Abstract
Protein tyrosine phosphatase (PTP) 1B has been implicated as a negative regulator of multiple signaling pathways downstream of receptor tyrosine kinases. Inhibition of this enzyme was initially thought to potentially lead to increased oncogenic signaling and tumorigenesis. Surprisingly, we show that platelet-derived growth factor-stimulated extracellular-regulated kinase signaling in PTP1B-deficient cells is not significantly hyperactivated. Moreover, these cells exhibit decreased Ras activity and reduced proliferation by way of previously uncharacterized pathways. On immortalization, PTP1B-deficient fibroblasts display increased expression of Ras GTPase-activating protein (p120RasGAP). Furthermore, we demonstrate that p62Dok (downstream of tyrosine kinase) is a putative substrate of PTP1B and that tyrosine phosphorylation of p62Dok is indeed increased in PTP1B-deficient cells. Consistent with the decreased Ras activity in cells lacking PTP1B, introduction of constitutively activated Ras restored extracellular-regulated kinase signaling and their proliferative potential to those of WT cells. These results indicate that loss of PTP1B can lead to decreased Ras signaling, despite enhanced signaling of other pathways. This finding may in part explain the absence of increased tumor incidence in PTP1B-deficient mice. Thus, PTP1B can positively regulate Ras activity by acting on pathways distal to those of receptor tyrosine kinases.
Protein tyrosine phosphatase (PTP) 1B is the prototype for the superfamily of PTPs and has been implicated in multiple signaling pathways (1). Of particular interest, gene-targeting studies in mice have established PTP1B as a critical physiological regulator of metabolism by attenuating insulin, leptin, and growth hormone signaling (2–6). PTP1B function seems to be dispensable for embryonic development. However, PTP1B-deficient mice exhibit resistance to diabetes and obesity, the two major metabolic diseases in industrialized societies. Not surprisingly, PTP1B is a highly regarded target of the pharmaceutical industry in the treatment of these disorders (7).
Because PTP1B is a negative regulator of multiple receptor tyrosine kinases (RTKs) (1), the concern is that PTP1B inhibition may lead to increased oncogenic signaling. Indeed, PTP1B-deficient fibroblasts display increased insulin-like growth factor I (IGF-I) receptor, epidermal growth factor receptor, and platelet-derived growth factor receptor (PDGFR) tyrosine phosphorylation (8, 9). Regardless of this potentially enhanced oncogenic signaling, PTP1B-deficient mice do not overtly undergo tumorigenesis. One possibility is that PTP1B may not regulate RTK signaling in all cell types, or that functional redundancy may exist. Alternatively, loss of PTP1B may affect the progression of a tumorigenic event, but not its rate of initiation. Finally, it is also possible that PTP1B may be involved in the activation of oncogenic pathways downstream of RTKs.
We decided to pursue the third alternative based on our previous studies with PTP1B in the IGF-I receptor pathway. Paradoxically, IGF-I-stimulated extracellular-regulated kinase (Erk) phosphorylation in PTP1B-deficient fibroblasts is significantly diminished. This finding could, in part, be explained by previous results suggesting that PTP1B is involved in the activation of Src (10, 11). Indeed, we demonstrated that adhesion-mediated Erk and Src activation in PTP1B-deficient fibroblasts are both impaired (12). However, most of our studies were performed with cells immortalized with the SV40 large T antigen (TAg). Importantly, TAg has been shown to abrogate the requirements of Src kinases during PDGF-induced mitogenesis (13), and PDGF-induced Erk activation in TAg-immortalized fibroblasts lacking Src kinases is relatively unchanged (14). This result suggested to us that there were additional mechanisms in PTP1B-deficient fibroblasts that were responsible for the diminished Erk activity.
In this study, we show that, although PTP1B-deficient cells exhibit increased PDGFR and AKT phosphorylation, Erk activation does not occur to the same extent. We show that loss of PTP1B results in diminished Ras activity and that this event occurs through increased p120RasGAP (Ras GTPase-activating protein) expression and p62Dok (downstream of tyrosine kinase) phosphorylation. Taken together, these results propose how PTP1B can act as a positive regulator of Ras signaling downstream of RTKs and may in part explain why PTP1B knockout mice do not present an increased incidence of tumors.
Methods
Antibodies. Rabbit polyclonal antibodies against PTP1B have been described (3). Additional antibodies were purchased from Cell Signaling Technology (Beverly, MA) (pan and phospho anti-AKT, anti-Erk); Upstate Biotechnology (Lake Placid, NY) (anti-phosphotyrosine 4G10); Santa Cruz Biotechnology (anti-PDGFRβ, anti-Dok1); Transduction Laboratories (Lexington, KY) (anti-p190RhoGAP, anti-p120RasGAP, anti-H-Ras, and anti-Shc); and BioSource International (Camarillo, CA) (anti-Src).
Cell Culture and Cell Lines. All cell lines were maintained in DMEM (Invitrogen) supplemented with 10% FBS (BioSource International) and antibiotics (5 mg/ml penicillin/streptomycin, Invitrogen). All spontaneously immortalized fibroblast cell lines derived from PTP1B or T cell PTP (TCPTP) knockout embryos have been described (9, 15). The SV40 TAg cells were rescued as described (9).
Generation of the V12Ras Clones. PTP1B WT or knockout spontaneously immortalized fibroblasts were transfected with a linearized V12Ras vector and a hygromycin selection vector (pMC1-HygR-pA) at a 10:1 ratio, respectively. Transfected cells were selected in DMEM containing 50–75 μg/ml hygromycin, and colonies were picked. Screening of positive clones was done by Western blotting by using anti-H-Ras antibody. In each case, at least 10 clones were isolated and further characterized.
Preparation of Cell Lysate and Immunoblotting. Cells were washed in ice-cold PBS and lysed in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl/50 mM Tris·HCl, pH 7.5/1% Nonidet P-40/0.25% sodium deoxycholate/1 mM Na3VO4/50 mM NaF) supplemented with Complete EDTA-free protease inhibitor mixture (Roche Molecular Biochemicals). Cell lysates were rotated end-over-end at 4°C for 10 min and cleared by centrifugation at 14,000 × g for 10 min at 4°C. The protein concentration was measured by the Bradford method (Bio-Rad). Protein samples were resolved by 8% SDS/PAGE and subjected to immunoblotting with the indicated antibodies.
Immunoprecipitation. Cell lysates were incubated with anti-p62Dok antibodies and 20 μl of protein G-Agarose (Invitrogen) at 4°C for 2 h. Precipitates were washed in lysis buffer, resuspended in SDS sample buffer, and resolved by 8% SDS/PAGE for immunoblot analysis.
Semiquantitative RT-PCR. Total RNA was isolated from cells by using TRIzol (Invitrogen), and first-strand cDNA synthesis was obtained from 1 μg of RNA with random hexamers by using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The cDNA was then used as a template for PCR with two sets of synthesized primers. Aliquots of 1 μl of the reverse transcription reaction were amplified by using 1 unit of AmpliTaq Gold (Applied Biosystems) under the following PCR conditions: 5 min at 95°C, then 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C for 30 cycles, 2 min at 72°C) in a 25-μl reaction mixture, by using 100 pmol each of the sense and antisense primers: 5′RasGAP, 5′-GGGTGTTTACAGAAATCAGTTC-3′; and 3′RasGAP, 5′-CTCATTGCTGAGTGTTCTCAG-3′. In parallel, a GAPDH PCR was performed to control for the RNA input in the RT-PCR: 5′GAPDH, 5′-AACGACCCCTTCATTGAC-3′; and 3′GAPDH, 5′-TCCACGACATACTCAGCAC-3′. Reaction products were separated by 1% agarose gel electrophoresis and detected with ethidium bromide staining.
Subtrate-Trapping Experiments. Details of the PTP1B WT and D181A constructs have been described (16). NIH 3T3 c-Src Y527F cells were transfected by Lipofectamine (Invitrogen) according to the manufacturer's instructions. Forty-eight hours posttransfection, cells were lysed in buffer containing 50 mM Hepes (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, and 10% glycerol (supplemented with Complete EDTA-free protease inhibitors). Crude lysates were then cleared by centrifugation at 14,000 × g. GST-tagged proteins were precipitated by using 25 μl of Glutathione Sepharose beads (Pharmacia), washed extensively in lysis buffer, and then resuspended in SDS sample buffer. Aliquots were resolved by SDS/PAGE and analyzed by immunoblotting with the indicated antibodies.
Soft Agar Assay. PTP1B WT and knockout cells stably expressing V12Ras were assessed for anchorage-independent growth by colony formation in soft agar. NIH 3T3 c-Src Y527F cells were used as positive control. The cells were plated at 3 × 103 cells per well in a six-well plate in triplicate, by using 0.35% low melting point agarose and grown in DMEM with 20% FBS. Media were changed every 3 days. Colony number was determined by scoring for those with a size >0.1 mm in size. Representative colonies were photographed in phase contrast from plates at day 10 of the assay (×10 magnification). Values are reported as the average of three experiments ± SE.
Ras-GTP Pull-Down Assay. Cells were lysed in MLB buffer (25 mM Hepes, pH 7.5/150 mM NaCl/1% Nonidet P-40/0.25% sodium deoxycholate/10% glycerol/25 nM NaF/1 mM Na3VO4/10 mM MgCl2/1 mM EDTA) supplemented with Complete EDTA-free protease inhibitor mixture. The level of Ras-GTP was determined by precipitation with a GST fusion protein of the Ras-binding domain on Raf1, which recognizes only active, GTP-bound Ras. Pull-downs were resolved by SDS PAGE and immunoblotted with an anti-Ras antibody to detect precipitated Ras-GTP.
Results
Differential Regulation of Downstream Pathways of the PDGFR by PTP1B. To investigate the mechanisms by which loss of PTP1B can attenuate Erk activation, we used the PDGF signaling system in TAg immortalized cells (TAg cells), for which Src function is dispensable (13, 14). Previous studies with PTP1B-deficient TAg cells showed that PDGF-stimulated Erk and AKT activation were not dramatically altered (8, 9). We used PDGF at a high concentration (50 ng/ml), allowing us to suspect that subtle changes by PTP1B may not be detectable in this case. Hence, in our studies, we used PDGF at much lower levels (10–20 ng/ml).
As expected, stimulation of TAg cells with low levels of PDGF resulted in increased cellular tyrosine phosphorylation and AKT activation, which was enhanced in PTP1B-deficient cells (Fig. 1). In contrast, PDGF-induced Erk phosphorylation was decreased in PTP1B knockout cells. Thus, PTP1B seems to differentially regulate signaling pathways diverging from the PDGFR.
Fig. 1.
Differential regulation of PDGF-signaling pathways by PTP1B. (a) Fibroblasts immortalized with SV40 TAg (+/+ PTP1B and –/– PTP1B) were serum-starved and stimulated with 10 ng/ml PDGF for indicated times or left unstimulated. The lysates were analyzed by Western blotting, and the membrane was probed with anti-phosphotyrosine antibodies. (b) The samples were analyzed for Erk and AKT proteins and phosphorylation levels by using phosphospecific antibodies.
PTP1B-Deficient Fibroblasts Display Decreased Growth and Ras Activity. Consistent with our data that IGF-I-(9) and PDGF-induced activation of Erk is impaired in PTP1B-deficient cells, we also observed that these cells display decreased monolayer growth compared with their WT counterparts (Fig. 2a). Similarly, although both WT and knockout cells were able to proliferate in 1% serum, PTP1B-deficient cells clearly present a diminished capacity to do so (Fig. 2b). Moreover, PTP1B-deficient TAg cells exhibit a consistent and reproducible elevation in PDGFR levels (Fig. 2c). This result is unlikely to be due to defective receptor internalization and degradation because stimulation with PDGF caused a rapid decrease in receptor levels in both PTP1B WT and knockout cells.
Fig. 2.
Decreased cell growth and Ras activity of SV40 TAg transformed fibroblasts lacking PTP1B. (a and b) Cells were seeded in a 24-well plate at a density of 1 × 104 cells per well in 10% or 1% FBS. At the indicated time points, the cells were trypsinized, and the total cell number per well was determined with a hemacytometer. Values are reported as the average of triplicate experiments ± SE. (c) PDGFR expression is increased in cells lacking PTP1B. Serum-starved cells were stimulated with PDGF for indicated times or left unstimulated. The lysates were analyzed by Western blotting for PDGFR expression. Equal loading was done by reprobing the membrane with antibodies against AKT/protein kinase B. (d) Serum-starved cells were stimulated with 20 ng/ml PDGF for 10 min or left unstimulated. Lysates were incubated with immobilized GST-Raf1-RBD (Raf-RBD) to precipitate active (GTP-bound) Ras. Ras-GTP was detected by using anti-H-Ras antibodies.
It has been established in fibroblasts that expression of the PDGFR is inversely proportional to Ras activity (17). Furthermore, Ras activity is necessary for full transformation by TAg (18). By using a Ras-GTP pull down assay, we indeed show that PTP1B-deficient TAg cells display diminished Ras-GTP levels (Fig. 2d). In addition, introduction of dominant active V12Ras in both PTP1B WT and knockout cells was able to suppress PDGFR levels (data not shown).
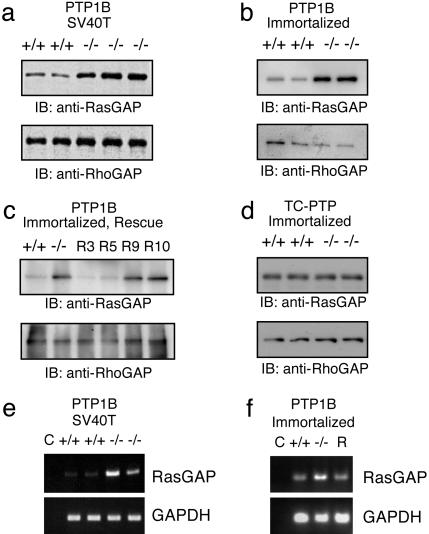
Immortalization Increases p120RasGAP Expression in PTP1B-Deficient Fibroblasts. To gain insight into the potential decrease in Ras activity in PTP1B-deficient cells, we first profiled several proteins upstream of the Erk signaling pathway. We found no significant alterations in the expression levels of the adapter proteins Shc or Grb2, or the kinases Src and Erk (data not shown). Importantly, however, we observed that the levels of p120RasGAP are elevated in PTP1B-deficient TAg cells (Fig. 3a). Yet, the levels of the related protein p190RhoGAP were not altered. The importance of this finding is underlined by the fact that p120RasGAP can attenuate Ras activity by promoting the intrinsic GTPase activity of Ras (19).
Fig. 3.
PTP1B-deficient cells display increased p120RasGAP expression. (a) Increased expression of p120RasGAP but not p190RhoGAP in TAg immortalized PTP1B-deficient fibroblasts. (b) Increased expression of RasGAP is also seen in spontaneously immortalized PTP1B-deficient fibroblasts. (c) Stable reexpression of PTP1B into PTP1B –/– cells decreases RasGAP levels. R3 and R5 are –/– cells that expressed PTP1B, and R9 and R10 are –/– cells mock transfected. (d) Expression level of RasGAP is not affected in TCPTP –/– immortalized fibroblasts. (e and f) Increased p120RasGAP mRNA in PTP1B-deficient fibroblasts assessed by RT-PCR. GAPDH was used as a loading control, and lane C (no DNA) was used as a negative control.
To exclude the possibility that this phenomenon was due to TAg, we also analyzed p120RasGAP levels in spontaneously immortalized cells, as well as primary fibroblasts. Similar to TAg cells, spontaneously immortalized PTP1B knockout cells also possess increased p120RasGAP levels compared with WT controls (Fig. 3b). Interestingly, however, this effect was not seen in primary cells, suggesting that an event during immortalization is required for this process (data not shown). To further exclude the possibility of clonal variation effects, reexpression of myc-tagged PTP1B into knockout cells was able to restore p120RasGAP expression (Fig. 3c). Finally, to show that the increase in p120RasGAP expression was specific for PTP1B knockout cells, we used TCPTP knockout cells as a control. Although TCPTP knockout cells also display decreased cell proliferation (15), p120RasGAP levels are not altered (Fig. 3d).
To determine whether there were increased levels of p120RasGAP mRNA in PTP1B-deficient fibroblasts, we performed semiquantitative RT-PCR analysis by using the GAPDH gene as a control. As shown in Fig. 3E, this is indeed the case in two independent PTP1B-deficient TAg cell lines compared with two WT controls. In Fig. 3f, reexpression of PTP1B into PTP1B-deficient spontaneously immortalized cells restores Ras-GAP mRNA levels. Although the mechanism by which this event occurs is unclear, our results suggest that PTP1B expression is required to suppress expression of the p120RasGAP during immortalization of fibroblasts.
p62Dok Is a Putative Substrate for PTP1B. Previous studies demonstrated that p120RasGAP is tyrosine phosphorylated in cells transformed by protein tyrosine kinases, including Src (20). Furthermore, tyrosine phosphorylation of p120RasGAP allows it to bind to other proteins to contribute to its ability to inhibit the Ras/mitogen-activated protein kinase (MAPK) pathway (19). To investigate whether p120RasGAP could be a potential substrate of PTP1B, we used a D181A mutant of PTP1B that was previously shown to possess diminished catalytic activity but retain binding ability, thus producing a “substrate trapping mutant” (21).
We coexpressed GST alone or GST-tagged PTP1B (WT or D181A) into NIH 3T3 cells stably expressing an activated Src mutant (Src Y527F cells) and examined whether we could detect tyrosine-phosphorylated proteins in a complex with PTP1B proteins. Pull-downs of lysates were performed with glutathione beads and then analyzed by immunoblotting with a panel of antibodies. From Fig. 4a, it is clear that neither GST alone nor GST-PTP1B (WT) could appreciably precipitate tyrosinephosphorylated proteins. In contrast, GST-PTP1B (D181A) was found to precipitate two such proteins of ≈60 and ≈130 kDa (pp60 and pp130). We were able to confirm the identity of pp130 as p130Cas, previously shown to be a potential substrate of PTP1B (22). In contrast, probing with p120RasGAP antibodies failed to demonstrate binding to PTP1B, suggesting that p120RasGAP is not a substrate of PTP1B (data not shown).
Fig. 4.
p62Dok is a putative substrate of PTP1B. (a) NIH 3T3 c-Src Y527F cells were transfected with GST-vector (V), GST-PTP1B WT (WT), and GST-PTP1B D181A (DA). Cell lysates were subjected to pull-down with glutathione beads and then analyzed by immunoblotting with the indicated antibodies. TCL, total cell lysate. (b and c) Increased p62Dok phosphorylation in fibroblasts lacking PTP1B. Serum-starved cells were stimulated with 20 ng/ml PDGF for 5 and 15 min or left unstimulated. p62Dok phosphorylation was analyzed by immunoprecipitating p62Dok, probing with anti-phosphotyrosine antibodies, and then reprobing with anti-Dok1 antibodies. Lane C is antibody and beads alone as a control. Rescued, PTP1B –/– cells that reexpressed PTP1B.
We next explored the identity of pp60. Using a candidate approach, we revealed that pp60 is the adaptor protein p62Dok. As controls, two other tyrosine-phosphorylated proteins ≈60 kDa, Shc and Src, were not found to be precipitated by the PTP1B D181A mutant. To further confirm that PTP1B is a substrate of PTP1B, we analyzed the amounts of tyrosinephosphorylated p62Dok in the PTP1B WT and knockout cells (Fig. 4b). As expected, p62Dok phosphorylation was increased in PTP1B-deficient TAg cells, and reexpression of myc-tagged PTP1B into these cells decreased p62Dok phosphorylation (Fig. 4c). Previous gene targeting studies have shown that p62Dok is a negative regulator of Erk signaling (23, 24). Furthermore, tyrosine phosphorylation of p62Dok can contribute to its inhibitory effect on Ras (25). Thus, this result provides another mechanism by which PTP1B may regulate Ras activity.
V12Ras Rescues the Proliferative Potential of PTP1B Knockout Cells. Our results demonstrate that, in immortalized PTP1B-deficient fibroblasts, both p120RasGAP expression and p62Dok phosphorylation are increased. These events can lead to the suppression of the Erk pathway and lie upstream of Ras activation. Thus, to further confirm our findings, we tested the effect of stably introducing a dominant active Ras mutant (V12Ras) in our PTP1B WT and knockout cells. This mutant is known to inhibit its intrinsic GTPase activity, thus stabilizing the active GTP-bound form of Ras (26). Furthermore, this modification renders V12Ras independent from the modulation by upstream signals, such as RTKs and GAPs.
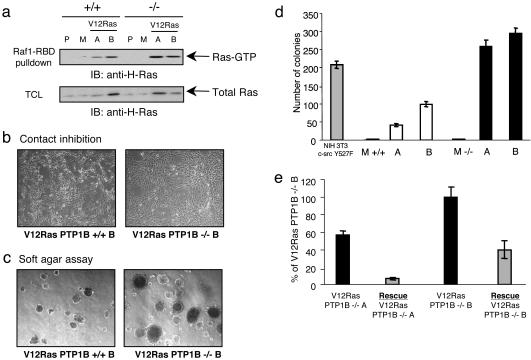
For our experiments, we chose three clones each for PTP1B WT and knockout cells. In this group, clones M (mock) do not express V12Ras, which we treated as our negative controls. Expression of V12Ras in the other clones (A +/+, B +/+, A –/–, B –/–) leads to an increase in Ras-GTP levels (Fig. 5a). Furthermore, both PTP1B WT and knockout cells were efficiently transformed by V12Ras, as judged by cell morphology and growth in soft agar (Fig. 5 b–d). In fact, the PTP1B-deficient clones even displayed enhanced soft agar growth, probably correlating with the slightly higher levels of Ras-GTP in these cells (Fig. 5a). Retroviral expression of PTP1B in these clones was able to suppress approximately half of the number of colonies (Fig. 5e), likely through its effects in down-regulating AKT activity (9). Thus, V12Ras is able to transform fibroblasts, even in the absence of PTP1B.
Fig. 5.
Transformation of PTP1B-deficient fibroblasts by activated Ras. (a) Expression of constitutively activated Ras leads to Ras activation. Cells were plated at 1 × 105 per 10-cm dish and cultured in supplemented DMEM until confluent. The lysates were incubated with immobilized GST-Raf1-RBD (Raf-RBD) to precipitate active (GTP-bound) Ras. Ras-GTP was detected with anti-H-Ras antibodies. (b) Stable cell lines expressing V12Ras were grown near confluence. Representative clones were photographed in phase contrast. (c) Anchorage-independent growth of PTP1B-deficient fibroblasts. Stable cell lines were grown in soft agar for 10 days. Colonies were counted under a microscope, and pictures of representative clones were taken in phase contrast. (d) Summary of the results obtained in c. Values are reported as the average of triplicate ± SE of three independent experiments. NIH 3T3 c-src Y527F were used as positive control. P, parental cell line; M, mock stable cell line (negative control); +/+V12Ras A and +/+V12Ras B, WT clones expressing V12Ras; –/–V12Ras A and –/–V12Ras B, PTP1B knockout clones expressing V12Ras. (e) Reexpression of PTP1B in V12Ras PTP1B –/– fibroblasts decreases the colonies' formation in soft agar. Both V12Ras PTP1B –/– cell lines were infected with a retroviral vector encoding PTP1B and grown in soft agar for 10 days. Colonies were counted as described in d.
Discussion
Overexpression and/or activating mutations of at least 30 protein tyrosine kinases have been linked to malignant transformation and cancer (27). In contrast, much less is known about the role of PTPs in human diseases. PTP1B is the prototypical PTP, and biochemical studies have implicated this enzyme in the dephosphorylation of several RTKs (1). We previously demonstrated that PTP1B-deficient fibroblasts display enhanced IGF-I-mediated receptor phosphorylation and AKT activation (9). Paradoxically, IGF-I-stimulated Erk activation was significantly impaired. Similarly, we also showed that PTP1B-deficient fibroblasts exhibit impaired adhesion-mediated Erk activation (12). In the present study, we provide evidence to explain how loss of PTP1B can lead to attenuation of Erk activation by way of impaired Ras signaling.
Stimulation of our TAg immortalized cells with PDGF resulted in enhanced tyrosine phosphorylation of cellular proteins in PTP1B knockout cells (Fig. 1a). Similarly, AKT phosphorylation was also increased (Fig. 1b), but PDGF-stimulated Erk activation was decreased in PTP1B knockout cells. In contrast, Haj et al. (8) reported only minor differences in Erk and AKT activation using independently established PTP1B knockout cell lines. One possible reason for this discrepancy is that those studies used high levels of PDGF (50 ng/ml), which may not allow for detecting subtle differences. Indeed, this situation has been shown for ShcA knockout cells (28). Thus, in our studies, we used lower amounts of PDGF (10 ng/ml). When we increased the levels of PDGF to 25 ng/ml, the differences we observed were diminished (data not shown).
Analysis of cell growth demonstrated that PTP1B is required for efficient proliferation of immortalized TAg fibroblasts (Fig. 2). Furthermore, we also noticed that PTP1B knockout cells had increased levels of the PDGFR (Fig. 3a), which suggested that Ras activity was lower in these cells (17). Consistent with this notion, Ras activity has been demonstrated to be essential for transformation by TAg in several model systems (18, 29, 30).
Profiling of key proteins involved in the Ras/MAPK pathway revealed an increased expression of p120RasGAP that was due to up-regulated transcription/message stability (Fig. 4). The significance of this finding is underscored by the ability of p120RasGAP to negatively regulate Ras activity by promoting the intrinsic GTPase activity of Ras (19). In addition, p62Dok, a binding partner of p120RasGAP, was found to be a potential substrate of PTP1B (Fig. 5). Previously, an unidentified phosphorylated 60-kDa protein was shown to be a potential substrate of PTP1B during epidermal growth factor signaling in COS cells (21). It is likely that this protein is also p62Dok. Importantly, p62Dok is a negative regulator of MAPK signaling (23, 24), and tyrosine phosphorylation of p62Dok can contribute to its inhibitory effect on Ras (25). Collectively, this finding suggested that loss of PTP1B in TAg cells leads to impaired Ras signaling.
In cells with high levels of Src activity, p62Dok has been shown to bind Csk, a negative regulator of Src kinases (31). The binding is thought to recruit Csk to cytoskeletal compartments to attenuate Src activity. Thus, it is intriguing to speculate that increased p62Dok phosphorylation in PTP1B knockout cells may contribute to decreased Src activity that was previously observed during fibronectin signaling (12). More studies will be required to verify this hypothesis.
If the impaired Ras signaling in PTP1B-deficient cells was due to p120RasGAP and p62Dok, then V12Ras should be able to transform these cells similar to that of WT controls. Indeed this is the case, and V12Ras-transformed PTP1B-deficient clones actually grow slightly better in soft agar (Fig. 5c). One possibility is that these clones possess Ras-GTP levels slightly higher than their WT counterparts (Fig. 5a). Alternatively, PTP1B-deficient cells also display enhanced AKT activity, which may promote a survival advantage. Nevertheless, our results show that V12Ras can transform cells in the absence of PTP1B.
Taken together, a model can be put forward to explain the impaired Erk activation seen in TAg immortalized PTP1B-deficient fibroblasts (Fig. 6). Loss of PTP1B leads to increased RTK phosphorylation and enhanced signaling of most downstream pathways. However, loss of PTP1B can also lead to cellular alterations that attenuate Erk signaling downstream of RTKs. For example, loss of PTP1B can lead to decreased Src activation by way of increased phosphorylation of the inhibitory site (12). In addition, loss of PTP1B leads to increased expression of p120RasGAP by way of an unidentified mechanism. Finally, p62Dok, a potential substrate of PTP1B, is hyperphosphorylated in PTP1B-deficient fibroblasts. All these events can contribute to attenuate Ras activity and thus Erk signaling. Consistent with this model, introduction of V12Ras into PTP1B-deficient cells can bypass these inhibitory events on Ras signaling and action.
Fig. 6.
Model of impaired Ras signaling in PTP1B-deficient fibroblasts. Loss of PTP1B leads to increased RTK phosphorylation and enhanced signaling of most downstream pathways. However, the absence of PTP1B can also lead to cellular alterations that attenuate MAPK signaling downstream of RTKs. For example, PTP1B deficiency can lead to decreased Src activation by way of increased phosphorylation of inhibitory site. In addition, loss of PTP1B leads to increased expression of p120RasGAP by way of an unidentified mechanism. Finally, p62Dok, a potential substrate of PTP1B, is hyperphosphorylated in PTP1B-deficient fibroblasts. All these events can contribute to attenuate Ras activity and thus MAPK signaling. Consistent with this model, introduction of activated Ras into PTP1B null cells can bypass these inhibitory events on Ras signaling and action.
In addition to the plasma membrane, a recent study demonstrated that Ras activation and signaling can occur at the endoplasmic reticulum (ER) (32). PTP1B predominantly localizes at the ER where it has been suggested to act on internalized PDGFRs and epidermal growth factor receptors (33, 34). It will be interesting to determine whether loss of PTP1B results in impaired Ras signaling globally, or in specific subcellular compartments. Targeted expression of PTP1B within knockout cells may provide further insight into this issue.
If loss of PTP1B leads to impaired Ras signaling, then what is the role of PTP1B during tumorigenesis? Approximately 30% of human cancers harbor activating mutations in the Ras gene (35). Because most of these mutations render Ras resistant to the actions of p120RasGAP, Src, and p62Dok, it is unlikely that loss of PTP1B would affect the Ras activity in this subset of cancer. However, loss of PTP1B does lead to increased IGF-I-induced AKT/protein kinase B activity (9), and, in this case, inhibition of PTP1B could offer an increased survival advantage to the transformed cells.
Breast cancer provides an interesting aspect in that rarely are Ras mutations found (36). In fact, most breast cancer cases are associated with increased expression of Src and members of the epidermal growth factor receptor family. Importantly, PTP1B has been identified as one of the major phosphatases that activate Src in breast cancer cells (11). Furthermore, increased expression of PTP1B has also been demonstrated in transformed human breast cells (37) and ovarian carcinomas (38). This result raises the intriguing possibility that PTP1B may positively contribute to the progression of these cancers by way of activation of Src. It will be interesting to determine the effects of introducing the PTP1B null background into transgenic models of breast cancer (39).
In conclusion, we have identified mechanisms by which PTP1B deficiency can actually lead to impaired Ras signaling and proliferation. Our results suggest that decreasing Ras activity through inhibition of PTP1B could even provide a means to treat a subset of cancers.
Acknowledgments
We thank Drs. Morag Park, Pankaj Tailor, Christophe Blanchetot, and Feng Gu for helpful discussions. N.D. and A.C. are recipients of a Canadian Institutes of Health Research doctoral research award and studentship, respectively. M.L.T. is a Canadian Institutes of Health Research Scientist. This work was supported by Cancer Research Society and Canadian Institute of Health Research (MOP-62887) operating grants to M.L.T.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IGF-I, insulin-like growth factor I; MAPK, mitogen-activated protein Erk, extracellular-regulated kinase; p62Dok, downstream of tyrosine kinase; Ras GTPase-activating protein; PDGFR, platelet-derived growth factor receptor; RTK, ceptor tyrosine kinase; TAg, SV40 large T antigen; PTP, protein tyrosine TCPTP, T cell PTP.
References
- 1.Ostman, A. & Bohmer, F. D. (2001) Trends Cell Biol. 11, 258–266. [DOI] [PubMed] [Google Scholar]
- 2.Cheng, A., Uetani, N., Simoncic, P. D., Chaubey, V. P., Lee-Loy, A., McGlade, C. J., Kennedy, B. P. & Tremblay, M. L. (2002) Dev. Cell 2, 497–503. [DOI] [PubMed] [Google Scholar]
- 3.Elchebly, M., Payette, P., Michaliszyn, E., Cromlish, W., Collins, S., Loy, A. L., Normandin, D., Cheng, A., Himms-Hagen, J., Chan, C. C., et al. (1999) Science 283, 1544–1548. [DOI] [PubMed] [Google Scholar]
- 4.Gu, F., Dubé, N., Kim, J. W., Cheng, A., Ibarra-Sanchez, M. d. J., Tremblay, M. L. & Boisclair, Y. R. (2003) Mol. Cell. Biol. 23, 3753–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klaman, L. D., Boss, O., Peroni, O. D., Kim, J. K., Martino, J. L., Zabolotny, J. M., Moghal, N., Lubkin, M., Kim, Y. B., Sharpe, A. H., et al. (2000) Mol. Cell. Biol. 20, 5479–5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zabolotny, J. M., Bence-Hanulec, K. K., Stricker-Krongrad, A., Haj, F., Wang, Y., Minokoshi, Y., Kim, Y. B., Elmquist, J. K., Tartaglia, L. A., Kahn, B. B. & Neel, B. G. (2002) Dev. Cell 2, 489–495. [DOI] [PubMed] [Google Scholar]
- 7.Johnson, T. O., Ermolieff, J. & Jirousek, M. R. (2002) Nat. Rev. Drug Discov. 1, 696–709. [DOI] [PubMed] [Google Scholar]
- 8.Haj, F. G., Markova, B., Klaman, L. D., Bohmer, F. D. & Neel, B. G. (2003) J. Biol. Chem. 278, 739–744. [DOI] [PubMed] [Google Scholar]
- 9.Buckley, D. A., Cheng, A., Kiely, P. A., Tremblay, M. L. & O'Connor, R. (2002) Mol. Cell. Biol. 22, 1998–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arregui, C. O., Balsamo, J. & Lilien, J. (1998) J. Cell Biol. 143, 861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjorge, J. D., Pang, A. & Fujita, D. J. (2000) J. Biol. Chem. 275, 41439–41446. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, A., Bal, G. S., Kennedy, B. P. & Tremblay, M. L. (2001) J. Biol. Chem. 276, 25848–25855. [DOI] [PubMed] [Google Scholar]
- 13.Broome, M. A. & Courtneidge, S. A. (2000) Oncogene 19, 2867–2869. [DOI] [PubMed] [Google Scholar]
- 14.Klinghoffer, R. A., Sachsenmaier, C., Cooper, J. A. & Soriano, P. (1999) EMBO J. 18, 2459–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibarra-Sanchez, M. J., Wagner, J., Ong, M. T., Lampron, C. & Tremblay, M. L. (2001) Oncogene 20, 4728–4739. [DOI] [PubMed] [Google Scholar]
- 16.Simoncic, P. D., Lee-Loy, A., Barber, D. L., Tremblay, M. L. & McGlade, C. J. (2002) Curr. Biol. 12, 446–453. [DOI] [PubMed] [Google Scholar]
- 17.Stice, L. L., Vaziri, C. & Faller, D. V. (1999) Front. Biosci. 4, D72–D86. [DOI] [PubMed] [Google Scholar]
- 18.Raptis, L., Brownell, H. L., Corbley, M. J., Wood, K. W., Wang, D. & Haliotis, T. (1997) Cell Growth Differ. 8, 891–901. [PubMed] [Google Scholar]
- 19.Donovan, S., Shannon, K. M. & Bollag, G. (2002) Biochim. Biophys. Acta 1602, 23–45. [DOI] [PubMed] [Google Scholar]
- 20.Ellis, C., Moran, M., McCormick, F. & Pawson, T. (1990) Nature 343, 377–381. [DOI] [PubMed] [Google Scholar]
- 21.Flint, A. J., Tiganis, T., Barford, D. & Tonks, N. K. (1997) Proc. Natl. Acad. Sci. USA 94, 1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, F., Hill, D. E. & Chernoff, J. (1996) J. Biol. Chem. 271, 31290–31295. [DOI] [PubMed] [Google Scholar]
- 23.Zhao, M., Schmitz, A. A., Qin, Y., Di Cristofano, A., Pandolfi, P. P. & Van Aelst, L. (2001) J. Exp. Med. 194, 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Cristofano, A., Niki, M., Zhao, M., Karnell, F. G., Clarkson, B., Pear, W. S., Van Aelst, L. & Pandolfi, P. P. (2001) J. Exp. Med. 194, 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wick, M. J., Dong, L. Q., Hu, D., Langlais, P. & Liu, F. (2001) J. Biol. Chem. 276, 42843–42850. [DOI] [PubMed] [Google Scholar]
- 26.Macaluso, M., Russo, G., Cinti, C., Bazan, V., Gebbia, N. & Russo, A. (2002) J. Cell Physiol. 192, 125–130. [DOI] [PubMed] [Google Scholar]
- 27.Blume-Jensen, P. & Hunter, T. (2001) Nature 411, 355–365. [DOI] [PubMed] [Google Scholar]
- 28.Lai, K. M. & Pawson, T. (2000) Genes Dev. 14, 1132–1145. [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas, M., Suwa, T., Yang, L., Zhao, L., Hawks, C. L. & Hornsby, P. J. (2002) Neoplasia 4, 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beachy, T. M., Cole, S. L., Cavender, J. F. & Tevethia, M. J. (2002) J. Virol. 76, 3145–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neet, K. & Hunter, T. (1995) Mol. Cell. Biol. 15, 4908–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiu, V. K., Bivona, T., Hach, A., Sajous, J. B., Silletti, J., Wiener, H., Johnson, R. L., 2nd, Cox, A. D. & Philips, M. R. (2002) Nat. Cell Biol. 4, 343–350. [DOI] [PubMed] [Google Scholar]
- 33.Haj, F. G., Verveer, P. J., Squire, A., Neel, B. G. & Bastiaens, P. I. (2002) Science 295, 1708–1711. [DOI] [PubMed] [Google Scholar]
- 34.Frangioni, J. V., Beahm, P. H., Shifrin, V., Jost, C. A. & Neel, B. G. (1992) Cell 68, 545–560. [DOI] [PubMed] [Google Scholar]
- 35.Bos, J. L. (1989) Cancer Res. 49, 4682–4689. [PubMed] [Google Scholar]
- 36.Andrechek, E. R. & Muller, W. J. (2000) Breast Cancer Res. 2, 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhai, Y. F., Beittenmiller, H., Wang, B., Gould, M. N., Oakley, C., Esselman, W. J. & Welsch, C. W. (1993) Cancer Res. 53, 2272–2278. [PubMed] [Google Scholar]
- 38.Wiener, J. R., Hurteau, J. A., Kerns, B. J., Whitaker, R. S., Conaway, M. R., Berchuck, A. & Bast, R. C., Jr. (1994) Am. J. Obstet. Gynecol. 170, 1177–1183. [DOI] [PubMed] [Google Scholar]
- 39.Hutchinson, J. N. & Muller, W. J. (2000) Oncogene 19, 6130–6137. [DOI] [PubMed] [Google Scholar]