Abstract
The transcription factor Nuclear Factor Kappa B (NF-κB) has been shown to be cardioprotective after permanent coronary occlusion (PO) and late ischemic preconditioning (IPC), and yet it is cell injurious after ischemia/reperfusion (I/R) in the heart. There is limited information regarding NF-κB-dependent cardioprotection, and the NF-κB-dependent genes that contribute to the cardioprotection after PO are completely unknown. The objective of the study was to identify NF-κB-dependent genes that contribute to cardioprotection after PO. Microarray analysis was used to delineate genes that potentially contribute to the NF-κB-dependent cardioprotection by determining the overlap between the set of PO regulated genes and genes regulated by NF-κB, using mice with genetic abrogation of NF-κB activation in the heart. This analysis identified 16 genes as candidates for NF-κB-dependent effects after PO. This set of genes overlaps with, but is significantly different from the set of genes we previously identified as regulated by NF-κB after IPC. The genes encoding heat shock protein 70.3 (hspa1a) and heat shock protein 70.1 (hspa1b) were the most significantly regulated genes after PO and were up-regulated by NF-κB. Results using knockout mice show that Hsp70.1 contributes to NF-κB-dependent cardioprotection after PO and likely underlies, at least in part, the NF-κB-dependent cardioprotective effect. Our previous results show that Hsp70.1 is injurious after I/R injury. This demonstrates that, like NF-κB itself, Hsp70.1 has antithetical effects on myocardial survival and suggests that this may underlie the similar antithetical effects of NF-κB after different ischemic stimuli. The significance of the research is that understanding the gene network regulated by NF-κB after ischemic insult may lead to identification of therapeutic targets more appropriate for clinical development.
Keywords: nuclear factor kappa B (NF-κB), heat shock protein 70 (Hsp70), permanent coronary occlusion (PO), myocardial infarction (MI), microarray, gene expression, cardioprotection
Introduction
One of the leading causes of death globally is cardiovascular disease, with most of these deaths being related to coronary heart disease, which includes acute myocardial ischemia [1]. Myocardial ischemia results in multiple biochemical and metabolic changes that include an imbalance between ATP/oxygen supply and demand, high intercellular calcium levels, cell death, and synthesis/release of cytokines [2–4]. These biochemical and metabolic changes occur during myocardial ischemia and reperfusion, result in the activation of transcription factors, for example, NF-κB [2–4].
Previous in vivo studies have shown that NF-κB activation in the heart after various stimuli (e.g. ischemia/reperfusion, I/R and cytokines) occurs mostly in the cardiomyocytes [4–7]. Studies using isolated rat cardiomyocytes induced by TNF-α resulted in NF-κB activation and expression of NF-κB-dependent proteins (e.g. protein A20) [8–10]. In contrast, cardiomyocytes transfected with mutated IκBα (S32A, S36A) or IκBΔN mutant (truncated IκBα, lacks IκBα phosphorylation sites) that block NF-κB activation, had an increase in cell death after TNF-α administration [8–10]. These results suggest that TNF-α activates NF-κB and provides cardioprotection. The mechanism behind NF-κB cardioprotection induced by TNF-α is unknown.
Both TNF-α and hypoxic stimuli are involved in the expression of stress-induced protein, e.g., heat shock protein 70 (Hsp72) in isolated adult feline cardiomyocytes [11,12]. Isolated adult cardiomyocytes treated with antisense (AS) hsp72 oligonucleotide resulted in an increase in injury, suggesting that the expression of Hsp72 protected cells from hypoxia injury [11]. TNF-α pretreated cells were resistant to subsequent hypoxic injury. However, AS hsp72 oligonucleotide failed to abrogate the TNF-α cardioprotection [12]. In contrast, mutated TNF-α ligands for TNF receptor 1 (TNFR1) and 2 (TNFR2) blocked the cardioprotective effects of TNF-α during hypoxia [12]. These results suggested that TNF-α cardioprotective effects after hypoxia are mediated through either or both TNFR1 and TNFR2 [12].
In 2000, Kurrelmeyer and colleagues subjected TNFR1/2 knockout (KO) mice to 24 hours coronary artery permanent occlusion, which resulted in a 40% increase in infarct size, compared to wild-type (WT) mice [13]. Interestingly, TNFR1 KO and TNFR2 single KO mice exhibited similar infarct size compared to WT mice after 24 hours PO, implying that TNFR1/2 are both required for the TNF-α dependent cardioprotection after PO [13]. The mechanisms behind TNFR1 and TNFR2 cardioprotection are unknown; however, one hypothesis is that NF-κB activation might contribute to the TNF-α cardioprotective effect [13]. In 2001, we showed that overexpression of a dominant-negative IκBα in the heart represses NF-κB activation [5]. Two years later IκBαΔN mice (cardiac-specific over expression of a dominant negative IκBα construct that inhibits NF-κB activation) were shown to have significantly increased infarct size after PO, suggesting that NF-κB is cardioprotective after PO [14]. In 2005, we showed, using genetic blockade of NF-κB, that NF-κB contributes to myocardial infarction after ischemia/reperfusion (I/R) injury [6], and in 2010, we elucidated the NF-κB-dependent gene network that underlies the cardioprotective effects of NF-κB after late ischemic preconditioning (IPC) [15].
As mentioned above, there is limited information regarding NF-κB-dependent cardioprotection in the PO model, and the NF-κB-dependent genes that contribute to this are unknown. An important question is whether the same genes underlie all NF-κB-dependent cardioprotective effects, for instance, after PO vs. after IPC. The objective of this study was to identify NF-κB-dependent genes that contribute to cardioprotection after PO.
We performed gene expression assays (microarrays and quantitative real-time RT-PCR, QRT-PCR) to determine genes that are up- or down-regulated after PO and dysregulated by genetic blockade of NF-κB. We identified 16 genes that likely constitute a NF-κB-dependent network activated after PO. Heat shock protein 1A (hspa1a/hsp70.3) and heat shock protein 1B (hspa1b/hsp70.1) were the most significantly regulated genes after PO and were regulated by NF-κB. Our results using knockout models showed that Hsp70.1 is a major contributor to NF-κB-dependent cardioprotection after PO. We discuss these findings in light of our recent discovery [15] that Hsp70.1 contributes to cell death after I/R injury. Delineation of NF-κB-dependent protective gene networks may result in identification of novel therapeutic targets for cardioprotective therapeutic strategies.
Materials and Methods
2.1 Animal models
All mice were bred and maintained in accordance with the University of Cincinnati Institutional Animal and Use Committee, and the Guide for the Care and Use of Laboratory Animals [16]. Cardiac-specific mutated IκBα (overexpressed the IκBαS32A, S36A cDNA in the C57B1/6J strain) dominant-negative mice (DN) have been fully characterized and demonstrated blockade of NF-κB after various insults [4–6]. The Hsp70.1 single knockout (Hsp70.1 KO) on the C57 background were obtained from Macrogen (S. Korea) and have been previously characterized [17,18]. Hsp70.1/70.3 double knockout mice (Hsp70.1/.3 KO) have been fully characterized and demonstrate a lack of functional hsp70.1 and hsp70.3 genes on the B6129SF2/J strain (B129) [19]. All experiments were controlled for age (10–16 weeks), sex (both males and females), and mouse strain (C57 mice for Hsp70.1 controls, non-transgenic siblings (WT) as controls for DN, and B129 for Hsp70.1/.3 KO controls). Statistical analysis post hoc was used to determine if there was an effect on gender. As previously described, this analysis showed no effect of gender upon the results of these studies [6].
2.2 Myocardial ischemia model
All mice were anesthetized with sodium pentobarbital (100mg/kg IP), intubated with polyethylene 90 tubing, and ventilated using a mini ventilator (Harvard Apparatus) to maintain the respiratory rate between 100 and 105 breaths/minute, as previously described [6,20]. Briefly, a lateral thoracotomy was performed by making a 1.5cm incision between the second and third ribs providing access to the left anterior descending coronary artery (LAD) [6,20]. Myocardial ischemia was achieved by tightening and tying a non-traumatic occluder to press against the coronary artery, and confirmed by visual observation (e.g. cyanosis) and continuous ECG monitoring [6,20]. In the permanent occlusion model, the LAD was permanently occluded and the mouse chest was then closed in layers. Sham mice were subjected to the same procedures without tightening the suture; therefore no occlusion occurred [6,20]. All mice were monitored during and after the surgery until they exhibited full consciousness.
2.3 Myocardial infarct analysis
Briefly, mice were euthanized, the aorta cannulated, and the heart perfused through the aortic root with 1% triphenyltetrazolium chloride (TTC) as previously described to determine the infarct tissue and risk region [6,20]. To distinguish the non-risk region, the suture was re-tied at the site of occlusion and the heart was perfused with a 5% phthalo blue dye solution [6,20]. Risk region, non-risk region and infarct region were measured using the method of Fishbein et al [21].
2.4 Tissue isolation
Hearts were removed and rinsed in ice-cold RNase free PBS. The ischemic tissue or analogues region (sham mice) (15–30mg of LV tissue) were isolated, quickly flash frozen (liquid nitrogen), and stored at −80°C. Nuclear and cytoplasmic extracts from frozen ischemic tissue or sham tissue were used for Western blots as described in detail in the supplement methods section [6].
2.5 Western blotting
Nuclear and cytoplasmic extract samples (20–30µg) were loaded in a 10–12% SDS-polyacrylamide gel. Proteins were transferred to 0.45-µm nitrocellulose (Amersham Bioscience Hybond-C Extra) or 0.45-µm PVDF membrane (Millipore), incubated with antibodies and developed using chemiluminescence (Perkin Elmer) as described in detail in the supplement.
2.6 RNA isolation
Total RNA was isolated from ischemic or sham tissue using an RNeasy Mini Kit (Qiagen, catalog # 74104) according to RNeasy Mini handbook (3rd edition; June 2001) Appendix C (Protocol for Isolation of Total RNA from Heart, Muscle, and Skin Tissue). In addition, Proteinase K (20mg/µl) was used to digest proteins and inactivate nucleases. To remove DNA contamination, the RNeasy mini columns were treated with Qiagen RNase-free DNase. RNA quantity and quality were assessed by optical density (Spectronic Unicam Genesys 10UV) at 260 nm, and optical density ratios of 260/280 nm and 260/230 nm ratios respectively.
2.7 Microarray Analysis
RNA samples (1µg/10µL) were submitted to the University of Cincinnati Microarray Core for Agilent Bioanalyzer/Nanodrop analysis (Agilent 2100 Bioanalyzer) and samples with RNA integrity number (RIN) greater then 7 and Nanodrop analysis ranging from 1.7 to 2.2 for the 260/280 ratios were used for microarrays. As previously described, competitive hybridization to microarrays of labeled cDNA targets generated from 4 separate samples per group was performed with two dye flips performed per group [15]. Additional details on microarray conditions and analysis are discussed in the supplement section.
Genes that were significantly up- and down-regulated (P<0.01 and >1.5 fold) by NF-κB and/or after PO were subjected to in silica functional analysis using CLustering Enrichment ANalysis (CLEAN) [22]. Briefly, CLEAN is an open-source R package that performs hierarchical clustering of genes based on their expression profiles and set of functional categories, e.g. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genome (KEGG) pathway) [22]. Functional enrichment for each functional category (GO and KEGG) was determined by performing Fisher’s Exact Test [22].
2.8 cDNA synthesis and Quantitative Real-Time RT-PCR (QRT-PCR)
Synthesis of cDNA was carried out using 0.5 µg of total RNA and using an RNA-to-cDNA kit (Applied Biosystems, P/N: 4387406) according to manufacturer’s instructions. cDNA quantity and quality were assessed by optical density (Spectronic Unicam Genesys 10UV) at 260 nm, and optical density ratios of 260/280 nm and 260/230 nm ratios respectively.
QRT-PCR was conducted with a Stratagene MX 3000P machine using a SYBR Green 2X RT-PCR master mix (Applied Biosystems, P/N: 430915) with a total reaction volume of 20µl. The thermocycling parameters were the following: 95° C for 10 minutes followed by 40 cycles of 90° C for 15 seconds, and 60°C for 60 seconds (with data collection at the end of 60°C step at each cycle) [15]. Primer sequences were determined by using Roche Applied Science Universal Probe Library Assay Design Software (ProbeFinder Version 2.45 Mouse) and were reconfirmed to be specific for the target gene by using a basic local alignment search tool (BLAST) (primer sequences in supplement section, Table 1S). All reactions were performed in triplicate on each plate with a minimum of three independent replicates, and gene expression values were calculated using the difference in target gene expression relative to 18S mRNA using the 2-ΔΔCt method [15,23].
2.9 Statistical Analysis
Results are reported as mean± standard error of mean (SEM). Unpaired student t-test using the Bonferroni correction and a one-way ANOVA test were used. Differences were considered significant at P≤0.05. Power analysis (power = 0.95; α = 0.05) was used to ensure proper sample size needed to determine significance.
Results
3.1 NF-κB cardioprotection after PO
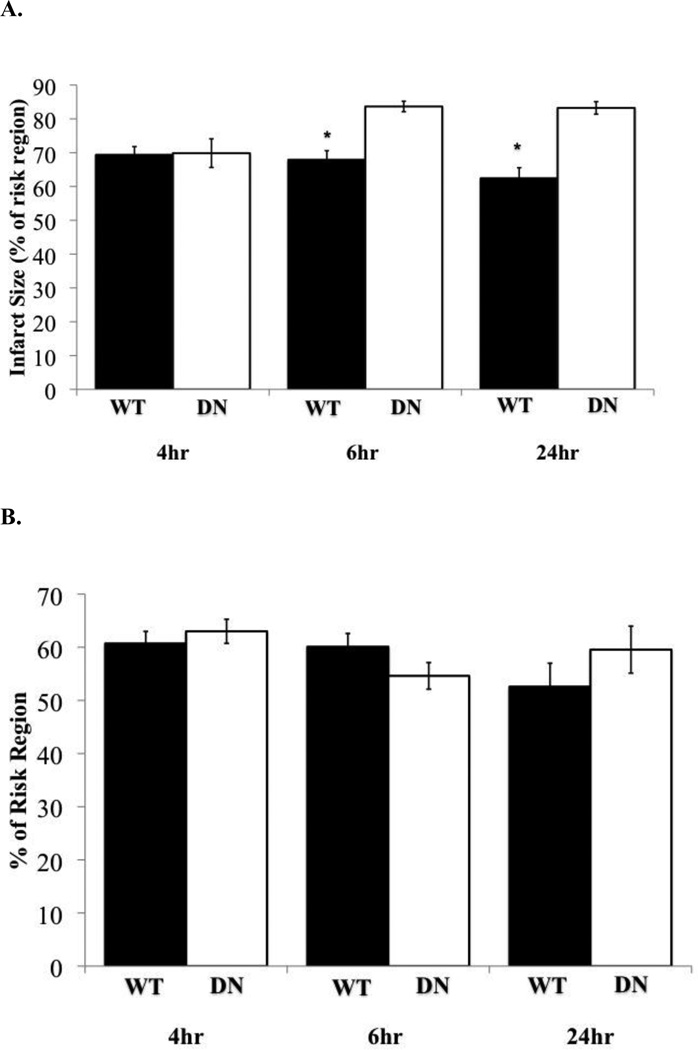
To identify the time period in which NF-κB-dependent cardioprotection occurs after PO, DN and WT mice were subjected to 4, 6 or 24 hours PO. After 4 hours PO, there were no significant differences in infarct size between DN mice (69.85%) and WT mice (69.35%) (Figure 1). However, DN mice subjected to 6 hours (83.66%) or 24 hours PO (83.23%) displayed a significantly (P≤0.05) larger infarct compared to WT mice 6 hours (67.89%) or 24 hours PO (62.42%) (Figure 1). DN mice exposed to 4 hours of PO (69.85%) exhibited a significantly (P≤0.05) smaller infarct compared to the 6 hours (83.66%) and 24 hours PO (83.23%) (Figure 1). There were no significant differences in infarct size after 4 hours (69.35%), 6 hours (67.89%), and 24 hours (62.42%) between WT PO groups (Figure 1). Our results suggest that NF-κB-dependent cardioprotection is initiated between 4 to 6 hours after PO.
Figure 1. NF-κB cardioprotection after PO.
(A) Assessment of infarct size (% of risk region) in WT mice (black bars) and DN (white bars) after 4, 6, and 24 hours (hr) PO. *P≤0.05 vs. corresponding DN, n=4–7. Bars are plotted as mean±SEM. (B) Percent of risk region was not significantly different (P>0.05) between all groups.
3.2 NF-κB translocation after PO
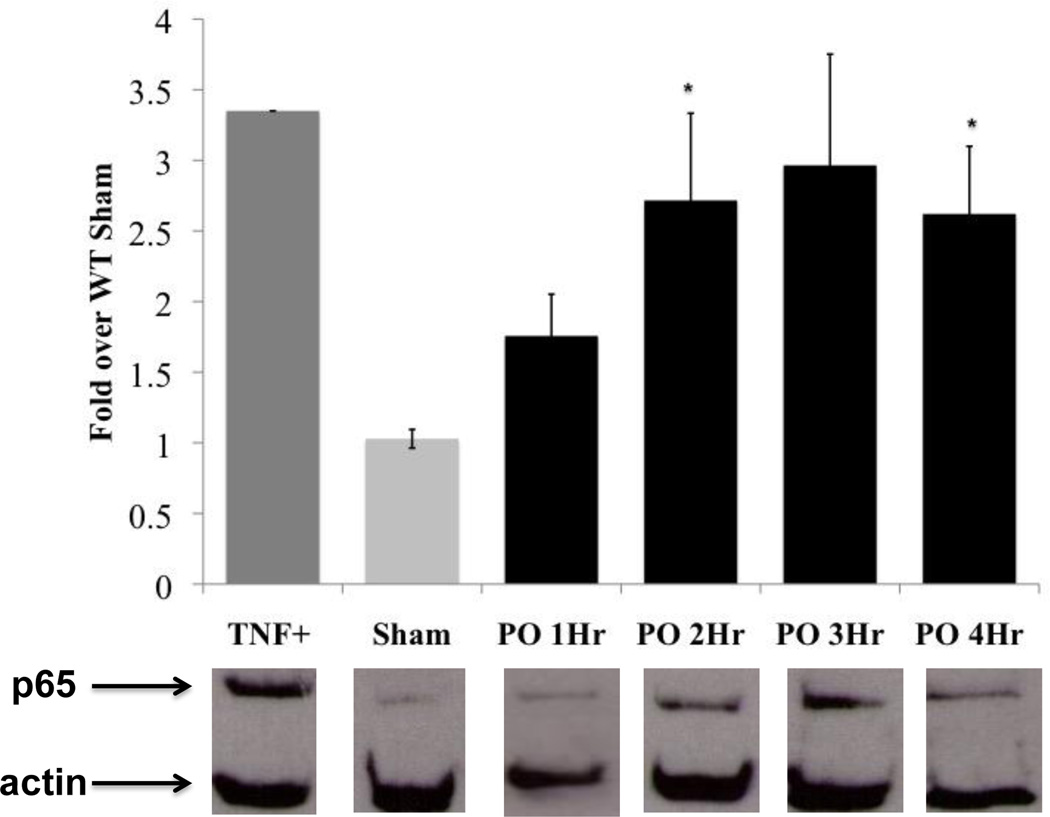
Quantitative analysis of p65 nuclear and cytoplasmic levels reveled a small, but not statistically significant increase of NF-κB (p65) in the nucleus relative to sham starting at 1 hour after occlusion (1.76 fold), followed by a significant (P≤0.05) increase at 2 hours (2.72 fold) (Figure 2). Steady levels of p65 remained in the nucleus up to 4 hours post coronary occlusion (3hr: 2.97 fold and 4hr: 2.62 fold) (Figure 2).
Figure 2. NF-κB translocation after PO.
Translocation of p65 into the nucleus is shown for the following groups (signal intensity in parentheses): TNF-α (positive control) (3.35), Sham (4 hour) (1.03±0.07), 1 hour PO (1.76±0.13), 2 hour PO (2.72±0.62), 3 hour PO (2.97±0.79), and 4 hour PO (2.62±0.48), with values plotted as fold over WT sham (sham average =1). Representative Western blots are below the graph, with p65 band at the top and actin below as a loading control. The actin signal was used and was used to normalized p65 signal and normalized signals were expressed as fold changes relative to sham. TNF-α control, WT Sham n=4, PO n=3–5. Bars represents mean ±SEM. *P≤0.05 vs. WT Sham. Additional details on Western blot analysis, confirmation of nuclear extracts (nuclear protein: TATA-TBP) and lack of any cytoplasmic contamination (no significant presence of cytoplasmic protein: GAPDH) in the nuclear extract are located in the supplement section.
3.3 Genes regulated during PO
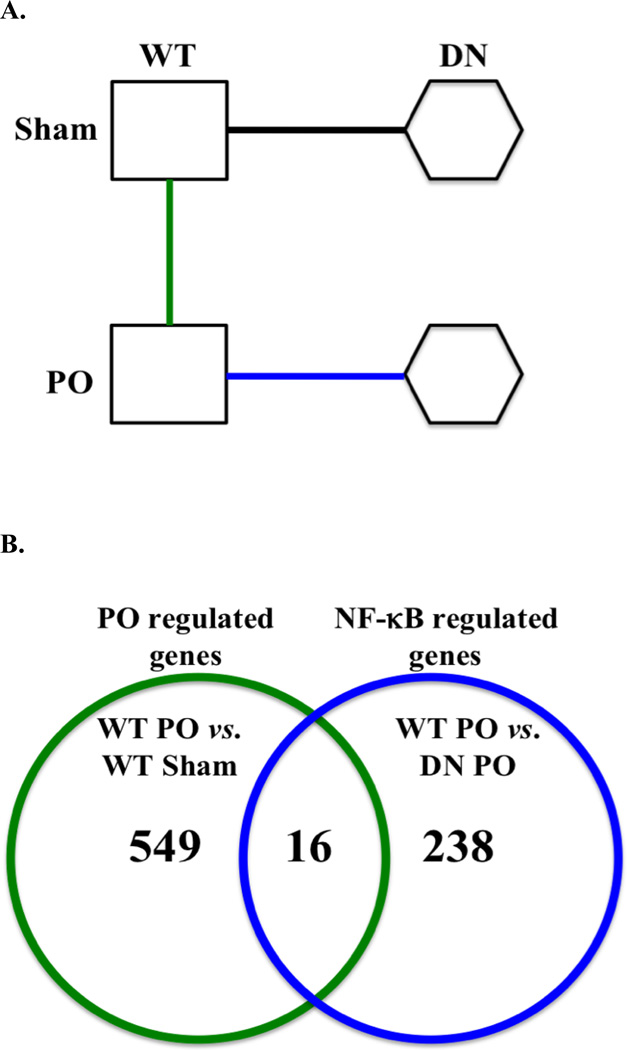
A microarray approach employing the DN mice was used to determine genes that are likely to underlie the NF-κB-dependent cardioprotection after PO (Figure 3A). Microarray analysis was used to determine the set of genes that are significantly up- and down-regulated (P<0.01 and >1.5 fold change) after PO (e.g. comparison between WT PO vs. WT Sham), and genes that are up- and down-regulated by NF-κB in response to PO (e.g. comparison between WT PO vs. DN PO) (Figure 3A). The two gene sets (e.g. PO regulated genes vs. NF-κB regulated genes) were overlapped to identify genes that most likely contribute to the NF-κB-dependent cardioprotection after PO (Figure 3B).
Figure 3. Microarray strategy and analysis.
(A) Microarray comparisons were made as shown by the sold lines between the WT PO, WT Sham (green line: WT Sham vs. WT PO) and DN PO (blue line: WT PO vs. DN PO). (B) Venn diagram represents the number of significantly up- and down-regulated genes between WT PO vs. WT Sham (green line circle: PO regulated genes) and WT PO vs. DN PO (blue line circle: NF-κB regulated genes). The 16 genes in the overlapping gene set were significantly up- and down-regulated by PO and by NF-κB. Statistical significances for the microarray analysis were P<0.01 and a fold change >1.5 fold change, n=4.
We found 565 genes (595 oligonucleotide probes) to be significantly regulated after PO (P<0.01 and fold change >1.5; WT PO vs. WT Sham) (Figure 3B). Out of the 565 genes, 435 were significantly up-regulated and 130 were down-regulated after PO. The most significantly up-regulated genes were heat shock proteins (e.g. hspa1a, hspa1b, and dnajb1) and transcription factors (e.g. egr1, fos, fosb, nr4a1, and atf3) (full list of genes see supplement section, Table S2). Functional annotation of the 565 genes according to GO term resulted in 255 significant GO categories (P<0.01) (see supplement section, Table S3). The most significantly regulated GO categories included: cell migration, response to stress, blood vessel development, inflammatory response, and cell adhesion (Table S3). There were 16 significantly regulated (P<0.01) KEGG pathways from the 565 genes. The most significantly regulated KEGG pathways included: MAPK signaling pathway, focal adhesion, and ECM-receptor interaction (Table S4).
3.4 Genes regulated by NF-κB in response to PO
Microarray comparisons were performed between WT PO and DN PO (e.g. WT PO vs. DN PO) to identify genes up- and down-regulated by NF-κB in response to PO (Figure 3A). This resulted in 254 genes (266 oligonucleotide probes) that were significantly regulated (P<0.01 and >1.5 fold change) by NF-κB (Figure 3B). There were 134 genes that were significantly up-regulated (e.g. car3, cxcl5) and 120 genes that were significantly down-regulated by NF-κB (e.g. cyp2b10, cdh22) (Table S5). In silico functional analysis of the 254 genes showed a significant enrichment (P<0.01) for 39 GO categories (Table S6). The most significantly regulated GO-term categories were regulation of endocytosis and regulation of phagocytosis (Table S6). There were only two significant (P<0.01) KEGG pathways, which were metabolism of xenobiotics by cytochrome P450 (P=7.57×10−5) and drug metabolism-cytochrome P450 (P=0.001) (Table S7).
3.5 NF-κB cardioprotective network
Genes that were significantly up- and down-regulated after PO (565 genes, section 3.3) were compared to genes that were significantly up- and down-regulated by NF-κB in response to PO (254 genes, section 3.4). This delineated 16 genes in the overlapping set (Figure 3B). These 16 genes are candidates for the NF-κB cardioprotective network after PO (Table 1). Out of these 16 genes, 11 genes were significantly up-regulated after PO, including hspa1a, hsp90aa1, car3, 2810474019rik, plscr1, igfbp3, ptx3 and dkk3 (Table 1). The three genes that were up-regulated after PO and were down-regulated by NF-κB were the following: edn2, ai605517, and myocd (Table 1). Five out of the 16 genes regulated after PO and by NF-κB were down-regulated after PO (Table 1). Four of these genes that were also down-regulated by NF-κB were ndufc1, sfil1, gm9783, and grhl2 (Table 1). There was only one gene that was down-regulated after PO, but was up-regulated by NF-κB; it was ankrd9 gene (Table 1).
Table 1.
Genes significantly regulated (P<0.01 and >1.5 fold change) after 5hr PO (WT PO vs. WT Sham) and by NF-κB (WT PO vs. DN PO). The fold change between WT PO vs. WT sham and WT PO vs. DN PO, are ratios of expression levels in the comparisons between the two groups.
| Gene ID |
Gene Name | Gene Symbol |
WT PO vs. WT Sham |
P-Values | WT PO vs. DN PO |
P-Values |
|---|---|---|---|---|---|---|
| 193740 | heat shock protein 1a | hspa1a (hsp70.3) | 5.86 | 1.06E-07 | 2.24 | 0.002 |
| 15519 | heat shock protein 90, alpha (cytosolic), class a member 1 | hsp90aa1 * | 3.91 | 3.33E-06 | 2.19 | 0.002 |
| 15519 | heat shock protein 90, alpha (cytosolic), class a member 1 | hsp90aa1 * | 2.86 | 8.55E0-5 | 2.06 | 0.001 |
| 12350 | carbonic anhydrase 3 | car3 | 1.74 | 0.002 | 2.13 | 0.0001 |
| 67246 | riken cdna 2810474o19 gene | 2810474o19rik | 1.88 | 0.004 | 1.69 | 0.003 |
| 22038 | phospholipid scramblase 1 | plscr1 | 2.10 | 0.004 | 1.85 | 0.006 |
| 16009 | insulin-like growth factor binding protein 3 | igfbp3 | 1.57 | 0.005 | 1.85 | 0.005 |
| 19288 | pentraxin related gene | ptx3 | 1.68 | 0.007 | 1.65 | 0.001 |
| 50781 | dickkopf homolog 3 (xenopus laevis) | dkk3 | 1.67 | 0.009 | 1.67 | 0.005 |
| 13615 | endothelin 2 | edn2 | 3.33 | 1.29E-06 | −2.66 | 0.0006 |
| 106622 | expressed sequence ai605517 | ai605517 | 2.14 | 0.0004 | −1.76 | 0.009 |
| 214384 | myocardin | myocd | 1.91 | 0.002 | −1.97 | 0.001 |
| 74251 | ankyrin repeat domain 9 | ankrd9 | −1.53 | 0.009 | 1.81 | 0.008 |
| 66377 | nadh dehydrogenase (ubiquinone) 1, subcomplex unknown, 1 | ndufc1 | −2.48 | 0.0002 | −1.84 | 0.008 |
| 78887 | sfi1 homolog, spindle assembly associated (yeast) | sfi1 | −1.76 | 0.004 | −1.97 | 0.001 |
| 69595 | predicted gene 9783 | gm9783 | −1.59 | 0.005 | −1.92 | 0.0006 |
| 252973 | grainyhead-like 2 (drosophila) | grhl2 | −1.50 | 0.008 | −1.58 | 0.008 |
Multiple oligonucleotide probes for the same genes.
The most significant (P=1.80E-05) GO-term category in the group of 16 genes was regulation of nitric oxide biosynthesis processes, which included the genes hsp90aa1 and ptx3 (Table 2A). The antigen processing and presentation pathway was the only KEGG pathway that was significantly regulated after PO and by NF-κB (Table 2B). There were two genes that were grouped into the antigen processing and presentation pathway; they were hsp90aa1 and hspa1a (Table 2B).
Table 2.
NF-κB cardioprotective gene program
(A) Significantly regulated gene ontology categories (P<0.01) of the 16 genes regulated by NF-κB and after PO.
(B) KEGG pathways that were significantly regulated (P<0.01). FDR= false discovery rate
| A | ||||
|---|---|---|---|---|
| GO Term ID | GO Term Category | P-Values | FDR | Gene symbol in category |
| GO:0045428 | Regulation of nitric oxide biosynthetic process | 1.80E-05 | 0.003342 | hsp90aa1, ptx3 |
| GO:0006809 | Nitric oxide biosynthetic process | 6.11E-05 | 0.003342 | hsp90aa1, ptx3 |
| GO:0046209 | Nitric oxide metabolic process | 6.11E-05 | 0.003342 | hsp90aa1, ptx3 |
| GO:0051171 | Regulation of nitrogen compound metabolic process | 6.82E-05 | 0.003342 | hsp90aa1, ptx3 |
| GO:0031328 | Positive regulation of cellular biosynthetic process | 0.000109 | 0.004308 | hsp90aa1, ptx3 |
| GO:0009607 | Response to biotic stimulus | 0.000300 | 0.009805 | hsp90aa1, ptx3, plscr1 |
| GO:0044271 | Nitrogen compound biosynthetic process | 0.001324 | 0.034026 | hsp90aa1, ptx3 |
| GO:0001558 | Regulation of cell growth | 0.001388 | 0.034026 | igfbp3, myocd |
| GO:0031326 | Regulation of cellular biosynthetic process | 0.001730 | 0.034793 | hsp90aa1, ptx3 |
| GO:0065008 | Regulation of biological quality | 0.001865 | 0.034793 | edn2, igfbp3, hspa1a, myocd |
| GO:0016049 | Cell growth | 0.001952 | 0.034793 | igfbp3, myocd |
| GO:0009891 | Positive regulation of biosynthetic process | 0.002182 | 0.035652 | hsp90aa1, ptx3, myocd |
| GO:0008361 | Regulation of cell size | 0.002435 | 0.036722 | igfbp3, myocd |
| GO:0031325 | Positive regulation of cellular metabolic process | 0.003087 | 0.041599 | hsp90aa1, ptx3, myocd |
| GO:0009893 | Positive regulation of metabolic process | 0.003183 | 0.041599 | hsp90aa1, ptx3, myocd |
| GO:0051707 | Response to other organism | 0.005105 | 0.062378 | ptx3, plscr1 |
| GO:0040008 | Regulation of growth | 0.005410 | 0.062378 | igfbp3, myocd |
| GO:0019229 | Regulation of vasoconstriction | 0.006526 | 0.065506 | edn2 |
| GO:0009620 | Response to fungus | 0.007826 | 0.065506 | ptx3 |
| GO:0001933 | Negative regulation of protein amino acid phosphorylation | 0.008476 | 0.065506 | igfbp3 |
| GO:0051147 | Regulation of muscle cell differentiation | 0.008476 | 0.065506 | myocd |
| GO:0050764 | Regulation of phagocytosis | 0.009125 | 0.065506 | ptx3 |
| GO:0050766 | Positive regulation of phagocytosis | 0.009125 | 0.065506 | ptx3 |
| GO:0002376 | Immune system process | 0.009451 | 0.065506 | hsp90aa1, ptx3, plscr1 |
| GO:0045595 | Regulation of cell differentiation | 0.009614 | 0.065506 | hsp90aa1, myocd |
| B | ||||
|---|---|---|---|---|
| KEGG ID | KEGG Pathway | P-Values | FDR | Gene symbol in category |
| mmu04612 | Antigen processing and presentation | 0.00145426 | 0.007271 | hsp90aa1, hspa1a |
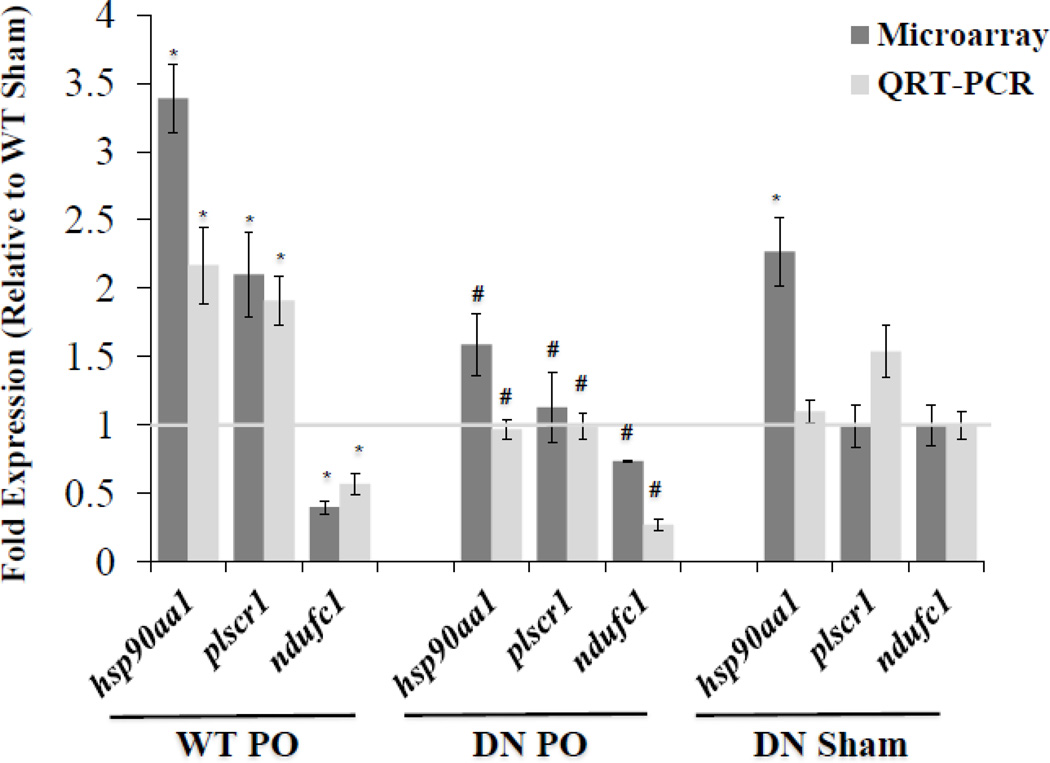
Gene expression patterns predicted by microarrays were quantitatively assessed using QRT-PCR to validate selected mRNA expression of two up-regulated genes plscr1, hsp90aa1, and one down-regulated gene, ndufc1 (Figure 4). QRT-PCR analysis confirmed the increased expression of plscr1 (1.91 fold) and hsp90aa1 (2.17 fold) during PO relative to sham (2.10 fold, and 3.39 fold receptivity predicted by microarrays) (Figure 4). In addition, NF-κB suppression (DN) resulted in basal expression of plscr1 and hsp90aa1, indicating these genes are positively regulated by NF-κB in response to PO (Figure 4). Microarrays (0.40 fold) and QRT-PCR (0.57 fold) analysis indicated that ndufc1 is down-regulated after PO compared to sham, but this was not reversed by NF-κB according to the QRT-PCR assay, in fact levels were further depressed by NF-κB blockade (Figure 4).
Figure 4. Gene expression confirmation by QRT-PCR.
Gene expression patterns predicted by microarrays were confirmed using QRT-PCR to validate selected mRNA expression of the following up-regulated genes: hsp90aa1, plscr1, and down-regulated gene ndufc1. Dark (microarray) and light bars (QRT-PCR) correspond to mRNA levels displayed as a fold change (mean±SEM) vs. WT Sham (represented by the light gray horizontal line at fold expression = 1). Microarray: *P<0.01 and >1.5 fold change vs. WT Sham, #P<0.01 and >1.5 fold change vs. WT PO, n=4. QRT-PCR: *P≤0.05 vs. WT Sham, #P≤0.05 vs. WT PO, n= 4–10.
3.6 Stress inducible heat shock protein 70 (hspa1a/hsp70.3 & hspa1b/hsp70.1)
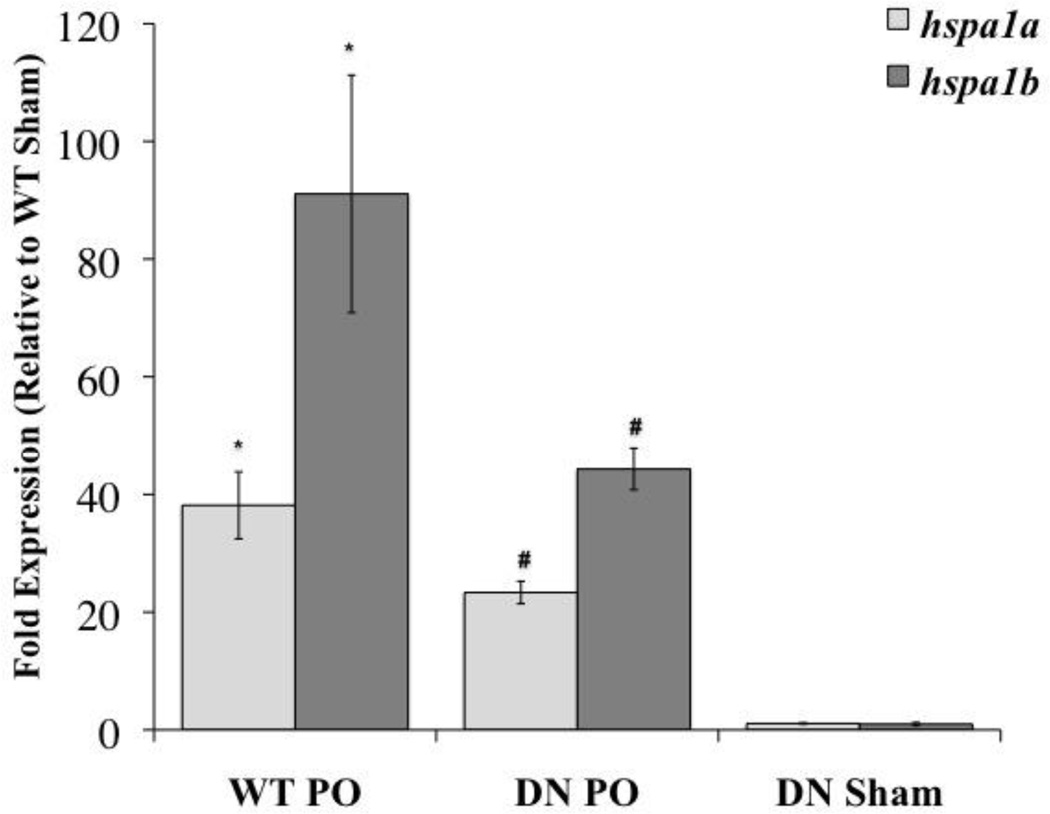
Our microarray analysis showed that hspa1b was the most significantly (P=1.88 ×10−11) and the highest regulated gene product after PO (59.59 fold compared to sham). Genetic blockade of NF-κB (DN mice) resulted in 2.01 fold reduction compared to WT PO (WT PO vs. DN PO, 29.56 fold compared to WT sham; P=0.04). QRT-PCR analysis showed that both hspa1a (38.16 fold over sham, P≤0.05) and hspa1b (91.07 fold over sham, P≤0.05) were highly induced after PO, and transcript levels (hspa1a: 23.34 fold over WT sham; hspa1b: 44.32 fold over WT sham) were significantly (P≤0.05) decreased in the DN mice compared to WT (Figure 5). Our results indicate hspa1a and hspa1b are both highly induced after PO and are regulated by NF-κB after PO.
Figure 5. QRT-PCR: hspa1a and hspa1b.
Both hspa1a (light bars) and hspa1b (dark bars) were highly induced after PO and transcript levels were significantly decreased in the DN mice. Dark and light bars correspond to mRNA levels displayed as a fold change (mean±SEM) vs. WT Sham (fold expression = 1). *P≤0.05 vs. WT Sham, #P≤0.05 vs. WT PO, n= 4–10.
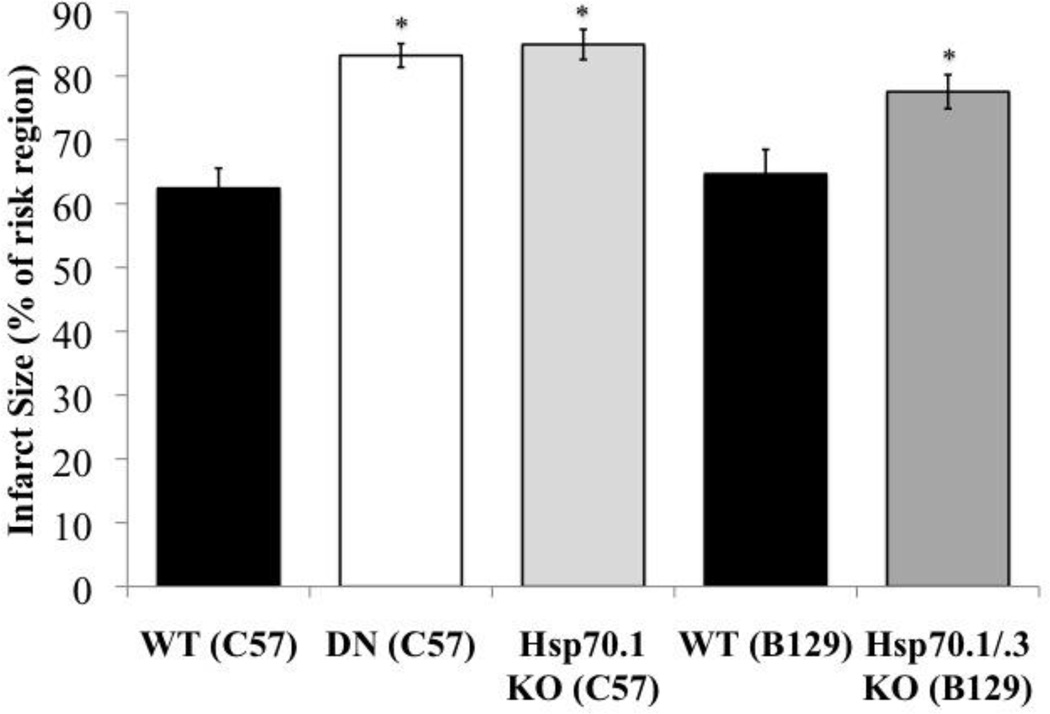
To assess the functional role of hspa1b gene product (Hsp70.1) during PO, Hsp70.1 KO and control mice were subjected to PO. Hsp70.1 KO had a significantly (P≤0.05) larger infarct (84.95%) compared to WT/C57 (62.42%) after PO (Figure 6). There was no difference in infarct size between Hsp70.1 KO (84.95%) and DN mice (83.23%) subjected to PO (Figure 6).
Figure 6. Functional studies: Hsp70.1 (hspa1b) and Hsp70.3 (hspa1a).
Assessment of infarct size (as a percent of risk region) after 24 hours PO in WT (C57) (black bar), DN (C57) (white bar), Hsp70.1 KO (C57) (light gray bar), WT (B129) (black bar) and Hsp70.1/.3 KO (B129) (dark gray bar). *P≤0.05 vs. corresponding WT, n=4–7. Bars are plotted as mean±SEM. Percent of risk region, percent of infarct/left ventricle, LV weight (mg), and survival rates are located in the supplement section (functional studies: Hsp70.1/hspa1b and Hsp70.3/hspa1a).
Since Hsp70.3 KO was not made available to us, we used the Hsp70.1/.3 double knockout mice to assess the functional role of hspa1a gene product (Hsp70.3). Hsp70.1/.3 KO mice had a significantly (P≤0.05) larger infarct (77.55%) compared to WT/B129 (64.66%) after 24 hours PO (Figure 6). There were no significant differences in infarct size between Hsp70.1 KO mice (84.95%) and Hsp70.1/.3 KO mice (77.55%) (Figure 6).
Discussion
There is controversy regarding the role of NF-κB in cell survival/cell death after ischemic insult. For example, NF-κB activation is cardioprotective after PO and after IPC [4,6,14,15]. However, NF-κB activation is also associated with cell death after I/R [4,6,14,15]. We previously have shown that the antithetical results of NF-κB are not due to specific mouse strains, differences in surgical models, or surgical technique [4]. From this data and the existing results in the literature, we developed the hypothesis that the regulation of different sets of NF-κB-dependent genes underlies the mechanistic basis of the differential effect of NF-κB upon cardioprotection vs. cell death after different ischemic stimuli [4]. Our objective was to identify the NF-κB-dependent gene network initiated after PO, to functionally validate components of this network, and to compare this with the NF-κB-dependent gene network that we already elucidated for late IPC [15].
Apoptotic and necrotic cell death is initiated within 2 to 6 hours after an initial ischemic event [13,14]. Misra et al., showed a significant increase in TUNEL positive cells in IκBαΔN mice compared to WT after 3 hours, with a further increase after 6 hours of PO [14]. These results suggest that NF-κB suppresses apoptotic cell death at 3 and 6 hours after PO and are consistent with our results that the major effect of cardiomyocyte-specific NF-κB abrogation upon infarct size occurs between 4 and 6 hours after coronary occlusion (Figure 1).
We hypothesized that NF-κB translocation into the nucleus occurs between 1 to 4 hours after PO, which, allowing time for primary gene expression, would explain the observation that NF-κB-dependent cardioprotection occurs within 4 to 6 hours after PO. NF-κB translocation to the nucleus was assessed by Western blot to quantify the amount of p65 in the nucleus after 1, 2, 3 and 4 hours after PO. There was a significant increase of p65 levels in the nucleus beginning 2 hours after PO with a steady level present to 4 hours PO (Figure 2). These results suggested that NF-κB is activated 2–4 hours after PO and this would allow time for transcription and translation of genes required to suppress cell death by 6 hours after PO. Increased levels of mRNA from NF-κB-dependent genes are known to be detectable as soon as 60 minutes following NF-κB activation (early response genes), while intermediate and late primary target genes are highly expressed by 3 hours [24,25]. In theory then, the peak regulation of the expression of the genes in question ought to occur between 4–6 hours after PO; therefore, we examined mRNA steady state levels after 5 hours PO in our model.
The main objective of this study was to identify genes that underlie the NF-κB-dependent cardioprotective effect after PO. Microarray comparison of mRNA levels between WT PO (5 hours) and WT sham (5 hours) mice resulted in 565 genes (595 oligonucleotide probes) significantly up- or down-regulated (P<0.01 and fold change of >1.5 fold) after PO (see supplement for details). Genes that were significantly up- and down-regulated after PO (565 genes) were compared to genes that were significantly up- and down-regulated by NF-κB in response to PO (254 genes). Comparison of the two sets of genes produced a set of 16 genes, candidates for contributing to NF-κB-dependent cardioprotection after PO (Figure 3).
Out of the 16 genes that were identified, four (4/16, 25%) (hsp90aa1, plscr1, ndufc1, and hspa1a) were chosen for validation of expression patterns predicted by our microarrays by QRT-PCR. These four genes were selected based upon the following criteria, representation of, 1) multiple expression patterns (both up- and down-regulated after PO and by NF-κB), 2) highly significant regulation after PO by NF-κB, and 3) likelihood of being involved in cardioprotection based on previous literature. In addition, we validated the mRNA expression level of hspa1b. The mRNA expression of, hsp90aa1, plscr1, hspa1a, and hspa1b were all confirmed by QRT-PCR to be significantly (P≤0.05) up-regulated after PO and by NF-κB (Figures 4 & 5). Therefore we concluded that the hspa1b mRNA is up-regulated by NF-κB after PO (P=1.88×10−11, fold over WT sham = 59.50), but was excluded by the array analysis as a false-negative. Expression levels of ndufc1 was confirmed by QRT-PCR to be significantly (P≤0.05) down-regulated after PO; however the QRT-PCR results suggested that, although levels dropped after PO, NF-κB significantly up-regulated ndufc1 after PO (Figure 4). This is likely due to the relatively small change in ndufc1 mRNA levels. Microarrays are considered to be accurate (up to 90%) in the identification of gene expression patterns (e.g. up or down expression level) [26]; however QRT-PCR is considered to be more accurate in measuring the quantitative expression levels of transcripts [26]. In general, where the two quantitative measures differ, we consider the QRT-PCR value the more accurate. We conclude that although ndufc1 mRNA levels are reduced after PO, that NF-κB seems to positively regulate the gene and does not mediate the reduction.
Of these four validated genes, several were previously implicated in cardioprotection. Hsp90, was shown to be cardioprotective after myocardial ischemia by enhancing nitric oxide synthesis [27,28,]. Our microarray analysis showed the most significant (P=1.80E-05) GO-term category was regulation of nitric oxide biosynthesis processes, which included the genes hsp90aa1 and ptx3. Both have been previously shown to be up-regulated by NF-κB [29–31], and are cardioprotective against myocardial ischemic injury [27,28,31]. The gene ptx3 is involved in the regulation of phagocytosis, one of the most significant GO term categories from our study. The genes hspa1a and plscr, were previously shown to be up-regulated by NF-κB (via cytokine induction) [12,32], and may contribute to cardioprotection, perhaps by regulating apoptosis [11,12,32,33]. The genes car3 and ndufc1, were not previoulsy known to be regulated by NF-κB or after ischemic stimuli. The ndufc1 gene, encodes an accessory subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase and is likely involved in transferring electrons from NADH to the respiratory chain [34]. The car3 gene encodes carbonic anhydrase, is involved in pH regulation in the heart, and was previously implicated in adaptation to ischemia and hypoxia [35–37].
Surprisingly, there were no bcl-2 anti-apoptotic gene family members that were significantly regulated by NF-κB after 5 hours PO. Misra et al. showed that Bcl-2 and c-IAP1 protein levels were reduced and suggested that this contributes to NF-κB-dependent cardioprotection after 1 hour PO. However, these investigators did not measure mRNA levels [14]. Perhaps these changes occur earlier, due to NF-κB inhibition, and are not due to NF-κB-dependent effects elicited by the ischemic stimulus.
Our microarray and QRT-PCR studies revealed that both stress inducible heat shock protein 70 (hspa1a and hspa1b) were highly expressed after PO and significantly (P≤0.05) up-regulated by NF-κB (Figure 5). Previous studies have shown that hspa1b (Rat hsp72) mRNA levels increase after PO [38]. In the mouse, rat and human, the stress inducible Hsp70 locus encodes hspa1a (encodes for Hsp70.3) and hspa1b (encodes for Hsp70.1), which are located in the major histocompatibility complex (MHC) III region, and are about 7kb apart in a head-to-tail orientation [41–43]. Hsp70.1 and Hsp70.3 are >99% identical to each other with two amino acid differences in the human proteins, whereas in the mouse, Hsp70.1 has one additional proline amino acid near the C-terminal end [15,17,39–41].
We employed Hsp70.1 KO and Hsp70.1/.3 KO mice (since, we were not able to obtain the Hsp70.3 KO) to investigate the individual role of Hsp70.1 and Hsp70.3 after PO. Hsp70.1 KO (C57) mice had a significantly larger infarct compared to WT (C57) mice; there were no significant differences in infarct size between the Hsp70.1 KO (C57) and DN (C57) mice (Figure 5). This result, taken together with the NF-κB-dependent regulation of the gene encoding Hsp70.1 (hspa1a), suggests that Hsp70.1 contributes to the NF-κB-dependent cardioprotection observed after PO. The Hsp70.1/.3 KO had a significantly larger infarct compared to WT (B129) mice, and infarct size was not significantly different between the Hsp70.1 and the Hsp70.1/.3 KOs (Figure 5). The relative contribution of Hsp70.3 after PO, though not directly measured, appears much less than Hsp70.1. However, the possibility of functional redundancy cannot be ruled out without the Hsp70.3 KO. Our results showed for the first time that Hsp70.1 (hspa1b) contributes to the NF-κB-dependent cardioprotective effect after PO.
Several studies, using various mouse models including Hsp70.1 KO, Hsp70.1/.3 KO and Hsp72 transgenic mice (Hsp70 TG) and various stressors, including myocardial ischemia, suggest that Hsp70 is protective [15,17,41–46]. For example, the double Hsp70 KO (Hsp70.1/.3 KO) has been shown previously to abrogate late ischemic preconditioning [15,41]. Hsp70.1 has been shown to be protective following ischemic stroke [17,45]. In this study we show that, after PO, activation of NF-κB and increased expression of Hsp70.1 contributes to cell survival, reducing infarct size after PO. We previously showed opposing roles for NF-κB activation in PO vs. I/R [4,6]. A recent study, by us, of NF-κB-dependent gene expression after late IPC demonstrated that Hsp70.3 plays a major cardioprotective role after late IPC, but that Hsp70.1 is pro-cell death after I/R without prior preconditioning [15,46]. Our results [15,46] suggest that the Hsp70.1 and Hsp70.3 are not simply redundant as has been assumed previously. The dissociation between mRNA levels and function that we observe may be due to post-transcriptional regulation, or to different relative avidities for binding partners. With regard to the latter, it is interesting to note that the single amino acid difference between the two Hsp70s is in the carboxy-terminus, which is known to be involved in some protein-protein interactions of Hsp70s [46]. However, due to the lack of antibodies specific for the various Hsp70s, we were unable to address this issue in the present study. Perhaps the antithetical action of Hsp70.1 in PO vs. I/R underlies the opposing effects of NF-κB (NF-κB paradox) in the ischemic models (PO vs. I/R) [46]. Nevertheless, the Hsp70s are unlikely to act alone in mediating the NF-κB effects after ischemic injury and there may indeed be post-transcriptional regulation of these gene programs that influences the functional roles of genes and gene combinations after these stressors.
In summary, our results were the following: 1) NF-κB acts after PO to limit infarct size and this myocardial salvage occurs between 4 to 6 hours after coronary ligation, 2) a significant amount of NF-κB is present in the nucleus starting at 2 hours and peaking at 3–4 hours after PO, 3) NF-κB-dependent up-regulation of Hsp70.1 is a major contributor to the NF-κB-dependent myocardial protection/salvage, and 4) microarray analysis resulted in the identification of 16 genes that are likely to be involved in NF-κB-dependent cardioprotection after PO. These 16 genes included both previously known NF-κB regulated genes (e.g. hspa1a, hsp90aa1, and plscr1) and genes not known to be regulated by NF-κB (e.g. car3, ankrd9, and ndufc1). We showed that NF-κB activation regulates different but overlapping gene programs depending upon the stimulus. The results of this study contribute significantly to our understanding the mechanism of NF-κB-induced myocardial salvage after PO, specifically, by the identification of a unique set of genes which may underlie it. The identification of NF-κB-dependent cardioprotective genes may result in the development of novel therapeutic targets to enhance cardioprotection during myocardial ischemia.
RESEARCH HIGHLIGHTS.
The transcription factor NF-κB is cardioprotective after permanent coronary ligation (PO)
NF-κB regulates different but overlapping gene expression networks after PO relative to late IPC
Hsp70.1 is up-regulated by NF-κB and is protective after PO, despite being proinfarct after I/R
The antithetical effect of Hsp70.1 may underlie that of NF-κB
Study of different gene sets regulated by NF-κB may identify new therapeutic targets
Supplementary Material
Acknowledgments
Acknowledgment and funding sources
This research was supported by NIH grants HL63034 (W.K. Jones), HL091478 (W. K. Jones). M.E. Wilhide was supported by National Research Service Award (NRSA) NIH Predoctoral F31 Fellowship (HL081923). We would like to thank Jackie Belew for assistance in mouse breeding and organizing procedures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None of the authors have disclosures to make relevant to this work.
References
- 1.Lloyd-Jones D, Adams R, Brown TM, Carnethon M, Dai S, De Simone G, et al. Heart disease and stroke statistics--2010 update: a report from the American Heart Association. Circulation. 2010;121(7):e46–e215. doi: 10.1161/CIRCULATIONAHA.109.192667. [DOI] [PubMed] [Google Scholar]
- 2.Zucchi R, Ronca F, Ronca-Testoni S. Modulation of sarcoplasmic reticulum function: a new strategy in cardioprotection? Pharmacol Ther. 2001;89(1):47–65. doi: 10.1016/s0163-7258(00)00103-0. [DOI] [PubMed] [Google Scholar]
- 3.Deten A, Volz HC, Briest W, Zimmer HG. Cardiac cytokine expression is upregulated in the acute phase after myocardial infarction. Experimental studies in rats. Cardiovasc Res. 2002;55(2):329–450. doi: 10.1016/s0008-6363(02)00413-3. [DOI] [PubMed] [Google Scholar]
- 4.Jones WK, Brown M, Wilhide M, He S, Ren X. NF-kappa B in Cardiovascular Disease: Diverse and Specific of a “General” Transcription Factor. Cardiovasc Toxicol. 2005;5(2):183–202. doi: 10.1385/ct:5:2:183. [DOI] [PubMed] [Google Scholar]
- 5.Dawn B, Xuan YT, Marian M, Flaherty MP, Murphree SS, Smith TL, et al. Cardiac-specific Abrogation of NF-KappaB activation in mice by transdominant expression of a mutant Ikappa-Balpha. J Mol Cell Cardiol. 2001;33:161–173. doi: 10.1006/jmcc.2000.1291. [DOI] [PubMed] [Google Scholar]
- 6.Brown M, McGuiness M, Wright T, Ren X, Wang Y, Boivin GP, et al. Cardiac-specific blockade of NF-kappaB in cardiac pathophysiology: differences between acute and chronic stimuli in vivo. Am J Physiol Heart Circ Physiol. 2005;289(1):H466–H476. doi: 10.1152/ajpheart.00170.2004. [DOI] [PubMed] [Google Scholar]
- 7.Haudek SB, Bryant DD, Giroir BP. Differential regulation of myocardial NF kappa B following acute or chronic TNF-alpha expression. J Mol Cell Cardiol. 2001;33:1263–1271. doi: 10.1006/jmcc.2001.1388. [DOI] [PubMed] [Google Scholar]
- 8.Higuchi Y, Chan TO, Brown MA, Zhang J, DeGeorge BR, Funakoshi H, et al. Cardioprotection afforded by NF-κB ablation is associated with activation of Akt in mice overexpressing TNF-α. Am J Physiol Heart Circ Physiol. 2006;290:H590–H598. doi: 10.1152/ajpheart.00379.2005. [DOI] [PubMed] [Google Scholar]
- 9.Mustapha S, Kirshner A, De Moissac D, Kirshenbaum LA. A direct requirement of nuclear factor-kappa B for suppression of apoptosis in ventricular myocytes. Am J Physiol Heart Circ Physiol. 2000;279(3):H939–H945. doi: 10.1152/ajpheart.2000.279.3.H939. [DOI] [PubMed] [Google Scholar]
- 10.Bergmann MW, Loser P, Dietz R, von Harsdorf R. Effect of NF-kappa B inhibition on TNF-alpha-induced apoptosis and downstream pathways in cardiomyocytes. J Mol. Cell Cardiol. 2001;33(6):1223–1232. doi: 10.1006/jmcc.2001.1385. [DOI] [PubMed] [Google Scholar]
- 11.Nakano M, Mann DL, Knowlton AA. Blocking the endogenous increase in HSP 72 increases susceptibility to hypoxia and reoxygenation in isolated adult feline cardiocytes. Circulation. 1997;95(6):1523–1531. doi: 10.1161/01.cir.95.6.1523. [DOI] [PubMed] [Google Scholar]
- 12.Nakano M, Knowlton AA, Dibbs Z, Mann DL. Tumor Necrosis factor-alpha confers resistance to hypoxic injury in the adult mammalian cardiac myocyte. Circulation. 1998;97(14):1392–1400. doi: 10.1161/01.cir.97.14.1392. [DOI] [PubMed] [Google Scholar]
- 13.Kurrelmeyer KM, Michael LH, Baumgarten G, Taffet GE, Peschon JJ, Sivasubramanian N, et al. Endogenous tumor necrosis factor protects the adult cardiac myocyte against ischemic-induced apoptosis in a murine model of acute myocardial infarction. Proc Natl Acad Sci USA. 2000;97(10):5456–5461. doi: 10.1073/pnas.070036297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Misra A, Haudek SB, Knuefermann P, Vallejo JG, Chen ZJ, Michael LH, et al. Nuclear factor-kappaB protects the adult cardiac myocyte against ischemia-induced apoptosis in a murine model of acute myocardial infarction. Circulation. 2003;108(25):3075–3078. doi: 10.1161/01.CIR.0000108929.93074.0B. [DOI] [PubMed] [Google Scholar]
- 15.Tranter M, Ren X, Forde T, Wilhide ME, Chen J, Sartor MA, et al. NF-κB driven cardioprotective gene programs; Hsp70.3 and cardioprotection after late ischemic precondition. J Mol Cell Cardiol. 2010;49(4):664–672. doi: 10.1016/j.yjmcc.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.REVISED GUIDE FOR THE CARE AND USE OF LABORATORY ANIMALS NIH GUIDE. 1996 Aug 16;Volume 25(Number 28) [Google Scholar]
- 17.Lee SH, Kim M, Yoon BW, Kim YJ, Ma SJ, Roh JK, et al. Targeted hsp70.1 Disruption Increases Infarction Volume After Focal Cerebral Ischemia in Mice. Stroke. 2001;32:2905–2912. doi: 10.1161/hs1201.099604. [DOI] [PubMed] [Google Scholar]
- 18.Shim EH, Kim J, 2nd, Bang ES, Heo JS, Lee JS, Kim EY, et al. Target disruption of hsp70.1 sensitizes to osmotic stress. EMBO Reports. 2002;3(9):857–861. doi: 10.1093/embo-reports/kvf175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt CR, Dix DJ, Sharma GG, Pandita RK, Gupta A, Funk M, et al. Genomic Instability and Enhanced Radiosensitivity in Hsp70.1- and Hsp70.3-Deficient Mice. Mol Cell Biol. 2004;24(2):889–911. doi: 10.1128/MCB.24.2.899-911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren X, Wang Y, Jones WK. TNF-alpha is required for late ischemic preconditioning but not for remote preconditioning of trauma. J. Surg. Res. 2004;121:120–129. doi: 10.1016/j.jss.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 21.Fishbein MC, Meerbaum S, Rit J, Lando U, Kanmatsuse K, Mercier JC, et al. Early phase acute myocardial infarct size quantification: validation of the triphenyl tetrazolium chloride tissue enzyme staining technique. Am Heart J. 1981;101:593–600. doi: 10.1016/0002-8703(81)90226-x. [DOI] [PubMed] [Google Scholar]
- 22.Freudenberg JM, Joshi VK, Hu Z, Medvedovic M. CLEAN: CLustering Enrichment ANalysis. BMC Bioinformatics. 2009;10:234. doi: 10.1186/1471-2105-10-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Sung MH, Salvatore L, De Lorenzi R, Indrawan A, Pasparakis M, Hager GL, et al. Sustained oscillations of NF-kappaB produce distinct genome scanning and gene expression profiles. PLoS One. 2009;4(9):e7163. doi: 10.1371/journal.pone.0007163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian B, Nowak DE, Brasier AR. A TNF-induced gene expression program under oscillatory NF-kappaB control. BMC Genomics. 2005;6:137. doi: 10.1186/1471-2164-6-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Draghic S, Khatri P, Eklund AC, Szallasi Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006;22(2):101–109. doi: 10.1016/j.tig.2005.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffin TM, Valdez TV, Mestril R. Radicocol activates heat shock protein expression and cardioprotection in neonatal rat cardiomyocytes. Am J Physiol Heart Circ Physiol. 2004;287(3):H1081–H1088. doi: 10.1152/ajpheart.00921.2003. [DOI] [PubMed] [Google Scholar]
- 28.Kupatt C, Dessy C, Hinkel R, Raake P, Daneau G, Bouzin C, et al. Heat shock protein 90 transfection reduces ischemia-reperfusion-induced myocardial dysfunction via reciprocal endothelial NO synthase serine 1177 phosphorylation and threonine 495 dephosphorylation. Arterioscler Thromb Vasc Biol. 2004;24(8):1435–1441. doi: 10.1161/01.ATV.0000134300.87476.d1. [DOI] [PubMed] [Google Scholar]
- 29.Altmeyer A, Klampfer L, Goodman AR, Vilcek J. Promoter structure and transcriptional activation of the murine TSG-14 gene encoding a tumor necrosis factor/interleukin-1-inducible pentraxin protein. J Biol Chem. 1995;270(43):25584–25590. doi: 10.1074/jbc.270.43.25584. [DOI] [PubMed] [Google Scholar]
- 30.Basile A, Sica A, d’Aniello E, Breviario F, Garrido G, Castellano M, et al. Characterization of the promoter for the human long pentraxin PTX3. Role of NF-kappaB in tumor necrosis factor-alpha and interleukin-1beta regulation. J Biol Chem. 1997;272(13):8172–8178. doi: 10.1074/jbc.272.13.8172. [DOI] [PubMed] [Google Scholar]
- 31.Sailo M, Chimenti S, De Angelis N, Molla F, Nebuloni M, Pasqualini F, et al. Cardioprotective function of the long pentraxin PTX3 in acute myocardial infarction. Circulation. 2008;117(8):1055–1064. doi: 10.1161/CIRCULATIONAHA.107.749234. [DOI] [PubMed] [Google Scholar]
- 32.Lu B, Sims PJ, Wiedmer T, Moser AH, Shigenaga JK, Grunfield C, et al. Expression of the phospholipid scramblase (PLSCR1) gene family during the acute phase response. Biochem Biophys Acta. 2007;1177(9):1777–1785. doi: 10.1016/j.bbalip.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Hamilton KL, Gupta S, Knowlton AA. Estrogen and regulation of heat shock protein expression in female cardiomyocytes: cross-talk with NF kappa B signaling. J Mol Cell Cardiol. 2004;36(4):577–584. doi: 10.1016/j.yjmcc.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Loeffen JL, Triepels RH, van den Heuvel LP, Schuelke M, Buskens CA, Smeets RJ, et al. cDNA of eight nuclear encoded subunits of NADH:ubiquinone oxidoreductase: human complex I cDNA characterization completed. Biochem Biophys Res Commun. 1998;253(2):415–422. doi: 10.1006/bbrc.1998.9786. [DOI] [PubMed] [Google Scholar]
- 35.Holotnakova T, Ziegelhoffer A, Ohradanova A, Hulikova A, Novakova M, Kopacek J, et al. Induction of carbonic anhydrase IX by hypoxia and chemical disruption of oxygen sensing in rat fibroblasts and cardiomyocytes. Pflugers Arch. 2008;456(2):323–337. doi: 10.1007/s00424-007-0400-6. [DOI] [PubMed] [Google Scholar]
- 36.Strnisková M, Ravingerová T, Neckár J, Kolár F, Pastoreková S, Barancík M. Changes in the expression and/or activation of regulatory proteins in rat hearts adapted to chronic hypoxia. Gen Physiol Biophys. 2006;25(1):25–41. [PubMed] [Google Scholar]
- 37.Vandenberg JI, Carter ND, Bethell HW, Nogradi A, Ridderstråle Y, Metcalfe JC, Grace AA. Carbonic anhydrase and cardiac pH regulation. Am J Physiol. 1996;271(6 PT 1):C1838–C1846. doi: 10.1152/ajpcell.1996.271.6.C1838. [DOI] [PubMed] [Google Scholar]
- 38.Yu H, Yokoyama M, Asano G. Time Course of Expression and Localization of Heat Shock Protein 72 in the Ischemic and Reperfused Rat Heart. Jpn Circ J. 1999;63:278–287. doi: 10.1253/jcj.63.278. [DOI] [PubMed] [Google Scholar]
- 39.Gorzowski JJ, Eckerley CA, Halgren RG, Mangurten AB, Phillips B. Methylation-associated Transcriptional Silencing of the Major Histocompatibility Complex-linked hsp70 Genes in Mouse Cell Lines. JBC. 1995;45(10):26940–26949. doi: 10.1074/jbc.270.45.26940. [DOI] [PubMed] [Google Scholar]
- 40.Milner CM, Campbell DR. Structure and expression of the three MHC-linked HSP70 genes. Immunogenetics. 1990;32:242–251. doi: 10.1007/BF00187095. [DOI] [PubMed] [Google Scholar]
- 41.Hampton CR, Shimamoto A, Rothnie CL, Griscavage-Ennis J, Chong A, Dix DJ, et al. HSP70.1 and -70.3 are required for late-phase protection induced by ischemic mouse hearts. Am J Physiol Heart Circ Physiol. 2003;285(2):H866–H874. doi: 10.1152/ajpheart.00596.2002. [DOI] [PubMed] [Google Scholar]
- 42.Kim YK, Suarez J, McDonough PM, Boer C, Dix DJ, Dillman WH. Deletion of the inducible 70-kDa heat shock protein genes in mice impairs cardiac contractile function and calcium handing associated with hypertrophy. Circulation. 2006;113(22):2589–2597. doi: 10.1161/CIRCULATIONAHA.105.598409. [DOI] [PubMed] [Google Scholar]
- 43.Plumier JC, Ross BM, Currie RW, Angelidis CE, Kazlaris H, Kollias G, et al. Transgenic mice expressing the human heat shock protein 70 have improved post-ischemic myocardial recovery. J Clin Invest. 1995;95:1854–1860. doi: 10.1172/JCI117865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuchiya D, Hong S, Matsumori Y, Shiina H, Kayama T, Swanson RA, et al. Overexpression of rat heat shock protein 70 is associated with reduction of early mitochondrial cytochrome C release and subsequent DNA fragmentation after permanent focal ischemia. J Cereb Blood Flow Metab. 2003;23(6):718–727. doi: 10.1097/01.WCB.0000054756.97390.F7. [DOI] [PubMed] [Google Scholar]
- 45.Lee SH, Kwon HM, Kim YJ, Lee KM, Yoon BW. Effects of hsp70.1 gene knockout on the mitochondrial apoptotic pathway after focal cerebral ischemia. Stroke. 2004;35(9):2195–2199. doi: 10.1161/01.STR.0000136150.73891.14. [DOI] [PubMed] [Google Scholar]
- 46.Wilhide ME. Hsp70.1 contributes to the NF-κB paradox after myocardial ischemic insults [dissertation] Cincinnati (OH): University of Cincinnati; 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.