Abstract
Angiosperms are among the major terrestrial radiations of life and a model group for studying patterns and processes of diversification. As a tool for future comparative studies, we compiled a supertree of angiosperm families from published phylogenetic studies. Sequence data from the plastid rbcL gene were used to estimate relative timing of branching events, calibrated by using robust fossil dates. The frequency of shifts in diversification rate is largely constant among time windows but with an apparent increase in diversification rates within the more recent time frames. Analyses of species numbers among families revealed that diversification rate is a labile attribute of lineages at all levels of the tree. An examination of the top 10 major shifts in diversification rates indicates they cannot easily be attributed to the action of a few key innovations but instead are consistent with a more complex process of diversification, reflecting the interactive effects of biological traits and the environment.
In a letter to J. D. Hooker dated July 22, 1879 (1), Charles Darwin described the rapid rise and early diversification within the angiosperms as “an abominable mystery.” Angiosperms are regarded as one of the greatest terrestrial radiations of recent geological times. The major lineages originated 130–90 million years ago (mya) (2, 3), followed by a dramatic rise to ecological dominance 100–70 mya (4). Approximately 250,000 extant species have been recognized (5), although estimates vary, and the final number might be double this (6). Within the group, sister clades can differ in species richness over several orders of magnitude. Darwin attempted to identify a single causal explanation for the rapid diversification of angiosperms but described his own efforts as “wretchedly poor” (1).
Subsequent attempts to understand angiosperm diversification have come from a variety of fields. Studies of the fossil record have explored the origin of angiosperms and the spatiotemporal patterns of their radiation (3, 7–9). A complementary approach has been the use of systematic data of living species to identify major trends in angiosperm evolution and their possible effects on diversification (10). For example, many authors have investigated the importance of biological traits, such as biotic pollination (2, 11, 12), biotic seed dispersal (13–15), and life history flexibility (16, 17), as putative key innovations. Increasingly, such studies rely on knowledge of phylogenetic relationships among higher taxa to estimate net diversification rates and pinpoint independent evolutionary events (18–21), thereby circumventing the problems associated with comparing higher taxa of different ages (22).
Recent advances in molecular phylogenetics have heralded a new era in plant phylogenetics. Since the molecular phylogenetic tree of angiosperms based on plastid rbcL sequence data by Chase et al. (23), a succession of large-scale angiosperm trees has appeared over the last decade (24–26). Increased sampling of taxa and the use of multiple genes (27–29) have led to increased resolution and confidence in angiosperm relationships (30). These data have become a major resource for comparative biology, but to date no single analysis has included all currently recognized angiosperm families.
Here we use a supertree approach to combine recent phylogenetic data into the first complete family-level phylogenetic tree of the angiosperms, a task that was described as “formidable” and “impossible to meet” just over a decade ago (18). We present this tree, together with dates calibrated by using the fossil record and estimated from molecular branch lengths, as a compilation of current knowledge and a tool for comparative plant biology. In addition, we use the supertree to present the first complete survey of diversification among familial angiosperm lineages. Our aim is to identify at which points on the tree major shifts occurred and use this information to guide the examination of factors that might explain the mystery of angiosperm diversification.
Methods
Supertree Construction. Supertree methods are being used increasingly to combine multiple sources of phylogenetic data into a single analysis. We used matrix representation with parsimony (MRP), which codes branching patterns of individual source trees as a binary matrix and missing taxa as question marks. The matrices for all of the trees are then combined, and a tree search is performed on the combined matrix using parsimony (31, 32). The best practice for supertree analyses is an active area of research (33), but MRP is widely recognized as one of the best current methods and has been successfully applied in a large number of studies (34–36).
Forty-six source trees were selected from published and unpublished work on the basis of either their comprehensive coverage or resolution of previously poorly understood relationships, with the aim of maximizing the number of families represented (a list of source trees is given in Table 2, which is published as supporting information on the PNAS web site). To take into account levels of support for relationships, we used bootstrap percentages for nodes in the source trees as character weights for the MRP binary matrix, following the method of Salamin et al. (34) (further details are provided in Supporting Methods, which is published as supporting information on the PNAS web site). Family delineations followed the Angiosperm Phylogeny Group (APG) classification (37, 38). For six families, we were unable to find published phylogenetic treatments (listed in Table 3, which is published as supporting information on the PNAS web site).
The supertr ee0.8b program [www.tcd.ie/Botany/NS/SuperTree.html (34)] was used to create a single binary matrix representing all of the relationships in the above trees. The binary matrix was analyzed with paup4.0b8 (39) by using weighted parsimony with the following heuristic search: 250 replicates of random taxon addition, subtree pruning–regrafting branch swapping, and holding 10 trees at each replicate. The saved trees were then used as the starting trees in another search using tree bisection–reconnection with a tree limit of 10,000 equally most parsimonious trees.
As estimation of divergence times and consequently diversification rates requires a completely bifurcating topology; all subsequent analysis was performed on one of the most parsimonious supertrees. To examine whether arbitrary resolutions may have biased our results, we repeated each subsequent analysis of diversification rates excluding nodes that collapsed in the strict consensus tree.
The topology of the supertree was compared to that of the three-gene (atpB, rbcL, and 18S rDNA) bootstrap tree generated from the matrix of Soltis et al. (28). Sampling both the plastid and nuclear genome and with broad taxonomic coverage, this tree is regarded as the best estimate of angiosperm phylogeny to date. Therefore, as quality control for the supertree, we checked whether strongly supported relationships in this source tree are also present in the supertree. We used a parsimony equivalent of the Shimodaira–Hasegawa test (40) to compare tree lengths for three-gene source data optimized onto each tree topology in turn by using 500 bootstrap replicates and 10,000 random trees. Second, we compared the number of nodes in common between the two trees by using the program treecorrect1.2b [www.tcd.ie/Botany/NS/software.html (41)].
Dating. We estimated the amount of molecular change along branches in the tree by using a matrix of rbcL sequences compiled from the source matrices or downloaded from GenBank (www.ncbi.nlm.nih.gov). The rbcL gene was chosen because it has been sequenced for most of the taxa in the supertree. Branch lengths were optimized onto the supertree by using maximum likelihood assuming an HKY85 + Γ DNA substitution model in paup4.0b8 (39). This model provides a compromise between model complexity and computational time (42). The phylogenetic tree was arranged with Amborellaceae as sister to the rest of the angiosperms (27, 28, 43–45). To correct for variation in substitution rate among lineages, we used nonparametric rate smoothing (46), as implemented in treeedit v1.0 a10 (http://evolve.zoo.ox.ac.uk/software/TreeEdit). A single family on the supertree, Triuridaceae, lacked rbcL sequence data and was placed arbitrarily halfway along the branch leading to its sister clade.
The tree was calibrated in units of millions of years by using the split between Fagales and Cucurbitales set to 84 mya [after Wikström et al. (47)]. To check consistency of date estimates, we also calibrated the tree, setting the stem lineage subtending the eudicot crown group set to 126 mya (48), and compared the alternative dates.
Measuring Diversification. Species numbers for families were taken from Watson and Dallwitz (refs. 50 and 51 and http://biodiversity.uno.edu/delta/angio). If the generic composition of a family differed from that currently accepted by the APG (37, 38), species richness was adjusted to be in agreement with the APG classification (see Supporting Methods). To determine whether there is significant variation in diversification rates among angiosperm lineages, we calculated the overall tree imbalance by using the mean tree imbalance measure of Fusco and Cronk (51) as modified by Purvis et al. (52) on the strict consensus tree, because arbitrary resolutions of polytomies have been shown to inflate imbalance (53, 54).
We used two complementary methods to pinpoint where diversification rates changed on the tree. First, we estimated net diversification rates for all clades on the tree using log(N)/t, where N is the number of species within a clade, and t is the time since the clade diverged from its sister clade on the dated tree. Changes in diversification rate on the tree were calculated by subtracting the rate for each clade from the rate of its immediate nesting clade. We refer to this measure as maximum likelihood estimate of shift in diversification rate (logN) rate shifts. Second, we compared the species richness of all sister clades on the tree by using the Slowinski–Guyer measure of imbalance (SG; ref. 56), which assigns a probability of observing an equal or greater difference in species numbers at each node under a general null model that diversification rates in the two daughter clades have been equal. Sister clades are the same age, and therefore this approach accounts for possible effects of different clade ages on current species richness using information on topology alone. Due to the nested nature of phylogenetic comparisons, families with a large or small number of species can influence the degree of imbalance at nodes nesting nearer the root (56). We corrected for this nonindependence by using a heuristic approach, described in Supporting Methods.
The distribution of shifts in diversification rate across the tree using the latter two measures were explored by using randomization tests to examine whether the diversification rate is phylogenetically conserved. First, we examined heritability of diversification rates among branches of the tree. Details of randomization test procedures are provided in Supporting Methods. Second, we looked for concentration of shifts in diversification rate in either particular time windows or particular angiosperm orders recognized by the APG.
Finally, for both measures of shifts in diversification rate, we identified the top 10 shifts found across the tree. Because the logN measure includes the direction of each shift as well as magnitude, we identified the top 10 increases and decreases in diversification rates separately. We then categorized the affected clades in terms of several factors previously proposed to influence diversification rates in angiosperms, ranging from pollination syndrome to geographic range (taken from Watson and Dallwitz's online database, http://biodiversity.uno.edu/delta/angio). Clades were labeled polymorphic if they exhibit a mixture of possible values. In addition, we used taxonomic descriptions to identify any other general features of the clades. The goal was not to perform a comprehensive test of correlates of diversification in angiosperms but rather to explore whether single factors or simple combinations might be associated with the major shifts in angiosperm radiation. We also recorded the level of support for the nodes: one explanation for large shifts might be phylogenetic error, for example, if a small family were mistakenly placed as sister to a larger clade. The nonindependence of characters within the MRP matrix violates the assumptions of the bootstrap; thus estimates of node support were inferred from the individual source trees.
Results and Discussion
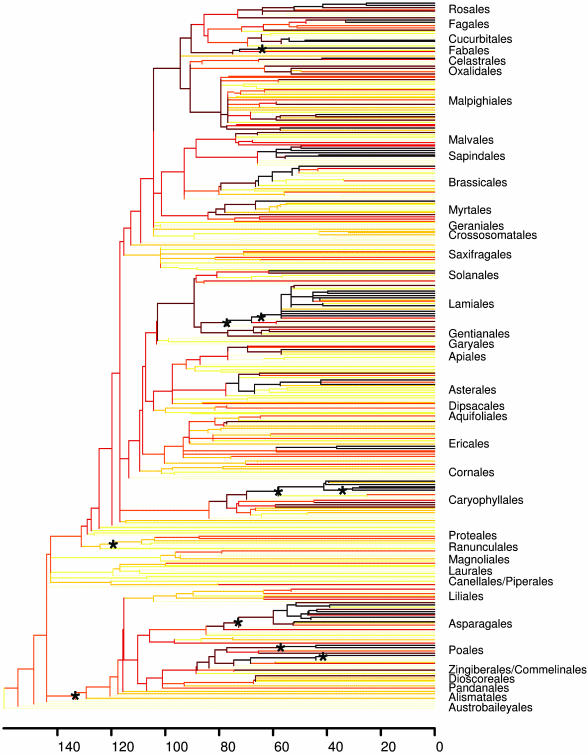
Supertree and Dates. Our MRP analysis generated 10,000 most parsimonious supertrees, one of which is summarized in Fig. 1 and presented in full in Fig. 2, which is published as supporting information on the PNAS web site; nodes collapsing in the strict consensus are indicated with an arrow on the latter. Although undoubtedly more equally most parsimonious trees could have been found with continued branch swapping, it may be reasonable to assume that those nodes liable to collapse in a strict consensus of all most parsimonious trees were identified by using the search implemented.
Fig. 1.
One of 10,000 most parsimonious supertrees with dates obtained by used nonparametric rate smoothing transformation of maximum likelihood branch lengths from rbcL sequence data. The time scale was calibrated by using the split between Fagales and Cucurbitales at 84 mya. The strength of shading reflects diversification rates estimated as log (number of species)/age since split from sister clade. See Fig. 2 for a larger figure showing names of all terminal taxa. Diversification rates vary from low (yellow to orange) to high (red to black). Asterisks indicate the top 10 most imbalanced nodes referred to in Table 1.
The final trees include 379 terminal taxa representing monophyletic clades, mostly families but also higher clades in cases for which recognized families are not monophyletic. Because the source trees were predominantly molecular, and all of the matrices included sequence data for rbcL, the supertree is inevitably biased toward the rbcL gene tree. We retained the input of rbcL data because the best estimate of relationships within the source trees is likely to come from combined analysis of all available markers (57).
Because the topology derives from existing phylogenetic hypotheses, we do not present an in-depth discussion of recovered relationships. As noted above, a few families do not appear monophyletic, most noticeable within Caryophyllales, despite our following the APG classification for circumscription of families. These families are known to be nonmonophyletic, for example Portulacaceae and Phytolaccaceae, but changes in circumscription were judged in the last APG classification to be premature until comprehensive studies are performed. The occurrence of nonmonophyletic families and polytomies within the supertree highlight areas in need of more rigorous analysis and more data.
Differences between the supertree and individual source trees could in principle be caused by hard incongruence among studies or by phylogenetic errors due to relationships with low levels of bootstrap support (58). Most conflicts between the supertree and the three-gene source tree are at nodes with weak support in the three-gene tree. Only 69% of nodes in the source tree are found in the supertree, but all nodes with bootstrap support >70% were present. The supertree was not significantly different from the three-gene source tree in its fit to the three-gene molecular data (modified Shimodaira–Hasegawa test, P = 0.58). These results indicate that the weighted MRP analysis accurately reproduced the relationships supported by the best sampled source tree.
There remains active debate over methods for calibrating phylogenetic trees (42, 47, 59), but many alternative methods are not applicable to such large data sets. The alternative calibration point, the origin of the eudicots, produced slightly younger estimates of divergence times, dating the split between Fagales and Cucurbitales ≈10 my younger than that suggested by the fossil record and leading to, on average, 89% younger dates than for the alternative calibration. More generally, there remain examples of inconsistencies in fossil and molecular dates for angiosperm lineages, with a tendency for molecular dates to overestimate deeper nodes, such as the origin of the eudicots, and underestimate more terminal nodes (e.g., Poaceae, Moraceae, and Salicaceae) (47). Discrepancies between molecular and fossil dates are frequent in all groups where comparisons have been made (60). Whether these differences relate to biases in molecular dating procedures, errors in fossil sampling and identification, or both remains to be investigated thoroughly. At present, we have no means to correct for these differences and therefore simply present our results as a comprehensive molecular estimate of branching events for all angiosperm families calibrated by fossil dates assumed to be robust. Because our later analyses rely predominantly on relative age estimates of different families, rather than absolute age, we discuss only results using the Fagales–Cucucurbitales calibration point.
Patterns of Diversification. Analysis of the supertree revealed significant imbalance in net diversification rates among angiosperm lineages compared to the null model that all lineages have an equal diversification rate (weighted mean I = 0.72, P < 0.001; I, tree imbalance). The comprehensive taxonomic sampling of the supertree allows increased confidence in these findings, which broadly correspond to previous estimates of phylogenetic imbalance within the angiosperms (51) and coincide with the general pattern found across a wide range of taxa (61, 62). Placing the six families not represented in the source trees in the final supertree based on published statements of their likely affinities (see Table 3) did not change our results; we discuss below only those results excluding these families.
The two methods of reconstructing shifts in diversification rate on the tree yielded mostly similar results. Nodes that exhibit a significant SG value tend to have a large logN rate shift. The few exceptions to this trend were cases in which two sister clades with balanced species numbers were joined by a relatively long stem branch. This led to reconstruction of a high rate in both sister clades compared to the rate expected for their nesting clade, a situation not recognizable from topology alone. Overall the measures give the same visual picture of diversification: frequent shifts in diversification rate have occurred across the tree (Fig. 1).
The randomization test found that diversification rates are significantly phylogenetically heritable between related lineages, but only marginally so (logN rate shifts, P = 0.040; SG values, P = 0.031). Hence, sister families are only marginally more likely to have similar species numbers than two families chosen at random, indicating that diversification rate is a labile attribute.
There was also only weak evidence that particular orders of angiosperms have experienced a greater frequency of shifts than others (randomization test, logN rate shifts, P > 0.1; SG values, P = 0.036), excluding collapsing nodes from the analysis further reduced significance in both analyses. However, the frequency of reconstructed shifts did vary among time windows, and the exact pattern differed between the SG and logN methods of assigning rate shifts (logN rate shifts, P = 0.024; SG values, P > 0.1; see Figs. 3 and 4, which are published as supporting information on the PNAS web site). Nodes in more recent time periods tended to display a greater logN rate shift (P < 0.001) than expected under the null model, associated with the observation of sister families with long stem branches outlined above. One possible explanation would be if diversification rates have increased uniformly across all lineages within very recent time periods. However, an alternative explanation is that this pattern reflects a bias due to the use of families as terminal taxa: shifts occurring within families can be reconstructed only as occurring in the entire family in our analyses. Reconstructed shifts in diversification rates at nodes deeper in the tree would be unaffected by any such bias; hence our overall results are not affected by the sampling of families as terminal taxa, providing all terminal clades are monophyletic, and we can assign all recognized species of angiosperms to one of the tips in the tree.
The top 10 most imbalanced nodes (SG measure) in the strict consensus supertree are shown in Table 1. Equivalent tables for the logN rate shifts are in Tables 2–8, which are published as supporting information on the PNAS web site. The exact membership of the tables varies with the measure of rate shifts used and whether we correct for nesting of species richness or not, but the general conclusions are unchanged. The top 10 nodes do not reflect poorly supported parts of the tree, rejecting phylogenetic inaccuracy as an explanation for their high imbalance. None of the biological traits stand out as unequivocal key innovations explaining the major shifts in diversification. As can be seen from Table 1 (see also Tables 4 and 5), clades with higher species richness tend to be more polymorphic in the traits considered and cover a wider geographical range, but whether this is a cause or an effect of increased species richness is difficult to evaluate at this level (e.g., see ref. 63). Similarly, major shifts near the root of the tree, such as those leading to the core eudicots and monocots, are characterized by species-rich clades that are polymorphic in all traits considered in this paper and have cosmopolitan distributions. In contrast, the species-poor sister lineages are polymorphic for only approximately a quarter of the traits considered and have typically much more restricted distributions.
Table 1. Top 10 most imbalanced nodes (SG) and their derived clades.
| Imbalance (SG) | Sister clades | Age, mya | Node support/(source ref.) | Pollination mode | Dispersal mode | Habit | Strictly dioecious | Chromosome no. | Geographic distribution | Lifestyle |
|---|---|---|---|---|---|---|---|---|---|---|
| 0.0002 | Lamiales I* | 77 | 53/(25) | Biotic/? | Poly | Poly | None | 6-96 | Cosmopolitan | Poly |
| Plocospermataceae | ? | Abiotic | Woody | None | ? | Central America | Perennial | |||
| 0.0002 | Poaceae | 41 | 97/(66) | Abiotic | Poly | Poly | Poly | 4-122 | Cosmopolitan | Poly |
| Ecdeiocoleaceae | Abiotic | ? | Herbaceous | None | ? | Australia | Perennial | |||
| 0.0004 | Monocots | 133 | 87/(26) | Poly | Poly | Poly | Poly | 4-180† | Cosmopolitan | Poly |
| Acoraceae | ? | Poly | Herbaceous | None | 24 | Old World and North America | Perennial | |||
| 0.0007 | Asparagales* | 72 | 69/(26) | Biotic/? | Poly | Poly | Poly | 6-180 | Cosmopolitan | Perennial |
| Xeronemataceae | ? | Biotic | Herbaceous | None | ? | New Zealand and New Caledonia | Annual | |||
| 0.0010 | Lamiales II* | 64 | 43/(25) | Biotic/? | Poly | Poly | None | 6-96 | Cosmopolitan | Poly |
| Tetrachondraceae | ? | Abiotic | Herbaceous | None | ? | New Zealand and Patagonia | Perennial | |||
| 0.0011 | Fabaceae | 64 | 54/(28) | Biotic | Poly | Poly | None | 10-112 | Cosmopolitan | Poly |
| Surianaceae | ? | Biotic | Woody | None | ? | Pan subtropical to tropical | Perennial | |||
| 0.0012 | Caryophyllales I* | 85 | 98/(28) | Poly | Poly | Poly | Poly | 12-144 | Cosmopolitan | Poly |
| Asteropeiaceae/Physenaceae | Abiotic/? | Abiotic | Woody | Poly | ? | Madagascar | Perennial | |||
| 0.0014 | Caryophyllales II* | 74 | 40/(25) | Poly | Poly | Poly | Poly | 12-144 | Cosmopolitan | Poly |
| Stegnospermaceae | ? | Abiotic | Woody | None | ? | North & central America | Perennial | |||
| 0.0015 | Ranunculales* | 119 | 88/(26) | Poly | Poly | Poly | Poly | 12-56† | Cosmopolitan | Poly |
| Eupteleaceae | Abiotic | Abiotic | Woody | None | 28 | East Asia | Perennial | |||
| 0.0015 | Cyperaceae/Juncaceae | 47 | 55/(28) | Poly | Abiotic | Poly | Poly | 12-112† | Cosmopolitan | Poly |
| Thumiaceae | Abiotic | Abiotic | Herbaceous | None | ? | North South America | ? |
Bold indicates the larger clade; the respective nodes are indicated in Fig. 1 by asterisks. Other than where stated, ecological data were derived from Watson and Dallwitz's online database (refs. 49 and 50 and http://biodiversity.uno.edu/delta/angio). Poly, polymorphic; ?, unknown.
Taxonomic description of clades is given in Table 6.
Values obtained from Plant DNA C-values Database 2.0 (www.rbgkew.org.uk/cval/homepage.html).
Conclusion
As a tool for comparative biology, we have reconstructed a dated supertree of angiosperm families with species numbers presented for the terminals. Our analyses revealed a strikingly labile pattern of diversification rate in the angiosperms. This pattern is not the result solely of phylogenetic inaccuracy and misplaced taxa, because many of the nodes with major shifts are strongly supported in the source trees.
Our results uphold Darwin's suspicions that simple explanations for the mystery of angiosperm diversification are inadequate. Our calibration of the diversification of the major angiosperm lineages does show an early rapid radiation of the basal lineages, and this could be taken to account for what Darwin considered to be the “rapid rise and early diversification” of the angiosperms, which was his “abominable mystery” (1). However, numerous other shifts in diversification rates have occurred throughout the history of angiosperms, including several large increases in rates in recent time periods. The pattern is not consistent with a simple model in which diversification is driven by a few major key innovations but rather argues for a more complex process in which propensity to diversify is highly labile: there are “winners” and “losers” at all levels, and shifts occur repeatedly. This conclusion is supported by our tabulation of characteristics of clades affected by the major shifts and previous studies on incomplete phylogenetic trees (21, 64). Traits that may characterize particular species-rich clades are not sufficient to guarantee phylogenetic success, because within all species-rich higher clades we observe several shifts to slower rates of diversification.
Together, these results have implications for future analyses on how the interaction between traits and the environment affects diversification: some traits convey success in some environments but not others. Phylogenetic studies of diversity rely on inferences from current species numbers in terminal clades. Therefore, patterns of diversification reconstructed onto phylogenetic trees depend on the age of lineages, their intrinsic attributes, and also the environments experienced since their origin, particularly recent conditions. Global environments have changed considerably during the history of angiosperm radiation: which lineages are diverse now depends on the match between traits and recent climates, e.g., the rise to dominance of grasses during the late Tertiary is linked to global cooling and drying (65). Ultimately, increasing phylogenetic resolution at the level of genera and below may be needed to produce detailed models of how these interacting effects influence diversification. Our supertree represents a step toward this goal.
Supplementary Material
Acknowledgments
We thank Nicolas Salamin and Andy Purvis for help with the supertree. This work was supported by a Natural Environment Research Council studentship (to T.J.D.), a Royal Society University Research Fellowship (to T.G.B.), a U.S.–U.K. Fulbright Distinguished Professorship (to D.E.S. and P.S.S.), and U.S. National Science Foundation Grant DEB-0090283 (to D.E.S., P.S.S., D. L. Dilcher, and P. S. Herendeen).
Abbreviations: mya, millions of years ago; SG, Slowinski–Guyer measure of imbalance; logN, maximum likelihood estimate of shift in diversification rate; APG, Angiosperm Phylogeny Group; MRP, matrix representation with parsimony.
References
- 1.Darwin, F. & Seward, A. C., eds. (1903) More Letters of Charles Darwin (John Murray, London), Vol. 2.
- 2.Labandeira, C. C., Dilcher, D. L., Davis, D. R. & Wagner, D. L. (1994) Proc. Natl. Acad. Sci. USA 91, 12278–12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crane, P. R., Friis, E. M. & Pedersen, K. J. (1995) Nature 374, 27–33. [Google Scholar]
- 4.Barrett, P. M. & Willis, K. J. (2001) Biol. Rev. 76, 411–447. [DOI] [PubMed] [Google Scholar]
- 5.Wilson, E. O. (1992) The Diversity of Life (Belknap, Harvard Univ. Press, Cambridge, MA).
- 6.Bramwell, D. (2002) Plant Talk 28, 32. [Google Scholar]
- 7.Axelrod, D. I. (1952) Evolution (Lawrence, Kans.) 6, 29–60. [Google Scholar]
- 8.Doyle, J. A. (1978) Annu. Rev. Ecol. Syst. 9, 365–392. [Google Scholar]
- 9.Niklas, K. J. & Tiffney, B. H. (1994) Philos. Trans. R. Soc. London B 345, 35–44. [Google Scholar]
- 10.Dilcher, D. L. (2000) Proc. Natl. Acad. Sci. USA 97, 7030–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ricklefs, R. E. & Renner, S. S. (1994) Evolution (Lawrence, Kans.) 48, 1619–1636. [DOI] [PubMed] [Google Scholar]
- 12.Crepet, W. L. (1984) Ann. Mo. Bot. Gard. 71, 607–630. [Google Scholar]
- 13.Eriksson, O. & Bremer, B. (1992) Evolution (Lawrence, Kans.) 46, 258–266. [DOI] [PubMed] [Google Scholar]
- 14.Herrera, C. M. (1989) Am. Nat. 133, 309–322. [Google Scholar]
- 15.Smith, J. F. (2001) Am. Nat. 157, 646–653. [DOI] [PubMed] [Google Scholar]
- 16.Midgely, J. J. & Bond, W. J. (1991) Philos. Trans. R. Soc. London B 333, 209–215. [Google Scholar]
- 17.Regal, P. J. (1997) Science 196, 622–629. [DOI] [PubMed] [Google Scholar]
- 18.Sanderson, M. J. & Donoghue, M. J. (1996) Trends Ecol. Evol. 11, 15–20. [DOI] [PubMed] [Google Scholar]
- 19.Dodd, M. E., Silvertown, J. & Chase, M. W. (1999) Evolution (Lawrence, Kans.) 53, 732–744. [DOI] [PubMed] [Google Scholar]
- 20.Barraclough, T. G. & Savolainen, V. (2001) Evolution (Lawrence, Kans.) 55, 677–683. [DOI] [PubMed] [Google Scholar]
- 21.Magallón, S. & Sanderson, M. J. (2001) Evolution (Lawrence, Kans.) 55, 1762–1780. [DOI] [PubMed] [Google Scholar]
- 22.Barraclough, T. G., Nee, S. & Harvey, P. H. (1998) Evol. Ecol. 12, 751–754. [Google Scholar]
- 23.Chase, M. W., Soltis, D. E., Olmstead, R. G., Morgan, D., Les, D. H., Mishler, B. D., Duvall, M. R., Price, R. A., Hills, H. G., Qiu, Y. L., et al. (1993) Ann. Mo. Bot. Gard. 80, 528–580. [Google Scholar]
- 24.Chase, M. W., Duvall, M. R., Hills, H. G., Conran, J. G., Cox, A. V., Caddick, L. R., Cameron, K. M. & Hoot, S. B. (1995) in Monocotyledons: Systematics and Evolution, eds. P. J. Rudall, P. J. Cribb, D. F. Cutler & Humphries, C. J. (Royal Botanic Gardens, Kew, U.K.), pp. 109–137.
- 25.Savolainen, V., Fay, M. F., Albach, D. C., Backlund, A., van der Bank, M., Cameron, K. M., Johnson, S. A., Lledo, M. D., Pintaud, J.-C., Powell, M., et al. (2000) Kew Bull. 55, 257–309. [Google Scholar]
- 26.Savolainen, V., Chase, M. W., Hoot, S. B., Morton, C. M., Soltis, D. E., Bayer, C., Fay, M. F., De Bruijn, A. Y., Sullivan, S. & Qiu, Y. L. (2000) Syst. Biol. 49, 306–362. [DOI] [PubMed] [Google Scholar]
- 27.Soltis, P. S., Soltis, D. E. & Chase, M. W. (1999) Nature 402, 402–404. [DOI] [PubMed] [Google Scholar]
- 28.Soltis, D. E., Soltis, P. S., Chase, M. W., Mort, M. E., Albach, D. C., Zanis, M., Savolainen, V., Hahn, W. H., Hoot, S. B., Fay, M. F., et al. (2000) Bot. J. Linn. Soc. 133, 381–461. [Google Scholar]
- 29.Graham, S. W. & Olmstead, R. G. (2000) Am. J. Bot. 87, 1712–1730. [PubMed] [Google Scholar]
- 30.Qiu, Y.-L., Lee, J. H., Bernasconi-Quadroni, F., Soltis, D. E., Soltis, P. S., Zanis, M., Zimmer, E. A., Chen, Z. D., Savolainen, V. & Chase, M. W. (1999) Nature 402, 404–407. [DOI] [PubMed] [Google Scholar]
- 31.Baum, B. R. (1992) Taxon 41, 3–10. [Google Scholar]
- 32.Ragan, M. A. (1992) Mol. Phylogenet. Evol. 1, 53–58. [DOI] [PubMed] [Google Scholar]
- 33.Bininda-Emonds, O. R. P., Gittleman, J. L. & Steel, M. A. (2002) Annu. Rev. Ecol. Syst. 33, 265–289. [Google Scholar]
- 34.Salamin, S., Hodkinson, T. R. & Savolainen, V. (2002) Syst. Biol. 51, 136–150. [DOI] [PubMed] [Google Scholar]
- 35.Purvis, A. (1995) Philos. Trans. R. Soc. London B 348, 405–421. [DOI] [PubMed] [Google Scholar]
- 36.Bininda-Emonds, O. R. P., Gittleman, J. L. & Purvis, A. (1999) Biol. Rev. Cambridge Philos. Soc. 74, 143–175. [DOI] [PubMed] [Google Scholar]
- 37.Angiosperm Phylogeny Group (1998) Ann. Mo. Bot. Gard. 85, 531–553. [Google Scholar]
- 38.Angiosperm Phylogeny Group II (2003) Bot. J. Linn. Soc. 141, 399–436. [Google Scholar]
- 39.Swofford, D. L. (2001) paup4.0b8 (Sinauer, Sunderland, MA).
- 40.Shimodaira, H. & Hasegawa, M. (1999) Mol. Biol. Evol. 16, 114–116.10331256 [Google Scholar]
- 41.Savolainen, V., Chase, M. W., Salamin, N., Soltis, D. E., Soltis, P. S., López, A. J., Fédrigo, O. & Naylor, G. J. P. (2002) Syst. Biol. 51, 638–647. [DOI] [PubMed] [Google Scholar]
- 42.Sanderson, M. J. & Doyle, J. A. (2001) Am. J. Bot. 88, 1499–1516. [PubMed] [Google Scholar]
- 43.Mathews, S. & Donoghue, M. J. (2000) Int. J. Plant Sci. 161, S41–S55. [Google Scholar]
- 44.Zanis, M. J., Soltis, D. E., Soltis, P. S., Mathews, S. & Donoghue, M. J. (2002) Proc. Natl. Acad. Sci. USA 99, 6848–6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu, Y.-L., Lee, J., Bernasconi-Quadroni, F., Soltis, D. E., Soltis, S. P., Zanis, M., Zimmers, E. A., Chen, Z., Savolainen, V. & Chase, M. W. (2000) Int. J. Plant Sci. 161, S3–S27. [Google Scholar]
- 46.Sanderson, M. J. (1997) Mol. Biol. Evol. 14, 1218–1231. [Google Scholar]
- 47.Wikström, N., Savolainen, V. & Chase, M. W. (2001) Proc. R. Soc. London B 268, 2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drinnan, A. N., Crane, P. R. & Hoot, S. B. (1994) Plant Syst. Evol. 8, 93–122 suppl. [Google Scholar]
- 49.Watson, L. & Dallwitz, M. J. (1991) Australian Syst. Bot. 4, 681–695. [Google Scholar]
- 50.Dallwitz, M. J. (1980) Taxon 29, 41–46. [Google Scholar]
- 51.Fusco, G. & Cronk, Q. C. B. (1995) J. Theor. Biol. 175, 235–243. [Google Scholar]
- 52.Purvis, A., Katzourakis, A. & Agapow, P. M. (2001) J. Theor. Biol. 214, 99–103. [DOI] [PubMed] [Google Scholar]
- 53.Mooers, A. Ø., Page, R. D. M., Purvis, A. & Harvey, P. H. (1995) Syst. Biol. 44, 332–342. [Google Scholar]
- 54.Heard, S. B. & Mooers, A. Ø. (1996) Syst. Biol. 45, 115–118. [Google Scholar]
- 55.Slowinski, J. B. & Guyer, C. (1993) Am. Nat. 142, 1019–1024. [DOI] [PubMed] [Google Scholar]
- 56.Sanderson, M. J. & Donoghue, M. J. (1994) Science 264, 1590–1593. [DOI] [PubMed] [Google Scholar]
- 57.Huelsenbeck, J. P., Bull, J. J. & Cunningham, W. (1996) Trends Ecol. Evol. 11, 152–158. [DOI] [PubMed] [Google Scholar]
- 58.Soltis, D. E., Soltis, P. S., Mort, M. E., Chase, M. W., Savolainen, V., Hoot, S. B. & Morton, C. M. (1998) Syst. Biol. 47, 32–42. [DOI] [PubMed] [Google Scholar]
- 59.Soltis, P. S., Soltis, D. E., Savolainen, V., Crane, P. R. & Barraclough, T. G. (2002) Proc. Natl. Acad. Sci. USA 99, 4430–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benton, M. J. & Ayala, F. J. (2003) Science 300, 1698–1700. [DOI] [PubMed] [Google Scholar]
- 61.Purvis, A. (1996) in New Uses for New Phylogenies, eds. Harvey, P. H., Brown, A. J. L., Smith, J. M. & Nee, S. (Oxford Univ. Press, Oxford), pp. 153–168.
- 62.Savolainen, V., Heard, S. B., Powell, M. P., Davies, T. J. & Mooers, A. Ø. (2002) Syst. Biol. 51, 1–9. [DOI] [PubMed] [Google Scholar]
- 63.Ricklefs, R. E. & Renner, S. S. (2000) Evolution (Lawrence, Kans.) 54, 1061–1065. [PubMed] [Google Scholar]
- 64.Sims, H. J. & McConway, K. J. (2003) Evolution (Lawrence, Kans.) 57, 460–479. [DOI] [PubMed] [Google Scholar]
- 65.Chapman, G. P. (1996) The Biology of Grasses (CAB International, Oxon, U.K.).
- 66.Bremer, K. (2002) Evolution (Lawrence, Kans.) 56, 1374–1387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.