Abstract
Amyloid-β precursor protein (APP), a type I membrane protein, is physiologically processed by α- or β-secretases that cleave APP N-terminal to the transmembrane region. Extracellular α-/β-cleavage of APP generates a large secreted N-terminal fragment, and a smaller cellular C-terminal fragment. Subsequent γ-secretase cleavage in the transmembrane region of the C-terminal fragment induces secretion of small extracellular peptides, including Aβ40 and Aβ42, which are instrumental in the pathogenesis of Alzheimer's disease, and intracellular release of a cytoplasmic tail fragment. Although APP resembles a cell-surface receptor, no functionally active extracellular ligand for APP that might regulate its proteolytic processing has been described. We now show that F-spondin, a secreted signaling molecule implicated in neuronal development and repair, binds to the conserved central extracellular domain of APP and inhibits β-secretase cleavage of APP. Our data indicate that F-spondin may be an endogenous regulator of APP cleavage, and suggest that the extracellular domains of APP are potential drug targets for interfering with β-secretase cleavage.
Keywords: Alzheimer's disease, α-secretase, β-secretase, γ-secretase
Amyloid-β precursor protein (APP) is a ubiquitous type I membrane protein that resembles a cell-surface receptor and is physiologically processed by site-specific proteolysis (1–4). The initial extracellular cleavage of APP by α- or β-secretases releases a large fragment called APPS that contains most of the extracellular sequences of APP. After α-/β-cleavage, the C-terminal fragments (CTFs) of APP remain in the membrane. The CTFs are composed of a small extracellular stub with different N termini, depending on the initial α-/β-secretase cleavage sites, the transmembrane region, and the cytoplasmic tail of APP. CTFs are recognized by another protease called γ-secretase, which cleaves the CTFs at multiple sites in the transmembrane region (5, 6). γ-Secretase cleavage results in the extracellular secretion of small peptides and the intracellular release of the cytoplasmic tail [APP intracellular domain (AICD)]. Once released, the AICD is thought to act as a transcriptional activator (7) and to perform other signaling roles (8–14). Different extracellular peptides are secreted after γ-cleavage, depending on whether the CTF was initially produced by α- or β-secretase cleavage. Especially important here is β-secretase cleavage in the brain that induces secretion of Aβ40 and Aβ42, peptides that are the major components of amyloid-β fibrils in Alzheimer's disease (1–4). APP is closely related to two mammalian proteins called APP-like proteins 1 and 2 (APLP1 and 2) (15–18). APLPs are also cleaved by secretases similar to APP (19–22), but their cleavage products do not appear to have a pathological role. Individual APP and APLP knockout mice are viable and fertile, but double APP/APLP2 or APLP1/APLP2 knockout mice do not survive postnatally (23, 24), suggesting that APP and APLPs are functionally redundant.
The site-specific proteolysis of APP and putative transcriptional signaling by the cytoplasmic tail of APP resembles that of Notch, a cell-surface protein that functions as a ligand-dependent regulator of cell fate (25, 26). These similarities suggested that cleavage of APP may also be regulated by ligands, but no functionally active ligands have been identified. F-spondin is a secreted neuronal protein that may be involved in regulating cell–cell interactions. Consistent with this notion, F-spondin is developmentally regulated, impairs binding of cells to the extracellular matrix, and is induced by neuronal injury (27–33). We now report that F-spondin binds to the central APP domain (CAPPD), and inhibits the initial α-/β-cleavage of APP. Our findings suggest that an endogenous ligand for APP can regulate cleavage, and may indicate a strategy for developing drugs that inhibit APP cleavage by binding to the CAPPD.
Materials and Methods
Plasmids. Vectors encoding various parts of human APP695 or F-spondin (American Type Culture Collection no. 2190694) were generated by subcloning the corresponding PCR fragments into pCMV-Ig9 (34), pGEX-KG, or pCMV5 (plasmid names with residue numbers from APP695: pCMVIg-APP.1 = residues 1–678; pCMVIg-APP.2 = 1–205; pGEX-CAPPD = 286–557, pCMV-APP as described (7), pCMV-APPΔ1 = deletion of residues 36–289 with insertion of Pro-Trp residues; pCMV-APPΔ2 = deletion of residues 288–493 with insertion of Thr-Arg residues; pCMVIg-F spondin.1 = residues 1–807 (full-length); pCMVIg-F spondin.2 = 1–501; pCMVIg-F spondin.3 = 1–614, pCMVIg-F spondin.4 = 1–754; pCMVIg-F spondin.5 = 1–225; pCMVIg-F spondin.6 = 1–442; pCMVIg-F spondin.7 = 443–807. Full-length myc-tagged F-spondin was generated by subcloning NotI–ClaI PCR fragments into pcDNA4-His/myc B. Vectors encoding human full-length Mindin [American Type Culture Collection no. 5183118 (35)] were generated by subcloning EcoRI–SalI fragment to pCMVIg9 vector and EcoRI–XhoI fragment to pcDNA4-His/myc A vector.
Generation of Brain Membrane Extracts. We homogenized 20 frozen rat brains (Pelfreeze, Rogers, AK) in 200 ml of 0.32 M sucrose, 5 mM Hepes-NaOH, pH 7.4, and 0.1 mM EDTA containing a standard protease inhibitor mix (0.1 g/liter PMSF/10 mg/liter leupeptin/aprotinin/1 mg/liter pepstatin A). The homogenate was centrifuged at low speed (800 × g for 15 min) to remove debris, and the supernatant was centrifuged (100,000 × g for 1 h) to yield a crude membrane pellet that was homogenized in buffer A (20 mM Hepes-NaOH, pH 7.4/150 mM NaCl/2 mM CaCl2/2 mM MgCl2 with the standard protease inhibitor mix). Subsequently, an equal volume of buffer B (buffer A containing 2% Triton X-100) was added for extraction (3 h at 4°C), and insoluble material was removed by centrifugation (100,000 × g for 1 h).
Affinity Chromatography on Immobilized GST- or Ig-Fusion Proteins. These procedures were performed essentially as described (36, 37). Brain membrane extract was precleared by incubation (2 h at 4°C) with glutathione agarose and incubated overnight at 4°C with immobilized GST-CAPPD on glutathione agarose beads preequilibrated with buffer B. Beads were washed with buffer B and were serially eluted with 2 ml of buffer B containing 0.3 M NaCl, 0.5 M NaCl, 1.0 M NaCl, or 1.0 M NaCl, 10 mM EGTA, and 5 mM EDTA (instead of 2 mM CaCl2). Eluted proteins were analyzed by SDS/PAGE and Coomassie blue staining. Bound proteins were identified by liquid chromatography/MS of tryptic fragments. For pull-down assays, the medium from COS cells transfected with pcDNA4-His/myc-F spondin or pcDNA-His/myc-Mindin (collected 48–72 h posttransfection) was adjusted to (final concentrations) 10 mM Hepes-NaOH, pH 7.4/1 mM EGTA/1% Triton X-100, proteinase inhibitors were added, and the supernatant was precleared. The treated medium was then incubated overnight at 4°C with GST or GST-CAPPD immobilized on glutathione agarose or with various Ig-APP fusion proteins immobilized on protein-A Sepharose. Glutathione agarose or Protein A beads were washed four to five times with buffer B, and were examined by SDS/PAGE and immunoblotting. COS cells that were transfected with pCMV-APP, pCMV-APPΔ1, pCMV-APPΔ2, or pCMV-APLPs were harvested in PBS 48 h posttransfection, membrane proteins were solubilized in buffer B, and the cell lysate was incubated overnight at 4°C with Protein A-Sepharose containing Ig-F spondins, Ig-Mindin, or Ig-C fusion protein. Protein A beads were washed with buffer B four to five times, and resuspended in SDS/PAGE sample buffer.
APP Cleavage in Transfected Cells by BACE 1. HEK293 cells were cotransfected in 12-well plates by using FuGENE reagent with APP alone, APP with BACE1, or combinations of APP and BACE1 with Ig-F spondin or Ig-C. APP fragments were examined by immunoblotting and quantitated by using 125I-labeled secondary antibodies (Amersham Pharmacia, Piscataway, NJ) with PhosphorImager (Molecular Dynamics) detection (38).
Transactivation Assays. HEK293 cells were cotransfected in 12-well plates by using Lipofectamine 2000 with pCMV-APP, pCMV-Tip60, pCMV-Fe65, and reporter plasmids pG5E1B-luc and pCMV-LacZ alone, or with Ig-F spondin, or Ig-neurexin 1β (Ig-N1β), or Ig-Mindin, or Ig-SynCAM, or Ig-N1β-3 or Ig-N1α-1, or Ig-C. Transactivation assays were performed as described (7, 39). The luciferase activity was standardized by the β-galactosidase activity as a control for transfection efficiency.
Results
Identification of F-spondin as a Potential APP Ligand. To search for APP ligands, we produced a recombinant GST-fusion protein containing the CAPPD of APP (Fig. 1A). Because the CAPPD has no cysteines, it lacks disulfide bonds and can be produced in bacteria. We used the CAPPD-GST fusion protein immobilized on glutathione-Sepharose for affinity chromatography experiments with membrane proteins that were solubilized from rat brain with 1% Triton X-100, and used immobilized GST as a negative control (data not shown). We eluted bound proteins with high salt, and identified by MS F-spondin as a major component of the proteins bound to CAPPD.
Fig. 1.
Binding of F-spondin to immobilized APP. (A) Domain structure of APP (Upper) and diagram of various APP vectors used for the present study (Lower). N-terminally, APP is composed of a signal peptide (SP), a CRD, a zinc-binding motif, acidic sequence regions, and a Kunitz domain. The center of APP is occupied by a large domain that contains no cysteine residues (referred to as CAPPD) and a short linker sequence that includes the cleavage sites for α- and β-secretases. C-terminally, APP contains a transmembrane region and a cytoplasmic tail. The constructs used here include Ig-fusion proteins of the entire extracellular region or CRD alone (Ig-APP.1 or Ig-APP.2 respectively), a GST-CAPPD fusion protein, and expression vectors that encode full-length APP, or APP in which the CRD or part of the CAPPD were deleted marked by dashed lines (pCMV-APPΔ1 or 2, respectively). Nonneuronal APP contains an alternatively spliced Kunitz domain that is absent from all APP constructs used here. (B) Affinity chromatography of secreted myc-tagged recombinant F-spondin pull-downs on immobilized APP proteins. Ig- and GST-fusion proteins of various fragments of APP, as indicated in A, were used to affinity-purify secreted myc-tagged F-spondin produced in the supernatant of transfected COS cells.
F-spondin is a secreted glycoprotein that contains an N-terminal reelin domain, a central F-spondin-specific sequence, and C-terminal thrombospondin domains (see Fig. 2A), and may function in axonal path finding, cell–cell interactions, and neural regeneration (27–33). To confirm the potential interaction of F-spondin with APP, we produced in transfected COS cells a fusion protein of the Fc-region of human Ig with the complete extracellular sequence of APP (Ig-APP.1; Fig. 1 A). We tested whether the purified Ig-APP fusion protein, immobilized on protein A-Sepharose, could pull down myc-tagged full-length F-spondin that was also produced in transfected COS cells (Fig. 1B). As a control, we used an Ig-fusion protein that contains only a few N-terminal residues of neurexin 1α as an irrelevant control, in addition to the Ig moiety (Ig-C; Fig. 1 A). We examined binding of recombinant secreted F-spondin to the immobilized Ig-APP fusion protein in the presence and absence of Ca2+, because many extracellular binding domains are stabilized by structural Ca2+ binding. We found that only Ig-APP but not Ig-C captured F-spondin, and that F-spondin was only bound in the presence of Ca2+ (Fig. 1B). We next investigated whether an Ig-fusion protein that includes only the N-terminal domains of APP (Ig-APP.2) also captures F-spondin (Fig. 1B), but detected no binding. However, when we tested the isolated CAPPD, immobilized as a bacterially expressed GST-fusion protein, we confirmed APP binding to F-spondin in a Ca2+-dependent manner similar to the Ig-fusion protein containing the full-length extracellular sequences of APP (Fig. 1B).
Fig. 2.
Binding of APP to immobilized F-spondin. (A) Domain structure of F-spondin (Upper) and constructs of F-spondin included in the various Ig-fusion vectors used for the present study (Lower). F-spondin is composed of an N-terminal signal peptide (SP), reelin-like and spondin domains, and six C-terminal thrombospondin repeats. The positions of the two N-glycosylation sites are indicated. (B) Pulldown of full-length APP695. APP695 was solubilized with 1% Triton X-100 from transfected COS cells and was bound to immobilized Ig-F spondin proteins containing full-length or parts of F-spondin (see A). (C) Pulldown of APP deletion mutants (see Fig. 1A for extent of the deletions) with full-length Ig-F spondin. (D) Comparison of the ability of immobilized full-length F-spondin to affinity-purified APP, APLP1, and APLP2 expressed in transfected COS cells and visualized with antibodies to the C termini of indicated proteins.
To validate the interaction of F-spondin with APP, we tested whether immobilized F-spondin can capture APP. We constructed a series of F-spondin Ig-fusion proteins that include different parts of F-spondin (Fig. 2 A), and performed pulldowns of recombinant full-length APP695 expressed in COS cells (Fig. 2B) or of endogenous brain APP (data not shown). We found that immobilized F-spondin specifically retained recombinant and native brain APP, and that the N-terminal reelin domain and the central spondin-specific region of F-spondin were essential for binding APP, whereas the C-terminal thrombospondin repeats were not required (Fig. 2B).
We then examined the specificity of APP binding by F-spondin. Expression of deletion constructs of APP revealed that deletion of the N-terminal cysteine-rich growth-factor-like domain (CRD; APPΔ1) did not abolish binding, whereas a partial deletion of the CAPPD (APPΔ2) blocked binding (Fig. 2C). APP is closely related to APLP1 and 2 (15–18), and the CAPPD is particularly well conserved among these proteins. If binding of F-spondin to APP was specific, one would expect the APLPs also to bind. Indeed, all three proteins were similarly captured by immobilized F-spondin (Fig. 2D). Viewed together, these findings indicate that the CAPPD of APP directly binds to F-spondin.
A protein related to F-spondin called Mindin has recently been characterized (32). Mindin contains a spondin-like sequence and a single thrombospondin repeat, but lacks a reelin domain (Fig. 3A). To test whether Mindin might bind to APP, we performed experiments similar to those described in Figs. 1 and 2 with myc-tagged Mindin (Fig. 3B) or with a Ig-Mindin fusion protein (Fig. 3C). In contrast to F-spondin, we did not observe Mindin binding in either assay configuration, suggesting that Mindin does not bind to APP.
Fig. 3.
Lack of an interaction of APP with Mindin. (A) Domain structure of F-spondin with Mindin. SP, signal peptide, a spondin-like domain; TSR, thrombospondin repeat. (B) Pulldown of myc-tagged Mindin with immobilized GST-CAPPD fusion protein. (C) Pulldown of APP with a Ig-Mindin fusion protein.
F-spondin Inhibits APP Cleavage by BACE 1, the Primary β-Secretase. A key feature of APP is that it is digested by α- and β-secretases that cleave APP at a site C-terminal of the CAPPD (Fig. 1 A). To test whether binding of F-spondin alters APP cleavage, we cotransfected BACE 1, the enzyme that mediates β-secretase activity (40–42), with APP. When we transfected only APP into HEK293 cells and analyzed APP cleavage by immunoblotting with an antibody to its C-terminal cytoplasmic tail, we could barely detect CTFs at a low steady-state level (Fig. 4A, lanes 1–3; experiments are carried out in triplicate for quantifications). However, when we cotransfected BACE 1 with APP, the steady-state level of CTFs dramatically increased (lanes 4–6). After BACE 1 cleavage, we observed two closely migrating CTFs on SDS/PAGE that may correspond to the two major BACE 1 cleavage sites in APP (41, 42). When we cotransfected Ig-C together with APP and BACE 1, we found a small decrease in both APP and the CTFs (Fig. 3A, lanes 7–9), probably because cotransfection of a third plasmid dilutes the cellular transcription/translation machinery. However, when we cotransfected full-length Ig-F spondin fusion protein with APP and BACE 1, we observed a dramatic decline in the level of CTFs, indicating that F-spondin inhibits BACE 1-dependent APP cleavage (lanes 10–12). The decrease in CTFs by F-spondin was confirmed in quantitations of full-length APP and the CTFs in the transfected cells using 125I-labeled secondary antibodies (Fig. 4B). These measurements demonstrated that F-spondin decreased the CTFs of APP by ≈70–80% (corrected for the amount of full-length APP present to control for cotransfection effects).
Fig. 4.
F-spondin inhibits cleavage of APP by BACE 1. (A) Immunoblot of HEK293 cells that were transfected without or with BACE 1, Ig-C, or Ig-F spondin as indicated. Experiments were carried out in triplicate to ensure reproducibility. Numbers on the left indicate positions of molecular mass markers. Note that BACE 1 cotransfection promotes production of two C-terminal fragments, termed CTFβ1 and CTFβ2. (B) Quantification of the results shown in A. Relative levels of full-length APP and of both CTFs were quantified using 125I-labeled secondary antibodies and PhosphorImager detection. Data shown are means ± SEM derived by dividing for each sample the signal for CTFβ1 or CTFβ2 by the APP signal.
We next asked whether the effect of F-spondin on APP cleavage was dose-dependent. We performed similar transfection experiments with increasing amounts of F-spondin, and quantified the level of F-spondin protein in addition to the levels of full-length APP proteins and of CTFs. As expected, transfection of increasing amounts of F-spondin plasmid led to a dose-dependent increase in F-spondin protein (Fig. 5A). In addition, a moderate decrease in full-length APP was observed, presumably because of competition between transfected plasmids for transcription. Transfection of <0.25 μg of F-spondin plasmid inhibited CTF production ≈75%, but had <20% effect on APP levels (Fig. 5A). Correcting the CTF levels for those of full-length APP confirmed that the drop in CTFs was not a simple reflection of the small decrease in APP, but was due to a large decline in APP cleavage by relatively low levels of F-spondin (Fig. 5B).
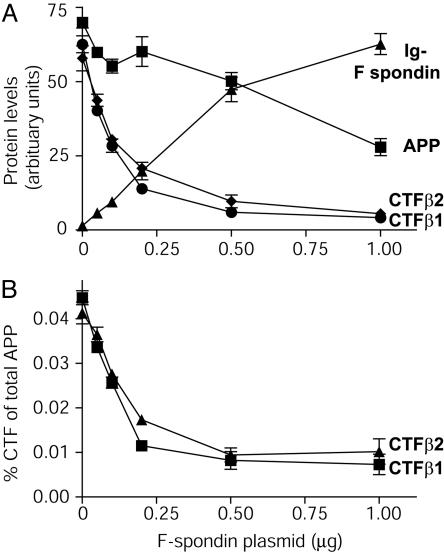
Fig. 5.
Titration of F-spondin-mediated inhibition of APP cleavage by BACE 1. (A) Relative levels of proteins expressed in an experiment similar to that described in Fig. 4. Increasing amounts of Ig-F spondin plasmid were cotransfected with constant amounts of APP and BACE 1. The levels of full-length APP and the CTFs of APP and of F-spondin were quantified by immunoblotting and are shown in arbitrary units. (B) Ratio of CTF to full-length APP as a function of increasing amount of F-spondin. CTF levels were corrected for APP expression. Data shown are means ± SEM from a representative experiment (n = 3) independently repeated multiple times.
F-spondin Impairs APP-Dependent Transactivation of Gal4-Tip60 Transcription. Previous studies (7) suggested that the AICD of APP functions in transcriptional activation by binding to the adaptor protein Fe65 that, in turn, binds to the chromosome remodeling factor Tip60. Unmodified APP strongly transactivates Gal4-Tip60-mediated transcription by a mechanism that depends on Fe65, probably because the AICD of APP (which binds to Fe65) is released by α-/β- and γ-cleavage of APP and cooperates with Fe65 in transcription. To test whether F-spondin alters the transcriptional activation mediated by APP as an additional, indirect assay for APP cleavage, we cotransfected increasing amounts of Ig-F spondin with a constant amount of APP and Fe65 into HEK293 cells (Fig. 6A). We found that APP alone greatly stimulated Gal4-Tip60-dependent transactivation as expected (7), but that even low amounts of cotransfected F-spondin plasmid (<100 ng) inhibited transactivation, consistent with an inhibition of cleavage by F-spondin.
Fig. 6.
Effect of F-spondin on APP-dependent transactivation of Gal4-Tip60-mediated transcription. (A) F-spondin inhibits APP-dependent transactivation. A constant amount of Gal4-Tip60, Fe65, and APP was cotransfected with increasing amounts of Ig-F spondin. Note that, without F-spondin, APP causes a strong stimulation of Gal4-Tip60-dependent transcription as described (7). F-spondin dramatically inhibits APP-dependent transactivation of transcription by Gal4-Tip60, such that even low concentrations of F-spondin (<100 ng of transfected plasmid) almost completely block the response. (B) Comparison of the effects of multiple Ig-fusion proteins on the APP-dependent transactivation of Gal4-Tip60. All proteins were expressed with 50 ng of cotransfected plasmids. (C) Increasing concentrations of APP are unable to rescue the F-spondin-dependent inhibition of APP-dependent transactivation of Gal4-Tip60. Constant amounts of Gal4-Tip60, Fe65, and Ig-C, Ig-N1β-1, or Ig-F spondin were cotransfected with increasing concentrations of APP. The bell-shaped dose–response curve under control conditions as reported (7) is probably due to dilution of transcription factors by increasing amounts of APP. Nevertheless, even at high concentrations of APP, F-spondin induces a relative inhibition of transactivation.
We next investigated the specificity of this inhibition. We cotransfected small amounts of plasmids (50 ng of DNA) encoding various unrelated Ig-fusion proteins (Ig-C, Ig-F spondin, Ig-Mindin, Ig-SynCAM, and three different Igneurexins) with Gal4-Tip60 and Fe65, and measured the relative level of transactivation by APP in the presence of these Ig-fusion proteins (Fig. 6B). Ig-F spondin potently inhibited transactivation, whereas Ig-SynCAM, Ig-N1β-1, and Ig-N1β-3 produced no inhibition of transactivation, and Ig-Mindin and Ig-N1α-1 caused an intermediate degree of inhibition. The intermediate inhibition of transactivation caused by Ig-Mindin and Ig-N1α-1, although significantly less than the inhibition mediated by F-spondin, raised the possibility that the F-spondin-dependent inhibition in this assay is not specific, but is an indirect effect. Ig-SynCAM, Ig-N1β-1, and Ig-N1β-3 may have been unable to inhibit because they were expressed in the wrong ratio with APP. To address this possibility, we cotransfected increasing amounts of APP with a constant amount of Fe65 and of either Ig-C, Ig-N1β-1, or Ig-F spondin. The design of this experiment aimed to control for potential nonspecific effects of the Ig moiety in the Ig-F spondin fusion protein, or for trafficking effects induced by expressing a neuronal cell-surface protein. Increasing concentrations of APP were tested to account for the possibility that a protein did not truly inhibit transactivation, but simply shifted the requirement for APP. Indeed, in the presence of Ig-C, APP potentiated transcription in a bell-shaped dose–response curve (Fig. 6C) as described (7). This bell-shaped dose–response curve is probably due to the fact that high concentrations of APP are less efficient in stimulating transcription because the overexpressed APP dilutes out expression of the other components. Ig-F spondin greatly inhibited transactivation at all APP levels, whereas Ig-N1β-1 had no effect (Fig. 6C). Together these data are consistent with the notion that F-spondin, by binding to the extracellular CAPPD of APP, inhibits APP processing and thereby impairs transcriptional transactivation.
Discussion
We report that F-spondin, a secreted protein that is thought to function in brain development and neuronal repair (26–32), can bind to the conserved CAPPD, and we show that F-spondin binding can control cleavage of APP by BACE 1, which cleaves APP C-terminal to the CAPPD. The evidence for these conclusions is based on four principal findings: (i) F-spondin is efficiently captured by Ig- and GST-fusion proteins of APP (Fig. 1); (ii) APP is affinity-purified on immobilized Ig-F spondin fusion proteins (Fig. 2); (iii) coexpression of Ig-F spondin with APP and BACE 1 inhibits cleavage of APP in a dose-dependent manner (Figs. 4 and 5); and (iv) cotransfection of Ig-F spondin impairs APP-dependent transactivation of transcription mediated by Gal4-Tip60 (Fig. 6). As controls, we show that APP does not bind to other proteins, including an Ig-fusion protein of Mindin that contains a subset of domains present in F-spondin (Fig. 3). Our data only addressed BACE 1-dependent APP cleavage directly, but it seems likely that α-secretase-dependent cleavage is also suppressed by F-spondin, because F-spondin inhibited APP-dependent transactivation in tissue culture cells, whereas most APP cleavage is likely mediated by α-secretase.
F-spondin is a secreted neuronal glycoprotein that was identified by subtractive hybridization, because F-spondin is abundantly expressed in the floor plate during spinal cord development (27). Neuronal expression of F-spondin appears to be high during embryonic development but diminishes after birth (31). Multiple developmental roles for F-spondin were shown. Recombinant F-spondin promotes neurite outgrowth (27), and endogenous F-spondin is involved in axonal path finding (28). In addition, F-spondin inhibits adhesion of neural crest cells to the extracellular matrix (30). In adult animals, axotomy of sciatic nerve causes a large increase in F-spondin expression distal to the lesion (31), suggesting a possible function for F-spondin in neuronal regeneration. F-spondin is also widely expressed outside of the brain (31). F-spondin was purified based on an in vitro assay as a vascular smooth muscle growth promoting factor that stimulates the growth of smooth muscle cells (32). Furthermore, F-spondin inhibits adhesion of endothelial cells to the extracellular matrix, and impairs endothelial cell migration (33). Also, when injected into the rat cornea, F-spondin impaired neovascularization (33). Together, these data suggest that F-spondin is a secreted extracellular matrix component that inhibits adhesion of several cell types, including neurons, may have a role in axonal path finding, and could be involved in injury repair in adult brain. The properties of F-spondin, especially its wide tissue distribution, up-regulation by nerve dissection, and secretion as a glycoprotein, are consistent with a functional interaction with APP and APLPs, which are also ubiquitously expressed extracellular proteins that have been implicated in similar processes (1–4). Despite considerable effort, the in vivo roles of F-spondin (and also of APP and APLPs) in brain and in peripheral tissues remain unclear. F-spondin has not been localized by immunocytochemistry, and no binding partners for F-spondin (apart from APP/APLPs reported here) have been identified. Moreover, no genetic model incorporating F-spondin mutations exist.
Our results show that F-spondin is a potential ligand for APP. Although we used several strategies to analyze the interaction of F-spondin with APP, our observations are subject to limitations that need to be considered. The data are based on in vitro binding reactions and transfected cells, and do not describe a physiological complex between endogenous APP and F-spondin. The significance of the F-spondin/APP complex will have to be tested in genetically modified animals, such as knockout and/or knock-in mice in which F-spondin and APP are altered. However, the fact that F-spondin binds to APLP1 and APLP2 (Fig. 2D), which exhibit functional redundancy with APP (23, 24), supports the notion that F-spondin binding performs a biologically significant role. In addition, Mindin was shown not to bind to APP although it is related protein to F-spondin, and also contains spondin domains and one thrombospondin repeat, but lacks the reelin domain.
Ligands for the extracellular domains of APP could have several functions: they regulate cleavage, act as receptors for the cleaved and secreted extracellular fragment (APPS), or mediate cell adhesion by attaching uncleaved cell-surface APP to another cell or the extracellular matrix. F-spondin selectively binds to the CAPPD of APP, suggesting that other domains of APP, especially the N-terminal CRD, could interact with other ligands. Furthermore, the developmental dynamics of F-spondin and APP appear to be different, raising the possibility that the CAPPD has also additional ligands. We report that F-spondin impairs APP cleavage and directly affects the transcriptional signal by APP. Coexpression of F-spondin with APP and BACE 1 inhibited the production of CTFs, thus indicating that F-spondin inhibits BACE 1-dependent APP cleavage. These experiments were independently confirmed with transactivation assays to assess the specificity for APP cleavage. We document that Ig-F spondin fusion protein inhibited transactivation consistent with an inhibition of cleavage by F-spondin in comparison to other cell-surface proteins that did not alter APP-dependent transactivation (Fig. 6). However, it is unclear whether APP binding and the effect on APP cleavage by F-spondin occur on the cell surface with extracellularly derived F-spondin, or during secretory transport in secretory vesicles. In the first case, F-spondin would be a paracrine agent, and in the second, an autocrine agent. Both scenarios do not exclude biological significance and cannot be evaluated until the physiological importance of APP cleavage is known. At this point, the biological significance of BACE 1 cleavage is obscure, given the finding that the BACE 1 knockout mice do not have an immediately obvious phenotype (43, 44). It should also be noted that inhibition of APP cleavage by F-spondin could represent an epiphenomenon, and that the real function of the F-spondin/APP interaction is a signaling role on the cell surface. It is possible, for example, that APP acts as a receptor for F-spondin, which mediates the inhibition of cell adhesion to substratum observed for recombinant F-spondin (30, 33).
Although the physiological significance of F-spondin binding to APP remains unclear, the potent effect of F-spondin on APP cleavage demonstrates that APP cleavage can in principle be regulated by ligands that bind to the CAPPD. It is unclear whether F-spondin binding to the CAPPD inhibits cleavage sterically, or whether F-spondin binding causes an allosteric change or an alteration of APP trafficking. Independent of the mechanism of inhibition, however, compounds binding to the CAPPD could mimic this effect and act as extracellular inhibitors of APP cleavage that potentially decrease Aβ production. APLPs are also cleaved by secretases, including BACE 1 (19–22), and it is likely that F-spondin binding to APLPs (Fig. 2) also inhibits the cleavage of APLPs. However, because there are significant sequence differences between the CAPPDs of APP and APLPs, it is possible that specific inhibitors of APP cleavage based on CAPPD binding can be isolated. Our findings therefore indicate an approach to developing inhibitors of Aβ production, suggesting that it may be possible to isolate small-molecule ligands that bind extracellularly to the CAPPD and are specific for APP as opposed to other BACE 1 substrates such as APLPs.
Acknowledgments
We thank I. Kornblum and E. Borowicz for excellent technical assistance; and Drs. X. Cao, Q. Li, M. Khvotchev, G. Yu, and T. Biederer for advice and reagents. This work was supported by National Institute of Mental Health Grant R01-MH69585-01 (to T.C.S.) and by National Institute on Aging Fellowship Award AG05844 (to A.H.).
Abbreviations: CTF, C-terminal fragment; APP, amyloid-β precursor protein; AICD, APP intracellular domain; APLP, APP-like protein; CAPPD, central APP domain; CRD, cysteinerich domain; Ig-C, control Ig; Ig-N1, Ig neurexin 1.
References
- 1.Price, D. L. & Sisodia, S. S. (1998) Annu. Rev. Neurosci. 21, 479-505. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe, D. J. (1998) Trends Cell Biol. 8, 447-453. [DOI] [PubMed] [Google Scholar]
- 3.Bayer, T. A., Cappai, R., Masters, C. L., Beyreuther, K. & Multhaup, G. (1999) Mol. Psychiatry 4, 524-528. [DOI] [PubMed] [Google Scholar]
- 4.Haass, C. & De Strooper, B. (1999) Science 286, 916-919. [DOI] [PubMed] [Google Scholar]
- 5.Sastre, M., Steiner, H., Fuchs, K., Capell, A., Multhaup, G., Condron, M. M., Teplow, D. B. & Haass, C. (2001) EMBO Rep. 2, 835-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weidemann, A., Eggert, S., Reinhard, F. B., Vogel, M., Paliga, K., Baier, G., Masters, C. L., Beyreuther, K. & Evin, G. (2002) Biochemistry 41, 2825-2835. [DOI] [PubMed] [Google Scholar]
- 7.Cao, X. & Südhof, T. C. (2001) Science 293, 115-120. [DOI] [PubMed] [Google Scholar]
- 8.Homayouni, R., Rice, D. S., Sheldon, M. & Curran, T. (1999) J. Neurosci. 19, 7507-7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howell, B. W., Lanier, L. M., Frank, R., Gertler, F. B. & Cooper, J. A. (1999) Mol. Cell. Biol. 19, 5179-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarr, P. E., Roncarati, R., Pelicci, G., Pelicci, P. G. & D'Adamio, L. (2002) J. Biol. Chem. 277, 16798-16804. [DOI] [PubMed] [Google Scholar]
- 11.Trommsdorff, M., Borg, J. P., Margolis, B. & Herz, J. (1998) J. Biol. Chem. 273, 33556-33560. [DOI] [PubMed] [Google Scholar]
- 12.Baek, S. H., Ohgi, K. A., Rose, D. W., Koo, E. H., Glass, C. K. & Rosenfeld, M. G. (2002) Cell 110, 55-67. [DOI] [PubMed] [Google Scholar]
- 13.Leissring, M. A., Murphy, M. P., Mead, T. R., Akbari, Y., Sugarman, M. C., Jannatipour, M., Anliker, B., Muller, U., Saftig, P., De Strooper, B., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 4697-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koo, E. H. (2002) Traffic 3, 763-770. [DOI] [PubMed] [Google Scholar]
- 15.Wasco, W., Bupp, K., Magendantz, M., Gusella, J. F., Tanzi, R. E. & Solomon, F. (1992) Proc. Natl. Acad. Sci. USA 89, 10758-10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wasco, W., Gurubhagavatula, S., Paradis, M. D., Romano, D. M., Sisodia, S. S., Hyman, B. T., Neve, R. L. & Tanzi, R. R. (1993) Nat. Genet. 5, 95-99. [DOI] [PubMed] [Google Scholar]
- 17.Sprecher, C. A., Morgenstern, K. A., Mathewes, S., Dahlen, J. R., Schrader, S. K., Foster, D. C. & Kisiel, W. (1993) Biochemistry 32, 4481-4486.8485127 [Google Scholar]
- 18.Slunt, H. H., Thinakaran, G., Von Koch, C., Lo, A. C., Tanzi, R. E. & Sisodia, S. S. (1994) J. Biol. Chem. 269, 2637-2644. [PubMed] [Google Scholar]
- 19.Naruse, S., Thinkaran, G., Luo, J. J., Kusiak, J. W., Tomita, T., Iwatsubo, T., Qian, X., Ginty, D. D., Price, D. L., Borchel, D. R., et al. (1998) Neuron 21, 1213-1221. [DOI] [PubMed] [Google Scholar]
- 20.Scheinfeld, M. H., Ghersi, E., Laky, K., Fowlkes, B. J. & D'Adamio, L. (2002) J. Biol. Chem. 277, 44195-44201. [DOI] [PubMed] [Google Scholar]
- 21.Walsh, D. M., Fadeeva, J. V., LaVoie, M. J., Paliga, K., Eggert, S., Kimberly, W. T., Wasco, W. & Selkoe, D. J. (2003) Biochemistry 42, 6664-6673. [DOI] [PubMed] [Google Scholar]
- 22.Li, Q. & Südhof, T. C. J. Biol. Chem., in press.
- 23.von Koch, C. S., Zheng, H., Chen, H., Trumbauer, M., Thinakaran, G., van der Ploeg, L. H., Price, D. L. & Sisodia, S. S. (1997) Neurobiol. Aging 18, 661-669. [DOI] [PubMed] [Google Scholar]
- 24.Heber, S., Herms, J., Gajic, V., Hainfellner, J., Aguzzi, A., Rulicke, T., von Kretzschmar, H., von Koch, C., Sisodia, S., Tremml, P., et al. (2000) J. Neurosci. 20, 7951-7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fortini, M. E. (2002) Nat. Rev. Mol. Cell Biol. 3, 673-684. [DOI] [PubMed] [Google Scholar]
- 26.Hartmann, D., Tournoy, J., Saftig, P., Annaert, W. & De Strooper, B. (2001) J. Mol. Neurosci. 17, 171-181. [DOI] [PubMed] [Google Scholar]
- 27.Klar, A., Baldassare, M. & Jessell, T. M. (1992) Cell 69, 95-110. [DOI] [PubMed] [Google Scholar]
- 28.Tzarfati-Majar, V., Burstyn-Cohen, T. & Klar, A. (2001) Proc. Natl. Acad. Sci. USA 98, 4722-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burstyn-Cohen, T., Tzarfaty, V., Frumkin, A., Feinstein, Y., Stoeckli, E. & Klar, A. (1999) Neuron 23, 233-246. [DOI] [PubMed] [Google Scholar]
- 30.Debby-Brafman, A., Burstyn-Cohen, T., Klar, A. & Kalcheim, C. (1999) Neuron 22, 475-488. [DOI] [PubMed] [Google Scholar]
- 31.Burstyn-Cohen, T., Frumkin, A., Xu, Y. T., Scherer, S. S. & Klar, A. (1998) J. Neurosci. 18, 8875-8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyamoto, K., Morishita, Y., Yamazaki, M., Minamino, N., Kangawa, K., Matsuo, H., Mizutani, T., Yamada, K. & Minegishi, T. (2001) Arch. Biochem. Biophys. 390, 93-100. [DOI] [PubMed] [Google Scholar]
- 33.Terai, Y., Abe, M., Miyamoto, K., Koike, M., Yamasaki, M., Ueda, M., Ueki, M. & Sato, Y. (2001) J. Cell Physiol. 188, 394-402. [DOI] [PubMed] [Google Scholar]
- 34.Ushkaryov, Y. A., Hata, Y., Ichtchenko, K., Moomaw, C., Afendis, S., Slaughter, C. A. & Südhof, T. C. (1994) J. Biol. Chem. 269, 11987-11992. [PubMed] [Google Scholar]
- 35.Feinstein, Y., Borrell, V., Garcia, C., Burstyn-Cohen, T., Tzarfaty, V., Frumkin, A., Nose, A., Okamoto, H., Higashijima, S., Soriano, E. & Klar, A. (1999) Development (Cambridge, U.K.) 126, 3637-3648. [DOI] [PubMed] [Google Scholar]
- 36.Ichtchenko, K., Hata, Y., Nguyen, T., Ullrich, B., Missler, M., Moomaw, C. & Südhof, T. C. (1995) Cell 81, 435-443. [DOI] [PubMed] [Google Scholar]
- 37.Hata, Y., Slaughter, C. A. & Südhof, T. C. (1993) Nature 366, 347-351. [DOI] [PubMed] [Google Scholar]
- 38.Rosahl, T. W., Spillane, D., Missler, M., Herz, J., Selig, D. K., Wolff, J. R., Hammer, R. E., Malenka, R. C. & Südhof, T. C. (1995) Nature 375, 488-493. [DOI] [PubMed] [Google Scholar]
- 39.Biederer, T., Cao, X., Südhof, T. C. & Liu, X. (2002) J. Neurosci. 22, 7340-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan, R., Bienkowski, M. J., Shuck, M. E., Miao, H., Tory, M. C., Pauley, A. M., Brashier, J. R., Stratman, N. C., Mathews, W. R. & Buhl, A. E. (1999) Nature 402, 533-537. [DOI] [PubMed] [Google Scholar]
- 41.Sinha, S., Anderson, J. P., Barbour, R., Basi, G. S., Caccavello, R., Davis, D., Doan, M., Dovey, H. F., Frigon, N. & Hong, J. (1999) Nature 402, 537-540. [DOI] [PubMed] [Google Scholar]
- 42.Vassar, R., Bennett, B. D., Babu-Khan, S., Kahn, S., Mendiaz, E. A., Denis, P., Teplow, D. B., Ross, S., Amarante, P. & Loeloff, R. (1999) Science, 286, 735-741. [DOI] [PubMed] [Google Scholar]
- 43.Cai, H., Wang, Y., McCarthy, D., Wen, H., Borchelt, D. R., Price, D. L. & Wong, P. C. (2001) Nat. Neurosci. 4, 233-234. [DOI] [PubMed] [Google Scholar]
- 44.Roberds, S. L., Anderson, J., Basi, G., Bienkowski, M. J., Branstetter, D. G., Chen, K. S., Freedman, S. B., Frigon, N. L., Games, D. & Hu, K. (2001) Hum. Mol. Genet. 10, 1317-1324. [DOI] [PubMed] [Google Scholar]