Abstract
RNA interference is a powerful genetic approach for efficiently silencing target genes. The existing method of gene suppression by the constitutive expression of short hairpin RNAs (shRNAs) allows analysis of the consequences of stably silencing genes but limits the analysis of genes essential for cell survival, cell cycle regulation, and cell development. We have developed an inducible U6 promoter for synthesis of shRNAs in both human and murine cells. Cells containing stably integrated shRNA expression constructs demonstrate stringent dosage- and time-dependent kinetics of induction with undetectable background expression in the absence of the inducer ecdysone. Inducible suppression of human p53 in glioblastoma cells shows striking morphological changes and defects in cell cycle arrest caused by DNA damage, as expected. Remarkably, the inducibility is reversible after withdrawal of the inducer, as observed by reappearance of the protein and a restoration of the original cell phenotype. Inducible and reversible regulation of RNA interference has broad applications in the areas of mammalian genetics and molecular therapeutics.
The ability to vary expression levels of an endogenous gene product at will and to monitor effects on cells or whole animals can provide powerful insights into its biological role. RNA interference (RNAi)-mediated gene silencing has emerged as a powerful approach to regulate levels of an endogenous protein within its physiological limits.
RNAi is a process of sequence-specific posttranscriptional gene silencing mediated by double-stranded RNA and is a powerful genetic approach to analyze gene function in many organisms (1, 2). The endogenous mediators of sequence-specific mRNA degradation are 21- and 22-nt short interfering RNAs (siRNAs) generated from longer double-stranded RNAs by the ribonuclease III activity of the evolutionary conserved dicer enzyme (3, 4). Gene-specific long double-stranded RNAs have been successfully used in worms and flies for RNAi-mediated gene silencing (5). However, in mammalian cells, double-stranded RNAs >30 bp trigger the antiviral/IFN pathways that results in global shut-down of protein synthesis (6, 7). Recently it was demonstrated that RNAi-mediated gene silencing can be obtained in cultured mammalian cells by delivery of chemically synthesized short (<30 nt) double-stranded siRNA molecules (3) or by endogenous expression of short hairpin RNAs (shRNAs) bearing a fold-back stem-loop structure (8–12).
Plasmid- and viral vector-based constitutive expression of shRNAs by RNA polymerase III U6 and H1 small nuclear RNA promoters (U6 or H1) often result in stable and efficient suppression of target genes (9, 10, 13). However, the inability to adjust levels of suppression has imposed limitations in the analysis of genes essential for cell survival, cell cycle regulation, and cell development. Besides, gross suppression of a gene for longer periods may result in nonphysiological responses. This problem can be circumvented by generating inducible regulation of RNAi in mammalian cells. The two most widely used inducible mammalian systems use tetracycline- or ecdysone-responsive transcriptional elements (14, 15). The chief drawback of the tetracycline-inducible system is a relatively high background of expression in the uninduced state in certain cell lines (15, 16). The ecdysone-inducible system is tightly regulated (15, 17), with no expression in the uninduced state and a rapid inductive response (15), and the components of the inducible system are inert with rapid clearance kinetics and, therefore, do not affect mammalian physiology.
To facilitate stable and inducible suppression of any gene we have developed an ecdysone-inducible synthesis of shRNAs under the control of a modified RNA polymerase III-specific U6 promoter. By using a retroviral delivery and fluorescence-activated cell sorting (FACS) analysis of enhanced GFP (EGFP) positive cells, stable cell lines were rapidly and efficiently established both in murine and human cells, thus alleviating the labor-intensive isolation and analysis of multiple independent clones. Significantly, RNAi inducibility follows a stringent dose- and time-dependent kinetics of induction with undetectable background expression in the absence of the inducer. We inducibly expressed shRNAs targeting the human tumor suppressor p53 gene in the human glioblastoma cell line and MyoD in the murine endothelial cell line. Dose- and time-dependent suppression of p53 gene expression was associated with changes in cell morphology and concomitant reduction in its downstream target p21. Furthermore, the suppression was specific because it could override p53-dependent cell cycle arrest caused by γ-irradiation. Remarkably, withdrawal of the inducer completely reversed the phenotype as indicated by reappearance of the protein, a restoration of original morphology, and a gain in the ability to undergo p53-mediated cell cycle arrest in response to γ-irradiation. An inducible regulation of RNAi with reversible properties has broad implications in the areas of mammalian genetics and molecular therapeutics.
Materials and Methods
Generation of Retroviral Constructs. Retroviral vectors expressing the two nuclear receptor/transcription factors, VgEcR and RXR, were constructed as described (17). To construct pEind-RNAi, a GAL4-Oct-2Q(Q → A) fragment was PCR-amplified from pCG-GAL4-Oct-2Q(Q → A) (18) by using a forward primer (ACGCCCGCGGATGAAGCTACTGTCTTCTATC) and reverse primer (CACCCTGAAGTTCTCAGGATCC), digested with SacII/BamHI, and inserted into the SacII/BamH1 site upstream of vector pIRES-EGFP (Clontech). Next, the GAL4-Oct-2Q(Q → A) internal ribosomal entry site-linked EGFP was PCR-amplified by using a forward primer (AGCTTTGTTTAAACCGAATTCTGCAGTCGACGGTA) and reverse primer CAGCTGATCATTA CTTGTACAGCTCGTCC, digested with PmeI/BclI, and inserted into the PmeI/BclI site of pI-TKHygro retroviral vector (17). Next, a BglII site was created downstream of XhoI in pI-TKHygro by site-directed mutagenesis (Stratagene) by using two primers: ACAGTGGCGGCCGCTCGAGATCTCTTGGAGTGGTGAATCCGTT (upper) and TGTCACCGCCGGCGAGCTCTAG AGAACCTCACCACTTAGGCAA (lower). The resulting construct was digested with BglII and blunt-ended with Klenow enzyme into which a gateway destination cassette ccdB (Invitrogen) was inserted.
To generate 4XGAL4 DNA-binding sites upstream of the U6 promoter-containing vector (19), a SacI site was created by site-directed mutagenesis (Stratagene) upstream of the Oct-1-binding site by using two primers, CAGGCTCCGCGGCCGCCGAGCTCACCGAGGGCCTATTTCCCATG (upper) and GTCCGAGGCGCCG GC GGCTCGAGTGGCTCCCGGATAAAGGGTAC (lower). The Oct-1 and staf binding elements were removed by digestion with SacI/NdeI and replaced with 4XGAL4-binding sites obtained by digesting vector pU6/-198-4XG17M (18) with SacI/NdeI.
Designing and Cloning of shRNAs. A majority of shRNA probes were designed by using computer software (www.cshl.org/public/SCIENCE/hannon.html). shRNA sequences (two complementary ≈83-nt DNA oligos) were annealed and cloned directly into a 4XGAL4 U6 promoter-containing vector by using a ligation-independent cloning method (19). The entire 4XGAL4 U6 promoter-shRNA cassette was transferred into pEind-RNAi by gateway clonase recombination reaction (Invitrogen). shRNA against the p53 gene was designed based on a published sequence (11). shRNA against firefly luciferase (9) served as a nonspecific control.
Cell Culture, Retroviral Transductions, and Selection of Stable Cells. U87MG (human glioblastoma-derived cells, ATTC HTB-14) was maintained in DMEM supplemented with 10% FBS, 0.1 mg/ml penicillin and 0.1 mg/ml streptomycin (Life Technologies, Rockville, MD). The murine endothelial cell line, mHEVc, was grown in RPMI medium 1640 supplemented with 10% FBS/1 mM Hepes/10 mM sodium bicarbonate/2 mM glutamine. U87MG stably expressing receptors VgEcR and RXR (17) was maintained in a puromycin (0.4 mg/ml) and G418 (1 μg/ml) selection, whereas mHEVc was maintained in puromycin (0.6 mg/ml) and G418 (6 μg/ml). Both of the host cell lines carrying pEind-RNAi were maintained in hygromycin at 0.1 mg/ml. For biosafety purposes the pEind-RNAi is a self-inactivating (sin) retrovirus that lacks U3 enhancers in the 3′ long terminal repeat (LTR). On proviral integration, this deletion flanks the retroviral insert, thereby removing the enhancers from both 5′ and 3′ LTRs minimizing the risk of generating replication-competent recombinants.
High-titer virus were produced by calcium phosphate transfection of retroviral DNA constructs into the LinX amphotrophic retroviral packaging cell line (17) followed by incubation with dexamethasone and butyrate (each 1 μM) of packagers at 32°C for 3 days. Infections were carried out by harvesting supernatants from LinX packagers, filtered through a 0.45 μM membrane (Millipore), followed by the addition of polybrene (8 μM) to supernatants, finally overlaying onto actively dividing host cells (70% confluent). Cells were gently spun at room temperature for 30 min and incubated at 32°C for 6 h. Recipients were infected one more time by using freshly prepared supernatant from the same packaging plate, at 8-h intervals. At 24 h after the final infection, cells were split to lower densities and antibiotic selection applied for 2–3 days. Cells were split on six-well plates and induced with 2 μM murA, and the top 3% of GFP+ cells were collected by FACS. Cells that expressed EGFP in the absence of inducer (≈2%) were removed by FACS. The GFP+ cells were expanded for further analysis. An equal amount of ethanol was added to the plates that were uninduced. Given the low percentage of “leaky” cells in the absence of the inducer, one round of FACS for isolating GFP+ cells should suffice for routine applications. However, in experiments requiring long-term cultures the removal of “leaky” cells may be necessary where constitutive suppression of a gene (for example, a tumor suppressor) may confer growth advantage to the cell in the absence of the inducer.
Western Blot Analysis and Antibodies. Stable cell lines inducibly expressing shRNAs targeting human p53 or murine MyoD mRNA were separately grown in six-well plates at a density of 0.3 × 105 per well. Cells were induced with the indicated concentrations of murA; and, 72 h after induction, cells were harvested and lysed with RIPA lysis buffer (150 mM NaCl/50 mM Tris, pH 8.0/0.5% sodium deoxycholate/0.1% SDS/1% Nonidet P-40) containing complete protease inhibitor (Roche Applied Science). Equal amounts of lysate were subjected to Western blot analysis (20) by using antibodies against p53 (1:1,000, NovoCastra, Newcastle, U.K.), MyoD (1:500), GAL4 (1:1,000), p21 (1:500), EGFP (1:200), and horseradish peroxidase-conjugated secondary antibody (1:5,000), obtained from Santa Cruz Biotechnology. The blots were stripped by incubating with 100 mM β-mercaptoethanol/2% SDS/62.5 mM Tris·HCl (pH 6.7) at 50°C for 20 min and reprobed with anti-β-tubulin (1:5,000, Sigma) primary antibody and horseradish peroxidase-conjugated secondary antibody (1:3,000, Amersham Biosciences) to show equal loading. The blots were developed by using ECL Plus (Amersham Biosciences) or West Femto (Pierce) blotting detection system. Signal intensities were determined after background corrections by using alpha-imager 2000 documentation and analysis software (Alpha Innotech, San Leandro, CA). The percentage reduction in band intensity for each concentration of murA was calculated relative to the uninduced samples and normalized against β-tubulin.
Northern Blot Analysis. Cells were plated at a density of 1 × 105 in a 10-cm dish and induced with 5 μM murA. After 72 h cells were harvested and total RNA was extracted by using a RNeasy kit (Qiagen, Valencia, CA). RNA (30 μg) was loaded on a 15% denaturing polyacrylamide-urea gel and Northern blot analysis was performed (21). A 21-nt sense strand of p53 (GACTCCAGTGGTAATCTACTT) was end-labeled with [γ-32P]ATP by using a KinaseMax kit (Ambion, Austin, TX). Hybridization was performed with a NorthernMax kit (Ambion). After hybridization, blots were washed and exposed to x-ray film at –70°C.
Immunofluoresence. U87MG cells stably and inducibly expressing human p53 shRNA or nonspecific shRNA were separately grown on glass coverslips in a 12-well plate at a density of 1 × 103 cells per well. Nonspecific shRNA was targeted against the Luciferase gene. Cells were induced with 5 μM murA for 72 h, fixed in 4% paraformaldehyde for 30 min, and washed three times for 5 min with PBS, followed by permeabilization in 0.2% Triton X-100 and 0.5% normal goat serum for 10 min. The permeabilized cells were blocked for 30 min in 10% normal goat serum, incubated with p53 antibody (1:200, NovoCastra) and β-tubulin (1:400, Sigma) for 1 h at room temperature, and washed three times for 5 min with PBS and 0.5% normal goat serum. Next, cells were incubated with secondary antibodies, Alexa 488 and Alexa 594 (Molecular Probes), for 1 h at room temperature and stained with 4′,6-diamidino-2-phenylindole (1 μg/ml). The coverslip was mounted on antifade mounting medium (Fluoromount-G, Southern Biotechnology Associates) and visualized under a fluorescent microscope. Images were captured by using a Zeiss AxioCam HRm camera at equal exposure time for all panels.
Flow Cytometric Analysis of Cell Cycle Distribution. U87MG cells stably and inducibly expressing human p53 shRNA were plated at a density of 1 × 105 cells per well in six-well plates and induced with 5 μM murA for 72 h followed by γ-irradiation at 20 Gy. In reversal experiments cells were trypsinized and replated to maintain appropriate densities. Twenty-four hours after γ-irradiation, adherent cells were harvested, washed once in PBS, and fixed in ice-cold 70% ethanol in distilled water. Cells were then washed twice in PBS supplemented with 1% BSA and resuspended in PBS containing 0.1% Triton X-100, 50 μg/ml propidium iodide, 5 mM sodium citrate, and 50 μg/ml RNase A. After incubation at room temperature for 20 min, cells were analyzed for cell cycle distribution with an LSRII flow cytometer (Becton Dickinson) and facsdiva software (Becton Dickinson). Red fluorescence (585 ± 42 nm) was evaluated on a linear scale, and pulse width analysis was used to exclude cell doublets and aggregates from the analysis. Cells with DNA content between 2N and 4N were designated as being in the G1, S, or G2/M phase of the cell cycle. The number of cells in each compartment of the cell cycle was expressed as a percentage of the total number of cells present.
Results and Discussion
Design and Characterization of the Retroviral-Based, Ecdysone-Inducible RNAi System. The U6 promoter is widely used for directing expression of shRNAs because it is active in all cell types and efficiently directs synthesis of small, noncoding transcripts bearing well defined ends. However, so far, attempts to generate robust inducible RNA polymerase III promoters have met with less than satisfactory results (22). To facilitate stable and inducible suppression of any gene we developed an ecdysone-inducible synthesis of shRNAs under the control of a modified U6 promoter. We accomplished this by replacing the natural U6 enhancers with heterologous GAL4-DNA-binding sites and tested GAL4-transactivator fusions (18), which activate transcription specifically from the wild-type U6 promoter but not from pol II mRNA, U1 small nuclear RNA, or U6 TATA– promoters. A synthetic transactivator, Oct-2Q(Q → A), specifically expressed shRNA with no background expression in the absence of the inducer and was, therefore, used for further analysis.
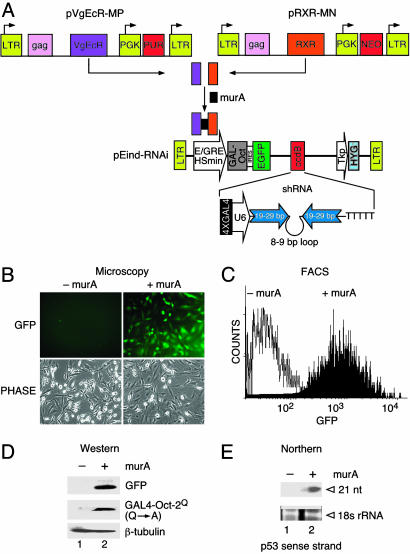
The ecdysone-inducible system is composed of three Moloney murine leukemia virus-based retroviral vectors: the two vectors expressing nuclear receptors/transcription factors VgEcR and RXR and a third construct, pEind-RNAi, expressing a chimeric transactivator GAL4-Oct-2Q(Q → A) and an internal ribosomal entry site-linked enhanced GFP (EGFP) under an inducible promoter hybrid ecdysone-responsive element/Hsmin (Fig. 1A). GFP expression permits enrichment of transduced cells by fluorescence-activated cell sorting (FACS). We incorporated a “gateway” site-specific acceptor, ccdB (Invitrogen), so that any DNA-encoded shRNA of interest can be readily transferred from a donor vector by recombination downstream of the U6 promoter. The enhancer of the U6 promoter comprises an octamer motif and an adjacent element that binds the transactivators Oct-1 and staf, respectively (23). We deleted the natural enhancer region and engineered four tandem GAL4-binding sites (4XGAL4) in its place. On induction with muristerone A (murA, an analogue of ecdysone), the two receptors/transcription factors dimerize and bind to the hybrid ecdysone-responsive element to activate GAL4-Oct-2Q(Q → A) expression. GAL4-Oct-2Q(Q → A) in turn binds to the 4XGAL4 DNA-binding sites and activates the U6 promoter, which drives expression of a shRNA (Fig. 1 A).
Fig. 1.
Design of retroviral vectors for ecdysone-inducible synthesis of shRNA and experimental validation in mammalian cells. (A) Vector descriptions. pVgEcR-MP, retroviral vector for constitutive expression of a modified Drosophila ecdysone receptor (containing the VP16 transactivating domain) from the LTR and marked with a puromycin resistance gene; pRXR-MN, retroviral vector for constitutive expression of the RXR from the LTR and marked with the neomycin (G418) resistance gene; pEind-RNAi, a self-inactivating retroviral ecdysone-inducible vector marked with hygromycin resistance gene; E/GRE, hybrid ecdysone response element; Hsmin, minimal heat shock promoter; IRES, internal ribosomal entry site; PUR, puromycin; NEO, G418 resistance gene; Tkp, enhancerless thymidine kinase promoter; PGK, phosphoglucokinase promoter; ccdB, Gateway system cassette. (B) Microscopic analysis of a representative cell line to show GFP+ cells in the absence of murA (–murA) and presence of 0.5 μM murA (+murA) at 72 h after induction. (C) FACS analysis to show GFP+ cells in the absence (–murA) and presence (+murA) of 5 μM murA at 72 h after induction. (D) Western blot analysis showing expression of EGFP and GAL4-Oct-2Q(Q → A) in uninduced cells (lane 1) and cells induced with 5 μM MurA (lane 2) for 72 h. (E) Northern blot analysis showing inducible expression of p53-specific siRNAs in cells treated with 5 μM murA for 72 h by probing with 32P-labeled p53 sense strand. 18S RNA served as an internal control to show equal loading.
Stable cell lines were generated as described in Materials and Methods (also see Fig. 5, which is published as supporting information on the PNAS web site). The use of a retroviral delivery and flow sorting of EGFP+ cells expedited rapid and efficient generation of stable cell lines both in murine and human cells. Analysis of sorted cells after induction with murA showed >95% EGFP+ cells as determined by fluorescent microscopy (Fig. 1B) and FACS (Fig. 1C). Western blot analysis demonstrated increased levels of GAL4-Oct-2Q(Q → A) and EGFP protein in samples treated with murA but not in untreated controls (Fig. 1D). Having established inducible expression of the activator GAL4-Oct-2Q(Q → A), we next demonstrated its ability to activate p53 shRNA expression from the modified U6 promoter. Northern blot analysis showed production of 21-nt siRNAs specifically in the cells expressing the activator (Fig. 1E). These results indicate that the GAL4-Oct-2Q(Q → A) induced formation of the stem-loop precursor transcript that was cleaved in the cell to produce a functional siRNA. Taken together, these data indicate that the induction is highly specific and lacks detectable background expression levels in the absence of the inducer.
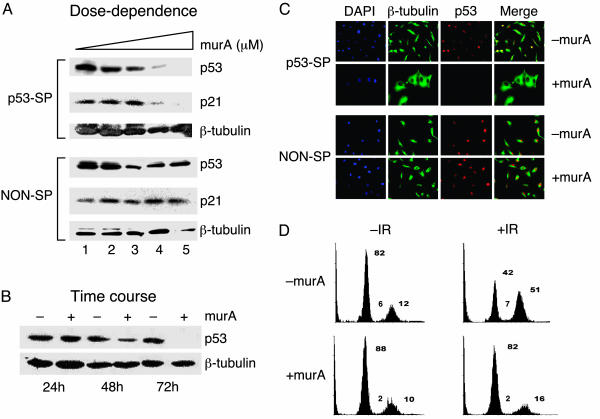
Stable and Efficient RNAi-Mediated Inducible Suppression of Human p53 Gene. The usefulness of the RNAi-inducible system in suppressing expression of a cognate target gene was evaluated next. As a first target we chose the human p53 gene because of detectable expression in mammalian cells, availability of reliable antibodies to monitor levels of the protein, and the presence of an effective shRNA against the p53 gene (11). A human glioblastoma cell line, U87MG, carrying a functional wild-type p53 (24) and stably expressing receptors VgEcR and RXR was transduced with a pEind-RNAi virus expressing an shRNA targeting the p53 gene. Stable cells treated with 0.5–5 μM murA showed a dose-dependent reduction in endogenous p53 level with almost 95% reduction in the presence of 5 μM murA (Fig. 2A Upper). The suppression of p53 was also time-dependent, showing partial reduction (60%) at 48 h and maximal reduction (>95%) at 72 h after induction (Fig. 2B). To ascertain that the induced shRNA targeted the p53 mRNA specifically, we measured the levels of cyclin-dependent kinase inhibitor p21, a well characterized transcriptional target of p53. As expected, the decrease in p21 levels correlated well with inducible suppression of p53 (Fig. 2 A Upper), relative to that obtained with a nonspecific shRNA (Fig. 2 A Lower). In contrast, levels of Cdk4 protein, an upstream component of the p53-signaling pathway, remained unchanged (data not shown). Taken together, these results suggest that the p53 shRNA synthesized by the inducible system is tightly regulated and exhibits high target specificity.
Fig. 2.
Stable and efficient RNAi-mediated inducible suppression of human p53 gene in U87MG cells. (A) Dose–response of ecdysone-inducible RNAi. Stable cell lines carrying p53-specific shRNA (p53-SP) and nonspecific shRNA (NON-SP) were induced with 0.0 (lane 1), 0.5 (lane 2), 2.0 (lane 3), 3.0 (lane 4), and 5.0 (lane 5) μM murA. Whole-cell extracts were prepared after 72 h and analyzed by Western blotting for p53, p21, and β-tubulin (control). Fold reduction in p53 protein level is 30% (lane 2), 64% (lane 3), 90% (lane 4), and >95% (lane 5) relative to uninduced sample (lane 1). (B) Time course of ecdysone-inducible RNAi. Stable cells carrying shRNA for p53 were induced with 5 μM murA and analyzed for p53 levels at indicated time points; fold reduction of p53 protein level is 60% (+murA at 48 h) and >95% (+murA at 96 h). Fold reduction of protein level was based on densitometric measurement. (C) Inducible suppression of p53 at a single-cell level by immunofluorescence, showing silencing of p53 gene by an inducible p53 shRNA in the presence of 5 μM murA (Upper, bottom row) but not in the absence of murA (Upper, top row), 72 h after induction. Staining with p53-specific antibody (red) and α-tubulin (green). 4′,6-Diamidino-2-phenylindole (DAPI) was used to stain nuclei (blue). (D) Cell cycle analysis of γ-irradiated U87MG cells carrying inducible p53 shRNA by FACS. Stable cell lines carrying p53 shRNA were either uninduced (–murA) or induced (+murA) with 5 μM murA for 72 h and subjected to 20 Gy of γ-irradiation. After 24 h FACS analysis showing distribution of cells in G1, S, and G2/M phases of the cell cycle is shown. IR, irradiation. This experiment is a representation of three independent data sets.
We next assessed the effects of inducing expression of p53 shRNA at a single-cell level by immunofluorescence experiments. The results showed the presence of nuclear p53 in the absence of induction (Fig. 2C Upper, top row), as expected. Induction of p53 shRNA expression resulted in a dramatic reduction in the levels of p53 (Fig. 2C Upper, bottom row), relative to that obtained with a nonspecific shRNA (Fig. 2C Lower). Cells with reduced levels of p53 were flat and large in contrast to normal long-spindle forms observed in the uninduced state (Fig. 2C Upper). The morphological change was specific to effects of p53 hairpin and not to the activity of either the inducer or the expression of GAL4-Oct-2Q(Q → A), EGFP, and nonspecific shRNA (Fig. 2C Lower). Although reports indicating morphology changes in HeLa cells as a consequence of p53 suppression by antisense RNA are available (25), it is likely that in the U87MG cells' suppression of p53 cooperates with the absence of PTEN (26, 27) to generate the observed phenotype.
After having established specific down-regulation of p53 gene expression by inducible RNAi, we next wondered whether the p53 suppression had a functional consequence. FACS analysis showed a dramatic increase in the cell number in G2/M phase (39%) and a concomitant decrease in G0/G1 phase (40%) in uninduced, irradiated samples (Fig. 2D Upper Right). In contrast, cells induced for p53 shRNA and exposed to γ-irradiation lost their ability to arrest at the G2/M phase of the cell cycle (Fig. 2D Lower Right). We were unable to observe G0/G1 arrest in multiple independent experiments, underscoring the importance of p53 in mediating G2/M arrest in response to DNA damage in U87MG cells consistent with published observations (28, 29). Indeed, cell cycle arrest in G2/M has been shown to depend on the cell type (30, 31).
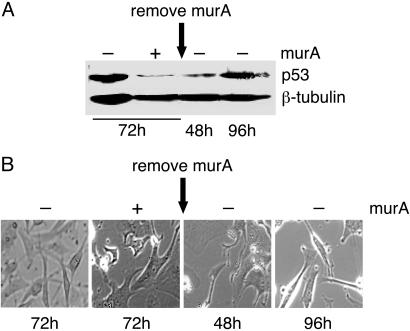
The RNAi-Mediated Inducible Gene Suppression Is Reversible. A major limitation of constitutive shRNA expression systems is the irreversible suppression of gene expression that could result in nonphysiological responses. We determined whether the gene suppression obtained by the RNAi-inducible system could be reversed after withdrawal of the inducer. In the presence of the inducer stable cells carrying p53 shRNA showed, as expected, reduction in endogenous p53 levels (Fig. 3A) and associated morphological changes at 72 h after induction (Fig. 3B). On removal of the inducer a partial recovery occurred in p53 protein levels at 48 h, with almost full recovery at 96 h (Fig. 3A). The recovery of p53 protein level was associated with restoration in the original cell morphology (Fig. 3B, compare large flat cells at 72 h in the presence of murA with spindle-shaped cells at 96 h after withdrawal of murA), and cell function as determined by γ-irradiation induced cell cycle arrest (see Fig. 6, which is published as supporting information on the PNAS web site), underscoring the swift clearance kinetics of murA that results in a rapid phenotypic switch. These studies also demonstrate that the p53 gene suppression does not lead to an irreversible cascade of molecular events.
Fig. 3.
RNAi-mediated inducible gene suppression is reversible. (A) Western blot analysis showing 93% fold reduction in p53 protein levels in cells induced with 5 μM murA (+murA) relative to uninduced (–murA) at 72 h. After murA removal, the fold recovery in p53 levels is 20% (48 h) and >90% (96 h). (B) Phase-contrast microscopy showing morphology of U87MG cells either uninduced (–murA) or induced with 5 μM murA (+murA) for 72 h and at 48 and 96 h after murA removal.
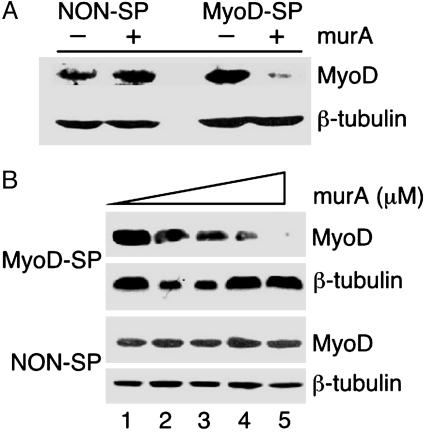
Inducible Suppression of MyoD in a Murine Cell line. The general utility of the RNAi-inducible system was determined by targeting suppression of a MyoD gene in a murine cell line. Murine endothelial cells, mHEVc (32), were transduced with viruses carrying the two receptors/transcription factors and pEind-RNAi expressing a MyoD shRNA, as described earlier. Efficient suppression of MyoD gene expression was observed at 72 h after induction (Fig. 4A). Furthermore, increasing concentrations of murA (0.5–5 μM) showed a dose-dependent reduction in endogenous MyoD levels with almost 95% reduction in the presence of 5 μM murA (Fig. 4B Upper), relative to cells stably expressing a nonspecific shRNA (NON-SP, Fig. 4B Lower).
Fig. 4.
Inducible suppression of MyoD gene expression in a murine endothelial cells. (A) Cells stably and inducibly expressing MyoD shRNA (MyoD-SP) and nonspecific shRNA (NON-SP) were treated with 5 μM murA. Whole-cell extracts were prepared at 72 h after induction and analyzed by Western blot for MyoD and β-tubulin. Fold reduction of protein level is >95% for MyoD (MyoD-SP) relative to nonspecific shRNA. (B) Stable cells carrying MyoD specific shRNA (MyoD-SP) and nonspecific shRNA (NON-SP) were induced with 0.0 (lane 1), 0.5 (lane 2), 1.0 (lane 3), 3.0 (lane 4), and 5.0 (lane 5) μM murA and subjected to Western blot for MyoD and β-tubulin. Fold reduction of MyoD protein level for shRNA (MyoD-SP) is 5% (lane 2), 25% (lane 3), 72% (lane 4), and >95% (lane 5) relative to uninduced sample (lane 1). Fold reduction in MyoD was normalized against β-tubulin for each band. shRNA used for the suppression of MyoD gene expression was identified in a high-throughput screen (19).
In summary, we provide a powerful approach to stably and inducibly suppress gene expression in mammalian cells. Given the success of the ecdysone-based inducible systems in animal models (15, 33) and germ-line transmission of RNAi (34), it is conceivable to generate transgenic animals inducibly expressing RNAi. A considerable improvement to the existing design would be to obtain tissue-specific regulation in vivo by either expressing the GAL4 activator under the control of a tissue-specific promoter or by using a “caged ecdysteroid” (35). Recently, doxycyclin-controllable suppression of genes in mammalian cells was reported (36, 37). However, it was not clear how much basal expression in the absence of the inducer these systems allow. Furthermore, doxycyclin is linked to toxicity problems, a major disadvantage for studies involving both cultured cells and animals. Ecdysone is more suitable for use in vivo because it is a naturally occurring lipophilic steroid that can penetrate tissues and is quickly metabolized and cleared (33).
A major application of the RNAi-inducible system would be in studies where partial down-regulation of gene expression is desired, in particular, in cases where partial suppression results in distinct phenotypes. For example, shRNAs showing varying levels of p53 suppression generated distinct tumor phenotypes in vivo (38). Partial suppression is also useful where lethality associated with complete suppression of essential genes is of concern. In our system, the levels of shRNA expression can be easily and finely controlled by simply varying the dosage of the inducer.
Supplementary Material
Acknowledgments
We thank Nouria Hernandez for comments on the manuscript and helpful discussions and Winship Herr, Zaher Nahle, Douglas Conklin, Linda Rodgers, Elisa De Stanchina, Lance Palmer, and Kimberly LaVine for reagents.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RNAi, RNA interference; siRNA, short interfering RNA; shRNA, short hairpin RNA; EGFP, enhanced GFP; FACS, fluorescence-activated cell sorting; LTR, long terminal repeat.
References
- 1.Hammond, S. M., Boettcher, S., Caudy, A. A., Kobayashi, R. & Hannon, G. J. (2001) Science 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- 2.Sharp, P. A. (1999) Genes Dev. 13, 139–141. [PubMed] [Google Scholar]
- 3.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. (2001) Nature 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 5.Paddison, P. J. & Hannon, G. J. (2002) Cancer Cell 2, 17–23. [DOI] [PubMed] [Google Scholar]
- 6.Marcus, P. I. & Sekellick, M. J. (2001) J. Interferon Cytokine Res. 21, 423–429. [DOI] [PubMed] [Google Scholar]
- 7.Gil, J. & Esteban, M. (2000) Apoptosis 5, 107–114. [DOI] [PubMed] [Google Scholar]
- 8.Elbashir, S. M., Harborth, J., Weber, K. & Tuschl, T. (2002) Methods 26, 199–213. [DOI] [PubMed] [Google Scholar]
- 9.Paddison, P. J., Caudy, A. A., Bernstein, E., Hannon, G. J. & Conklin, D. S. (2002) Genes Dev. 16, 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sui, G., Soohoo, C., Affar el, B., Gay, F., Shi, Y. & Forrester, W. C. (2002) Proc. Natl. Acad. Sci. USA 99, 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 12.Paul, C. P., Good, P. D., Winer, I. & Engelke, D. R. (2002) Nat. Biotechnol. 20, 505–508. [DOI] [PubMed] [Google Scholar]
- 13.Rubinson, D. A., Dillon, C. P., Kwiatkowski, A. V., Sievers, C., Yang, L., Kopinja, J., Zhang, M., McManus, M. T., Gertler, F. B., Scott, M. L. & Van Parijs, L. (2003) Nat. Genet. 33, 401–406. [DOI] [PubMed] [Google Scholar]
- 14.Gossen, M., Freundlieb, S., Bender, G., Muller, G., Hillen, W. & Bujard, H. (1995) Science 268, 1766–1769. [DOI] [PubMed] [Google Scholar]
- 15.No, D., Yao, T. P. & Evans, R. M. (1996) Proc. Natl. Acad. Sci. USA 93, 3346–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Craenenbroeck, K., Vanhoenacker, P., Leysen, J. E. & Haegeman, G. (2001) Eur. J. Neurosci. 14, 968–976. [DOI] [PubMed] [Google Scholar]
- 17.Stolarov, J., Chang, K., Reiner, A., Rodgers, L., Hannon, G. J., Wigler, M. H. & Mittal, V. (2001) Proc. Natl. Acad. Sci. USA 98, 13043–13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das, G., Hinkley, C. S. & Herr, W. (1995) Nature 374, 657–660. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, R., Conklin, D. S. & Mittal, V. (2003) Genome Res. 13, 2333–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harlow, E. & Lane, D. (1988) Antibodies, A Laboratory Manual. (Cold Spring Harbor Lab. Press, Plainview, NY).
- 21.Dalmay, T., Hamilton, A., Rudd, S., Angell, S. & Baulcombe, D. C. (2000) Cell 101, 543–553. [DOI] [PubMed] [Google Scholar]
- 22.Meissner, W., Rothfels, H., Schafer, B. & Seifart, K. (2001) Nucleic Acids Res. 29, 1672–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaub, M., Myslinski, E., Schuster, C., Krol, A. & Carbon, P. (1997) EMBO J. 16, 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Meir, E. G., Kikuchi, T., Tada, M., Li, H., Diserens, A. C., Wojcik, B. E., Huang, H. J., Friedmann, T., de Tribolet, N. & Cavenee, W. K. (1994) Cancer Res. 54, 649–652. [PubMed] [Google Scholar]
- 25.Iotsova, V. & Stehelin, D. (1995) Eur J. Cell Biol. 68, 122–132. [PubMed] [Google Scholar]
- 26.Mayo, L. D., Dixon, J. E., Durden, D. L., Tonks, N. K. & Donner, D. B. (2002) J. Biol. Chem. 277, 5484–5489. [DOI] [PubMed] [Google Scholar]
- 27.Stambolic, V., MacPherson, D., Sas, D., Lin, Y., Snow, B., Jang, Y., Benchimol, S. & Mak, T. W. (2001) Mol. Cell 8, 317–325. [DOI] [PubMed] [Google Scholar]
- 28.Badie, B., Goh, C. S., Klaver, J., Herweijer, H. & Boothman, D. A. (1999) Cancer Gene Ther. 6, 155–162. [DOI] [PubMed] [Google Scholar]
- 29.Hirose, Y., Berger, M. S. & Pieper, R. O. (2001) Cancer Res. 61, 1957–1963. [PubMed] [Google Scholar]
- 30.Taylor, W. R. & Stark, G. R. (2001) Oncogene 20, 1803–1815. [DOI] [PubMed] [Google Scholar]
- 31.Bunz, F., Dutriaux, A., Lengauer, C., Waldman, T., Zhou, S., Brown, J. P., Sedivy, J. M., Kinzler, K. W. & Vogelstein, B. (1998) Science 282, 1497–1501. [DOI] [PubMed] [Google Scholar]
- 32.Tudor, K. S., Deem, T. L. & Cook-Mills, J. M. (2000) Biochem. Cell Biol. 78, 99–113. [PubMed] [Google Scholar]
- 33.Karns, L. R., Kisielewski, A., Gulding, K. M., Seraj, J. M. & Theodorescu, D. (2001) BMC Biotechnol. 1, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carmell, M. A., Zhang, L., Conklin, D. S., Hannon, G. J. & Rosenquist, T. A. (2003) Nat. Struct. Biol 10, 91–92. [DOI] [PubMed] [Google Scholar]
- 35.Lin, W., Albanese, C., Pestell, R. G. & Lawrence, D. S. (2002) Chem. Biol. 9, 1347–1353. [DOI] [PubMed] [Google Scholar]
- 36.Wiznerowicz, M. & Trono, D. (2003) J. Virol. 77, 8957–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van de Wetering, M., Oving, I., Muncan, V., Pon Fong, M. T., Brantjes, H., van Leenen, D., Holstege, F. C., Brummelkamp, T. R., Agami, R. & Clevers, H. (2003) EMBO Rep. 4, 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hemann, M. T., Fridman, J. S., Zilfou, J. T., Hernando, E., Paddison, P. J., Cordon-Cardo, C., Hannon, G. J. & Lowe, S. W. (2003) Nat. Genet. 33, 396–400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.