isogenic TnphoA insertion mutant 33A9, which carries an RL003 gene mutation, exhibits reduced plant and mouse pathogenicity (21). Because RL003 is absent from PAO1, suggesting that it might occur within a P. aeruginosa pathogenicity island, we screened a PA14 cosmid library with a 300-bp RL003 probe. Initial results with the cosmids pA113, pB104, pI48, pH44, and pG68 showed that only pA113, pB104, and pI48 overlap. Although both borders of pH44 and pG68 contain PAO1 sequences, only the left borders of pA113, pB104, and pI48 carry PAO1 DNA, indicating that RL003 occurs in at least two sites in the PA14 genome, one of which includes a large genomic block.
To further define this block, we performed a progressive cosmid walk, starting with a pI48 probe that contains the PAO1/PA14 left junction, and identified a cosmid carrying the right PA14/PAO1 junction (Fig. 4A, which is published as supporting information on the PNAS web site). A set of five cosmid clones, pI48, pG22, pSK91, pSK24, and pF62, were assembled to define a contiguous 150-kb region, designated PAPI-1, found in PA14 and absent from PAO1 (Figs. 1A and 4A). Similarly, we used probes that correspond to the right and left borders of pH44 to confirm that this cosmid does not overlap PAPI-1, demonstrating that a second copy of RL003 occurs on a smaller PA14 genomic block, designated PAPI-2 (Figs. 1B and 4B).
Fig. 1.
Correspondence of the PA14 PAPI-1 (A) and PAPI-2 (B) elements with the PAO1 genome. Gene designations and linear coordinates (bp) are presented above and below the lines, respectively. Light and dark gray shading represent the conserved left and right boundaries, respectively. The figure is not drawn to scale.
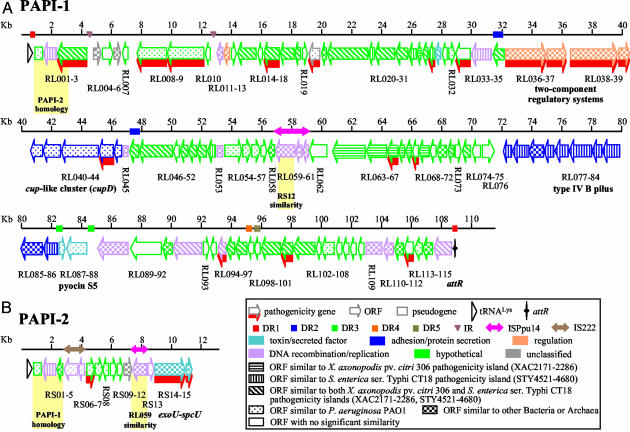
PAPI-1 Is a Pathogenicity Island. Comparison of the nucleotide sequence of the region defined by the five PAPI-1 cosmids (annotated in Fig. 2A and in Table 2, which is published as supporting information on the PNAS web site) with the PAO1 genome (26), shows that the 20-kb left end of pI48 (GenBank accession no. AY273871) is collinear with the PAO1 genome, whereas the internal 107,899 bp are unique to PA14 (Figs. 1 A and 2 A). This 108-kb region has all of the features of a genomic island: it occupies a block absent from several P. aeruginosa strains); its G+C content (59.7%) is different from that of the core genome (66.6%); it is associated with tRNA genes, because a tRNAAsn, tRNAPro, and tRNALys gene cluster (annotated as PA4541.1–3 in PAO1) occurs at its leftward PAO1/PA14 junction, and a 58-bp direct repeat of the 3′ half of the tRNALys gene, designated attR, occurs just within its right border, so that it is bounded by 58-bp direct repeats (Figs. 1 and 2 A); it contains seven mobility factor genes that encode integrases and transposases, plus four related pseudogenes and direct and inverted repeat sequences (Table 3, which is published as supporting information on the PNAS web site); it appears to have undergone deletions in different P. aeruginosa strains and/or additional insertions have occurred in PA14 (Fig. 3 and described below); and finally, it carries at least 19 virulence factors that occur on genomic islands found in a wide spectrum of other pathogenic bacteria (Tables 1 and 2).
Fig. 2.
The organization of PAPI-1 (A) and PAPI-2 (B). The boxes with arrows represent individual ORFs and their corresponding transcriptional orientations. Empty boxes represent pseudogenes; triangles represent tRNA genes; and the marked vertical line represents the presumptive attR “attachment” site. The numbered lines represent size (kb), and the coincident rectangles and single or double-headed arrows on the line respectively correspond to direct repeats (DR1–5), inverted repeat (IR), and insertion sequences. ORF color and pattern, respectively, correspond to the predicted protein function and the bacterial species that it is most related to, according to the key. Pathogenesis-related ORFs are indicated by red shading. Gene clusters functions are marked and correspond to the ORFs above the notations. The shaded regions show the homology between PAPI-1 and PAPI-2.
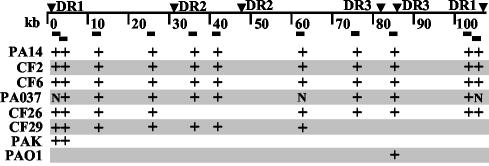
Fig. 3.
Presence of PAPI-1 in P. aeruginosa clinical isolates. The thick black line represents the PAPI-1 coordinates (kb). The arrowheads indicate the position of the DR. The black rectangles correspond to the probes used for hybridization. + denotes positive hybridization; N denotes experiment not done. All strains giving positive hybridization are shown.
Table 1. Mutations in PAPI ORFs produce attenuated animal and plant pathogenicity versus the PA14 parent strain.
| Strain name* | % Mouse mortality†§ | Growth in Arabidopsis leaf‡§ | Closest published homologue (organism/GenBank accession no.) |
|---|---|---|---|
| PA14 | 100 | 4.9 × 106 | Wild–type |
| RL003¶ | 41 | 2.3 × 105 | P. syringae pv. tomato DC3000/AAO54371 |
| RL003R | 95 | 4.4 × 106 | The mutated RL003 locus was replaced with its wild-type copy |
| RL008¶ | 38 | 4.1 × 106 | Methanosarcina acetivorans C2A/AAM05538 and P. aeruginosa PAO1/AAG05323 |
| RL009¶ | 31 | 1.3 × 104 | P. aeruginosa PAO1/AAG05327 |
| RL009R | 100 | 4.0 × 106 | The mutated RL009 locus was replaced with its wild-type copy |
| RL016¶ | 100 | 2.8 × 104 | P. syringae pv. tomato DC3000/AAO54383 |
| RL020∥ | 50 | 3.4 × 105 | protein-disulfide isomerase, P. aeruginosa PAO1/AAG04371 |
| RL022¶ | 88 | 3.3 × 106 | P. syringae pv. tomato DC3000/AAO54394 |
| RL029∥ | 38 | 9.4 × 104 | P. aeruginosa C/AAN62148 |
| RL033∥ | 25 | 4.9 × 104 | No significant similarity |
| RL036¶ | 44 | 1.9 × 105 | Two-component sensor P. aeruginosa PA14/AAM15532 |
| RL037¶ | 43 | 1.2 × 105 | Two-component regulator pvrR, P. aeruginosa PA14 AAM15533 |
| RL038¶ | 31 | 4.4 × 104 | Two-component sensor rcsC, S. typhimurium LT2/AAL21172 |
| RL039¶ | 31 | 2.7 × 105 | Two-component regulator rcsB, E. coli O157:H7 EDL933/AAG57352 |
| RL043∥ | 75 | 1.7 × 106 | Probable pili assembly chaperone cupA2, P. aeruginosa PAO1/AAG05517 |
| RL054∥ | 63 | NT | P. aeruginosa PAO1/AAG05610 |
| RL062∥ | 78 | NT | No significant similarity |
| RL065∥ | 63 | 4.5 × 105 | X. axonopodis pv. citri 306/AAM37094 |
| RL068∥ | 56 | 2.6 × 105 | No significant similarity |
| RL090∥ | 67 | 2.7 × 104 | No significant similarity |
| RL092¶ | 88 | 3.9 × 106 | Topoisomerase I TopA, Xyella fastidiosa 9a5c (plasmid pXF51)/AAF85572 |
| RL095∥ | 50 | 5.3 × 105 | Single-stranded DNA binding protein Ssb, P. aeruginosa C/AAN62318 |
| RL101∥ | 38 | 1.8 × 106 | Pseudomonas sp. B13/CAD60668 |
| RL112∥ | 38 | 1.6 × 104 | No significant similarity |
| RS06¶ | 100 | 1.8 × 105 | P. aeruginosa PAO1/AAG04369 |
NT, not tested.
Strain designations correspond to the mutated ORFs (see Fig. 2)
Mice were infected with 5 × 105 bacterial cells as described, and animal mortality (7) was scored for up to 10 days. Eight to 16 mice were used per experiment
To assess plant pathogenicity, Arabidopsis leaves were inoculated with 3.3 × 105 bacterial cells and assayed 4 days after infection for bacterial CFU/cm2. Four different leaves were sampled. All experiments were performed twice
Statistical significance for mortality data and bacterial growth in Arabidopsis leaves was determined by the t test. Differences between groups were considered statistically significant at P ≤ 0.05 and are shown in bold
In-frame deletion mutants
Transposon-generated mutants
Functional Organization and Predicted ORFs of PAPI-1. Data in Fig. 2 and in Table 2 illustrate the highly modular organization and complex origin of PAPI-1, which is inserted in a hypervariable region of the P. aeruginosa genome near the pilA gene (14, 15, 18). Remarkably, >80% of its DNA sequence is unique and has no similarity with any GenBank sequences. Furthermore, 75 of its 115 predicted ORFs are unrelated to any previously identified proteins or functional domains, and thus cannot be assigned any function by homology.
Conversely, 40 PAPI-1 ORF-translated sequences show homology to proteins from several bacterial species, demonstrating the modular evolution of PAPI-1. For instance, 18 PAPI-1 genes display significant homology to pathogenicity-related genes, including a putative type III effector (RL030), a type IVB-like pilus gene cluster (RL077–86), and a chaperone/usher pathway (cup) gene cluster (RL040–44) (Fig. 2 and Table 2, and described below). The majority of the remaining related PAPI-1 genes encode functions related to DNA mobilization, integration, and partition activities. Many PAPI-1 predicted proteins are related to sequences found in Salmonella, pathogenic E. coli, Haemophilus somnus, Yersinia pestis, P. aeruginosa, Pseudomonas syringae, Pseudomonas fluorescens, Xylella fastidiosa, Burkholderia fungorum, and Xanthomonas (Table 2 and Table 4, which is published as supporting information on the PNAS web site). Also, 26 PAPI-1 ORF translated sequences are similar to predicted proteins on both the 134-kb island of the mammalian pathogen Salmonella enterica (STY4521–4680) (8), and the 130-kb island of the phytopathogen Xanthomonas axonopodis (XAC2171–2286) (12) (Fig. 2 and Tables 2 and 4). Moreover, 21 additional PAPI-1 ORFs show similarity with ORFs from only one of these pathogenicity islands, 14 with S. enterica (RL052, 72, 77–79, 81–85) and 7 with X. axonopodis (RL020, 35, 63–65, 67). This abundant array of putative pathogenicity-related genes likely plays a role in the broad host range of PA14.
Interspersed with its ORFs, PAPI-1 carries at least five pairs of direct repeats (DR), a pair of inverted repeats, and an insertion sequence also found in Pseudomonas putida (28) (Fig. 2and Tables 3 and 5, which is published as supporting information on the PNAS web site). The 63-bp DR1 repeats, which border the entire PAPI-1 element and are part of the tRNALys gene, include the 58-bp attR sequence. A hairpin-like structure thought necessary for DNA insertion (29) occurs downstream of the right DR1, and this sequence might correspond to the actual PAPI-1 integration site, generating the attR and attL sequences. We note that DR1-like sequences also occur in P. aeruginosa strains C and SG17M genomic islands, and in Xylella fastidiosa (17).
The 662-bp DR2 repeats encode two proteins of unknown function (RL035 and RL046) and may have served as a DNA integration site. The 15-kb region found between these repeats, which encodes nine predicted ORFs, includes bacterial genes not previously associated with genomic islands, including two pairs of two-component regulatory systems (RL036–37 and RL038–39). RL037 is the pvrR gene (30), whereas RL039 and RL038 share domains with rcsB and rcsC, which encode a response regulator and a sensor protein of animal and plant pathogenic bacteria, respectively (31, 32). rcsC is involved in Salmonella virulence (33). Downstream of these regulatory systems lies a putative fimbrial chaperone-usher gene cluster (RL040–44) that is both related and distinct to cup clusters in P. aeruginosa strains PAO1 and PAK (34), and to similar clusters from S. enterica and Yersinia pestis (10, 35). We designate the PAPI-1 cluster cupD, because its predicted products are <70% identical with those of other cup clusters. cup genes assemble adhesive organelles expressed by many pathogenic bacteria that mediate attachment to epithelial cells (36) and contribute to initiation of biofilm formation (34).
The 248-bp DR3 repeats prescribe a 2.5-kb region of 46.4% G+C, indicating its foreign origin (Fig. 2 and Table 3). This region contains the RL087–8 genes, which are homologues of PA0984–5 that encode a bacteriocin, pyocin S5, and its immunity protein (37). We note that the RL087–8 genes are nearly identical homologues of the pyocin S5 genes found on the PAO1 island (Table 4) corresponding to PAPI-2. A pilus biogenesis system (RL077–86) is located just upstream of the left DR3 (Fig. 2). This system resembles type IV group B pili clusters found in other pathogenic bacteria, including the enteropathogenic E. coli bundle-forming pilus, the S. enterica CT18 type IVB pilus, and the Vibrio cholerae toxin-coregulated pilus (8, 38–40). Interestingly, the type IV pilus biogenesis machinery is highly homologous to the type II secretion pathways (Table 6, which is published as supporting information on the PNAS web site).
Both DR4 and DR5 consist of two consecutive direct repeats. These repeats are adjacent to the RL092, RL095, RL102, RL109–11, and RL114 ORFs, which are related to plasmid-encoded replication and recombination functions, suggesting that portions of PAPI-1 might be plasmid-derived. In contrast, only two PAPI-1 ORFs (RL103 and RL110) are phage-related (Table 2). Interestingly, the integrase RL002 and the chromosome-partitioning protein Soj RL115 genes are located at the ends of the island, similar to the P. aeruginosa clone C islands, suggesting that this island may have had an intermediate circular form that once integrated into tRNA sequences (19).
Finally, Fig. 2 and Table 2 demonstrate that genomic “shuffling” has also contributed to PAPI-1 organization, as the RL001, RL020, RL087, and RL088 ORFs are closely related to the PA0977–87 genes, which are located on a PAO1 genomic island. RL054–56 and RL113 share homology with the PAO1 genes PA2221–8, which also occur in a region having atypical G+C content (26).
Characterization of the PAPI-2 Pathogenicity Island. Sequencing of pH44 and pG68, which carry a second copy of RL003, reveals a 10,722-bp region, designated PAPI-2, located near the phnAB genes (Figs. 1B and 2B), a hypervariable region of the P. aeruginosa genome (14, 15, 18). Fig. 2B illustrates the organization of the 15 PAPI-2 predicted ORFs, seven of which correspond to hypothetical proteins of unknown function (Table 7, which is published as supporting information on the PNAS web site). PAPI-2 exhibits features of a genomic island, with a G+C content of 56.4%. It contains multiple predicted mobility functions, including one integrase gene, four transposase genes, and one related pseudogene; and at its left border has an almost complete IS222 element, plus a portion of ISPpu14, a putative transposase gene (Fig. 2B and Table 5).
Half of PAPI-2 is homologous to PA0977–0987, an 8.9-kb PAO1 genomic island (19), which encodes 11 predicted ORFs (Fig. 2B and Table 8, which is published as supporting information on the PNAS web site). Unlike PA0977–0987, PAPI-2 is not associated with an attR site, although it is located at the same position in the P. aeruginosa core genome (Fig. 1B). Furthermore, these two islands share upstream and downstream sequences, and six ORFs (Figs. 1B and 2B and Table 7). Although the PAO1 island, unlike PAPI-2, does not contain the entire RS02 integrase gene, it does have an intact tRNALys (attL) at its left border and a corresponding 22-bp direct repeat at its right border (attR) (Fig. 1B). The RS03 predicted product shares homology with the N terminus of RL003, the product of the 33A9 locus (Table 7). Interestingly, the 2.5-kb left end of PAPI-2 is identical to the 2.5-kb left end of PAPI-1, from the tRNALys gene to RL003 and RS03, respectively (Fig. 2). Finally, the PAO1 pyocin genes PA0984–85 are replaced in PAPI-2 by the cytotoxin exoU gene and its chaperone spcU (RS14–15) (Table 8). exoU encodes a type III effector that plays an important role in pathogenesis (41). Its presence on PAPI-2 defines this block as a pathogenicity island.
PAPI Island ORFs Encode Pathogenicity-Related Functions. We generated and analyzed 23 mutant strains (Table 1), including 11 in-frame deletion mutants and 12 TnphoA transposon insertion mutants, to assess whether the PAPI-1 and PAPI-2 ORFs that encode hypothetical/unknown functions promote P. aeruginosa pathogenesis. None of the mutants are defective for growth in liquid culture or for the extracellular production of pyocyanin and pyoverdine and for protease activity. Because some of the known PAPI-1 genes are involved in adhesion and/or motility, we also evaluated the mutants for colony morphology, in vitro adhesion, and swimming, twitching, and swarming motilities. All of the mutants behave like the wild-type parent, indicating that these activities do not depend on the mutated gene under our experimental conditions (data not shown).
Virulence was assessed in plants and animals by using the Arabidopsis leaf infiltration and the mouse thermal injury models (21). Table 1 shows that 19 of the 23 mutants exhibit attenuated virulence phenotype in at least one of the hosts, with 11 attenuated in both. We note that caution must be taken when assigning a virulent phenotype to a particular mutated gene, as some of the mutations in our pool of TnphoA insertion mutants (Table 1) may be polar and thus also effect loss-of-function for downstream genes. Indeed an example of this affect is observed with two different mutations in the RL003 locus, the TnphoA generated mutant 33A9 (21) and the in-frame deletion mutant RL003, which are weakly and moderately virulent, respectively.
Importantly, 15 of the mutants that exhibit attenuated virulence phenotype correspond to recently discovered genes and one to a known gene (pvrR) that has not been previously shown to be involved in virulence. Of the mutated ORFs, RL016 and RL022 occur within a large region (RL012–30) found in several other phytopathogenic bacteria, and RL036–37 and RL038–39 encode two-component regulatory systems, suggesting that pathogenicity activities regulated by these systems are evolutionarily conserved. To test whether two of the mutations that we generated are indeed responsible for the Arabidopsis and mice attenuated-in-virulence phenotypes, we replaced the mutated loci in the in-frame mutant strains RL003 and RL009 with the corresponding wild-type loci to generate the strains RL003R and RL009R. As predicted, these strains exhibit restored wild-type virulence in plants and mice (Table 1).
Presence of PAPI-1 in Other P. aeruginosa Clinical Isolates. We used 11 hybridization probes spanning PAPI-1 to assess the occurrence of this island in 14 P. aeruginosa pathogenic strains, 12 of which were isolated from CF patients (Fig. 3) (15, 16). Whereas CF1, CF3–5, CF27–28, CF30, and CF32 do not hybridize with any of the probes used, PAO37, CF2, and CF6 hybridize with all of them, suggesting that these strains carry the entire 108-kb PAPI-1 island. In contrast, other strains hybridize to a subset of probes. CF26 appears to carry the complete island, except for the region found between the DR2 sequences, whereas CF29 only carries its leftward half. Both PAK and PAO1 harbor only a small segment of PAPI-1, with PAK carrying its left end, and PAO1 carrying a 1.7-kb region that encodes the pyocin S5 and immunity genes, which occupy different chromosomal locations in PAO1 versus PA14.
Discussion
Different P. aeruginosa strains have the remarkable ability to inhabit diverse environments and infect a range of organisms, from amoeba to humans (1, 2). This environmental and pathogenic promiscuity is due in part to the large and genetically diverse P. aeruginosa genome. Sequencing of the PAO1 reference strain reveals that it encodes an abundant repertoire of specialized genes that promote success in various environments (26). We propose that the pathogenic promiscuity of certain P. aeruginosa strains is further brought about via factors carried by pathogenicity islands. Indeed, our results for PAPI-1 and PAPI-2 provide observations and implications for pathogenicity island modular structure and evolution, relatedness to other bacterial species, and contribution to PA14 pathogenic promiscuity and the generation of pathogenic variants.
PAPI-1 has a highly mosaic structure. It harbors blocks of ORFs related to virulence functions in other human and phytopathogenic bacteria, and ORFs similar to genes found in Archaea and phages, illustrating its diverse foreign origin. The PAPI-1 border regions exemplify this mosaicism. Whereas the right border contains ORFs unrelated to any GenBank sequences, the left border carries ORFs found in Archaea species and in other P. aeruginosa strains. Interestingly, one of these ORFs, RL008, encodes a putative helicase fused to sequences homologous to a PAO1 gene that encodes an unknown function (42). By mutation analysis, this hybrid ORF encodes a mammalian virulence factor, and thus might represent a previously undescribed pathogenic function generated via gene fusion.
The highly modular organization of PAPI-1 demonstrates that it was generated by multiple recombination events, as it carries several unrelated genes and gene clusters. Indeed, a large portion of PAPI-1 is similar to ORF clusters found in the genomes of the phytopathogen X. axonopodis pv. citri (12), and the human pathogen S. enterica ser. Typhi (8). This region is interrupted by unrelated ORFs located between repeat sequences, suggesting that a fragment homologous to the X. axonopodis and S. enterica gene blocks may have been acquired by P. aeruginosa as a complete DNA fragment, and later interrupted by the insertion of unrelated fragments. Interestingly, one of these secondary regions, RL036–39, contains two pairs of two-component regulatory systems, which we show affect plant and mammalian pathogenesis. It will be interesting to determine whether these systems regulate genes located on PAPI-1 or on the core genome. Acquisition of regulatory systems and virulence genes from other microorganisms may have contributed to the evolution of P. aeruginosa pathogenic variants to thrive in diverse environments. For instance, the PAPI-1 group B type IV pili genes are related to virulence factors that promote pathogen attachment to host cells. Acquisition of these genes could increase P. aeruginosa host range by promoting attachment to different surfaces, such as different epithelial cells.
That several PAPI-1 and PAPI-2 genes have known relatives in the genomes of plant-, soil-, animal-, and human-associated bacterial species is not surprising, because P. aeruginosa inhabits soil and water environments, and is associated with several hosts. It is likely that, during its evolution, P. aeruginosa has encountered a diverse array of bacterial species that have donated, and continue to donate, foreign DNA fragments. In turn, these fragments have affected the evolution of P. aeruginosa pathogenic variants to colonize even more environments. Presumably, this gene flow is bidirectional, such that P. aeruginosa virulence genes have spread to other bacterial species, to generate virulent strains in these species as well.
PAPI-1 and PAPI-2 mutational analyses demonstrate that both islands carry genes that allow this pathogen to thrive on evolutionary diverse hosts, including plants and mammals. Indeed, of 23 mutated ORFs, 19 encode functions necessary for Arabidopsis or mice virulence, with 11 required for full “wild-type” virulence in both hosts. Although the majority of these genes encode products of unknown function, their presence in P. aeruginosa clinical isolates, including those from CF patients, suggests that they direct conserved functions important for fitness and survival. Future characterization of these pathogenicity factors should provide insights into broad host pathogenic and defense mechanisms. Completion of the PA14 genome sequence might also lead to the identification of additional PAPI blocks and virulence genes.
Perhaps our most interesting results concern the identification of 16 virulence factors and the sequence of PAPI-1. Remarkably, >80% of its DNA sequence is unique, and 75 of its 115 predicted ORFs are unrelated to any previously identified proteins or functional domains. Significantly, many of these ORFs act in pathogenesis. They likely represent a previously undescribed repertoire of virulence activities functioning beyond the large set of virulence and regulatory genes already present on the P. aeruginosa core genome, apparently to provide promiscuous pathogenic variants with an enhanced bag of tricks with which to infect more host species.
Supplementary Material
Acknowledgments
We thank S. Lory for providing the CF strains, S. Stachel for comments and editing, and S. Gopalan for helpful discussion on the statistical analysis. This work was supported in part by a grant from Aventis S.A. (to L.G.R.). E.D. is supported by a postdoctoral fellowship from Canadian Institutes of Health Research. R.L.B. was partially supported by a fellowship from the Fundação de Amparo à Pesquisa do Estado de São Paulo, Brazil. N.T.L., D.L., and J.U. were supported in part by National Heart, Lung, and Blood Institute Grant U01 HL66678 (to F. M. Ausubel).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CF, cystic fibrosis; DR, direct repeats.
References
- 1.Pukatzki, S., Kessin, R. H. & Mekalanos, J. J. (2002) Proc. Natl. Acad. Sci. USA 99, 3159-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahme, L. G., Ausubel, F. M., Cao, H., Drenkard, E., Goumnerov, B. C., Lau, G. W., Mahajan-Miklos, S., Plotnikova, J., Tan, M. W., Tsongalis, J., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 8815-8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Govan, J. R. W. & Deretic, V. (1996) Microbiol. Rev. 60, 539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyczak, J. B., Cannon, C. L. & Pier, G. B. (2000) Microbes Infect. 2, 1051-1060. [DOI] [PubMed] [Google Scholar]
- 5.Lau, G. W., Goumnerov, B. C., Walendziewicz, C. L., Hewitson, J., Xiao, W., Mahajan-Miklos, S., Tompkins, R., Perkins, L. A. & Rahme, L. G. (2003) Infect. Immun. 71, 4059-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahme, L. G., Stevens, E. J., Wolfort, S. F., Shao, J., Tompkins, R. G. & Ausubel, F. M. (1995) Science 268, 1899-1902. [DOI] [PubMed] [Google Scholar]
- 7.Hacker, J. & Kaper, J. B. (2000) Annu. Rev. Microbiol. 54, 641-679. [DOI] [PubMed] [Google Scholar]
- 8.Parkhill, J., Gougan, G., James, K. D., Thomson, N. R., Pickard, D., Wain, J., Churcher, C., Mungall, K. L., Bentley, S. D., Holden, M. T. G., et al. (2001) Nature 413, 848-852. [DOI] [PubMed] [Google Scholar]
- 9.Welch, R. A., Burland, V., Plunkett, G., III, Redford, P., Roesch, P., Rasko, D., Buckles, E. L., Liou, S. R., Boutin, A., Hackett, J., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parkhill, J., Wren, B. W., Thomson, N. R., Titball, R. W., Holden, M. T., Prentice, M. B., Sebaihia, M., James, K. D., Churcher, C., Mungall, K. L., et al. (2001) Nature 413, 523-527. [DOI] [PubMed] [Google Scholar]
- 11.Perna, N. T., Plunkett, G., III, Burland, V., Mau. B., Glasner, J. D., Rose, D. J., Mayhew, G. F., Evans, P. S., Gregor, J., Kirkpatrick, H. A., et al. (2001) Nature 409, 529-533. [DOI] [PubMed] [Google Scholar]
- 12.da Silva, A. C., Ferro, J. A., Reinach, F. C., Farah, C. S., Furlan, L. R., Quaggio, R. B., Monteiro-Vitorello, C. B., Van Sluys, M. A., Almeida, N. F., Alves, L. M., et al. (2002) Nature 417, 459-463. [DOI] [PubMed] [Google Scholar]
- 13.Censini, S., Lange, C., Xiang, Z., Crabtree, J. E., Ghiara, P., Borodovsky, M., Rappuoli, R. & Covacci, A. (1996) Proc. Natl. Acad. Sci. USA 93, 14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spencer, D. H., Kas, A., Smith, E. E., Raymond, C. K., Sims, E. H., Hastings, M., Burns, J. L., Kaul, R. & Olson, M. V. (2003) J. Bacteriol. 185, 1316-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfgang, M. C., Kulasekara, B. R., Liang, X., Boyd, D., Wu, K., Yang, Q., Miyada, C. G. & Lory, S. (2003) Proc. Natl. Acad. Sci. USA 100, 8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang, X. & Pham, X. Q., Olson, M. V. & Lory, S. (2001) J. Bacteriol. 183, 843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larbig, K. D., Christmann, A., Johann, A., Klockgether, J., Hartsch, T., Merkl, R., Wiehlmann, L., Fritz, H. J. & Tummler, B. (2002) J. Bacteriol. 184, 6665-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romling, U., Schmidt, K. D. & Tummler, B. (1997) J. Mol. Biol. 271, 386-404. [DOI] [PubMed] [Google Scholar]
- 19.Kiewitz, C., Larbig, K., Klockgether, J., Weinel, C. & Tummler, B. (2000) Microbiology 146, 2365-2373. [DOI] [PubMed] [Google Scholar]
- 20.Arora, S. K., Bangera, M., Lory, S. & Ramphal, R. (2001) Proc. Natl. Acad. Sci. USA 98, 9342-9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahme, L. G., Tan, M. W., Le, L., Wong, S. M., Tompkins, R. G., Calderwood, S. B. & Ausubel, F. M. (1997) Proc. Natl. Acad. Sci. USA 94, 13245-13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoang, T., Karkhoff-Schweizer, R., Kutchma, A. & Schweizer, H. (1998) Gene 212, 77-86. [DOI] [PubMed] [Google Scholar]
- 23.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (1998) Current Protocols in Molecular Biology (Wiley, New York).
- 24.Sokol, P. A., Ohman, D. E. & Iglewski, B. H. (1979) J. Clin. Microbiol. 9, 538-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Déziel, E., Comeau, Y. & Villemur, R. (2001) J. Bacteriol. 183, 1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., Brinkman, F. S., Hufnagle, W. O., Kowalik, D. J., Lagrou, M., et al. (2000) Nature 406, 959-964. [DOI] [PubMed] [Google Scholar]
- 27.Lukashin, A. & Borodovsky, M. (1998) Nucleic Acids Res. 26, 1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson, K. E., Weinel, C., Paulsen, I. T., Dodson, R. J., Hilbert, H., Martins dos Santos, V. A., Fouts, D. E., Gill, S. R., Pop, M., Holmes, M., et al. (2002) Environ. Microbiol. 4, 799-808. [DOI] [PubMed] [Google Scholar]
- 29.van der Meer, J. R., Ravatn, R. & Sentchilo, V. (2001) Arch. Microbiol. 175, 79-85. [DOI] [PubMed] [Google Scholar]
- 30.Drenkard, E. & Ausubel, F. M. (2002) Nature 416, 740-743. [DOI] [PubMed] [Google Scholar]
- 31.Gottesman, S. & Stout, V. (1991) Mol. Microbiol. 5, 1599-1606. [DOI] [PubMed] [Google Scholar]
- 32.Virlogeux, I., Waxin, H., Ecobichon, C., Lee, J. O. & Popoff, M. Y. (1996) J. Bacteriol. 178, 1691-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Detweiler, C. S., Monack, D. M., Brodsky, I. E., Mathew, H. & Falkow, S. (2003) Mol. Microbiol. 48, 385-400. [DOI] [PubMed] [Google Scholar]
- 34.Vallet, I., Olson, J. W., Lory, S., Lazdunski, A. & Filloux, A. (2001) Proc. Natl. Acad. Sci. USA 98, 6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Townsend, S. M., Kramer, N. E., Edwards, R., Baker, S., Hamlin, N., Simmonds, M., Stevens, K., Maloy, S., Parkhill, J., Dougan, G., et al. (2001) Infect. Immun. 69, 2894-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soto, G. E. & Hultgren, S. J. (1999) J. Bacteriol. 181, 1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michel-Briand, Y. & Baysse, C. (2002) Biochimie 84, 499-510. [DOI] [PubMed] [Google Scholar]
- 38.Attridge, S. R., Voss, E. & Manning, P. A. (1999) J. Bacteriol. 73, 109-117. [DOI] [PubMed] [Google Scholar]
- 39.Donnenberg, M. S., Zhang, H. Z. & Stone, K. D. (1997) Gene 192, 33-38. [DOI] [PubMed] [Google Scholar]
- 40.Giron, J. A., Gomez-Duarte, O. G., Jarvis, K. G. & Kaper, J. B. (1997) Gene 192, 39-43. [DOI] [PubMed] [Google Scholar]
- 41.Miyata, S., Casey, M., Frank, D. W., Ausubel, F. M. & Drenkard, E. (2003) Infect. Immun. 71, 2404-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi, J. Y., Sifri, C. D., Goumnerov, B. C., Rahme, L. G., Ausubel, F. M. & Calderwood, S. B. (2002) J. Bacteriol. 184, 952-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.